

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140104.6 - phase: 0
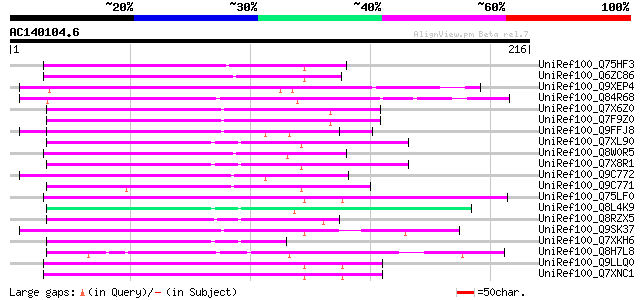
(216 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q75HF3 Hypothetical protein OSJNBa0024F18.8 [Oryza sat... 89 1e-16
UniRef100_Q6ZC86 Hypothetical protein P0702G08.10 [Oryza sativa] 80 3e-14
UniRef100_Q9XEP4 Hypothetical protein [Sorghum bicolor] 80 4e-14
UniRef100_Q84R68 Hypothetical protein OSJNBb0113I20.10 [Oryza sa... 73 4e-12
UniRef100_Q7X6Z0 OSJNBb0043H09.1 protein [Oryza sativa] 70 4e-11
UniRef100_Q7F9Z0 OSJNBa0079C19.27 protein [Oryza sativa] 70 4e-11
UniRef100_Q9FFJ8 Arabidopsis thaliana genomic DNA, chromosome 5,... 68 1e-10
UniRef100_Q7XL90 OSJNBb0115I09.22 protein [Oryza sativa] 63 6e-09
UniRef100_Q8W0R5 Hypothetical protein [Sorghum bicolor] 62 1e-08
UniRef100_Q7X8R1 OSJNBa0004L19.4 protein [Oryza sativa] 62 1e-08
UniRef100_Q9C772 Hypothetical protein F11B9.20 [Arabidopsis thal... 60 4e-08
UniRef100_Q9C771 Hypothetical protein F11B9.21 [Arabidopsis thal... 57 3e-07
UniRef100_Q75LF0 Hypothetical protein OSJNBb0079B16.19 [Oryza sa... 55 1e-06
UniRef100_Q8L4K9 Hypothetical protein OSJNBa0079H13.17 [Oryza sa... 55 1e-06
UniRef100_Q8RZX5 En/Spm-like transposon-like protein [Oryza sativa] 54 2e-06
UniRef100_Q9SK37 Hypothetical protein At2g24960 [Arabidopsis tha... 54 2e-06
UniRef100_Q7XKH6 OSJNBb0033P05.11 protein [Oryza sativa] 54 3e-06
UniRef100_Q8H7L8 Hypothetical protein OSJNBb0014I10.8 [Oryza sat... 50 3e-05
UniRef100_Q9LLQ0 Hypothetical protein DUPR11.2 [Oryza sativa] 50 4e-05
UniRef100_Q7XNC1 OSJNBa0029C04.7 protein [Oryza sativa] 50 4e-05
>UniRef100_Q75HF3 Hypothetical protein OSJNBa0024F18.8 [Oryza sativa]
Length = 483
Score = 88.6 bits (218), Expect = 1e-16
Identities = 45/127 (35%), Positives = 70/127 (54%), Gaps = 2/127 (1%)
Query: 15 GKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRM 74
G A WD TK+ LDIC+ E K W+ + + ++ KNY KQL+N+
Sbjct: 4 GHAYWDDRLTKIFLDICIAEKEKLNYNKKGLTKVGWQNVYRNFREQTCKNYDSKQLQNKF 63
Query: 75 DTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAK-FRFQGLEFCDEL 133
+TL+ + ++WK+L K +G GW+++ G I D WW+ +I + AK FR + L DEL
Sbjct: 64 NTLKRQHSLWKKLKNK-SGAGWDNNTGTIRCDDDWWEDRIEEDREAKQFRHKPLAHEDEL 122
Query: 134 EFIFGEI 140
+FG +
Sbjct: 123 TILFGSM 129
>UniRef100_Q6ZC86 Hypothetical protein P0702G08.10 [Oryza sativa]
Length = 172
Score = 80.5 bits (197), Expect = 3e-14
Identities = 42/125 (33%), Positives = 66/125 (52%), Gaps = 2/125 (1%)
