

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144502.11 + phase: 0
(675 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
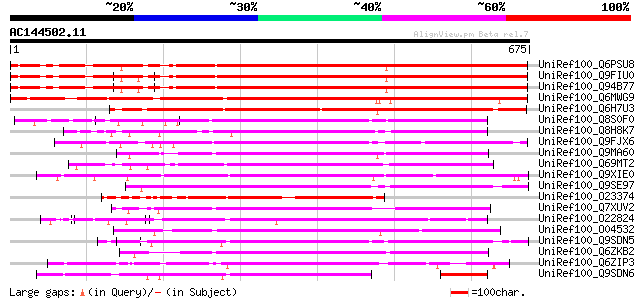
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6PSU8 Formin homology 2 domain-containing protein 5 [... 689 0.0
UniRef100_Q9FIU0 Similarity to unknown protein [Arabidopsis thal... 689 0.0
UniRef100_Q94B77 Hypothetical protein MRB17.15 [Arabidopsis thal... 687 0.0
UniRef100_Q6MWG9 B1160F02.7 protein [Oryza sativa] 575 e-162
UniRef100_Q6H7U3 Putative formin I2I isoform [Oryza sativa] 531 e-149
UniRef100_Q8S0F0 Putative formin-like protein AHF1 [Oryza sativa] 448 e-124
UniRef100_Q8H8K7 Putative formin-like protein [Oryza sativa] 426 e-117
UniRef100_Q9FJX6 Formin-like protein AtFH6 [Arabidopsis thaliana] 407 e-112
UniRef100_Q9MA60 F22F7.8 protein [Arabidopsis thaliana] 403 e-111
UniRef100_Q69MT2 Diaphanous protein-like [Oryza sativa] 400 e-110
UniRef100_Q9XIE0 F23H11.22 protein [Arabidopsis thaliana] 398 e-109
UniRef100_Q9SE97 Formin-like protein AHF1 [Arabidopsis thaliana] 395 e-108
UniRef100_O23374 P140mDia like protein [Arabidopsis thaliana] 386 e-105
UniRef100_Q7XUV2 OSJNBa0072F16.14 protein [Oryza sativa] 372 e-101
UniRef100_O22824 Hypothetical protein At2g43800 [Arabidopsis tha... 364 5e-99
UniRef100_O04532 F20P5.14 protein [Arabidopsis thaliana] 357 8e-97
UniRef100_Q9SDN5 FH protein NFH2 [Nicotiana tabacum] 356 1e-96
UniRef100_Q6ZKB2 Putative formin homology(FH)protein [Oryza sativa] 352 3e-95
UniRef100_Q6ZIP3 Putative formin homology(FH) domain-containing ... 338 4e-91
UniRef100_Q9SDN6 FH protein NFH1 [Nicotiana tabacum] 305 3e-81
>UniRef100_Q6PSU8 Formin homology 2 domain-containing protein 5 [Arabidopsis
thaliana]
Length = 900
Score = 689 bits (1779), Expect = 0.0
Identities = 377/683 (55%), Positives = 463/683 (67%), Gaps = 53/683 (7%)
Query: 2 NDYSFGSSSNNLGDNKKTSLQGNQSMGAFAVVGSPFELNPPPGRVGTIHSGMPPLRPPPG 61
+DYS GSS N G + K QG+QS ++ G + G+ L
Sbjct: 260 SDYSVGSSIN-YGGSVKGDKQGHQSFNIYSNQGKMSSFD------GSNSDTSDSLE---- 308
Query: 62 RMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPP 121
L HE N + PPP R + V S + S +P+PP PP
Sbjct: 309 --ERLSHEGLRNNSITNHGLPPLKPPPGRTASVLSGK---------SFSGKVEPLPPEPP 357
Query: 122 LHPALPGAKPSPPPPPAPLGAKP---GPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRP 178
+ K S PPPP P P GPP PPPPAPP GP PPPPP P G +P
Sbjct: 358 KFLKVSSKKASAPPPPVPAPPMPSSAGPPRPPPPAPPPGSGGPKPPPPPGPKGPRP---- 413
Query: 179 PPPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSM 238
PPP G PRPP GP A+A + D K KLKPFFWDKV AN + SM
Sbjct: 414 -PPPMSLGPKAPRPPSGP----------ADALDD---DAPKTKLKPFFWDKVQANPEHSM 459
Query: 239 VWNQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLL 298
VWN I+SGSFQFNEEMIE+LFGY A +KN + K+ SS Q PQ++QI++ KK QNL
Sbjct: 460 VWNDIRSGSFQFNEEMIESLFGYAAADKN--KNDKKGSSGQAALPQFVQILEPKKGQNLS 517
Query: 299 ILLRALNVTMEEVCDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRF 358
ILLRALN T EEVCDAL EGNELP EF+QTLLKMAPT +EELKLRL+ G+++QLG A+RF
Sbjct: 518 ILLRALNATTEEVCDALREGNELPVEFIQTLLKMAPTPEEELKLRLYCGEIAQLGSAERF 577
Query: 359 LKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKT 418
LKA+VDIP AFKR+E LLFMCT EE+ ESF LEVACKELR SRLF KLLEAVLKT
Sbjct: 578 LKAVVDIPFAFKRLEALLFMCTLHEEMAFVKESFQTLEVACKELRGSRLFLKLLEAVLKT 637
Query: 419 GNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQ 478
GNRMNDGT+RGGAQAFKLDTLLKL+DVKGTDGKTTLLHFVVQEIIR+EG++AAR ++SQ
Sbjct: 638 GNRMNDGTFRGGAQAFKLDTLLKLADVKGTDGKTTLLHFVVQEIIRTEGVRAARTIRESQ 697
Query: 479 SLSNIKTDEL------HETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLG 532
S S++KT++L E+E++YR LGLE VS LS+ELE+VK+ + +DAD LT T +K+G
Sbjct: 698 SFSSVKTEDLLVEETSEESEENYRNLGLEKVSGLSSELEHVKKSANIDADGLTGTVLKMG 757
Query: 533 HGLVKAKDILNKNLKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFH 592
H L KA+D +N +K+ ++ GFRE +E F++NAE + +LE+EK+IMALVKSTGDYFH
Sbjct: 758 HALSKARDFVNSEMKSSGEESGFREALEDFIQNAEGSIMSILEEEKRIMALVKSTGDYFH 817
Query: 593 GNATKDDGLRLFVVVRDFLIMLDKVCKEVRDAQKKPAKPIKQETSRGLSSSDT-RPSPS- 650
G A KD+GLRLFV+VRDFLI+LDK CKEVR+A+ +P + +++ S +SS+T R +PS
Sbjct: 818 GKAGKDEGLRLFVIVRDFLIILDKSCKEVREARGRPVRMARKQGSTASASSETPRQTPSL 877
Query: 651 DFRQRLFPAIAERRIDDDSSDEE 673
D RQ+LFPAI ERR+D SSD +
Sbjct: 878 DPRQKLFPAITERRVDQSSSDSD 900
Score = 47.0 bits (110), Expect = 0.002
Identities = 27/77 (35%), Positives = 32/77 (41%), Gaps = 6/77 (7%)
Query: 136 PPAPLGAKPGPPPPPPPA-PPSAKPGPPPPP-----PPAPSGAKPGPRPPPPPPKSGVAP 189
P L KPG P P P+ PP GPP PP PP + P R PPPPP A
Sbjct: 134 PRRNLATKPGSSPSPSPSRPPKRSRGPPRPPTRPKSPPPRKASFPPSRSPPPPPAKKNAS 193
Query: 190 PRPPIGPKAGGPKATEN 206
P + K ++
Sbjct: 194 KNSTSAPVSPAKKKEDH 210
Score = 43.9 bits (102), Expect = 0.016
Identities = 25/65 (38%), Positives = 31/65 (47%), Gaps = 8/65 (12%)
Query: 127 PGAKPSPPPPPAPLGAKPGPPPPP--PPAPPSAKPG-----PPPPPPPAPSGAKPGPRPP 179
PG+ PSP P P ++ GPP PP P +PP K PPPPP + +K P
Sbjct: 142 PGSSPSPSPSRPPKRSR-GPPRPPTRPKSPPPRKASFPPSRSPPPPPAKKNASKNSTSAP 200
Query: 180 PPPPK 184
P K
Sbjct: 201 VSPAK 205
Score = 41.6 bits (96), Expect = 0.078
Identities = 21/59 (35%), Positives = 27/59 (45%), Gaps = 6/59 (10%)
Query: 130 KPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVA 188
KP P P+P ++P PP PP+ P PPP + P PPPPP K +
Sbjct: 141 KPGSSPSPSP--SRPPKRSRGPPRPPTR----PKSPPPRKASFPPSRSPPPPPAKKNAS 193
Score = 40.0 bits (92), Expect = 0.23
Identities = 23/73 (31%), Positives = 30/73 (40%), Gaps = 7/73 (9%)
Query: 96 SARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPP 155
+ +P ++PSP S PPRPP +P PPP PPPPP
Sbjct: 139 ATKPGSSPSPSPSRPPKRSRGPPRPP-------TRPKSPPPRKASFPPSRSPPPPPAKKN 191
Query: 156 SAKPGPPPPPPPA 168
++K P PA
Sbjct: 192 ASKNSTSAPVSPA 204
>UniRef100_Q9FIU0 Similarity to unknown protein [Arabidopsis thaliana]
Length = 900
Score = 689 bits (1777), Expect = 0.0
Identities = 377/683 (55%), Positives = 463/683 (67%), Gaps = 53/683 (7%)
Query: 2 NDYSFGSSSNNLGDNKKTSLQGNQSMGAFAVVGSPFELNPPPGRVGTIHSGMPPLRPPPG 61
+DYS GSS N G + K QG+QS ++ G + G+ L
Sbjct: 260 SDYSVGSSIN-YGGSVKGDKQGHQSFNIYSNQGKMSSFD------GSNSDTSDSLE---- 308
Query: 62 RMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPP 121
L HE N + PPP R + V S + S +P+PP PP
Sbjct: 309 --ERLSHEGLRNNSITNHGLPPLKPPPGRTASVLSGK---------SFSGKVEPLPPEPP 357
Query: 122 LHPALPGAKPSPPPPPAPLGAKP---GPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRP 178
+ K S PPPP P P GPP PPPPAPP GP PPPPP P G +P
Sbjct: 358 KFLKVSSKKASAPPPPVPAPQMPSSAGPPRPPPPAPPPGSGGPKPPPPPGPKGPRP---- 413
Query: 179 PPPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSM 238
PPP G PRPP GP A+A + D K KLKPFFWDKV AN + SM
Sbjct: 414 -PPPMSLGPKAPRPPSGP----------ADALDD---DAPKTKLKPFFWDKVQANPEHSM 459
Query: 239 VWNQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLL 298
VWN I+SGSFQFNEEMIE+LFGY A +KN + K+ SS Q PQ++QI++ KK QNL
Sbjct: 460 VWNDIRSGSFQFNEEMIESLFGYAAADKN--KNDKKGSSGQAALPQFVQILEPKKGQNLS 517
Query: 299 ILLRALNVTMEEVCDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRF 358
ILLRALN T EEVCDAL EGNELP EF+QTLLKMAPT +EELKLRL+ G+++QLG A+RF
Sbjct: 518 ILLRALNATTEEVCDALREGNELPVEFIQTLLKMAPTPEEELKLRLYCGEIAQLGSAERF 577
Query: 359 LKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKT 418
LKA+VDIP AFKR+E LLFMCT EE+ ESF LEVACKELR SRLF KLLEAVLKT
Sbjct: 578 LKAVVDIPFAFKRLEALLFMCTLHEEMAFVKESFQKLEVACKELRGSRLFLKLLEAVLKT 637
Query: 419 GNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQ 478
