

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139344.7 + phase: 1 /pseudo
(478 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
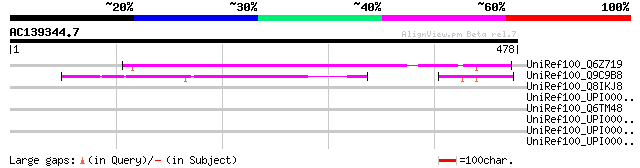
UniRef100_Q6Z719 Hypothetical protein P0575F10.13 [Oryza sativa] 276 9e-73
UniRef100_Q9C9B8 Hypothetical protein F2P9.16 [Arabidopsis thali... 169 2e-40
UniRef100_Q8IKJ8 Hypothetical protein [Plasmodium falciparum] 36 2.1
UniRef100_UPI000032A68E UPI000032A68E UniRef100 entry 35 3.6
UniRef100_Q6TM48 Hypothetical protein orf54 [Bacteriophage D3112] 35 3.6
UniRef100_UPI000043740D UPI000043740D UniRef100 entry 34 8.1
UniRef100_UPI00003055F4 UPI00003055F4 UniRef100 entry 34 8.1
UniRef100_UPI00002F8790 UPI00002F8790 UniRef100 entry 34 8.1
>UniRef100_Q6Z719 Hypothetical protein P0575F10.13 [Oryza sativa]
Length = 749
Score = 276 bits (706), Expect = 9e-73
Identities = 158/385 (41%), Positives = 228/385 (59%), Gaps = 31/385 (8%)
Query: 107 RTLRFRNL-----GNDLQDR-VLLQCVTLGMTRTISFSSHSSLFVCLGLSLLTRILPLPR 160
RT+RF G + DR +LLQCV LG+T+ + H S+ C+ ++LL +LPLP
Sbjct: 295 RTIRFAAEKAVLEGKHVDDRRLLLQCVALGLTQCGQVTPHESVLRCVCMALLEELLPLPD 354
Query: 161 LYE-SVFELSPSSGGLKVNEIKEHPDNILFKEAGAVTGIFCNLYVLADEENKNIVENLIW 219
L + SV +S + N +K+H D++LFKEAG V GI CN Y A ++ K VE +W
Sbjct: 355 LLKMSVQCPDGNSPEIVKNRVKQHLDSVLFKEAGPVAGILCNQYSFASDKAKTSVETCVW 414
Query: 220 EYYRDIYFGHRKVVMDLKGKEDELLTNFEKTAESAFLMVVVFALSVTKHKLSSTFAQEIQ 279
EY + +Y R V+ +GK+D+L+T+ EK AE+AFLMVVVF+ VTKH+L++ ++ Q
Sbjct: 415 EYAQVLYCHLRAAVILHQGKQDDLITDIEKIAEAAFLMVVVFSAEVTKHRLNAKSSEGFQ 474
Query: 280 TEISLKILVSLSCVEYFRHVRLPEYMETIRKVTAIVKKNENACTFFVNSIPSYGDLTNGP 339
++++KILVS SC+E+ R +RLPEY E +R+ + ++N F+ SIPSY +LTN
Sbjct: 475 PDVAVKILVSFSCLEHLRRLRLPEYTEAVRRAVLVNQENAAVAALFIESIPSYAELTNLL 534
Query: 340 D-QKTKYFWSKDEVQTARVLLYLRVIPTLIECLRGPVFGDMVAPTMFLYMEHPNGKVARA 398
T+Y W D VQT+R+L YLR+ +C + Y++H N KV RA
Sbjct: 535 TLDGTRYIWHGDVVQTSRILFYLRIFDIKKKCPYHKI---------VRYIQHSNEKVTRA 585
Query: 399 SHSVFTAFISMGKESEKIDSLIEGEARFLSLSIIILPNTL----------GMASGVVGMV 448
SHSV +F+S G +++ D + E L+ + TL G+ASGV +
Sbjct: 586 SHSVVVSFLSSGNDTDPDDRMALKE----QLAFYYIKRTLEAYPGVTPFEGLASGVAALA 641
Query: 449 RHLPAGSPTTFYCIHSLVEKANQLC 473
RHLPAGSP T +CIH+LV KA LC
Sbjct: 642 RHLPAGSPATLFCIHNLVVKAKDLC 666
>UniRef100_Q9C9B8 Hypothetical protein F2P9.16 [Arabidopsis thaliana]
Length = 699
Score = 169 bits (428), Expect = 2e-40
Identities = 109/291 (37%), Positives = 158/291 (53%), Gaps = 43/291 (14%)
Query: 50 ESFKENYAPFVVFMSGIGVLRVTDRYASSTGMKVDVLTRMRTSAIVRVEALVSDLVSRTL 109
E+ KE YA F VFM+ GV+R + SS +++ +++R SA R+E + LVS
Sbjct: 264 ETSKEKYAVFAVFMAAAGVVRASTAGFSSGAQSLEI-SKLRNSAEKRIEFVAQILVSNG- 321