Query: 15 GKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRM 74
G+ WD TK+ LD+C+ E K N W+ + + +++ KNY KQL+N+
Sbjct: 4 GRTYWDKALTKIFLDLCIAEKTKRNHNKKGRTNIGWQNLYRNFREQSGKNYDSKQLQNKF 63
Query: 75 DTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAK-FRFQGLEFCDEL 133
T + ++ +WK L K G GW+++ I D WW+ +I + A+ FR + LE DEL
Sbjct: 64 STFKRQYKLWKSLKNKSGG-GWDNNSSTIRCDDDWWEDRIEENRDARQFRGKPLEHEDEL 122
Query: 134 EFIFG 138
FG
Sbjct: 123 TTHFG 127
>UniRef100_Q9XEP4 Hypothetical protein [Sorghum bicolor]
Length = 1001
Score = 80.1 bits (196), Expect = 4e-14
Identities = 56/215 (26%), Positives = 89/215 (41%), Gaps = 34/215 (15%)
Query: 5 ESLPNSSENLG--------------KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKW 50
+ LP S +LG KA W + L D C EI + F W
Sbjct: 195 DRLPKGSRSLGRKIEIFLFGSSMGDKADWSDRFLRNLFDACKEEIDAGNRPMGIFTATGW 254
Query: 51 EEIRDEYNKRANKNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWW 110
+ + ++ ++ T+KQLKN++D L+ E+T++ + TGLGW+ +D WW
Sbjct: 255 KNVVSKFGDKSGDKRTKKQLKNKLDILKKEYTMFMEFKNFATGLGWDAENKTVDCPKEWW 314
Query: 111 D---AKIRN------VKYAKFRFQGLEFCDELEFIFGEIVATSQCACAPAMGVPLESTGK 161
D A+ N + KFR G +F +++ IFG+ A P + + T
Sbjct: 315 DEHLARCNNPEKGIKCSHVKFRKHGPKFLEDMHVIFGKAHVDGSTATCPG-DISSDDTSD 373
Query: 162 NTTADVAREIIESDDELNIDELSPMENTQSKNKRK 196
A+V + E + +N Q K KRK
Sbjct: 374 EDVAEVPKPTKEKEKTVN----------QGKRKRK 398
>UniRef100_Q84R68 Hypothetical protein OSJNBb0113I20.10 [Oryza sativa]
Length = 298
Score = 73.2 bits (178), Expect = 4e-12
Identities = 56/207 (27%), Positives = 104/207 (50%), Gaps = 12/207 (5%)
Query: 5 ESLPNSSENL--GKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRAN 62
E++P+ S + A W+ ++T+++ +I ++ + N ++E+ +++ +
Sbjct: 3 ENVPSKSVKMPIDYADWNDYNTRVVCEIFADQVVAGNRPNTHLSNSGYDEVIEQFASKTG 62
Query: 63 KNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVK-YAK 121
YT+ Q+KN+ D LR E+ WK+ L +TGLGW+ + A +A W ++ K
Sbjct: 63 LRYTRLQIKNKWDKLRIEYNCWKK-LKSQTGLGWDDARQTVTATAARWKQLKEDIAGCLK 121
Query: 122 FRFQGLEFCDELEFIFGEIVATSQCACAPAMGVPLESTGKNTTADVAREIIESDDELNID 181
FR GL+ + LE +F +I T + G + S N T +A ++ E DDE + D
Sbjct: 122 FRKAGLQNEELLEKMFEDIRNTGADHWSLGQGT-IPSATTNGTIQIA-DVNEIDDETDED 179
Query: 182 ELSPMENTQSKNKRKVSPRMRKQQRVR 208
E S +K KR S ++ K ++ +
Sbjct: 180 EPS------AKRKRGDSSKVDKLKKTK 200
>UniRef100_Q7X6Z0 OSJNBb0043H09.1 protein [Oryza sativa]
Length = 408
Score = 70.1 bits (170), Expect = 4e-11
Identities = 39/140 (27%), Positives = 69/140 (48%), Gaps = 2/140 (1%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