GNRMNDGT+RGGAQAFKLDTLLKL+DVKGTDGKTTLLHFVVQEIIR+EG++AAR ++SQ
Sbjct: 638 GNRMNDGTFRGGAQAFKLDTLLKLADVKGTDGKTTLLHFVVQEIIRTEGVRAARTIRESQ 697
Query: 479 SLSNIKTDEL------HETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLG 532
S S++KT++L E+E++YR LGLE VS LS+ELE+VK+ + +DAD LT T +K+G
Sbjct: 698 SFSSVKTEDLLVEETSEESEENYRNLGLEKVSGLSSELEHVKKSANIDADGLTGTVLKMG 757
Query: 533 HGLVKAKDILNKNLKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFH 592
H L KA+D +N +K+ ++ GFRE +E F++NAE + +LE+EK+IMALVKSTGDYFH
Sbjct: 758 HALSKARDFVNSEMKSSGEESGFREALEDFIQNAEGSIMSILEEEKRIMALVKSTGDYFH 817
Query: 593 GNATKDDGLRLFVVVRDFLIMLDKVCKEVRDAQKKPAKPIKQETSRGLSSSDT-RPSPS- 650
G A KD+GLRLFV+VRDFLI+LDK CKEVR+A+ +P + +++ S +SS+T R +PS
Sbjct: 818 GKAGKDEGLRLFVIVRDFLIILDKSCKEVREARGRPVRMARKQGSTASASSETPRQTPSL 877
Query: 651 DFRQRLFPAIAERRIDDDSSDEE 673
D RQ+LFPAI ERR+D SSD +
Sbjct: 878 DPRQKLFPAITERRVDQSSSDSD 900
Score = 48.1 bits (113), Expect = 8e-04
Identities = 25/60 (41%), Positives = 27/60 (44%), Gaps = 7/60 (11%)
Query: 136 PPAPLGAKPGPPPPPPPA-PPSAKPGPPPPP------PPAPSGAKPGPRPPPPPPKSGVA 188
P L KPG P P P+ PP GPP PP PP S P PPPPP K +
Sbjct: 134 PRRNLATKPGSSPSPSPSRPPKRSRGPPRPPTRPKSPPPRKSSFPPSRSPPPPPAKKNAS 193
Score = 47.0 bits (110), Expect = 0.002
Identities = 24/62 (38%), Positives = 27/62 (42%), Gaps = 5/62 (8%)
Query: 127 PGAKPSPPPPPAPLGAKPGPPPP-----PPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPP 181
PG+ PSP P P ++ P PP PPP S P PPPPPA A P
Sbjct: 142 PGSSPSPSPSRPPKRSRGPPRPPTRPKSPPPRKSSFPPSRSPPPPPAKKNASKNSTSAPV 201
Query: 182 PP 183
P
Sbjct: 202 SP 203
Score = 40.0 bits (92), Expect = 0.23
Identities = 23/73 (31%), Positives = 30/73 (40%), Gaps = 7/73 (9%)
Query: 96 SARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPP 155
+ +P ++PSP S PPRPP +P PPP PPPPP
Sbjct: 139 ATKPGSSPSPSPSRPPKRSRGPPRPP-------TRPKSPPPRKSSFPPSRSPPPPPAKKN 191
Query: 156 SAKPGPPPPPPPA 168
++K P PA
Sbjct: 192 ASKNSTSAPVSPA 204
>UniRef100_Q94B77 Hypothetical protein MRB17.15 [Arabidopsis thaliana]
Length = 900
Score = 687 bits (1774), Expect = 0.0
Identities = 377/683 (55%), Positives = 462/683 (67%), Gaps = 53/683 (7%)
Query: 2 NDYSFGSSSNNLGDNKKTSLQGNQSMGAFAVVGSPFELNPPPGRVGTIHSGMPPLRPPPG 61
+DYS GSS N G + K QG+QS ++ G + G+ L
Sbjct: 260 SDYSVGSSIN-YGGSVKGDKQGHQSFNIYSNQGKMSSFD------GSNSDTSDSLE---- 308
Query: 62 RMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPP 121
L HE N + PPP R + V S + S +P+PP PP
Sbjct: 309 --ERLSHEGLRNNSITNHGLPPLKPPPGRTASVLSGK---------SFSGKVEPLPPEPP 357
Query: 122 LHPALPGAKPSPPPPPAPLGAKP---GPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRP 178
+ K S PPPP P P GPP PPPPAPP GP PPPPP P G +P
Sbjct: 358 KFLKVSSKKASAPPPPVPAPQMPSSAGPPRPPPPAPPPGSGGPKPPPPPGPKGPRP---- 413
Query: 179 PPPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSM 238
PPP G PRPP GP A+A + D K KLKPFFWDKV AN + SM
Sbjct: 414 -PPPMSLGPKAPRPPSGP----------ADALDD---DAPKTKLKPFFWDKVQANPEHSM 459
Query: 239 VWNQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLL 298
VWN I+SGSFQFNEEMIE+LFGY A +KN + K+ SS Q PQ++QI++ KK QNL
Sbjct: 460 VWNDIRSGSFQFNEEMIESLFGYAAADKN--KNDKKGSSGQAALPQFVQILEPKKGQNLS 517
Query: 299 ILLRALNVTMEEVCDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRF 358
ILLRALN T EEVCDAL EGNELP EF+QTLLKMAPT +EELKLRL+ G+++QLG A+RF
Sbjct: 518 ILLRALNATTEEVCDALREGNELPVEFIQTLLKMAPTPEEELKLRLYCGEIAQLGSAERF 577
Query: 359 LKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKT 418
LKA+VDIP AFKR+E LLFMCT EE+ ESF LEVACKELR SRLF KLLEAVLKT
Sbjct: 578 LKAVVDIPFAFKRLEALLFMCTLHEEMAFVKESFQKLEVACKELRGSRLFLKLLEAVLKT 637
Query: 419 GNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQ 478
GNRMNDGT+RGGAQAFKLDTLLKL+DVKGTDGKTTLLHFVVQEIIR+EG++AAR ++SQ
Sbjct: 638 GNRMNDGTFRGGAQAFKLDTLLKLADVKGTDGKTTLLHFVVQEIIRTEGVRAARTIRESQ 697
Query: 479 SLSNIKTDEL------HETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLG 532
S S++KT++L E+E++YR LGLE VS LS+ELE+VK+ + +DAD LT T +K+G
Sbjct: 698 SFSSVKTEDLLVEETSEESEENYRNLGLEKVSGLSSELEHVKKSANIDADGLTGTVLKMG 757
Query: 533 HGLVKAKDILNKNLKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFH 592
H L KA+D +N +K+ + GFRE +E F++NAE + +LE+EK+IMALVKSTGDYFH
Sbjct: 758 HALSKARDFVNSEMKSSGEGSGFREALEDFIQNAEGSIMSILEEEKRIMALVKSTGDYFH 817
Query: 593 GNATKDDGLRLFVVVRDFLIMLDKVCKEVRDAQKKPAKPIKQETSRGLSSSDT-RPSPS- 650
G A KD+GLRLFV+VRDFLI+LDK CKEVR+A+ +P + +++ S +SS+T R +PS
Sbjct: 818 GKAGKDEGLRLFVIVRDFLIILDKSCKEVREARGRPVRMARKQGSTASASSETPRQTPSL 877
Query: 651 DFRQRLFPAIAERRIDDDSSDEE 673
D RQ+LFPAI ERR+D SSD +
Sbjct: 878 DPRQKLFPAITERRVDQSSSDSD 900
Score = 48.1 bits (113), Expect = 8e-04
Identities = 25/60 (41%), Positives = 27/60 (44%), Gaps = 7/60 (11%)
Query: 136 PPAPLGAKPGPPPPPPPA-PPSAKPGPPPPP------PPAPSGAKPGPRPPPPPPKSGVA 188
P L KPG P P P+ PP GPP PP PP S P PPPPP K +
Sbjct: 134 PRRNLATKPGSSPSPSPSRPPKRSRGPPRPPTRPKSPPPRKSSFPPSRSPPPPPAKKNAS 193
Score = 47.0 bits (110), Expect = 0.002
Identities = 24/62 (38%), Positives = 27/62 (42%), Gaps = 5/62 (8%)
Query: 127 PGAKPSPPPPPAPLGAKPGPPPP-----PPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPP 181
PG+ PSP P P ++ P PP PPP S P PPPPPA A P
Sbjct: 142 PGSSPSPSPSRPPKRSRGPPRPPTRPKSPPPRKSSFPPSRSPPPPPAKKNASKNSTSAPV 201
Query: 182 PP 183
P
Sbjct: 202 SP 203
Score = 40.0 bits (92), Expect = 0.23
Identities = 23/73 (31%), Positives = 30/73 (40%), Gaps = 7/73 (9%)
Query: 96 SARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPP 155
+ +P ++PSP S PPRPP +P PPP PPPPP
Sbjct: 139 ATKPGSSPSPSPSRPPKRSRGPPRPP-------TRPKSPPPRKSSFPPSRSPPPPPAKKN 191
Query: 156 SAKPGPPPPPPPA 168
++K P PA
Sbjct: 192 ASKNSTSAPVSPA 204
>UniRef100_Q6MWG9 B1160F02.7 protein [Oryza sativa]
Length = 906
Score = 575 bits (1483), Expect = e-162
Identities = 328/713 (46%), Positives = 430/713 (60%), Gaps = 69/713 (9%)
Query: 1 MNDYSFGSSSNNLGDNKKTSLQGNQSMGAFAVVGSPFELNPPPGRVGTIHSGMPPLRPPP 60
MN S S + + ++ T++ G G S + V + G P PPP
Sbjct: 222 MNKVSMQSQAMRMSSHEITTIAG---AGRVENKVSTIAPSAAAAAVASAGGGQVPAAPPP 278
Query: 61 GRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRP 120
P P PP PP PP P PP + PP P
Sbjct: 279 PAGPPPPAPPPL--------------PPSHHHHHGHHPPPPHPLPPGAGAGAGTGAPPPP 324
Query: 121 PLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAK--PGPPPPPPPAPSGAKPGPRP 178
P HPA P A P P P P+ GA GPPPPPPPA P+A PGP P PPP P A G
Sbjct: 325 PAHPAAP-APPPPAPSPSAAGAGSGPPPPPPPAAPAAPRPPGPGPGPPPPPGAAGRGGGG 383
Query: 179 PPPPPKSGVAPPRPPIGPKAGGPKATENAE--AGAEGGADTSKAKLKPFFWDKVPANSDQ 236
PPPP G GP+A GP + + A A AD +KAKLKPFFWDKV AN +Q
Sbjct: 384 PPPPALPG--------GPRARGPPPFKKSPGAAAAAAQADPNKAKLKPFFWDKVTANPNQ 435
Query: 237 SMVWNQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQN 296
+MVW+QIK+GSFQFNEEMIE+LFG + K + +KES + Q+++I+D KKAQN
Sbjct: 436 AMVWDQIKAGSFQFNEEMIESLFGAQSTEKKSTDAKKESGKE---ATQFVRILDPKKAQN 492
Query: 297 LLILLRALNVTMEEVCDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPAD 356
L I L+AL+V+ E+V A+ EG++LP + +QTL++ +PTSDEEL+LRL+ G+ +QLGPA+
Sbjct: 493 LAISLKALSVSAEQVRAAVMEGHDLPPDLIQTLVRWSPTSDEELRLRLYAGEPAQLGPAE 552
Query: 357 RFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVL 416
+F++A++D+P ++R++ LLFM EE +SFA LEVAC+ELR SRLF KLLEAVL
Sbjct: 553 QFMRAIIDVPYLYQRLDALLFMAALPEEAAAVEQSFATLEVACEELRGSRLFKKLLEAVL 612
Query: 417 KTGNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAK- 475
KTGNRMNDGT+RGGAQAFKLDTLLKL+DVKG DGKTTLLHFVVQEIIRSEG++AARAA
Sbjct: 613 KTGNRMNDGTFRGGAQAFKLDTLLKLADVKGVDGKTTLLHFVVQEIIRSEGVRAARAASG 672
Query: 476 -------------------DSQS-------LSNIKTDELHETED---HYRELGLEMVSHL 506
