

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140104.12 - phase: 0
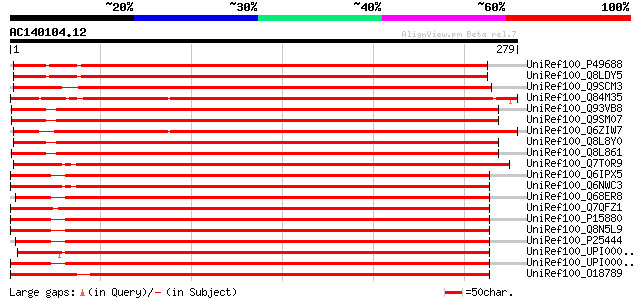
(279 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P49688 40S ribosomal protein S2 [Arabidopsis thaliana] 472 e-132
UniRef100_Q8LDY5 40S ribosomal protein S2 [Arabidopsis thaliana] 469 e-131
UniRef100_Q9SCM3 40S ribosomal protein S2 homolog [Arabidopsis t... 465 e-130
UniRef100_Q84M35 Putative 40S ribosomal protein S2 [Oryza sativa] 462 e-129
UniRef100_Q93VB8 Ribosomal protein S2, putative [Arabidopsis tha... 459 e-128
UniRef100_Q9SM07 XW6 protein [Arabidopsis thaliana] 459 e-128
UniRef100_Q6ZIW7 Putative 40S ribosomal protein S2 [Oryza sativa] 458 e-128
UniRef100_Q8L8Y0 Ribosomal protein S2, putative [Arabidopsis tha... 457 e-127
UniRef100_Q8L861 Ribosomal protein S2, putative [Arabidopsis tha... 457 e-127
UniRef100_Q7T0R9 Sop-prov protein [Xenopus laevis] 397 e-109
UniRef100_Q6IPX5 Ribosomal protein S2 [Homo sapiens] 394 e-108
UniRef100_Q6NWC3 Zgc:85824 [Brachydanio rerio] 392 e-108
UniRef100_Q68ER8 MGC89305 protein [Xenopus tropicalis] 392 e-108
UniRef100_Q7QFZ1 ENSANGP00000015322 [Anopheles gambiae str. PEST] 392 e-108
UniRef100_P15880 40S ribosomal protein S2 [Homo sapiens] 392 e-108
UniRef100_Q8N5L9 Ribosomal protein S2 [Homo sapiens] 391 e-107
UniRef100_P25444 40S ribosomal protein S2 [Mus musculus] 391 e-107
UniRef100_UPI00002513F3 UPI00002513F3 UniRef100 entry 390 e-107
UniRef100_UPI000013EEA4 UPI000013EEA4 UniRef100 entry 390 e-107
UniRef100_O18789 40S ribosomal protein S2 [Bos taurus] 388 e-107
>UniRef100_P49688 40S ribosomal protein S2 [Arabidopsis thaliana]
Length = 285
Score = 472 bits (1214), Expect = e-132
Identities = 233/261 (89%), Positives = 243/261 (92%), Gaps = 3/261 (1%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
ERGG+RG FG GFGGRGG RG RGGRG GRGGRR GGR EEEKWVPVTKLGRLV G
Sbjct: 11 ERGGERGDFGRGFGGRGG-RGDRGGRGRGGRGGRR--GGRASEEEKWVPVTKLGRLVAAG 67
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
I+ +EQIYLHSLP+KE+QIID L+GPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD
Sbjct: 68 HIKQIEQIYLHSLPVKEYQIIDMLIGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDG 127
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSVTV
Sbjct: 128 NGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 187
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFLTP
Sbjct: 188 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFLTP 247
Query: 243 EFWKETRFSKSPFQEYTDLLA 263
EFWKETRFS+SP+QE+TD LA
Sbjct: 248 EFWKETRFSRSPYQEHTDFLA 268
>UniRef100_Q8LDY5 40S ribosomal protein S2 [Arabidopsis thaliana]
Length = 285
Score = 469 bits (1207), Expect = e-131
Identities = 232/261 (88%), Positives = 242/261 (91%), Gaps = 3/261 (1%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
ERGG+RG FG GFGGRGG RG RGGRG GRGGRR GGR EEEKWVPVTKLGR V G
Sbjct: 11 ERGGERGDFGRGFGGRGG-RGDRGGRGRGGRGGRR--GGRASEEEKWVPVTKLGRHVAAG 67
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
I+ +EQIYLHSLP+KE+QIID L+GPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD
Sbjct: 68 HIKQIEQIYLHSLPVKEYQIIDMLIGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDG 127
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSVTV
Sbjct: 128 NGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 187
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFLTP
Sbjct: 188 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFLTP 247
Query: 243 EFWKETRFSKSPFQEYTDLLA 263
EFWKETRFS+SP+QE+TD LA
Sbjct: 248 EFWKETRFSRSPYQEHTDFLA 268
>UniRef100_Q9SCM3 40S ribosomal protein S2 homolog [Arabidopsis thaliana]
Length = 276
Score = 465 bits (1196), Expect = e-130