Query: 110 RFRNLGNDLQDRVLLQCVTLGMTRTISFSSHSSLFVCLGLSLLTRILPLPRLYES---VF 166
L ++ LL+C + + R S SS + L +CL +LLT++ PL ++YES F
Sbjct: 322 NVVTLPTTQREGPLLKCFAIALARCGSVSSSAPLLLCLTSALLTQVFPLGQIYESFCNAF 381
Query: 167 ELSPSSGGLKVNEIKEHPDNILFKEAGAVTGIFCNLYVLADEENKNIVENLIWEYYRDIY 226
P G ++ ++EH ++LFKE+GA++G FCN Y A EENK IVEN+IW++ +++Y
Sbjct: 382 GKEPI--GPRLIWVREHLSDVLFKESGAISGAFCNQYSSASEENKYIVENMIWDFCQNLY 439
Query: 227 FGHRKVVMDLKGKEDELLTNFEKTAESAFLMVVVFALSVTKHKLSSTFAQEIQTEISLKI 286
HR++ M L G ED LL + EK AES+FLMVVVFAL+VTK L ++E + E
Sbjct: 440 LQHRQIAMLLCGIEDTLLGDIEKIAESSFLMVVVFALAVTKQWLKPIVSKERKME----- 494
Query: 287 LVSLSCVEYFRHVRLPEYMETIRKVTAIVKKNENACTFFVNSIPSYGDLTN 337
N+ C FV SIP+Y LTN
Sbjct: 495 -------------------------------NDAPCVSFVESIPAYDSLTN 514
Score = 56.6 bits (135), Expect = 2e-06
Identities = 33/77 (42%), Positives = 46/77 (58%), Gaps = 6/77 (7%)
Query: 405 AFISMGKESEKIDSLIEGEAR---FLSLSIIILPNTL---GMASGVVGMVRHLPAGSPTT 458
AF+S KESE+ + E ++ S+ + P G+ASGV +V+HLPAGSP
Sbjct: 517 AFLSSAKESEEDERTQFKEQLVFYYMQRSLEVYPEITPFEGLASGVATLVQHLPAGSPAI 576
Query: 459 FYCIHSLVEKANQLCSE 475
FY +HSLVEKA+ +E
Sbjct: 577 FYSVHSLVEKASTFSTE 593
>UniRef100_Q8IKJ8 Hypothetical protein [Plasmodium falciparum]
Length = 1068
Score = 36.2 bits (82), Expect = 2.1
Identities = 32/90 (35%), Positives = 46/90 (50%), Gaps = 9/90 (10%)
Query: 175 LKVNEIKEHPD-NILFKEAGAVTGIFCNLYVLADEENKNIVENLIWEYYRDIYFGHRKVV 233
L+ N IKE D + L K + GI N++ A E K I++N +E FG
Sbjct: 375 LRRNNIKEIFDVDDLVKNIKSFLGIKSNIHE-AFENQKLIIKNCNYES-----FGPELCS 428
Query: 234 MDLKGKEDELLTNFEKTAESAFLMVVVFAL 263
+D K KE +L +EK SAFL +++F L
Sbjct: 429 VDEKAKE--MLWKYEKKKNSAFLFIILFTL 456
>UniRef100_UPI000032A68E UPI000032A68E UniRef100 entry
Length = 260
Score = 35.4 bits (80), Expect = 3.6
Identities = 28/93 (30%), Positives = 48/93 (51%), Gaps = 8/93 (8%)
Query: 176 KVNEIKEHPDNILFKEAGAVTGIFCNLYVLADEENKNIVENLIWEYYRDIYFGHRKVVMD 235
K+N + N++F E T IF + E+ KN+ + IW++Y D H + + +
Sbjct: 119 KININMDKQSNVIFCE----TSIFGRRAM--SEKIKNLSFSDIWKFYIDNKISHLESI-N 171
Query: 236 LKGKEDELLTN-FEKTAESAFLMVVVFALSVTK 267
+ G+ D+LL N F +SA +++F VTK
Sbjct: 172 IDGETDKLLKNKFTLDKQSAISTILIFGDIVTK 204
>UniRef100_Q6TM48 Hypothetical protein orf54 [Bacteriophage D3112]
Length = 383
Score = 35.4 bits (80), Expect = 3.6
Identities = 24/89 (26%), Positives = 41/89 (45%), Gaps = 12/89 (13%)
Query: 384 MFLYMEHPNGKVARASHSVFTAFISMGKESEKIDSLIEGEARFLSLSIIILPNTLGMASG 443
+ + M P+G +AR +TA ++ G + S ++G R+L ++ + L +A G
Sbjct: 221 LLVAMGRPSGSLARYGVEFYTALVTAGPNATTGSSGLDGTTRYLQMTNV--SRALFIADG 278
Query: 444 ----------VVGMVRHLPAGSPTTFYCI 462
G + +PAG PTT Y I
Sbjct: 279 WSTAVLWVRAETGSLHFMPAGVPTTDYRI 307
>UniRef100_UPI000043740D UPI000043740D UniRef100 entry