SQS S++ L + +D YR+LGL +VS L
Sbjct: 673 GGGGSSISSISSSDDLILLQSQSSIGSNSGRSSVDASSLEQEQDETERYRQLGLGVVSSL 732
Query: 507 STELENVKRGSVLDADSLTATTIKLGHGLVKAKDILNKNLKNVEDDRGFRETVESFVKNA 566
+L+NV++ + DAD+LT T LGH LVKA + L+ ++++E+D GF+ + SFV+ +
Sbjct: 733 GDDLQNVRKAASFDADALTITVASLGHRLVKANEFLSTGMRSLEEDSGFQRRLASFVQQS 792
Query: 567 EADVKKLLEDEKKIMALVKSTGDYFHGNATKDDGLRLFVVVRDFLIMLDKVCKEVRDAQK 626
+ V +LLEDEK++ +LV++T DYFHG+ KD+GLRLFVVVRDFL +LDKVC+EV++
Sbjct: 793 QEQVTRLLEDEKRLRSLVRATVDYFHGSTGKDEGLRLFVVVRDFLGILDKVCREVKEQAA 852
Query: 627 KPAKPIKQE------TSRGLSSSDTRPSPSDFRQRLFPAIAERRIDDDSSDEE 673
AK KQ+ SR S S R + R A++ SSD +
Sbjct: 853 ANAKAKKQQQPTPAPRSRQSSQSSFRDPRQQIQDRRAAALSRNNSSSSSSDSD 905
>UniRef100_Q6H7U3 Putative formin I2I isoform [Oryza sativa]
Length = 881
Score = 531 bits (1367), Expect = e-149
Identities = 285/553 (51%), Positives = 382/553 (68%), Gaps = 31/553 (5%)
Query: 130 KPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAP 189
K PPPPP PPPPPPP PP P PPPPPPP GA PPP PPK+ +A
Sbjct: 350 KLMPPPPP--------PPPPPPPPPPPPPPRPPPPPPPIKKGA-----PPPAPPKATMAR 396
Query: 190 -PR--PPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSG 246
P+ P ++ A+E A +E + +AKL+PF+WDKV AN DQSM W+ IK G
Sbjct: 397 FPKLSPTESSRSEESSASELASESSETEVNAPRAKLRPFYWDKVLANPDQSMAWHDIKFG 456
Query: 247 SFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNV 306
SF NEEMIE LFGY A N+NN + KE S + DPSPQ++ ++D KK+ NL ++ +A+NV
Sbjct: 457 SFHVNEEMIEELFGYGAGNQNN-VKDKEISIA-DPSPQHVSLLDVKKSCNLAVVFKAMNV 514
Query: 307 TMEEVCDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRFLKAMVDIP 366
EE+ DAL EGNELP L+T+L+M PT +EE KLRL+NGD SQLG A++ +KA++DIP
Sbjct: 515 RAEEIHDALVEGNELPRLLLETILRMKPTDEEEQKLRLYNGDCSQLGLAEQVMKALIDIP 574
Query: 367 SAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGT 426
AF+R+ LLFM + +E+ ++ ESF LE AC EL++ RLF KLLEA+LKTGNR+NDGT
Sbjct: 575 FAFERIRALLFMSSLQEDASSLRESFLQLEAACGELKH-RLFLKLLEAILKTGNRLNDGT 633
Query: 427 YRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDS------QSL 480
+RGGA AFKLDTLLKLSDVKG DGKTTLLHFVVQEIIRSEG++ AR A ++ S
Sbjct: 634 FRGGANAFKLDTLLKLSDVKGADGKTTLLHFVVQEIIRSEGVREARLAMENGRSPPFPST 693
Query: 481 SNIKTDE-LHETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAK 539
S+ ++E L E ++Y LGL++VS LS EL+NVKR + LDAD+L+ + L H L++AK
Sbjct: 694 SDDNSNESLQEDGNYYSNLGLKIVSGLSNELDNVKRVAALDADALSTSVANLRHELLRAK 753
Query: 540 DILNKNLKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDD 599
+ LN ++ ++E++ GF ++ESF+++AE + LL+++K++ LVK T YFHGN KDD
Sbjct: 754 EFLNSDMASLEENSGFHRSLESFIEHAETETNFLLKEDKRLRMLVKRTIRYFHGNDEKDD 813
Query: 600 GLRLFVVVRDFLIMLDKVCKEVRDAQKKPAKPIKQETSRGLSSSDTRPSPSDFRQRLFPA 659
G RLFV+VRDFL+MLDK CKEV +QKK S+ +S+ S S+ +++ FPA
Sbjct: 814 GFRLFVIVRDFLVMLDKACKEVGASQKKAT-----NKSQANGNSNNPSSQSNPQEQQFPA 868
Query: 660 IAERRIDDDSSDE 672
+ + D S++
Sbjct: 869 VLDHHFDSSDSND 881
>UniRef100_Q8S0F0 Putative formin-like protein AHF1 [Oryza sativa]
Length = 960
Score = 448 bits (1152), Expect = e-124
Identities = 290/676 (42%), Positives = 384/676 (55%), Gaps = 83/676 (12%)
Query: 7 GSSSNNLGDNKKTSLQGNQSMGAF----------AVVGSPFELNPPPGRVGTIHSGMPPL 56
G +S + GD + S QG+ M AV +P PG + + P
Sbjct: 257 GFASPSSGDEEFYSPQGSSKMSTSHRTLAAAVEAAVAARDRSKSPSPGSIVST----PSY 312
Query: 57 RPPPGR-MNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVAS-ARPPATPSPPQSLLAGAK 114
PG M+P P PP F+ G S R V + +PPA P PP A
Sbjct: 313 PSSPGATMSPAPASPPLFSSPGQ---SGRRSVKSRSDSVRTFGQPPAPPPPPP--FAPTL 367
Query: 115 PVPPRPPLHPALPGAKPSPPPPPAPL-----------------GAKPGPPPPPPPAPPSA 157
P PP P P PSP PP +PL P PPPPP
Sbjct: 368 PPPPPPRRKP------PSPSPPSSPLIENTSALRSTTTTDTTIPRNPFVQPPPPPTHTHG 421
Query: 158 KPGPPPPPPPAPSG------AKPGPRPPPPPPKSGVAPPRPPIGPKAGGP---------- 201
P PPPPPPP P G KPG ++PP K+G P
Sbjct: 422 PPPPPPPPPPPPVGYWESRVRKPGTGTSKETRSPALSPPPQAASFKSGLPTDAFPGRLAD 481
Query: 202 KATENAEAGAEGGAD-----TSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIE 256
A A A A GG D T + KLKP WDKV A+SD+ MVW+Q+KS SFQ NEEMIE
Sbjct: 482 NADHAAAAAAGGGGDKSEETTPRPKLKPLHWDKVRASSDRVMVWDQLKSSSFQVNEEMIE 541
Query: 257 TLFGYNAVNKNNGQRQKESSSSQD--PSPQYI-QIVDKKKAQNLLILLRALNVTMEEVCD 313
TLF N N KE ++ + P+P+ +++D KK+QN+ ILLRALNV+ E+VCD
Sbjct: 542 TLF---ICNPANSAPPKEPATRRPVLPTPKTDNKVLDPKKSQNIAILLRALNVSKEQVCD 598
Query: 314 ALYEGN--ELPSEFLQTLLKMAPTSDEELKLRLFNGDLS--QLGPADRFLKAMVDIPSAF 369
AL EGN +E L+TLLKMAPT +EE+KLR F + S +LGPA++FLKA++DIP AF
Sbjct: 599 ALCEGNTENFGAELLETLLKMAPTKEEEIKLREFKEETSPIKLGPAEKFLKAVLDIPFAF 658
Query: 370 KRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRG 429
KR++ +L++ F+ E+ +SF LE AC ELRNSRLF KLLEAVLKTGNRMN GT RG
Sbjct: 659 KRVDAMLYIANFESEVNYLKKSFETLETACDELRNSRLFLKLLEAVLKTGNRMNVGTNRG 718
Query: 430 GAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSLSNIKTDELH 489
A AFKLDTLLKL DVKGTDGKTTLLHFVVQEIIR+EG + + +QS + + L
Sbjct: 719 DAHAFKLDTLLKLVDVKGTDGKTTLLHFVVQEIIRTEG---SHLSASNQSTPRTQANPLR 775
Query: 490 ETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDI--LNKNLK 547
+ E ++LGL++V+ L EL NVK+ + +D+D L++ KL G+ K ++ LN+ +K
Sbjct: 776 D-ELECKKLGLQVVAGLGNELSNVKKAAAMDSDVLSSYVSKLAGGIEKITEVLRLNEEVK 834
Query: 548 NVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDG--LRLFV 605
+ ED F ++++ F+K A+ D+ ++ E ++LVK +YFHG++ K++ R+F+
Sbjct: 835 SREDAWRFHDSMQKFLKRADDDIIRVQAQESVALSLVKEITEYFHGDSAKEEAHPFRIFM 894
Query: 606 VVRDFLIMLDKVCKEV 621
VVRDFL +LD+VCKEV
Sbjct: 895 VVRDFLSVLDQVCKEV 910
Score = 50.8 bits (120), Expect = 1e-04
Identities = 38/110 (34%), Positives = 45/110 (40%), Gaps = 21/110 (19%)
Query: 112 GAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSG 171
G + R LH + S PP PAP PGP PP PA P PPPPPA +G
Sbjct: 26 GLEVAVARRQLHQPFFPDQSSSPPTPAP----PGPAPPFFPALPV------PPPPPATAG 75
Query: 172 AKPGPRPPPPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSKAK 221
+ P P +G AGG AT + G GG K+K
Sbjct: 76 QEQPTYPALVLPNTG-----------AGGAAATAAPDGGGGGGGGARKSK 114
Score = 35.8 bits (81), Expect = 4.3
Identities = 26/76 (34%), Positives = 29/76 (37%), Gaps = 15/76 (19%)
Query: 100 PATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKP 159
P TP+PP P PP PALP PPPPPA G + P P P +
Sbjct: 48 PPTPAPPG----------PAPPFFPALP----VPPPPPATAGQEQ-PTYPALVLPNTGAG 92
Query: 160 GPPPPPPPAPSGAKPG 175
G P G G
Sbjct: 93 GAAATAAPDGGGGGGG 108
>UniRef100_Q8H8K7 Putative formin-like protein [Oryza sativa]
Length = 849
Score = 426 bits (1094), Expect = e-117
Identities = 269/574 (46%), Positives = 348/574 (59%), Gaps = 58/574 (10%)
Query: 71 PSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAK 130
PS T AAAP P + A+A PPA P+P ++ PPR A GA+
Sbjct: 266 PSLTSDFFPTTPAAAPVP---APAAAAPPPAPPAPR------SRRTPPRTRFS-AGSGAE 315
Query: 131 -----PSPP--PPPAPLGAKPGPPPPPPPAP---PSAKPGPPPPPPPAPSGAKPGPRPPP 180
SPP PPPAP PPPPPPP+ + KP PPPPPP P+G R
Sbjct: 316 MNKQMASPPSNPPPAP------PPPPPPPSRFNNTTPKPPPPPPPPEPPTGPVSARRLLR 369
Query: 181 PPPKSG--VAPPRPP---IGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSD 235
P P G + PR P + AT + E D + KLKP WDKV +SD
Sbjct: 370 PLPAEGPSIVIPRAPAMAVTKDNDATAATMSVRTRGEAAGDEPRPKLKPLHWDKVRTSSD 429
Query: 236 QSMVWNQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQ---IVDKK 292
+ MVW+++K +E+MIE LF +N + + + + PQ+ Q ++D K
Sbjct: 430 RDMVWDRLK-----LDEDMIEVLF----MNNSTAVAPRMDNPKKVGMPQFKQEERVLDPK 480
Query: 293 KAQNLLILLRALNVTMEEVCDALYEGNE--LPSEFLQTLLKMAPTSDEELKLRLFNGDLS 350
KAQN+ ILLRALNVT+EEV DAL +GN L +E L+TL+KMAPT +EELKLR F GDLS