KA+W T +T++ ++C+ +I + + ++ I +Y ++ +++ QLKNR D
Sbjct: 145 KAEWSTTNTRIFCELCVEQIEAGNRPIGIMTTRGYQNIAVKYLQKTGLRHSKMQLKNRWD 204
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAKFRFQGLEFCDE-LE 134
L+ ++ W LL K+TGLGW+ G + A +W + K G C++ L
Sbjct: 205 ILKALYSFWLGLL-KDTGLGWDSTQGTVSASDKYWKQVTKGHSEWKKLQHGPPDCEDLLS 263
Query: 135 FIFGEIVATSQCACAPAMGV 154
+F E+ ACAP V
Sbjct: 264 QMFAEVAVDGSSACAPGEDV 283
>UniRef100_Q7F9Z0 OSJNBa0079C19.27 protein [Oryza sativa]
Length = 728
Score = 70.1 bits (170), Expect = 4e-11
Identities = 39/140 (27%), Positives = 69/140 (48%), Gaps = 2/140 (1%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
KA+W T +T++ ++C+ +I + + ++ I +Y ++ +++ QLKNR D
Sbjct: 190 KAEWSTTNTRIFCELCVEQIEAGNRPIGIMTTRGYQNIAVKYLQKTGLRHSKMQLKNRWD 249
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAKFRFQGLEFCDE-LE 134
L+ ++ W LL K+TGLGW+ G + A +W + K G C++ L
Sbjct: 250 ILKALYSFWLGLL-KDTGLGWDSTQGTVSASDKYWKQVTKGHSEWKKLQHGPPDCEDLLS 308
Query: 135 FIFGEIVATSQCACAPAMGV 154
+F E+ ACAP V
Sbjct: 309 QMFAEVAVDGSSACAPGEDV 328
>UniRef100_Q9FFJ8 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MJJ3
[Arabidopsis thaliana]
Length = 449
Score = 68.2 bits (165), Expect = 1e-10
Identities = 41/148 (27%), Positives = 67/148 (44%), Gaps = 2/148 (1%)
Query: 5 ESLPNSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKN 64
+S+ +SS K W + K+ LD+ + E K + F + W+ I N+
Sbjct: 156 QSMCSSSNPQTKGYWSPSTHKLFLDLLVQETLKGNRPDTHFNKEGWKTILGTINENTGLG 215
Query: 65 YTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIR-NVKYAKFR 123
YT+ QLKN D R W IW QL+G + + W+ + A W IR N + +FR
Sbjct: 216 YTRPQLKNHWDCTRKAWKIWCQLVG-ASSMKWDPESRSFGATEEEWRIYIRENPRAGQFR 274
Query: 124 FQGLEFCDELEFIFGEIVATSQCACAPA 151
+ + D+L IF ++ + P+
Sbjct: 275 HKEVPHADQLAIIFNGVIEPGETYTPPS 302
Score = 55.1 bits (131), Expect = 1e-06
Identities = 31/123 (25%), Positives = 56/123 (45%), Gaps = 2/123 (1%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
KA W+ ++ +D+C+ + K G F + W I + ++ Y + QLKN D
Sbjct: 4 KAVWEPEYHRVFVDLCVEQTMLGNKPGTHFSKEGWRNILISFQEQTGAMYDRMQLKNHWD 63
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDA-DSAWWDAKIRNVKYAKFRFQGLEFCDELE 134
T+ +W IW++L+ + + + WN A D W + N ++R +LE
Sbjct: 64 TMSRQWKIWRRLV-ETSFMNWNPESNRFRATDDDWANYLQENPDAGQYRLSVPHDLKKLE 122
Query: 135 FIF 137
+F
Sbjct: 123 ILF 125
>UniRef100_Q7XL90 OSJNBb0115I09.22 protein [Oryza sativa]
Length = 804
Score = 62.8 bits (151), Expect = 6e-09