Identities = 229/263 (87%), Positives = 239/263 (90%), Gaps = 8/263 (3%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
ERGGDRG FG GFGGRGG RGG GRG R GR EEEKWVPVTKLGRLVKEG
Sbjct: 7 ERGGDRGDFGRGFGGRGGGRGGPRGRG--------RRAGRAPEEEKWVPVTKLGRLVKEG 58
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
KI +EQIYLHSLP+KE+QIID LVGP+LKDEVMKIMPVQKQTRAGQRTRFKAF+VVGD+
Sbjct: 59 KITKIEQIYLHSLPVKEYQIIDLLVGPSLKDEVMKIMPVQKQTRAGQRTRFKAFIVVGDS 118
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
NGHVGLGVKCSKEVATAIRGAIILAKLSV+P+RRGYWGNKIGKPHTVPCKVTGKCGSVTV
Sbjct: 119 NGHVGLGVKCSKEVATAIRGAIILAKLSVVPIRRGYWGNKIGKPHTVPCKVTGKCGSVTV 178
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFLTP
Sbjct: 179 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFLTP 238
Query: 243 EFWKETRFSKSPFQEYTDLLAKP 265
EFWKETRFSKSP+QE+TD L P
Sbjct: 239 EFWKETRFSKSPYQEHTDFLLIP 261
>UniRef100_Q84M35 Putative 40S ribosomal protein S2 [Oryza sativa]
Length = 274
Score = 462 bits (1188), Expect = e-129
Identities = 237/281 (84%), Positives = 256/281 (90%), Gaps = 9/281 (3%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
MAERGG+RGG GFG RG RGGRG RG R RGGRR G R+EEEKWVPVTKLGRLVK
Sbjct: 1 MAERGGERGGERGGFG-RGFGRGGRGDRGGR-RGGRR---GPRQEEEKWVPVTKLGRLVK 55
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
EG+ +E+IYLHSLP+KEHQI++TLV P LKDEVMKI PVQKQTRAGQRTRFKAFVVVG
Sbjct: 56 EGRFSKIEEIYLHSLPVKEHQIVETLV-PGLKDEVMKITPVQKQTRAGQRTRFKAFVVVG 114
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
DNNGHVGLGVKC+KEVATAIRGAIILAKLSV+PVRRGYWGNKIG+PHTVPCKVTGKCGSV
Sbjct: 115 DNNGHVGLGVKCAKEVATAIRGAIILAKLSVVPVRRGYWGNKIGQPHTVPCKVTGKCGSV 174
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGI+DVFTSSRGSTKTLGNFVKATFDCLMKTYGFL
Sbjct: 175 TVRMVPAPRGSGIVAARVPKKVLQFAGIEDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 234
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKALILEE--ERVEA 279
TP+FW++T+F KSPFQEYTDLLAKPT KAL+++ E VEA
Sbjct: 235 TPDFWRDTKFVKSPFQEYTDLLAKPT-KALMIDAPVENVEA 274
>UniRef100_Q93VB8 Ribosomal protein S2, putative [Arabidopsis thaliana]
Length = 284
Score = 459 bits (1181), Expect = e-128
Identities = 231/269 (85%), Positives = 239/269 (87%), Gaps = 6/269 (2%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
AERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV
Sbjct: 10 AERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVA 64
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 65 DNKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 124
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 125 DGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSV 184
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFL
Sbjct: 185 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFL 244
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKA 269
TPEFWKETRFS+SP+QE+TD L+ A
Sbjct: 245 TPEFWKETRFSRSPYQEHTDFLSTKAVSA 273
>UniRef100_Q9SM07 XW6 protein [Arabidopsis thaliana]
Length = 284
Score = 459 bits (1181), Expect = e-128
Identities = 231/269 (85%), Positives = 239/269 (87%), Gaps = 6/269 (2%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
AERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV
Sbjct: 10 AERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVA 64
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 65 DNKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 124
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 125 DGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSV 184
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFL
Sbjct: 185 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFL 244
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKA 269