Sbjct: 481 KAQNIAILLRALNVTLEEVTDALLDGNAECLGAELLETLVKMAPTKEEELKLRDFTGDLS 540
Query: 351 QLGPADRFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHK 410
+LG A+RFLKA++DIP AFKR++V+L+ F+ E+ +SF LE AC +L+ SRLF K
Sbjct: 541 KLGSAERFLKAVLDIPFAFKRVDVMLYRANFENEVNYLRKSFQTLEAACDDLKGSRLFLK 600
Query: 411 LLEAVLKTGNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKA 470
LLEAVL+TGNRMN GT RG A+AFKLDTLLKL+DVKG DGKTTLLHFVVQEI+RSE K+
Sbjct: 601 LLEAVLRTGNRMNVGTNRGEAKAFKLDTLLKLADVKGADGKTTLLHFVVQEIVRSEDAKS 660
Query: 471 ARAAKDSQSLSNI-KTDELHETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTI 529
+A ++ ++NI K ++L R GL++VS LSTEL NVKR + +D D L
Sbjct: 661 EKAPEN--HITNIAKVEQL-------RRQGLKVVSGLSTELGNVKRAATMDFDVLHGYVS 711
Query: 530 KLGHGLVKAKDILNKNLKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGD 589
KL GL K K +L K F T+ F+K AE +++++ DEK + VK +
Sbjct: 712 KLEAGLGKIKSVLQLE-KQCSQGVNFFATMREFLKEAEQEIEQVRHDEKAALGRVKEITE 770
Query: 590 YFHGNATKDDG--LRLFVVVRDFLIMLDKVCKEV 621
YFHGNA K++ LR+F+VVRDFL MLD VC+EV
Sbjct: 771 YFHGNAVKEEAHPLRIFMVVRDFLSMLDHVCREV 804
Score = 34.7 bits (78), Expect = 9.5
Identities = 32/104 (30%), Positives = 41/104 (38%), Gaps = 20/104 (19%)
Query: 119 RPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRP 178
R LH L + +PPP A P PP+P + P P P +G GP
Sbjct: 26 RRVLHEPLFPIEWTPPPSTAS---------PSPPSPDFSS-DPSTPATPVDNG---GPAL 72
Query: 179 PPPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSKAKL 222
PPPP + VA + GP + A GG T KA +
Sbjct: 73 LPPPPPNTVA---ADVSSSRSGP----DPRARGGGGGGTPKAAI 109
>UniRef100_Q9FJX6 Formin-like protein AtFH6 [Arabidopsis thaliana]
Length = 899
Score = 407 bits (1047), Expect = e-112
Identities = 267/657 (40%), Positives = 363/657 (54%), Gaps = 77/657 (11%)
Query: 59 PPGRMNPLPHEPPSF--TPFGNTAVSAAAPPPPRQS----GVASARPPATPSPPQSLLAG 112
P LPH + + FG+ +AA+ P + + PP PP L
Sbjct: 271 PRSANGSLPHSKRTSPRSKFGSAPTTAASRSPEMKHVIIPSIKQKLPPPVQPPPLRGLES 330
Query: 113 AKPVPPRPPLHPALPGAKPSPPPPPAPLGA---KPGPPPPPPPAPPSAKPGPPPPPPPAP 169
+ P P ++P PPP A A + P PPP +PP + PPPPPPP
Sbjct: 331 DEQELPYSQNKPKF--SQPPPPPNRAAFQAITQEKSPVPPPRRSPPPLQTPPPPPPPP-- 386
Query: 170 SGAKPGPRPPPPPPK--------------------SGVAPPRPPI----GPKAGGPKATE 205
P PPPPP+ S +P R PK +
Sbjct: 387 ------PLAPPPPPQKRPRDFQMLRKVTNSEATTNSTTSPSRKQAFKTPSPKTKAVEEVN 440
Query: 206 NAEAGA-----EGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIETLFG 260
+ AG+ +G D SK KLKP WDKV A+SD++ VW+Q+KS SFQ NE+ +E LFG
Sbjct: 441 SVSAGSLEKSGDGDTDPSKPKLKPLHWDKVRASSDRATVWDQLKSSSFQLNEDRMEHLFG 500
Query: 261 YNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGN- 319
N+ ++ ++ S + +++D KK+QN+ ILLRALNVT EEV +AL +GN
Sbjct: 501 CNS--GSSAPKEPVRRSVIPLAENENRVLDPKKSQNIAILLRALNVTREEVSEALTDGNP 558
Query: 320 -ELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRFLKAMVDIPSAFKRMEVLLFM 378
L +E L+TL+KMAPT +EE+KLR ++GD+S+LG A+RFLK ++DIP AFKR+E +L+
Sbjct: 559 ESLGAELLETLVKMAPTKEEEIKLREYSGDVSKLGTAERFLKTILDIPFAFKRVEAMLYR 618
Query: 379 CTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFKLDT 438
F E+ SF LE A EL+ SRLF KLLEAVL TGNRMN GT RG A AFKLDT
Sbjct: 619 ANFDAEVKYLRNSFQTLEEASLELKASRLFLKLLEAVLMTGNRMNVGTNRGDAIAFKLDT 678
Query: 439 LLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSLSNIKTDELHETEDHYREL 498
LLKL D+KG DGKTTLLHFVVQEI RSEG KD LH D +R+
Sbjct: 679 LLKLVDIKGVDGKTTLLHFVVQEITRSEG---TTTTKDETI--------LHGNNDGFRKQ 727
Query: 499 GLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDILNKNLKNVEDDRGFRET 558
GL++V+ LS +L NVK+ + +D D L++ KL GL D L LK F ++
Sbjct: 728 GLQVVAGLSRDLVNVKKSAGMDFDVLSSYVTKLEMGL----DKLRSFLKTETTQGRFFDS 783
Query: 559 VESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDG--LRLFVVVRDFLIMLDK 616
+++F+K AE +++K+ E+K +++VK +YFHGNA +++ LR+F+VVRDFL +LD
Sbjct: 784 MKTFLKEAEEEIRKIKGGERKALSMVKEVTEYFHGNAAREEAHPLRIFMVVRDFLGVLDN 843
Query: 617 VCKEVRDAQKKPAKPIKQETSRGLSSSDTRPSPSDFRQRLFPAIAERRIDDDSSDEE 673
VCKEV+ Q+ + + ++R S T P R + R DD SSD E
Sbjct: 844 VCKEVKTMQEM-STSMGSASARSFRISATASLPVLHRYK-------ARQDDTSSDSE 892
Score = 39.3 bits (90), Expect = 0.39
Identities = 23/65 (35%), Positives = 28/65 (42%), Gaps = 5/65 (7%)
Query: 101 ATPSPPQSLLAGAKPVPPRP-----PLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPP 155
+TP PP + P+P P P +P+ P PPPPP G P P
Sbjct: 40 STPPPPDFQSTPSPPLPDTPDQPFFPENPSTPQQTLFPPPPPPVSADVNGGLPIPTATTQ 99
Query: 156 SAKPG 160
SAKPG
Sbjct: 100 SAKPG 104
Score = 34.7 bits (78), Expect = 9.5
Identities = 20/51 (39%), Positives = 23/51 (44%), Gaps = 4/51 (7%)
Query: 139 PLGAKPGPPPPP----PPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKS 185
PL + PPPP P+PP P P PS + PPPPPP S
Sbjct: 34 PLFPESSTPPPPDFQSTPSPPLPDTPDQPFFPENPSTPQQTLFPPPPPPVS 84
>UniRef100_Q9MA60 F22F7.8 protein [Arabidopsis thaliana]
Length = 884
Score = 403 bits (1036), Expect = e-111
Identities = 231/530 (43%), Positives = 317/530 (59%), Gaps = 72/530 (13%)
Query: 140 LGAKPGPPPPPPPAP------------------------------------------PSA 157
+ A P PPPPPPP P P++
Sbjct: 365 VSAPPPPPPPPPPLPQFSNKRIHTLSSPETANLQTLSSQLCEKLCASSSKTSFPINVPNS 424
Query: 158 KPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADT 217
+P PPPPPPP G PPPP S R P+G K G P
Sbjct: 425 QPRPPPPPPPPQQLQVAGINKTPPPPLSLDFSERRPLG-KDGAP---------------- 467
Query: 218 SKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSS 277
KLKP WDKV A D++MVW+++++ SF+ +EEMIE+LFGY + E
Sbjct: 468 -LPKLKPLHWDKVRATPDRTMVWDKLRTSSFELDEEMIESLFGYTM----QSSTKNEEGK 522
Query: 278 SQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGNELPSEFLQTLLKMAPTSD 337
S+ PSP +++ K+ QN ILL+ALN T +++C AL +G L + L+ L+KM PT +
Sbjct: 523 SKTPSPGK-HLLEPKRLQNFTILLKALNATADQICSALGKGEGLCLQQLEALVKMVPTKE 581
Query: 338 EELKLRLFNGDLSQLGPADRFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEV 397
EELKLR + G + +LG A++FL+A+V +P AF+R E +L+ TF++E+ SF++LE
Sbjct: 582 EELKLRSYKGAVDELGSAEKFLRALVGVPFAFQRAEAMLYRETFEDEVVHLRNSFSMLEE 641
Query: 398 ACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHF 457
ACKEL++SRLF KLLEAVLKTGNRMN GT RGGA+AFKLD LLKLSDVKGTDGKTTLLHF
Sbjct: 642 ACKELKSSRLFLKLLEAVLKTGNRMNVGTIRGGAKAFKLDALLKLSDVKGTDGKTTLLHF 701
Query: 458 VVQEIIRSEGIKAARAAKD---SQSLSNIKTDELHETEDHYRELGLEMVSHLSTELENVK 514
VVQEI RSEGI+ + + +Q + +T E E E+ YR +GL++VS L+TEL NVK
Sbjct: 702 VVQEISRSEGIRVSDSIMGRIMNQRSNKNRTPE--EKEEDYRRMGLDLVSGLNTELRNVK 759
Query: 515 RGSVLDADSLTATTIKLGHGLVKAKDILNKNLKNVEDDRGFRETVESFVKNAEADVKKLL 574
+ + +D + L + L GL + + ++ LK E++R F ++ SF++ E +++L
Sbjct: 760 KTATIDLEGLVTSVSNLRDGLGQLSCLASEKLKGDEENRAFVSSMSSFLRYGEKSLEELR 819
Query: 575 EDEKKIMALVKSTGDYFHGNATKDD--GLRLFVVVRDFLIMLDKVCKEVR 622
EDEK+IM V +YFHG+ D+ LR+FV+VRDFL MLD VC+E+R
Sbjct: 820 EDEKRIMERVGEIAEYFHGDVRGDEKNPLRIFVIVRDFLGMLDHVCRELR 869
>UniRef100_Q69MT2 Diaphanous protein-like [Oryza sativa]
Length = 788
Score = 400 bits (1028), Expect = e-110
Identities = 245/582 (42%), Positives = 340/582 (58%), Gaps = 61/582 (10%)
Query: 77 GNTAVSAAA---------PPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALP 127
G+TA +A++ PP PRQ P+T A K PP + P
Sbjct: 208 GSTATAASSASSPELRPMPPLPRQFQQTRTSMPSTSQTIHEAGAEDKRAPPPQSVRP--- 264
Query: 128 GAKPSPPPPPAPLGAKPGPPPPPPPAPP-------SAKPGPPPPPPPAPSGAKPGPR--- 177
PPPPP PPPPPPP PP A P PPPP P A +G+ PR
Sbjct: 265 -----PPPPP--------PPPPPPPMPPRTDNASTQAAPAPPPPLPRAGNGSGWLPRRYT 311
Query: 178 --PPPPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKV-PANS 234
P ++ P P P+ E A+ A + KLKP WDKV PA+S
Sbjct: 312 ERAAPTVIRASAGAVHPEESPARASPE-----EKAADAAA---RPKLKPLHWDKVRPASS 363