Identities = 46/152 (30%), Positives = 68/152 (44%), Gaps = 3/152 (1%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
KA W T L +I EI A+ + ++ ++ +Y +RAN ++ KQ+KNR
Sbjct: 185 KANWTEEKTYTLCEIACEEIRDGHCPNGAWTTRGYQNLKAKYFQRANLRHSTKQIKNRFT 244
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYA-KFRFQGLEFCDELE 134
L+ + W L ++TG G G I A +AWW+ KI+ A KFR E+ D L
Sbjct: 245 HLKNYYIAW-VWLNQQTGAG-RSASGEIIASNAWWEKKIKEKGDAKKFRHGNPEYLDFLI 302
Query: 135 FIFGEIVATSQCACAPAMGVPLESTGKNTTAD 166
IF + A P E G+ D
Sbjct: 303 EIFQGVAVDGSTAYIPGFEDEDEEEGEEEAGD 334
>UniRef100_Q8W0R5 Hypothetical protein [Sorghum bicolor]
Length = 284
Score = 62.0 bits (149), Expect = 1e-08
Identities = 36/127 (28%), Positives = 59/127 (46%), Gaps = 2/127 (1%)
Query: 15 GKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRM 74
G+A WD +T+M LD+C+ E K W + + + + Y KQL+N+
Sbjct: 4 GRAHWDDHTTRMFLDLCIAEKEKENYNSKGLTKIGWHNLYRNFKEHTRRVYDTKQLQNKF 63
Query: 75 DTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKI-RNVKYAKFRFQGLEFCDEL 133
++L+ + +W+Q K G GW+ + I D+ D I N FR + L D L
Sbjct: 64 NSLKRMYNLWRQQNDKTRG-GWDKNSSTITQDADACDNHITENSAEEDFRGKALAHEDAL 122
Query: 134 EFIFGEI 140
+FG +
Sbjct: 123 TILFGSM 129
>UniRef100_Q7X8R1 OSJNBa0004L19.4 protein [Oryza sativa]
Length = 823
Score = 61.6 bits (148), Expect = 1e-08
Identities = 46/152 (30%), Positives = 68/152 (44%), Gaps = 3/152 (1%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
KA W T L +I EI A+ + ++ ++ +Y +RAN ++ KQ+KNR
Sbjct: 206 KANWTEEKTYTLCEIACEEIRDGHCPNGAWTTRGYQNLKAKYFQRANLRHSTKQIKNRFT 265
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYA-KFRFQGLEFCDELE 134
L+ + W L ++TG G G I A +AWW+ KI+ A KFR E+ D L
Sbjct: 266 HLKNYYIAW-VWLNQQTGAG-RSASGEIIASNAWWEKKIKEKGDAKKFRHGNPEYLDFLI 323
Query: 135 FIFGEIVATSQCACAPAMGVPLESTGKNTTAD 166
IF + A P E G+ D
Sbjct: 324 EIFQGVAVDGSTAYIPGFEDEDEEEGEEEAWD 355
>UniRef100_Q9C772 Hypothetical protein F11B9.20 [Arabidopsis thaliana]
Length = 459
Score = 60.1 bits (144), Expect = 4e-08
Identities = 38/138 (27%), Positives = 62/138 (44%), Gaps = 2/138 (1%)
Query: 5 ESLPNSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKN 64
+S N S K W S ++ +D+ E K + + + W+ I + N+ K+
Sbjct: 153 QSASNFSSPQSKGYWSPSSHELFVDLLFQEALKGNRPDSHYPKETWKMILETINQNTGKS 212
Query: 65 YTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDA-DSAWWDAKIRNVKYAKFR 123
+T+ QLKN D R W IW Q++G + W+ A D W + N + A FR
Sbjct: 213 FTRPQLKNHWDCTRKSWKIWCQVIGAPV-MKWDATSRTFGATDEDWKNYLKENHRAAPFR 271
Query: 124 FQGLEFCDELEFIFGEIV 141
+ L D+L IF ++
Sbjct: 272 RKQLPHADKLATIFKGLI 289