TPEFWKETRFS+SP+QE+TD L+ A
Sbjct: 245 TPEFWKETRFSRSPYQEHTDFLSTKAVSA 273
>UniRef100_Q6ZIW7 Putative 40S ribosomal protein S2 [Oryza sativa]
Length = 276
Score = 458 bits (1179), Expect = e-128
Identities = 227/277 (81%), Positives = 245/277 (87%), Gaps = 9/277 (3%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
ERGG+RGGFG GFG RGGRGDRGRGGR G R+EEEKWVPVTKLGRLVKE
Sbjct: 9 ERGGERGGFGRGFG--------RGGRGDRGRGGRGGRRGPRQEEEKWVPVTKLGRLVKEN 60
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
KI +E+IYLHSLP+KEHQI++ LV P LKDEVMKI PVQKQTRAGQRTRFKAFVVVGD
Sbjct: 61 KIHKIEEIYLHSLPVKEHQIVEQLV-PGLKDEVMKITPVQKQTRAGQRTRFKAFVVVGDG 119
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
+GHVGLGVKC+KEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSVTV
Sbjct: 120 DGHVGLGVKCAKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 179
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
RMVPAPRGSGIVAA VPKKVLQFAGI+DVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP
Sbjct: 180 RMVPAPRGSGIVAAHVPKKVLQFAGIEDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 239
Query: 243 EFWKETRFSKSPFQEYTDLLAKPTAKALILEEERVEA 279
+FW+ETRF K+PFQEYTDLLA+P + E++EA
Sbjct: 240 DFWRETRFIKTPFQEYTDLLARPKGLVIEAPAEKIEA 276
>UniRef100_Q8L8Y0 Ribosomal protein S2, putative [Arabidopsis thaliana]
Length = 284
Score = 457 bits (1177), Expect = e-127
Identities = 230/268 (85%), Positives = 238/268 (87%), Gaps = 6/268 (2%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVKE 61
ERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV +
Sbjct: 11 ERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVAD 65
Query: 62 GKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD 121
KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD
Sbjct: 66 NKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD 125
Query: 122 NNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVT 181
NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSVT
Sbjct: 126 GNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSVT 185
Query: 182 VRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLT 241
VRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFLT
Sbjct: 186 VRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFLT 245
Query: 242 PEFWKETRFSKSPFQEYTDLLAKPTAKA 269
PEFWKETRFS+SP+QE+TD L+ A
Sbjct: 246 PEFWKETRFSRSPYQEHTDFLSTKAVSA 273
>UniRef100_Q8L861 Ribosomal protein S2, putative [Arabidopsis thaliana]
Length = 284
Score = 457 bits (1175), Expect = e-127
Identities = 230/269 (85%), Positives = 238/269 (87%), Gaps = 6/269 (2%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
AERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV
Sbjct: 10 AERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVA 64
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 65 DNKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 124
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 125 DGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSV 184
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLG FVKATFDCL KTYGFL
Sbjct: 185 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGKFVKATFDCLQKTYGFL 244
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKA 269
TPEFWKETRFS+SP+QE+TD L+ A
Sbjct: 245 TPEFWKETRFSRSPYQEHTDFLSTKAVSA 273
>UniRef100_Q7T0R9 Sop-prov protein [Xenopus laevis]
Length = 281
Score = 397 bits (1019), Expect = e-109
Identities = 196/273 (71%), Positives = 227/273 (82%), Gaps = 3/273 (1%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
+ GG+RGGF GFGG G RG GRG RGRG R A G + ++++WVPVTKLGRLVK+
Sbjct: 4 DAGGNRGGFRGGFGGGGRGRGRGRGRG-RGRG--RGARGGKADDKEWVPVTKLGRLVKDM 60
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