Query: 235 DQSMVWNQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKA 294
+ VW+Q+K+ SF+ NEEMIETLF N+ + + KE++++ Q +++D KK+
Sbjct: 364 GRPTVWDQLKASSFRVNEEMIETLFVSNSTRRASKNGVKEANAAC--CNQENKVLDPKKS 421
Query: 295 QNLLILLRALNVTMEEVCDALYEGN--ELPSEFLQTLLKMAPTSDEELKLRLFNGD-LSQ 351
QN+ I+LRAL+ T EEVC AL +G L +E L+TLLKMAP+ +EE+KL+ F D +S+
Sbjct: 422 QNIAIMLRALDATKEEVCKALLDGQAESLGTELLETLLKMAPSREEEIKLKEFREDAVSK 481
Query: 352 LGPADRFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKL 411
LGPA+ FLKA++ IP AFKR+E +L++ F E+ SF LE AC+ELR SRLFHK+
Sbjct: 482 LGPAESFLKAVLAIPFAFKRVEAMLYIANFDSEVDYLKTSFKTLEAACEELRGSRLFHKI 541
Query: 412 LEAVLKTGNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAA 471
L+AVLKTGNRMN GT RG A AFKLD LLKL DVKG DGKTTLLHFV++EI++SEG
Sbjct: 542 LDAVLKTGNRMNTGTNRGNASAFKLDALLKLVDVKGADGKTTLLHFVIEEIVKSEGASIL 601
Query: 472 RAAKDSQSLSNIKTDELHETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKL 531
+ S S I D +++GL +V+ L EL NVK+ + +D+D+L + KL
Sbjct: 602 ATGQTSNQGSAIADD------FQCKKVGLRIVASLGGELGNVKKAAGMDSDTLASCVAKL 655
Query: 532 GHGLVKAKDILNKNLKNVEDD--RGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGD 589
G+ K + L N + DD + FR ++ F++ AEA++ + E ++LV+ T +
Sbjct: 656 SAGVSKISEALQLNQQLGSDDHCKRFRASIGEFLQKAEAEITAVQAQESLALSLVRETTE 715
Query: 590 YFHGNATKDDG--LRLFVVVRDFLIMLDKVCKEVRDAQKKPA 629
+FHG++ K++G LR+F+VVRDFL +LD VCK+V ++ A
Sbjct: 716 FFHGDSVKEEGHPLRIFMVVRDFLTVLDHVCKDVGRMNERTA 757
Score = 39.7 bits (91), Expect = 0.29
Identities = 24/48 (50%), Positives = 25/48 (52%), Gaps = 11/48 (22%)
Query: 124 PALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSG 171
PALP P PPPPPAP P P APP PPP PAP+G
Sbjct: 52 PALP---PPPPPPPAPFF-----PFLPDSAPPQL---PPPVTTPAPAG 88
Score = 38.9 bits (89), Expect = 0.50
Identities = 29/91 (31%), Positives = 34/91 (36%), Gaps = 14/91 (15%)
Query: 128 GAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGV 187
G++ P PL P PPPPPP P P P P P+ PPP V
Sbjct: 36 GSRRELHEPLFPLENAPALPPPPPPPPAPFFPFLPDSAP---------PQLPPP-----V 81
Query: 188 APPRPPIGPKAGGPKATENAEAGAEGGADTS 218
P P G GG A A A + +S
Sbjct: 82 TTPAPAGGAGDGGTDAGAAATGDASSSSSSS 112
>UniRef100_Q9XIE0 F23H11.22 protein [Arabidopsis thaliana]
Length = 929
Score = 398 bits (1023), Expect = e-109
Identities = 262/681 (38%), Positives = 363/681 (52%), Gaps = 53/681 (7%)
Query: 36 PFELNPPPGRVGTIHSGMPPLRPPP-----------GRMNPLPHEPPSFTPFGNTAVSAA 84
P PPPG+ ++ P PPP PLP P P N +S +
Sbjct: 260 PSSTQPPPGQYMAGNASFPSSTPPPPGQYMAGNAPFSSSTPLP---PGQYPAVNAQLSTS 316
Query: 85 APPPPRQSGVASA--RPPATPSPPQSL-----LAGAKPVPPRPPLHPALPGAKPSPPPPP 137
AP P G +A P +T + P SL + G + PL P +PP PP
Sbjct: 317 APSVPLPPGQYTAVNAPFSTSTQPVSLPPGQYMPGNAALSASTPLTPGQFTTANAPPAPP 376
Query: 138 APLGAKPGPPPPPP-----PAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRP 192
P PPPPPP P PP K GP PPPP P G K G PPPPPP S PP+P
Sbjct: 377 GPANQTSPPPPPPPSAAAPPPPPPPKKGPAAPPPPPPPGKK-GAGPPPPPPMSKKGPPKP 435
Query: 193 PIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNE 252
P PK GP + D ++ KLKP WDK+ ++ +SMVW++I GSF F+
Sbjct: 436 PGNPK--GPTKSGETSLAVGKTEDPTQPKLKPLHWDKMNPDASRSMVWHKIDGGSFNFDG 493
Query: 253 EMIETLFGYNA--VNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEE 310
+++E LFGY A +++N Q ++ S+ P Q I+D +K+QN I+L++L +T EE
Sbjct: 494 DLMEALFGYVARKPSESNSVPQNQTVSNSVPHNQ-TYILDPRKSQNKAIVLKSLGMTKEE 552
Query: 311 VCDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRFLKAMVD-IPSAF 369
+ D L EG++ S+ L+ L +APT +E+ ++ F+G+ L AD L ++ +PSAF
Sbjct: 553 IIDLLTEGHDAESDTLEKLAGIAPTPEEQTEIIDFDGEPMTLAYADSLLFHILKAVPSAF 612
Query: 370 KRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRG 429
R V+LF + E+ S LE AC ELR LF KLLEA+LK GNRMN GT RG
Sbjct: 613 NRFNVMLFKINYGSEVAQQKGSLLTLESACNELRARGLFMKLLEAILKAGNRMNAGTARG 672
Query: 430 GAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAA----RAAKDSQSLSNIKT 485
AQAF L L KLSDVK D KTTLLHFVV+E++RSEG +AA + D+ S N
Sbjct: 673 NAQAFNLTALRKLSDVKSVDAKTTLLHFVVEEVVRSEGKRAAMNKNMMSSDNGSGENADM 732
Query: 486 DELHETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDILNKN 545
E E + ++GL ++ LS+E NVK+ + +D DS ATT+ LG + + K +L+++
Sbjct: 733 SR-EEQEIEFIKMGLPIIGGLSSEFTNVKKAAGIDYDSFVATTLALGTRVKETKRLLDQS 791
Query: 546 LKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDGL-RLF 604
K ED G + SF ++AE ++K + E++ +IM LVK T +Y+ A K+ L +LF
Sbjct: 792 -KGKED--GCLTKLRSFFESAEEELKVITEEQLRIMELVKKTTNYYQAGALKERNLFQLF 848
Query: 605 VVVRDFLIMLDKVCKEVRDAQKKPAKPIKQETSRGLSSSDTR-PSPSDFRQR---LFPAI 660
V++RDFL M+D C E+ Q+K + T G SSS PS + QR FP +
Sbjct: 849 VIIRDFLGMVDNACSEIARNQRKQQQQRPATTVAGASSSPAETPSVAAAPQRNAVRFPIL 908
Query: 661 -------AERRIDDDSSDEES 674
+ R SD ES
Sbjct: 909 PPNFMSESSRYSSSSDSDSES 929
>UniRef100_Q9SE97 Formin-like protein AHF1 [Arabidopsis thaliana]
Length = 1051
Score = 395 bits (1015), Expect = e-108
Identities = 236/542 (43%), Positives = 325/542 (59%), Gaps = 42/542 (7%)
Query: 151 PPAPPSAKPGPPPPPPPAP---------SGAKPGPRPPP-PPPKSGVAPPRPPIGPKAGG 200
P S P PPPPPPP P + A RPP PP P + P
Sbjct: 518 PQILQSRVPPPPPPPPPLPLWGRRSQVTTKADTISRPPSLTPPSHPFVIPSENL-PVTSS 576
Query: 201 PKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIETLFG 260
P T +E +T K KLK WDKV A+SD+ MVW+ ++S SF+ +EEMIETLF
Sbjct: 577 PMETPETVCASEAAEETPKPKLKALHWDKVRASSDREMVWDHLRSSSFKLDEEMIETLFV 636
Query: 261 YNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGNE 320
++N Q Q Q +++D KKAQN+ ILLRALNVT+EEVC+AL EGN
Sbjct: 637 AKSLNNKPNQSQTTPRCVLPSPNQENRVLDPKKAQNIAILLRALNVTIEEVCEALLEGNA 696
Query: 321 --LPSEFLQTLLKMAPTSDEELKLRLFNGDLS-QLGPADRFLKAMVDIPSAFKRMEVLLF 377
L +E L++LLKMAPT +EE KL+ +N D +LG A++FLKAM+DIP AFKR++ +L+
Sbjct: 697 DTLGTELLESLLKMAPTKEEERKLKAYNDDSPVKLGHAEKFLKAMLDIPFAFKRVDAMLY 756
Query: 378 MCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFKLD 437
+ F+ E+ +SF LE AC+ELRNSR+F KLLEAVLKTGNRMN GT RG A AFKLD
Sbjct: 757 VANFESEVEYLKKSFETLEAACEELRNSRMFLKLLEAVLKTGNRMNVGTNRGDAHAFKLD 816
Query: 438 TLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSLSNIKTDELHETEDHYRE 497
TLLKL DVKG DGKTTLLHFVVQEIIR+EG + S +N +TD++ R+
Sbjct: 817 TLLKLVDVKGADGKTTLLHFVVQEIIRAEGTRL--------SGNNTQTDDI-----KCRK 863
Query: 498 LGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDIL--NKNLKNVEDDRGF 555
LGL++VS L +EL NVK+ + +D++ L++ KL G+ K + + + + + F
Sbjct: 864 LGLQVVSSLCSELSNVKKAAAMDSEVLSSYVSKLSQGIAKINEAIQVQSTITEESNSQRF 923
Query: 556 RETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDG--LRLFVVVRDFLIM 613
E++++F+K AE ++ ++ E ++LVK +YFHGN+ K++ R+F+VVRDFL +
Sbjct: 924 SESMKTFLKRAEEEIIRVQAQESVALSLVKEITEYFHGNSAKEEAHPFRIFLVVRDFLGV 983
Query: 614 LDKVCKEVRDAQKKPAKPIKQETSRGLSSSDTRPSP-SDFRQRLFPAIAERRIDDDSSDE 672
+D+VCKEV ++ +SS+ P P + + P + RR SS
Sbjct: 984 VDRVCKEVGMINERTM----------VSSAHKFPVPVNPMMPQPLPGLVGRRQSSSSSSS 1033
Query: 673 ES 674
S
Sbjct: 1034 SS 1035
Score = 37.4 bits (85), Expect = 1.5
Identities = 29/96 (30%), Positives = 41/96 (42%), Gaps = 21/96 (21%)
Query: 68 HEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALP 127
HEP F P + S +PPP + +S PP++ P S P PL+P+ P
Sbjct: 27 HEP--FFPIDSPPPSPPSPPPLPKLPFSSTTPPSSSDPNAS---------PFFPLYPSSP 75
Query: 128 GAKPSPPPPPAPLGAKPGP-----PPPPPPAPPSAK 158
PPP PA + P P +PP++K
Sbjct: 76 -----PPPSPASFASFPANISSLIVPHATKSPPNSK 106
Score = 37.0 bits (84), Expect = 1.9
Identities = 23/62 (37%), Positives = 26/62 (41%), Gaps = 8/62 (12%)
Query: 131 PSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKP--GPRPPPPPPKSGVA 188
P PPP+P P PPP P P + PP P S P PPPP P S +