Score = 45.1 bits (105), Expect = 0.001
Identities = 30/123 (24%), Positives = 57/123 (45%), Gaps = 9/123 (7%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
KA W+ ++ +D+C+ + + G + I + +R +T+ QLKN D
Sbjct: 3 KAAWEPEYHRVFVDLCVEQKMLGNQPGT-------QHILKPFLQRTGARFTRNQLKNHWD 55
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDA-DSAWWDAKIRNVKYAKFRFQGLEFCDELE 134
T+ +W IW +L+ + + + W+ A D W + N + ++R F ++LE
Sbjct: 56 TMIKQWKIWCRLV-QCSDMQWDPQTNTFGANDQDWANYLHVNPEAGQYRLNPPSFLEKLE 114
Query: 135 FIF 137
IF
Sbjct: 115 LIF 117
>UniRef100_Q9C771 Hypothetical protein F11B9.21 [Arabidopsis thaliana]
Length = 539
Score = 57.4 bits (137), Expect = 3e-07
Identities = 40/141 (28%), Positives = 64/141 (45%), Gaps = 7/141 (4%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRN-----KKWEEIRDEYNKRANKNYTQKQL 70
KA W + S ++ +D+ E K + A RN + W + + +N++ YT+KQL
Sbjct: 173 KAYWSSSSHEIFVDLLFTESLKENRPKPARRNGYYAKETWNMMVESFNQKTGLRYTRKQL 232
Query: 71 KNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYA-KFRFQGLEF 129
KN + R W W Q +G L W+ + A S W+ + K A +FR + +
Sbjct: 233 KNHWNITRDAWRRWCQAVGSPL-LKWDANTKTFGATSEDWENYSKENKRAEQFRLKHIPH 291
Query: 130 CDELEFIFGEIVATSQCACAP 150
D+L IF V + A P
Sbjct: 292 ADKLAIIFKGHVEPGKTALRP 312
Score = 42.0 bits (97), Expect = 0.011
Identities = 41/177 (23%), Positives = 75/177 (42%), Gaps = 21/177 (11%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
K W+ K+ +D+C+ + K+ + FR I + + + +T+ QLKN D
Sbjct: 5 KVMWEPELHKVFVDLCVEQ-----KM-LGFRLPGLNRIWESFVQNTGARFTRDQLKNHWD 58
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIR-NVKYAKFRFQGL--EFCDE 132
T+ W W +L+ + + + W+ A + W R N K ++RF+ F +
Sbjct: 59 TMLRLWRAWCRLV-ECSEMKWDPQTKKFGASTEVWTNYFRVNPKAKQYRFRSSPPPFLKD 117
Query: 133 LEFIF-----GEIVATSQCACAPAMGVPLESTGKNTTADVAREIIESDDELNIDELS 184
L+ IF G+ TS C +P N T D + + D+ D+++
Sbjct: 118 LKMIFEGTDLGDEEGTS---CGKRKRIP---DADNDTGDEDNDTGDDDNYTGDDDIT 168
>UniRef100_Q75LF0 Hypothetical protein OSJNBb0079B16.19 [Oryza sativa]
Length = 693
Score = 55.5 bits (132), Expect = 1e-06
Identities = 43/195 (22%), Positives = 80/195 (40%), Gaps = 2/195 (1%)
Query: 15 GKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRM 74
GK +W + L+ C+ E+ + + ++ + + +R +N T+ Q+K
Sbjct: 4 GKIRWSDDEIGIFLEACLEELSAMTITSTCPKPQGYKNLIQKMKERIGRNLTKDQVKYIW 63
Query: 75 DTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAK-FRFQGLEFCDEL 133
R W +W L K TG+G + I AD WW+ K + A+ F+ L+ D
Sbjct: 64 CQCRKRWMLWTWLESKATGIGRDPITQAIVADDNWWEDKDQKKDGARVFKDSPLKHIDLH 123