KI+SLE+IYL SLPIKE +IID +G TLKDEV+KIMPVQKQTRAGQRTRFKAFV +GD
Sbjct: 61 KIKSLEEIYLFSLPIKESEIIDFFLGSTLKDEVLKIMPVQKQTRAGQRTRFKAFVAIGDY 120
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV V
Sbjct: 121 NGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSVLV 180
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
R++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +LTP
Sbjct: 181 RLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYLTP 240
Query: 243 EFWKETRFSKSPFQEYTDLLAKPTAKALILEEE 275
+ WKET F+KSP+QEYTD LAK + + E
Sbjct: 241 DLWKETVFAKSPYQEYTDHLAKTHTRVSVQRTE 273
>UniRef100_Q6IPX5 Ribosomal protein S2 [Homo sapiens]
Length = 293
Score = 394 bits (1011), Expect = e-108
Identities = 193/264 (73%), Positives = 222/264 (83%), Gaps = 7/264 (2%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
M RGG RGGFGSG GRG RG RGRG R A G + E+++W+PVTKLGRLVK
Sbjct: 19 MGNRGGFRGGFGSGIRGRGRGRG-------RGRGRGRGARGGKAEDKEWMPVTKLGRLVK 71
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 72 DMKIKSLEEIYLFSLPIKESEIIDFFLGASLKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 131
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 132 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSV 191
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 192 LVRLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 251
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD LAK
Sbjct: 252 TPDLWKETVFTKSPYQEFTDHLAK 275
>UniRef100_Q6NWC3 Zgc:85824 [Brachydanio rerio]
Length = 280
Score = 392 bits (1008), Expect = e-108
Identities = 195/264 (73%), Positives = 223/264 (83%), Gaps = 3/264 (1%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
MA+ G RGGF GFG G RG GRG RGRG R A G + E+++WVPVTKLGRLVK
Sbjct: 1 MADDAGGRGGFRGGFGTGGRGRGRGRGRG-RGRG--RGARGGKSEDKEWVPVTKLGRLVK 57
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL+SLPIKE +IID +G LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 58 DMKIKSLEEIYLYSLPIKESEIIDFFLGSALKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 117
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS+IPVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 118 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIIPVRRGYWGNKIGKPHTVPCKVTGRCGSV 177
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 178 LVRLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 237
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD LAK
Sbjct: 238 TPDLWKETVFTKSPYQEFTDHLAK 261
>UniRef100_Q68ER8 MGC89305 protein [Xenopus tropicalis]
Length = 281
Score = 392 bits (1007), Expect = e-108
Identities = 193/261 (73%), Positives = 221/261 (83%), Gaps = 7/261 (2%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEGK 63
RGG RGGFGSG GRG RG RGRG R A G + ++++WVPVTKLGRLVK+ K
Sbjct: 9 RGGFRGGFGSGGRGRGRGRG-------RGRGRGRGARGGKADDKEWVPVTKLGRLVKDMK 61
Query: 64 IRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNN 123
I+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +GD N
Sbjct: 62 IKSLEEIYLFSLPIKESEIIDFFLGSSLKDEVLKIMPVQKQTRAGQRTRFKAFVAIGDYN 121
Query: 124 GHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTVR 183
GHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV VR
Sbjct: 122 GHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSVLVR 181
Query: 184 MVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTPE 243
++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +LTP+
Sbjct: 182 LIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYLTPD 241
Query: 244 FWKETRFSKSPFQEYTDLLAK 264
WKET F+KSP+QEYTD LAK
Sbjct: 242 LWKETVFTKSPYQEYTDHLAK 262
>UniRef100_Q7QFZ1 ENSANGP00000015322 [Anopheles gambiae str. PEST]
Length = 271