Sbjct: 32 PIDSPPPSP------PSPPPLPKLPFSSTTPPSSSDPNASPFFPLYPSSPPPPSPASFAS 85
Query: 189 PP 190
P
Sbjct: 86 FP 87
Score = 37.0 bits (84), Expect = 1.9
Identities = 24/69 (34%), Positives = 27/69 (38%), Gaps = 10/69 (14%)
Query: 127 PGAKPSPPP-PPAPLGAKPGPPPPPPPAPP--SAKPGPPPPPPPAPSGAKPG-------P 176
P + PSPPP P P + P P A P P PPPP PA + P P
Sbjct: 37 PPSPPSPPPLPKLPFSSTTPPSSSDPNASPFFPLYPSSPPPPSPASFASFPANISSLIVP 96
Query: 177 RPPPPPPKS 185
PP S
Sbjct: 97 HATKSPPNS 105
Score = 36.6 bits (83), Expect = 2.5
Identities = 19/50 (38%), Positives = 24/50 (48%), Gaps = 5/50 (10%)
Query: 115 PVPPRPPLHPALPGAKPSPPPPPAPLGAK-----PGPPPPPPPAPPSAKP 159
P PP PP P LP + +PP P + P PPPP PA ++ P
Sbjct: 38 PSPPSPPPLPKLPFSSTTPPSSSDPNASPFFPLYPSSPPPPSPASFASFP 87
>UniRef100_O23374 P140mDia like protein [Arabidopsis thaliana]
Length = 645
Score = 386 bits (991), Expect = e-105
Identities = 223/369 (60%), Positives = 258/369 (69%), Gaps = 46/369 (12%)
Query: 120 PPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPP 179
PPL LP + +PPPPPA PPP PPP PP P P PPPPP + RPP
Sbjct: 299 PPLK--LPPGRSAPPPPPAA-----APPPQPPPPPP---PKPQPPPPPKIA------RPP 342
Query: 180 PPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMV 239
P PPK G AP R G + G + ++E GA K KLKPFFWDK+ AN DQ MV
Sbjct: 343 PAPPK-GAAPKRQ--GNTSSGDASDVDSETGA------PKTKLKPFFWDKM-ANPDQKMV 392
Query: 240 WNQIKSGSFQFNEEMIETLFGYNAVNKN-NGQRQKESSSSQDPSPQYIQIVDKKKAQNLL 298
W++I +GSFQFNEE +E+LFGYN NKN NGQ+ +SS + P QYIQI+D +KAQNL
Sbjct: 393 WHEISAGSFQFNEEAMESLFGYNDGNKNKNGQKSTDSSLRESPL-QYIQIIDTRKAQNLS 451
Query: 299 ILLRALNVTMEEVCDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRF 358
ILLRALNVT EEV DA+ EGNELP E LQTLLKMAPTS+EELKLRL++GDL LGP
Sbjct: 452 ILLRALNVTTEEVVDAIKEGNELPVELLQTLLKMAPTSEEELKLRLYSGDLHLLGP---- 507
Query: 359 LKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKT 418
R E LLFM + +EE++ E+ LEVACK+LRNSRLF KLLEAVLKT
Sbjct: 508 ------------RSESLLFMISLQEEVSGLKEALGTLEVACKKLRNSRLFLKLLEAVLKT 555
Query: 419 GNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQ 478
GNRMN GT+RG AQAFKLDTLLKLSDVKGTDGKTTLLHFVV EIIRSEG++A R S+
Sbjct: 556 GNRMNVGTFRGDAQAFKLDTLLKLSDVKGTDGKTTLLHFVVLEIIRSEGVRALRL--QSR 613
Query: 479 SLSNIKTDE 487
S S++KTD+
Sbjct: 614 SFSSVKTDD 622
Score = 42.0 bits (97), Expect = 0.059
Identities = 25/64 (39%), Positives = 27/64 (42%), Gaps = 9/64 (14%)
Query: 127 PGAKPSPPPPPAP-------LGAKPGPPPPPPPAPPSAKPGPPPPPPP--APSGAKPGPR 177
PG PS P PAP L P PP P + P P P APS + PGP
Sbjct: 124 PGPGPSFAPGPAPNPRSYDWLAPASSPNEPPAETPDESSPSPSEETPSVVAPSQSVPGPP 183
Query: 178 PPPP 181
PPP
Sbjct: 184 RPPP 187
Score = 38.5 bits (88), Expect = 0.66
Identities = 29/72 (40%), Positives = 31/72 (42%), Gaps = 8/72 (11%)
Query: 128 GAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPA--PSGAKPGPRPPPP---- 181
G PS P P P A PGP P P A P P PPA P + P P P
Sbjct: 117 GPAPSFAPGPGPSFA-PGPAPNPRSYDWLA-PASSPNEPPAETPDESSPSPSEETPSVVA 174
Query: 182 PPKSGVAPPRPP 193
P +S PPRPP
Sbjct: 175 PSQSVPGPPRPP 186
>UniRef100_Q7XUV2 OSJNBa0072F16.14 protein [Oryza sativa]
Length = 833
Score = 372 bits (954), Expect = e-101
Identities = 219/506 (43%), Positives = 300/506 (59%), Gaps = 64/506 (12%)
Query: 133 PPPPPAPLGAKPGPPPPPPPA-------PPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKS 185
PPPPP P PPPPPPPA S P PPPPPP P PPPPP
Sbjct: 365 PPPPPPP------PPPPPPPAVTQQQDVKTSCGPAVPPPPPPTP---------PPPPPL- 408
Query: 186 GVAPPRP----PIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWN 241
+AP + PI P A P A A G KLKP WDKV A ++ MVW+
Sbjct: 409 -LAPKQQSSGGPILPPAPAPPPLFRPWAPAVGKNGAPLPKLKPLHWDKVRAAPNRRMVWD 467
Query: 242 QIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILL 301
+I+S SF+ +E+MIE+LFGYNA + E S+ PS + ++D K+ QN IL+
Sbjct: 468 RIRSSSFELDEKMIESLFGYNA----RCSTKHEEVQSRSPSLGH-HVLDTKRLQNFTILM 522
Query: 302 RALNVTMEEVCDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRFLKA 361
+A++ T E++ AL GN L ++ L+ L+KMAP DE KL ++GD+ L PA+R LK
Sbjct: 523 KAVSATAEQIFAALLHGNGLSAQQLEALIKMAPAKDEADKLSAYDGDVDGLVPAERLLKV 582
Query: 362 MVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNR 421
++ IP AF R+E +L+ TF +E+ +SF +LE AC+EL +S+LF KLLEAVLKTGNR
Sbjct: 583 VLTIPCAFARVEAMLYRETFADEVGHIRKSFEMLEEACRELMSSKLFLKLLEAVLKTGNR 642
Query: 422 MNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSLS 481
MN GT RGGA AFKLD LLKL+DVKGTDGKTTLLHFVVQE+ RS +AA
Sbjct: 643 MNVGTARGGAMAFKLDALLKLADVKGTDGKTTLLHFVVQEMTRSRAAEAA---------- 692
Query: 482 NIKTDELHETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDI 541
++ + L EL NV++ + +D D LT + L HGL + K++
Sbjct: 693 -------------------DIAAGLGAELTNVRKTATVDLDVLTTSVSGLSHGLSRIKEL 733
Query: 542 LNKNLKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDG- 600
+ +L E ++ F + FV +A +++L + E++++A V+ +Y+HG+ KD+
Sbjct: 734 VGSDLSGDERNQCFVAFMAPFVAHAGEVIRELEDGERRVLAHVREITEYYHGDVGKDEAS 793
Query: 601 -LRLFVVVRDFLIMLDKVCKEVRDAQ 625
LR+FV+VRDFL ML++VCKEVR A+
Sbjct: 794 PLRIFVIVRDFLGMLERVCKEVRGAK 819
>UniRef100_O22824 Hypothetical protein At2g43800 [Arabidopsis thaliana]
Length = 894
Score = 364 bits (934), Expect = 5e-99
Identities = 245/606 (40%), Positives = 330/606 (54%), Gaps = 51/606 (8%)
Query: 41 PPPGRVGTIHS-------GMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSG 93
PPP + S G P LRP P PLP + SFTP S P RQ
Sbjct: 266 PPPASSSSSSSYSQYHKLGSPELRPLP----PLP-KLQSFTPVYK---STEQLNPKRQDF 317
Query: 94 VASARPPATPSPPQSLLAGAKPVPPRPPLHPALP-------GAKPSPPPPPAP-LGAKPG 145
P+ +G K P R + G+ P AP L A PG
Sbjct: 318 DGDDNENDEFFSPRGS-SGRKQSPTRVSDVDQIDNRSINGSGSNSCSPTNFAPSLNASPG 376
Query: 146 PPPPP----PPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPIGPKAGGP 201
P PP ++ P A+P P PPPPP S V A
Sbjct: 377 TSLKPKSISPPVSLHSQISSNNGIPKRLCPARPPPPPPPPPQVSEVP---------ATMS 427
Query: 202 KATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIETLFGY 261
+ ++ E +T K KLK WDKV A+S + MVW+QIKS SFQ NEEMIETLF
Sbjct: 428 HSLPGDDSDPEKKVETMKPKLKTLHWDKVRASSSRVMVWDQIKSNSFQVNEEMIETLFKV 487
Query: 262 NAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGNE- 320
N +S S ++ + +D +K+ N+ ILLRALNVT +EVC+AL EGN
Sbjct: 488 NDPTSRTRDGVVQSVSQEN------RFLDPRKSHNIAILLRALNVTADEVCEALIEGNSD 541
Query: 321 -LPSEFLQTLLKMAPTSDEELKLRLF----NGDLSQLGPADRFLKAMVDIPSAFKRMEVL 375
L E L+ LLKMAPT +EE KL+ +G S++GPA++FLKA+++IP AFKR++ +
Sbjct: 542 TLGPELLECLLKMAPTKEEEDKLKELKDDDDGSPSKIGPAEKFLKALLNIPFAFKRIDAM 601
Query: 376 LFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFK 435
L++ F+ E+ SF LE A EL+N+R+F KLLEAVLKTGNRMN GT RG A AFK
Sbjct: 602 LYIVKFESEIEYLNRSFDTLEAATGELKNTRMFLKLLEAVLKTGNRMNIGTNRGDAHAFK 661
Query: 436 LDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSLSNIKTDELHETEDHY 495
LDTLLKL D+KG DGKTTLLHFVVQEII+ EG + S N+ + +
Sbjct: 662 LDTLLKLVDIKGADGKTTLLHFVVQEIIKFEGARVPFTPSQSHIGDNMAEQSAFQDDLEL 721
Query: 496 RELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDILNKNLKNVEDDRGF 555
++LGL++VS LS++L NVK+ + +D++SL T ++ G+ K K+++ + LK F
Sbjct: 722 KKLGLQVVSGLSSQLINVKKAAAMDSNSLINETAEIARGIAKVKEVITE-LKQETGVERF 780
Query: 556 RETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDGLRLFVVVRDFLIMLD 615
E++ SF+ E ++ +L +M +VK +YFHGN ++ R+F VVRDFL +LD
Sbjct: 781 LESMNSFLNKGEKEITELQSHGDNVMKMVKEVTEYFHGN-SETHPFRIFAVVRDFLTILD 839
Query: 616 KVCKEV 621
+VCKEV
Sbjct: 840 QVCKEV 845
Score = 51.6 bits (122), Expect = 8e-05
Identities = 33/101 (32%), Positives = 41/101 (39%), Gaps = 17/101 (16%)
Query: 81 VSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPL 140