Query: 134 EFIF-GEIVATSQCACAPAMGVPLESTGKNTTADVAREIIESDDELNIDELSPMENTQSK 192
IF G V + A A A P + ++++ + + + + E E + +
Sbjct: 124 CAIFSGRTVVGNHSAVAGAAPAPPQQPTPRPNINISQLLAQQQNIATLGEFQVQEKARGQ 183
Query: 193 NKRKVSPRMRKQQRV 207
+ R Q+RV
Sbjct: 184 QMKLALRGDRLQRRV 198
>UniRef100_Q8L4K9 Hypothetical protein OSJNBa0079H13.17 [Oryza sativa]
Length = 916
Score = 55.1 bits (131), Expect = 1e-06
Identities = 51/203 (25%), Positives = 75/203 (36%), Gaps = 28/203 (13%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
KA W T L +I EI A+ + ++ ++ +Y +RAN ++ KQ+KNR
Sbjct: 206 KANWTEEKTYTLCEIACEEIRDGHCPNGAWTTRGYQNLKAKYFQRANLRHSTKQIKNRFT 265
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNV------------------ 117
L+ ++ W L ++TG G G I A +AWW+ K+R
Sbjct: 266 HLKNYYSAW-VWLNQQTGAG-RSASGEIIASNAWWEKKLRYAVVKNIASLHDFRVFCDID 323
Query: 118 --------KYAKFRFQGLEFCDELEFIFGEIVATSQCACAPAMGVPLESTGKNTTADVAR 169
KFR E+ D L IF + A P E G+ D
Sbjct: 324 LVSIQEKGDAKKFRHGNPEYLDFLIEIFQGVAVDGSTAYIPGFEDEDEEEGEEEAGDAGE 383
Query: 170 EIIESDDELNIDELSPMENTQSK 192
+ E SPM N K
Sbjct: 384 AARGERAVGDGYENSPMSNNSRK 406
>UniRef100_Q8RZX5 En/Spm-like transposon-like protein [Oryza sativa]
Length = 844
Score = 54.3 bits (129), Expect = 2e-06
Identities = 34/123 (27%), Positives = 61/123 (48%), Gaps = 3/123 (2%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
+A W + +LL +C+ ++ G + + I + Y+ + +++ QLKN++
Sbjct: 216 RANWTDRNNSILLALCIEQVRAGHYNGSQMNSDGYNAIAEGYHAKTGLMHSRLQLKNQIG 275
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAKFRFQGLEF-CDELE 134
L+ ++ W+ L TGLG G +DADS +W + I Y K +GL D+LE
Sbjct: 276 ILKSTYSFWR-YLQTHTGLG-RKPDGTVDADSDFWSSHIEGKPYLKKVLKGLPANLDQLE 333
Query: 135 FIF 137
+F
Sbjct: 334 EMF 336
>UniRef100_Q9SK37 Hypothetical protein At2g24960 [Arabidopsis thaliana]
Length = 797
Score = 54.3 bits (129), Expect = 2e-06
Identities = 45/185 (24%), Positives = 82/185 (44%), Gaps = 12/185 (6%)
Query: 5 ESLPNSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKN 64
ES+ S + K +W + ++I + +I + K G AF + W ++ +N R +
Sbjct: 158 ESVVLSGKESSKTEWTLEMDQYFVEIMVDQIGRGNKTGNAFSKQAWIDMLVLFNARFSGQ 217
Query: 65 YTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAK-FR 123
Y ++ L++R + L + + +L KE G W+ I AD A WD+ I++ A+ +R
Sbjct: 218 YGKRVLRHRYNKLLKYYKDMEAIL-KEDGFSWDETRLMISADDAVWDSYIKDHPLARTYR 276
Query: 124 FQGLEFCDELEFIFGEIVATSQCACAPAMGVPLESTGKNT-TADVAREIIESDDELNIDE 182
+ L ++L+ IF AC G G T++ ++ D I
Sbjct: 277 MKSLPSYNDLDTIF---------ACQAEQGTDHRDDGSAAQTSETKASQEQNSDRTRIFW 327