Length = 1285
Score = 34.3 bits (77), Expect = 8.1
Identities = 62/330 (18%), Positives = 125/330 (37%), Gaps = 65/330 (19%)
Query: 66 IGVLRVTDRYASSTGMKVDVLTRMRTSAIVRVEALVSDLVSRTLRFRNLGNDLQDRVLLQ 125
+GV+ +T +S G +V+ + S + R D S+ N ++ +L +
Sbjct: 816 LGVI-ITKNLSSLYGANYNVINQSIKSDMERWSTYPLDFASKI-------NVIKTNILPR 867
Query: 126 CVTLGMTRTISFSSHSSLFVCLGLSLLTRIL---PLPRLYESVFELSPSSGGLKVNEIKE 182
+ L + H + L++R + PR+ + +LS GG+ + +K+
Sbjct: 868 LLYLFQALPVEIPKHQ---FTIWNKLISRFIWDGKKPRIRYTTLQLSKQKGGMALPNLKD 924
Query: 183 HPDNILFKEAGAVTGIFCNLYVLADEENKNIVENLIWEYYRDIYFGHRKVVMDLKGKED- 241
+ F+ ++CN A + +E Y + G +++ +K + D
Sbjct: 925 YFYAAQFRP----LFLWCNNNYFARWKE---IETYTVGYQIQSFLGEKEIPSVVKSQLDS 977
Query: 242 ----------ELLTNFEKTAESAFLMVVVFALSVTKHKLSSTFAQ--------------- 276
E F+ + + L + F + +TF Q
Sbjct: 978 ITSGAVEFWFEFARQFKLSKDQKVLRWIAFDTQFKPGESDATFKQWATKGISAICTVLEN 1037
Query: 277 -EIQTEISLKILVSLSCVEYFRHVRLPEY------METIRKVTAIVK------KNENACT 323
E+QT +LK +SL+ + FR ++ +Y MET +++ +VK K+++ C
Sbjct: 1038 KELQTFQTLKEKLSLTNRDLFRFIQFRDYFYREIKMETSHEISPVVKIMMNAYKSDSKCA 1097
Query: 324 FFVNSIPSYGDLTNGPDQKTKYF---WSKD 350
++S Y + T Y W K+
Sbjct: 1098 KIISSF--YVSIMESKTNSTLYLKSRWEKE 1125
>UniRef100_UPI00003055F4 UPI00003055F4 UniRef100 entry
Length = 197
Score = 34.3 bits (77), Expect = 8.1
Identities = 17/43 (39%), Positives = 25/43 (57%), Gaps = 3/43 (6%)
Query: 174 GLKVNEIKEHPDNILFKEAGAVTGIFCNLYVLADEENKNIVEN 216
G+K+ E + N++FK A+ G+F L DEE K+I EN
Sbjct: 145 GIKLKEFPKLDSNLMFK---AIGGLFTGLIFFGDEEKKSISEN 184
>UniRef100_UPI00002F8790 UPI00002F8790 UniRef100 entry
Length = 171
Score = 34.3 bits (77), Expect = 8.1
Identities = 29/94 (30%), Positives = 41/94 (42%), Gaps = 1/94 (1%)
Query: 253 SAFLMVVVFALSVTKHKLSSTFAQEIQTEISLKILVSLSCVEYFRHVRLPEYMETIRKVT 312
S +L L VT L A E SL +L L CV Y + LPE+++ + K
Sbjct: 54 SLYLFGTTLGLLVTSGLLRFGPAFENTAWSSLFLLAPLGCVYYPLSI-LPEWLQILAKGL 112
Query: 313 AIVKKNENACTFFVNSIPSYGDLTNGPDQKTKYF 346
+V E A + VN+I Y ++ N YF
Sbjct: 113 PLVYIFEEARSILVNNIVDYKNIMNALALNLFYF 146
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.140 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 752,066,920
Number of Sequences: 2790947
Number of extensions: 30154228
Number of successful extensions: 93615
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 93603
Number of HSP's gapped (non-prelim): 10
length of query: 478
length of database: 848,049,833
effective HSP length: 131
effective length of query: 347
effective length of database: 482,435,776
effective search space: 167405214272
effective search space used: 167405214272
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC139344.7