V AAPPP +PP + PP P H +PPP L
Sbjct: 37 VVTAAPPP--------YQPPVSSQPPSP--------SPHTHHHHKKHLTTTTPPPHEKHL 80
Query: 141 GAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPP 181
+ PPPPPP+PP P P P + + + P P PPPP
Sbjct: 81 FSSVANPPPPPPSPPHPNPFFPSSDPTS-TASHPPPAPPPP 120
Score = 48.5 bits (114), Expect = 6e-04
Identities = 29/92 (31%), Positives = 36/92 (38%), Gaps = 4/92 (4%)
Query: 85 APPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKP 144
A PPP Q V+S P +P P P PPPPP+P P
Sbjct: 40 AAPPPYQPPVSSQPPSPSPHTHHHHKKHLTTTTPPPHEKHLFSSVANPPPPPPSP----P 95
Query: 145 GPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGP 176
P P P + P++ PPP PP P+ P
Sbjct: 96 HPNPFFPSSDPTSTASHPPPAPPPPASLPTFP 127
Score = 44.7 bits (104), Expect = 0.009
Identities = 29/97 (29%), Positives = 34/97 (34%), Gaps = 14/97 (14%)
Query: 124 PALPGAKPSPPPPPAPLGAKPGPPPP--------------PPPAPPSAKPGPPPPPPPAP 169
P P +PPP P+ ++P P P PPP PPPP P
Sbjct: 33 PFFPVVTAAPPPYQPPVSSQPPSPSPHTHHHHKKHLTTTTPPPHEKHLFSSVANPPPPPP 92
Query: 170 SGAKPGPRPPPPPPKSGVAPPRPPIGPKAGGPKATEN 206
S P P P P S + P P P A P N
Sbjct: 93 SPPHPNPFFPSSDPTSTASHPPPAPPPPASLPTFPAN 129
Score = 39.3 bits (90), Expect = 0.39
Identities = 31/104 (29%), Positives = 42/104 (39%), Gaps = 23/104 (22%)
Query: 59 PPGRMNPLPHEPPSFTPFGN----TAVSAAAPPPPRQSGVAS-ARPPATPSPPQSLLAGA 113
PP P+ +PPS +P + ++ PPP + +S A PP P
Sbjct: 42 PPPYQPPVSSQPPSPSPHTHHHHKKHLTTTTPPPHEKHLFSSVANPPPPP---------- 91
Query: 114 KPVPPRP-PLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPS 156
P PP P P P+ + PPPAP PPP P P+
Sbjct: 92 -PSPPHPNPFFPSSDPTSTASHPPPAP------PPPASLPTFPA 128
>UniRef100_O04532 F20P5.14 protein [Arabidopsis thaliana]
Length = 760
Score = 357 bits (915), Expect = 8e-97
Identities = 213/526 (40%), Positives = 303/526 (57%), Gaps = 42/526 (7%)
Query: 135 PPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGV---APPR 191
PPP ++P PPPPPP P PPPP G+ P P PPPP K G + +
Sbjct: 227 PPPQVKQSEPTPPPPPPSIAVKQSAPTPSPPPPIKKGSSPSPPPPPPVKKVGALSSSASK 286
Query: 192 PPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFN 251
PP P G A GG + + KLKP WDKV +SD SMVW++I GSF F+
Sbjct: 287 PPPAPVRG-----------ASGGETSKQVKLKPLHWDKVNPDSDHSMVWDKIDRGSFSFD 335
Query: 252 EEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEV 311
+++E LFGY AV G++ E ++P I I+D +K+QN I+L++L +T EE+
Sbjct: 336 GDLMEALFGYVAV----GKKSPEQGDEKNPKSTQIFILDPRKSQNTAIVLKSLGMTREEL 391
Query: 312 CDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRFLKAMV-DIPSAFK 370
++L EGN+ + L+ L ++APT +E+ + F+GD ++L A+ FL ++ +P+AF
Sbjct: 392 VESLIEGNDFVPDTLERLARIAPTKEEQSAILEFDGDTAKLADAETFLFHLLKSVPTAFT 451
Query: 371 RMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGG 430
R+ LF + E+ + L++ACKELR+ LF KLLEA+LK GNRMN GT RG
Sbjct: 452 RLNAFLFRANYYPEMAHHSKCLQTLDLACKELRSRGLFVKLLEAILKAGNRMNAGTARGN 511
Query: 431 AQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSL---------- 480
AQAF L LLKLSDVK DGKT+LL+FVV+E++RSEG K + S SL
Sbjct: 512 AQAFNLTALLKLSDVKSVDGKTSLLNFVVEEVVRSEG-KRCVMNRRSHSLTRSGSSNYNG 570
Query: 481 --SNIKTDELHETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKA 538
S+++ E E Y +LGL +V LS+E NVK+ + +D +++ AT L V+A
Sbjct: 571 GNSSLQVMSKEEQEKEYLKLGLPVVGGLSSEFSNVKKAACVDYETVVATCSALA---VRA 627
Query: 539 KDILNKNLKNVEDDRG--FRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNAT 596
KD + ED G F +T+ +F+ + E +VK +E+K+M LVK T DY+ A
Sbjct: 628 KD-AKTVIGECEDGEGGRFVKTMMTFLDSVEEEVKIAKGEERKVMELVKRTTDYYQAGAV 686
Query: 597 K--DDGLRLFVVVRDFLIMLDKVCKEV-RDAQ-KKPAKPIKQETSR 638
+ L LFV+VRDFL M+DKVC ++ R+ Q +K PI + R
Sbjct: 687 TKGKNPLHLFVIVRDFLAMVDKVCLDIMRNMQRRKVGSPISPSSQR 732
Score = 43.1 bits (100), Expect = 0.027
Identities = 28/80 (35%), Positives = 35/80 (43%), Gaps = 8/80 (10%)
Query: 68 HEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALP 127
H+PP S PPPP S +A + TPSPP + G+ P PP PP +
Sbjct: 225 HQPPP-----QVKQSEPTPPPPPPS-IAVKQSAPTPSPPPPIKKGSSPSPPPPPPVKKVG 278
Query: 128 G--AKPSPPPPPAPLGAKPG 145
+ S PPP GA G
Sbjct: 279 ALSSSASKPPPAPVRGASGG 298
Score = 37.4 bits (85), Expect = 1.5
Identities = 27/89 (30%), Positives = 39/89 (43%), Gaps = 5/89 (5%)
Query: 44 GRVGTIHSGM--PPLRPPPG--RMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARP 99
GR T HS + +PPP + P P PP +A + + PPPP + G + + P
Sbjct: 211 GRSSTSHSVIHNEDHQPPPQVKQSEPTPPPPPPSIAVKQSAPTPS-PPPPIKKGSSPSPP 269
Query: 100 PATPSPPQSLLAGAKPVPPRPPLHPALPG 128
P P L+ + PP P+ A G
Sbjct: 270 PPPPVKKVGALSSSASKPPPAPVRGASGG 298
Score = 34.7 bits (78), Expect = 9.5
Identities = 19/57 (33%), Positives = 25/57 (43%), Gaps = 2/57 (3%)
Query: 41 PPPGRVGTIHSGMPPLRPPPGR--MNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVA 95
PPP + S P PPP + +P P PP G + SA+ PPP G +
Sbjct: 240 PPPPSIAVKQSAPTPSPPPPIKKGSSPSPPPPPPVKKVGALSSSASKPPPAPVRGAS 296
>UniRef100_Q9SDN5 FH protein NFH2 [Nicotiana tabacum]
Length = 835
Score = 356 bits (914), Expect = 1e-96
Identities = 234/557 (42%), Positives = 320/557 (57%), Gaps = 52/557 (9%)
Query: 139 PLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPP----PKSGVAPPRPP- 193
P+ P P P PPPP P PPPPP P+ V P
Sbjct: 307 PVSVSPKRESCSRPKSPELIAVVTPPPPQRPP-------PPPPPFVHGPQVKVTANESPV 359
Query: 194 -IGPKAGGPKATEN--AEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQF 250
I P + EN E E + K KLK WDKV A+SD MVW+Q+KS SF+
Sbjct: 360 LISPMEKNDQNVENHSIEKNEEKSEEILKPKLKTLHWDKVRASSDCEMVWDQLKSSSFKL 419
Query: 251 NEEMIETLFGYNAVNKNNGQRQKE---SSSSQDPSPQYIQIVDKKKAQNLLILLRALNVT 307
NEEMIETLF N K SS SQ+ +++D KK+QN+ ILLR LN T
Sbjct: 420 NEEMIETLFVVKNPTLNTSATAKHFVVSSMSQEN-----RVLDPKKSQNIAILLRVLNGT 474
Query: 308 MEEVCDALYEGN--ELPSEFLQTLLKMAPTSDEELKLRLFNGDLS-QLGPADRFLKAMVD 364
EE+C+A EGN + +E L+ LLKMAP+ +EE KL+ + D +LGPA++FLKA++D
Sbjct: 475 TEEICEAFLEGNAENIGTELLEILLKMAPSKEEERKLKEYKDDSPFKLGPAEKFLKAVLD 534
Query: 365 IPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMND 424
IP AFKR++ +L++ F E+ SF LE AC+ELR+SR+F KLLEAVLKTGNRMN
Sbjct: 535 IPFAFKRIDAMLYISNFDYEVDYLGNSFETLEAACEELRSSRMFLKLLEAVLKTGNRMNV 594
Query: 425 GTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSLSNIK 484
GT RG A AFKLDTLLKL DVKG DGKTTLLHFVVQEII+SEG AR + +Q+
Sbjct: 595 GTNRGDAHAFKLDTLLKLVDVKGADGKTTLLHFVVQEIIKSEG---ARLSGGNQNHQQST 651
Query: 485 TDELHETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDILNK 544
T++ + ++LGL++VS++S+EL NVK+ + +D++ L +KL G+ +++
Sbjct: 652 TND----DAKCKKLGLQVVSNISSELINVKKSAAMDSEVLHNDVLKLSKGIQNIAEVVRS 707
Query: 545 -NLKNVEDD--RGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDG- 600
+E+ + F E++ F+K AE + +L E M+LVK +Y HG++ +++
Sbjct: 708 IEAVGLEESSIKRFSESMNRFMKVAEEKILRLQAQETLAMSLVKEITEYVHGDSAREEAH 767
Query: 601 -LRLFVVVRDFLIMLDKVCKEVRDAQKKPAKPIKQETSRGLSSSDTRPSPSDFRQRLFPA 659
R+F+VV+DFL++LD VCKEV ++ +SS+ P P L P
Sbjct: 768 PFRIFMVVKDFLMILDCVCKEVGTINERTI----------VSSAQKFPVP--VNPNLQPV 815
Query: 660 IAERRID--DDSSDEES 674
I+ R SSDEES
Sbjct: 816 ISGFRAKRLHSSSDEES 832
Score = 51.6 bits (122), Expect = 8e-05
Identities = 23/56 (41%), Positives = 30/56 (53%), Gaps = 4/56 (7%)
Query: 115 PVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPS 170
P+ PP P +P +PP PP P +P P P P+ + PPPPPPP+PS
Sbjct: 38 PIDSPPPSQPPIP----APPAPPTPYPFQPSTPDNNNPFFPTYRSPPPPPPPPSPS 89
Score = 47.4 bits (111), Expect = 0.001
Identities = 27/64 (42%), Positives = 31/64 (48%), Gaps = 10/64 (15%)
Query: 131 PSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPP----PPAPSGAKPGPRPPPPPPKSG 186
P PPP+ PP P PPAPP+ P P P P P+ P P PPPP P S