Score = 392 bits (1007), Expect = e-108
Identities = 190/265 (71%), Positives = 224/265 (83%), Gaps = 3/265 (1%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRG-GRRRAGGRREEEEKWVPVTKLGRLV 59
MA+ RGGF GFG RGG RGG GRG RGRG GR R G +E+ ++WVPVTKLGRLV
Sbjct: 1 MADAAPARGGFRGGFGSRGGARGG--GRGGRGRGRGRGRGRGGKEDSKEWVPVTKLGRLV 58
Query: 60 KEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVV 119
K+GKIR+LE+IYL+SLPIKE +IID + LKDEV+KIMPVQKQTRAGQRTRFKAFV +
Sbjct: 59 KDGKIRTLEEIYLYSLPIKEFEIIDFFLNRLLKDEVLKIMPVQKQTRAGQRTRFKAFVAI 118
Query: 120 GDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGS 179
GD+NGH+GLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIG+PHTVPCKV+GKCGS
Sbjct: 119 GDSNGHIGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGRPHTVPCKVSGKCGS 178
Query: 180 VTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGF 239
V VR++PAPRG+GIV+A VPKK+LQ AGIDD +T++RGST TLGNF KAT+ + KTY F
Sbjct: 179 VLVRLIPAPRGTGIVSAPVPKKLLQMAGIDDCYTTTRGSTTTLGNFAKATYAAIAKTYAF 238
Query: 240 LTPEFWKETRFSKSPFQEYTDLLAK 264
LTP+ WK+ +K+P+QE+ D L K
Sbjct: 239 LTPDLWKDLPLNKTPYQEFADFLDK 263
>UniRef100_P15880 40S ribosomal protein S2 [Homo sapiens]
Length = 293
Score = 392 bits (1007), Expect = e-108
Identities = 192/264 (72%), Positives = 221/264 (82%), Gaps = 7/264 (2%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
M RGG RGGFGSG GRG RG RGRG R A G + E+++W+PVTKLGRLVK
Sbjct: 19 MGNRGGFRGGFGSGIRGRGRGRG-------RGRGRGRGARGGKAEDKEWMPVTKLGRLVK 71
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 72 DMKIKSLEEIYLFSLPIKESEIIDFFLGASLKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 131
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 132 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSV 191
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 192 LVRLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 251
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD L K
Sbjct: 252 TPDLWKETVFTKSPYQEFTDHLVK 275
>UniRef100_Q8N5L9 Ribosomal protein S2 [Homo sapiens]
Length = 293
Score = 391 bits (1004), Expect = e-107
Identities = 192/264 (72%), Positives = 220/264 (82%), Gaps = 7/264 (2%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
M RGG RGGFGSG GRG RG RGRG R A G + E+++W+PVTKLGRLVK
Sbjct: 19 MGNRGGFRGGFGSGIRGRGRGRG-------RGRGRGRGARGGKAEDKEWMPVTKLGRLVK 71
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 72 DMKIKSLEEIYLFSLPIKESEIIDFFLGASLKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 131
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 132 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSV 191
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 192 LVRLIPAPRGIGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 251
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD L K
Sbjct: 252 TPDLWKETVFTKSPYQEFTDHLVK 275
>UniRef100_P25444 40S ribosomal protein S2 [Mus musculus]
Length = 293
Score = 391 bits (1004), Expect = e-107
Identities = 191/261 (73%), Positives = 220/261 (84%), Gaps = 7/261 (2%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEGK 63
RGG RGGFGSG GRG RG RGRG R A G + E+++W+PVTKLGRLVK+ K
Sbjct: 22 RGGFRGGFGSGLRGRGRGRG-------RGRGRGRGARGGKAEDKEWIPVTKLGRLVKDMK 74
Query: 64 IRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNN 123
I+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +GD N
Sbjct: 75 IKSLEEIYLFSLPIKESEIIDFFLGASLKDEVLKIMPVQKQTRAGQRTRFKAFVAIGDYN 134
Query: 124 GHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTVR 183
GHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV VR
Sbjct: 135 GHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSVLVR 194
Query: 184 MVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTPE 243