Query: 183 LSPME 187
PM+
Sbjct: 328 TPPMD 332
Score = 39.7 bits (91), Expect = 0.055
Identities = 26/125 (20%), Positives = 53/125 (41%), Gaps = 2/125 (1%)
Query: 19 WDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMDTLR 78
W + +D+ + +H+ + G F + W E+ +N + Y + LK+R L
Sbjct: 15 WTPTMERFFIDLMLEHLHRGNRTGHTFNKQAWNEMLTVFNSKFGSQYDKDVLKSRYTNLW 74
Query: 79 GEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAK-FRFQGLEFCDELEFIF 137
++ K LL G W+ + D + W ++ A+ ++ + + +L I+
Sbjct: 75 KQYNDVKCLL-DHGGFVWDQTHQTVIGDDSLWSLYLKAHPEARVYKTKPVLNFSDLCLIY 133
Query: 138 GEIVA 142
G VA
Sbjct: 134 GYTVA 138
Score = 39.3 bits (90), Expect = 0.072
Identities = 22/89 (24%), Positives = 42/89 (46%), Gaps = 1/89 (1%)
Query: 27 LLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMDTLRGEWTIWKQ 86
L+D+ + +++ ++G F W E+ +N + + + LKNR LR + K
Sbjct: 335 LIDLLVEQVNNGNRVGQTFITSAWNEMVTAFNAKFGSQHNKDVLKNRYKHLRRLYNDIKF 394
Query: 87 LLGKETGLGWNHHIGNIDADSAWWDAKIR 115
LL ++ G W+ + AD W+ I+
Sbjct: 395 LL-EQNGFSWDARRDMVIADDDIWNTYIQ 422
Score = 38.9 bits (89), Expect = 0.094
Identities = 24/112 (21%), Positives = 47/112 (41%), Gaps = 1/112 (0%)
Query: 4 KESLPNSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANK 63
K P S+ +W L+D+ + ++ + K+G F + W ++ + +N +
Sbjct: 519 KNDYPCSNIGPPCIEWTRVMDHCLIDLMLEQVSRGNKIGETFTEQAWADMAESFNAKFGL 578
Query: 64 NYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIR 115
L+NR L E +L + G W+ I A+ +W+A I+
Sbjct: 579 QTDMFMLENRYILLMKERDDINNILNLD-GFTWDVEKQTIVAEDEYWEAYIK 629
>UniRef100_Q7XKH6 OSJNBb0033P05.11 protein [Oryza sativa]
Length = 877
Score = 53.9 bits (128), Expect = 3e-06
Identities = 32/100 (32%), Positives = 51/100 (51%), Gaps = 2/100 (2%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
KA W T L +I EI A+ + ++ ++ +Y +RAN ++ KQ+KNR
Sbjct: 197 KANWTEEKTYTLCEIACEEIRDGHCPNGAWTTRGYQNLKAKYFQRANLRHSTKQIKNRFT 256
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIR 115
L+ + W L ++TG G G I A +AWW+ K+R
Sbjct: 257 HLKNYYMAW-VWLNQQTGAG-RSASGEIIASNAWWEKKLR 294
>UniRef100_Q8H7L8 Hypothetical protein OSJNBb0014I10.8 [Oryza sativa]
Length = 807
Score = 50.4 bits (119), Expect = 3e-05
Identities = 49/205 (23%), Positives = 84/205 (40%), Gaps = 28/205 (13%)
Query: 16 KAKWDTFSTKMLLDIC--MVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNR 73
KA W +T+ I ++I C + GI +N W +I Y + + ++Q R
Sbjct: 190 KANWSARNTRTFCSIWCHQMDIGNCVR-GIMQKNG-WRDIIKRYFQATGLVHNREQFMGR 247