Sbjct: 38 PIDSPPPSQ------PPIPAPPAPPTPYPFQPSTPDNNNPFFPTYRSPPPPPPPPSPSSL 91
Query: 187 VAPP 190
V+ P
Sbjct: 92 VSFP 95
>UniRef100_Q6ZKB2 Putative formin homology(FH)protein [Oryza sativa]
Length = 882
Score = 352 bits (902), Expect = 3e-95
Identities = 197/488 (40%), Positives = 291/488 (59%), Gaps = 55/488 (11%)
Query: 143 KPGPPPPPPPAPPSAKPGP----PPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPIGPKA 198
KP PPPPP PP G PPPPPP P + G P P
Sbjct: 427 KPAPPPPPQKNPPPNLKGQCYGQPPPPPPLPLQIQVGKDGSPLP---------------- 470
Query: 199 GGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGS--FQFNEEMIE 256
+LKP WDKV A ++SMVWN I+S S F+F+E+MI+
Sbjct: 471 ----------------------RLKPLHWDKVRAAPNRSMVWNDIRSSSFEFEFDEQMIK 508
Query: 257 TLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVCDALY 316
+LF YN G + E + ++ S +++ + QN ILL+ LN +VC+++
Sbjct: 509 SLFAYNL----QGSMKDEEAMNKTASTTK-HVIEHHRLQNTTILLKTLNANTSQVCNSVI 563
Query: 317 EGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRFLKAMVDIPSAFKRMEVLL 376
+GN L + L+ L+KM PT +EE KL ++GD++ L PA+ F+K ++ IP AF RMEV+L
Sbjct: 564 QGNGLSVQQLEALVKMKPTKEEEEKLLNYDGDINMLDPAENFVKVLLTIPMAFPRMEVML 623
Query: 377 FMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFKL 436
+ F +E+ SFA++E AC EL++S+LF +LLEAVLKTGNRMN GT RGGA AFKL
Sbjct: 624 YKENFDDEVAHIKMSFAMIEGACTELKSSKLFLRLLEAVLKTGNRMNVGTLRGGASAFKL 683
Query: 437 DTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSLSNIKTDELHETEDHYR 496
D LLKL+D++GTDGKTTLLHFVV+E+ RS+G+KA ++ S + E E Y
Sbjct: 684 DALLKLADIRGTDGKTTLLHFVVKEMARSKGLKALEKLNETPSSCHDTPTEREE----YS 739
Query: 497 ELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDILNKNLKNVEDDRGFR 556
+G E VS LS EL NVK+ + +D D+L + L GL + ++++ K+L + + + F
Sbjct: 740 SMGTEFVSELSNELGNVKKVASIDLDTLRNSISNLSCGLAQLRNLVEKDLASDDKNNNFL 799
Query: 557 ETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDG--LRLFVVVRDFLIML 614
+ ++SF+ +AE ++ L DE +++ V+ +Y+HG +KD+ L++F++V+DFL +L
Sbjct: 800 QCMKSFLNHAENTMQGLKADEAQVLLNVRELTEYYHGEVSKDESNLLQIFIIVKDFLGLL 859
Query: 615 DKVCKEVR 622
DKVC+E+R
Sbjct: 860 DKVCREMR 867
>UniRef100_Q6ZIP3 Putative formin homology(FH) domain-containing protein [Oryza
sativa]
Length = 753
Score = 338 bits (866), Expect = 4e-91
Identities = 236/632 (37%), Positives = 335/632 (52%), Gaps = 85/632 (13%)
Query: 50 HSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSL 109
H P LRP P P P P P S PPPP + + +
Sbjct: 176 HEKSPDLRPLPLLKQPSPDLRP--LPPLKRPESQPPPPPPSTPPLTTTGYSTDEEDQATY 233
Query: 110 LAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAP 169
K + PP A P PP PP P +P PPPPPPP
Sbjct: 234 YTAPKTAMSSFSRSTSQHSTLEQTAMPPM---AAPAPPQTNPPRP--VRP-PPPPPPPRQ 287
Query: 170 SGAKPGP-RPPPPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSK-AKLKPFFW 227
+P P PPP + + P+ P A E+ G A K LKP W
Sbjct: 288 RLLRPLPAESPPPAALANLELTGSPVKP------AVEDRGGENSGAARPPKPPHLKPLHW 341
Query: 228 DKVPANSDQSMVWNQIK-SGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPS---- 282
DK+ A S ++ VW+Q+K S +F+ +EE +E+LF +N G + S DP+
Sbjct: 342 DKLRAISGRTTVWDQVKNSDTFRVDEEAMESLF----LNSGGG-----GAGSSDPAARRG 392
Query: 283 ---PQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGN--ELPSEFLQTLLKMAPTSD 337
Q +++D K+ QN+ I+L++LNV +EV AL GN +L SEF +TL KMAPT +
Sbjct: 393 GSGKQERRLLDPKRLQNVAIMLKSLNVAADEVIGALVRGNPEDLGSEFYETLAKMAPTKE 452
Query: 338 EELKLRLFNGDLSQLGPADRFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEV 397
EELKL+ ++GDLS++ PA+RFLK ++ +P AF+R++ +L+ F E+ +SF LE
Sbjct: 453 EELKLKGYSGDLSKIDPAERFLKDVLGVPFAFERVDAMLYRANFDNEVNYLRKSFGTLEA 512
Query: 398 ACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHF 457
AC+ELR+S+LF KLL+AVLKTGNRMNDGT RG A+AFKLDTLLKL+D+K TDG+TTLLHF
Sbjct: 513 ACEELRSSKLFLKLLDAVLKTGNRMNDGTNRGEARAFKLDTLLKLADIKSTDGRTTLLHF 572
Query: 458 VVQEIIRSEGIKAARAAKDSQSLSNIKTDELHETEDHYRELGLEMVSHLSTELENVKRGS 517
VV+EIIRSEG + ++A + S S ++ ++ GL++++ LS+EL NVKR +
Sbjct: 573 VVKEIIRSEGFDSDQSAVNPGSGS----------KEQFKRDGLKLLAGLSSELSNVKRAA 622
Query: 518 VLDADSLTATTIKLGHGLVKAKDILNKNLKNVEDDRG----FRETVESFVKNAEADVKKL 573
L+ D+L+ ++L L K K +L LK D+G F + + F++ AEA++K +
Sbjct: 623 TLEMDTLSGNILRLEADLEKVKLVL--QLKETCSDQGASENFFQAMVVFLRRAEAEIKNM 680
Query: 574 LEDEKKIMALVKSTGDYFHGNATKDDGLRLFVVVRDFLIMLDKVCKEVRDAQKK------ 627
A + LR+FVVV +FL++LD+VC++V ++
Sbjct: 681 -------------------KTAEEPHPLRIFVVVDEFLLILDRVCRDVGRTPERVMMGSG 721
Query: 628 -----PA----KPIKQETSRGLSSSDTRPSPS 650
PA P + E R LSSSD S S
Sbjct: 722 KSFRVPAGTSLPPHRNENRRVLSSSDEDSSSS 753
>UniRef100_Q9SDN6 FH protein NFH1 [Nicotiana tabacum]
Length = 868
Score = 305 bits (781), Expect = 3e-81
Identities = 189/454 (41%), Positives = 257/454 (55%), Gaps = 33/454 (7%)
Query: 35 SPFELNPPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGV 94
SP L P + IH+ PP PP PLP +F P + +P PP S
Sbjct: 339 SPKSLMPKSPELVAIHTAPPPQYSPPPPPPPLPPRA-NFVPILVMGNESDSPSPPSSSSP 397
Query: 95 ASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAP 154
+ S P+S + + + + +P + PPPPPPP
Sbjct: 398 ERYSSRSIDSSPRSFNVWDQNLESPARITNQIQQIEPV---------SVASPPPPPPPLS 448
Query: 155 PSAKPGPPPPPPPAPSGAKPGPR---PPPPPPKSGVAPPRPP-------IGPKAGGPKAT 204
S PPPPPP P P+ PP P V P RP I P P +
Sbjct: 449 ISIPASVPPPPPPPPCKNWDSPKTLTPPTSKPPVLVTPLRPIALESPVLISPMDQLPSNS 508
Query: 205 ENAEAGAEG--GADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIETLFGYN 262
E E + +T K KLK WDKV A+SD+ VW+Q+KS SF+ +EEMIETLF
Sbjct: 509 EPIEKNEQKIENEETPKPKLKTLHWDKVRASSDRETVWDQLKSSSFKLDEEMIETLFVVK 568
Query: 263 AVNKNNGQRQKES---SSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGN 319
N + + + S SQ+ +++D KK+QN+ I LRAL+VT+EEVC+AL EGN
Sbjct: 569 TPTSNPKETTRRAVLPSQSQEN-----RVLDPKKSQNISIQLRALSVTVEEVCEALLEGN 623
Query: 320 E--LPSEFLQTLLKMAPTSDEELKLRLFNGDLS-QLGPADRFLKAMVDIPSAFKRMEVLL 376
L +E L++LLKMAP+ +EE KL+ + D +LGPA++FLKA++DIP AFKR++ +L
Sbjct: 624 ADALGTELLESLLKMAPSKEEERKLKEYKDDSPFKLGPAEKFLKAVLDIPFAFKRVDAML 683
Query: 377 FMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFKL 436
++ F E+ +SF LE +C+ELR++R+F KL+EAVLKTGNR+N GT RG A AFK+
Sbjct: 684 YISNFDSEVDYLKKSFETLEASCEELRSNRMFLKLVEAVLKTGNRLNVGTNRGDAHAFKV 743
Query: 437 DTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKA 470
DTLLKL+DVKG DGKT+ LHFVVQEIIR + +++
Sbjct: 744 DTLLKLADVKGADGKTSFLHFVVQEIIRLQAVES 777
Score = 57.4 bits (137), Expect = 1e-06
Identities = 28/63 (44%), Positives = 43/63 (67%), Gaps = 2/63 (3%)
Query: 561 SFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDG--LRLFVVVRDFLIMLDKVC 618
SF+ ++ +L E M+LVK +YFHGN+ +++ R+F+VVRDFL++LD+VC
Sbjct: 760 SFLHFVVQEIIRLQAVESVAMSLVKEITEYFHGNSAREEAHPFRIFMVVRDFLMVLDRVC 819
Query: 619 KEV 621
KEV
Sbjct: 820 KEV 822
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.312 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,327,973,075
Number of Sequences: 2790947
Number of extensions: 77241207
Number of successful extensions: 2199690
Number of sequences better than 10.0: 30251
Number of HSP's better than 10.0 without gapping: 14727
Number of HSP's successfully gapped in prelim test: 16864
Number of HSP's that attempted gapping in prelim test: 647212
Number of HSP's gapped (non-prelim): 307982
length of query: 675
length of database: 848,049,833
effective HSP length: 134
effective length of query: 541
effective length of database: 474,062,935
effective search space: 256468047835
effective search space used: 256468047835
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 78 (34.7 bits)
Medicago: description of AC144502.11