++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +LTP+
Sbjct: 195 LIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYLTPD 254
Query: 244 FWKETRFSKSPFQEYTDLLAK 264
WKET F+KSP+QE+TD L K
Sbjct: 255 LWKETVFTKSPYQEFTDHLVK 275
>UniRef100_UPI00002513F3 UPI00002513F3 UniRef100 entry
Length = 289
Score = 390 bits (1003), Expect = e-107
Identities = 193/264 (73%), Positives = 222/264 (83%), Gaps = 5/264 (1%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRG----GRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
GG G G G GGRGG RGG G GRG RGRG R A G + E+++W+PVTKLGRLVK
Sbjct: 9 GGPGGPGGPGLGGRGGFRGGFGSGLRGRG-RGRGRGRGARGGKAEDKEWIPVTKLGRLVK 67
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 68 DMKIKSLEEIYLFSLPIKESEIIDFFLGASLKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 127
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 128 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSV 187
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 188 LVRLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 247
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD L K
Sbjct: 248 TPDLWKETVFTKSPYQEFTDHLVK 271
>UniRef100_UPI000013EEA4 UPI000013EEA4 UniRef100 entry
Length = 293
Score = 390 bits (1001), Expect = e-107
Identities = 191/264 (72%), Positives = 220/264 (82%), Gaps = 7/264 (2%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
M RGG RGGFGSG GRG RG RGRG R A G + E+++W+PVTKLGRLVK
Sbjct: 19 MGNRGGFRGGFGSGIRGRGRGRG-------RGRGRGRGARGGKAEDKEWMPVTKLGRLVK 71
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRA QRTRFKAFV +G
Sbjct: 72 DMKIKSLEEIYLFSLPIKESEIIDFFLGASLKDEVLKIMPVQKQTRASQRTRFKAFVAIG 131
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 132 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSV 191
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 192 LVRLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 251
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD L K
Sbjct: 252 TPDLWKETVFTKSPYQEFTDHLVK 275
>UniRef100_O18789 40S ribosomal protein S2 [Bos taurus]
Length = 286
Score = 388 bits (997), Expect = e-107
Identities = 190/264 (71%), Positives = 219/264 (81%), Gaps = 7/264 (2%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
M RGG RGGFGSG GRG RG GRG RGG+ E+++W+PVTKLGRLVK
Sbjct: 12 MGGRGGFRGGFGSGVRGRGRGRGRGRGRGGGARGGKA-------EDKEWLPVTKLGRLVK 64
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 65 DMKIKSLEEIYLFSLPIKESEIIDFFLGASLKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 124
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 125 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSV 184
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++P PRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 185 LVRLIPPPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 244
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD L K
Sbjct: 245 TPDLWKETVFTKSPYQEFTDHLVK 268
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 488,641,166
Number of Sequences: 2790947
Number of extensions: 22542039
Number of successful extensions: 275928
Number of sequences better than 10.0: 3949
Number of HSP's better than 10.0 without gapping: 2775
Number of HSP's successfully gapped in prelim test: 1263
Number of HSP's that attempted gapping in prelim test: 168460
Number of HSP's gapped (non-prelim): 51405
length of query: 279
length of database: 848,049,833
effective HSP length: 126
effective length of query: 153
effective length of database: 496,390,511
effective search space: 75947748183
effective search space used: 75947748183
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC140104.12