Query: 74 MDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIR-NVKYAKFRFQGLEFCDE 132
LR +W + L +GLG N G + A+ WW A + + + KFR E+ E
Sbjct: 248 HRQLRKQWAFCNK-LRSSSGLGRNPD-GTVVAEDDWWAANTKGHPDWKKFRKGLPEYLPE 305
Query: 133 LEFIFGEIVATSQCACAPAMGVPLESTGKNTTADVAREIIESDDELNIDELSPME----- 187
++ +F + + P+E + E+DD+ + DEL+P+
Sbjct: 306 MDRMFEGVAVDGSTSFVATADQPVECDSSD----------EADDDEDDDELTPLSVDNKR 355
Query: 188 ------NTQSKNKRKVSPRMRKQQR 206
S +K+ SP +R R
Sbjct: 356 TSSTSTTASSPSKKSRSPAVRVMDR 380
>UniRef100_Q9LLQ0 Hypothetical protein DUPR11.2 [Oryza sativa]
Length = 809
Score = 50.1 bits (118), Expect = 4e-05
Identities = 37/143 (25%), Positives = 61/143 (41%), Gaps = 2/143 (1%)
Query: 15 GKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRM 74
GK +W + L+ C+ E+ + + ++ + + +R +N T+ Q+K
Sbjct: 115 GKIRWSDDEIGIFLEACLEELSAMTITSTCPKPQGYKNLIQKMKERIGRNLTKDQVKYIW 174
Query: 75 DTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAK-FRFQGLEFCDEL 133
R W +W L K TG+G + I AD WW+ K + A+ F+ L+ D
Sbjct: 175 RQCRKRWMLWTWLESKATGIGRDPITQAIVADDDWWEDKDQKKDGARVFKDSPLKHIDLH 234
Query: 134 EFIF-GEIVATSQCACAPAMGVP 155
IF G V + A A A P
Sbjct: 235 CAIFSGRTVVGNHSAVAGAAPAP 257
>UniRef100_Q7XNC1 OSJNBa0029C04.7 protein [Oryza sativa]
Length = 758
Score = 50.1 bits (118), Expect = 4e-05
Identities = 37/143 (25%), Positives = 61/143 (41%), Gaps = 2/143 (1%)
Query: 15 GKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRM 74
GK +W + L+ C+ E+ + + ++ + + +R +N T+ Q+K
Sbjct: 4 GKIRWSDDEIGIFLEACLEELSAMTITSTCPKPQGYKNLIQKMKERIGRNLTKDQVKYIW 63
Query: 75 DTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAK-FRFQGLEFCDEL 133
R W +W L K TG+G + I AD WW+ K + A+ F+ L+ D
Sbjct: 64 RQCRKRWMLWTWLESKATGIGRDPITQAIVADDDWWEDKDQKKDGARVFKDSPLKHIDLH 123
Query: 134 EFIF-GEIVATSQCACAPAMGVP 155
IF G V + A A A P
Sbjct: 124 CAIFSGRTVVGNHSAVAGAAPAP 146
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.132 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 364,964,703
Number of Sequences: 2790947
Number of extensions: 14737492
Number of successful extensions: 45705
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 45627
Number of HSP's gapped (non-prelim): 89
length of query: 216
length of database: 848,049,833
effective HSP length: 122
effective length of query: 94
effective length of database: 507,554,299
effective search space: 47710104106
effective search space used: 47710104106
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 72 (32.3 bits)
Medicago: description of AC140104.6


