

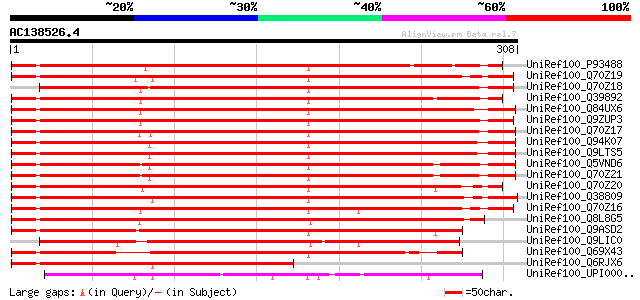
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138526.4 + phase: 0
(308 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P93488 NAP1Ps [Pisum sativum] 371 e-101
UniRef100_Q70Z19 Nucleosome assembly protein 1-like protein 1 [N... 367 e-100
UniRef100_Q70Z18 Nucleosome assembly protein 1-like protein 2 [N... 367 e-100
UniRef100_Q39892 Nucleosome assembly protein 1 [Glycine max] 363 4e-99
UniRef100_Q84UX6 Nucleosome/chromatin assembly factor group A [Z... 362 9e-99
UniRef100_Q9ZUP3 Putative nucleosome assembly protein [Arabidops... 362 9e-99
UniRef100_Q70Z17 Nucleosome assembly protein 1-like protein 3 [N... 359 6e-98
UniRef100_Q94K07 Nucleosome assembly protein [Arabidopsis thaliana] 358 1e-97
UniRef100_Q9LTS5 Nucleosome assembly protein [Arabidopsis thaliana] 358 1e-97
UniRef100_Q5VND6 Putative nucleosome assembly protein 1 [Oryza s... 357 2e-97
UniRef100_Q70Z21 Nucleosome assembly protein 1-like protein 1 [O... 356 4e-97
UniRef100_Q70Z20 Nucleosome assembly protein 1-like protein 2 [O... 350 3e-95
UniRef100_Q38809 Arabidopsis thaliana Col-0 nucleosome assembly ... 346 5e-94
UniRef100_Q70Z16 Nucleosome assembly protein 1-like protein 4 [N... 340 3e-92
UniRef100_Q8L8G5 Nucleosome/chromatin assembly factor group A [Z... 334 2e-90
UniRef100_Q9ASD2 Putative nucleosome assembly protein 1 [Oryza s... 320 2e-86
UniRef100_Q9LIC0 Nucleosome assembly protein-like [Arabidopsis t... 256 5e-67
UniRef100_Q69X43 Putative nucleosome assembly protein 1 [Oryza s... 252 1e-65
UniRef100_Q6RJX6 Induced stolon tip protein NAP1Ps [Capsicum ann... 231 1e-59
UniRef100_UPI000023CCD5 UPI000023CCD5 UniRef100 entry 201 2e-50
>UniRef100_P93488 NAP1Ps [Pisum sativum]
Length = 366
Score = 371 bits (953), Expect = e-101
Identities = 196/311 (63%), Positives = 239/311 (76%), Gaps = 22/311 (7%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
IQ+L Q L+ L SP+V KR+EVL+EIQ EHD+L+AKF EERAALEAKYQ+LYQPL
Sbjct: 33 IQSLAGQHSDVLESL--SPVVRKRVEVLREIQGEHDELEAKFLEERAALEAKYQILYQPL 90
Query: 62 YTKRYEIVNGLAEVEGADID-----EPEVKGVPYFWLVALQNNDEIADEITERDEDALKY 116
YTKRY+IVNG+AEV G ++ E + KGVP FWL A++NND +EITERDE ALK+
Sbjct: 91 YTKRYDIVNGVAEVVGVRVETAVAEEDKEKGVPSFWLNAMKNNDVGGEEITERDEGALKF 150
Query: 117 LKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEWLPGK 176
LKDIK+TR EP+GFKLEFFFDSNPYFSN++LTK YHMVDEDE I+EKAIGTEI+WLPGK
Sbjct: 151 LKDIKWTRIEEPKGFKLEFFFDSNPYFSNSVLTKIYHMVDEDEPILEKAIGTEIQWLPGK 210
Query: 177 SLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQNEMEH 229
LT+ KK AK +TE +SFFNFFN E+P+D +DE+ EELQN+ME
Sbjct: 211 CLTQKVLKKKPKKGAKNAKPITKTETCESFFNFFNPPEVPEDDEDIDEDMAEELQNQMEQ 270
Query: 230 DYDIGSTIRDKIIPNAVSWYTGEASDGES-GDLDDDDDDEDEEDDEEEVEVEKEEEDDNY 288
DYDIGSTIRDKIIP +SW+TGEA+ GE GDLDD+D+DED +D EE+ E E E+EDD
Sbjct: 271 DYDIGSTIRDKIIP--MSWFTGEAAQGEEFGDLDDEDEDED-DDAEEDDEEEDEDEDD-- 325
Query: 289 GLDDDEEDDNE 299
DD+EE++ +
Sbjct: 326 --DDEEEEETK 334
>UniRef100_Q70Z19 Nucleosome assembly protein 1-like protein 1 [Nicotiana tabacum]
Length = 374
Score = 367 bits (943), Expect = e-100
Identities = 187/322 (58%), Positives = 244/322 (75%), Gaps = 26/322 (8%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q +T + L+ L SP V KR+EVLKEIQ +HD+L+AKF+EERA LEAKYQ LYQPL
Sbjct: 33 LQDITGKPTNVLECL--SPNVRKRVEVLKEIQSQHDELEAKFYEERAVLEAKYQKLYQPL 90
Query: 62 YTKRYEIVNGLAEV-----EGADIDEPEV-----KGVPYFWLVALQNNDEIADEITERDE 111
YTKR++IVNG+ EV E A +D+ E KGVP FWL+A++NND +++EITERDE
Sbjct: 91 YTKRFDIVNGVVEVNTSETEAAAMDQDEDEDAVGKGVPDFWLIAMKNNDVLSEEITERDE 150
Query: 112 DALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIE 171
ALK+LKDIK+ + P+GFKLEFFFD+NPYF+NT+LTKTYHM+DEDE I+EKA+GTEIE
Sbjct: 151 GALKFLKDIKWAKIDNPKGFKLEFFFDTNPYFTNTVLTKTYHMIDEDEPILEKALGTEIE 210
Query: 172 WLPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQ 224
W PGK LT+ KK AK +TE+ +SFFNFF+ ++P+D +DE+A EELQ
Sbjct: 211 WYPGKCLTQKILKKKPKKGSKNAKPITKTEQCESFFNFFSPPQVPEDEEDIDEDAAEELQ 270
Query: 225 NEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEE 284
+ ME DYDIGSTIRDKII +AVSW+TGEA++ + DL+DDDDD++E+DD+E ++EEE
Sbjct: 271 SLMEQDYDIGSTIRDKIISHAVSWFTGEAAEDDFADLEDDDDDDEEDDDDE----DEEEE 326
Query: 285 DDNYGLDDDEEDDNEDVNNPKK 306
DD DD++E+D +D N KK
Sbjct: 327 DDE---DDEDEEDEDDTNTKKK 345
>UniRef100_Q70Z18 Nucleosome assembly protein 1-like protein 2 [Nicotiana tabacum]
Length = 377
Score = 367 bits (941), Expect = e-100
Identities = 185/307 (60%), Positives = 237/307 (76%), Gaps = 25/307 (8%)
Query: 19 SPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPLYTKRYEIVNGLAEVEGA 78
SP V KR+E L+EIQ EHD+L+AKFFEERAALEAKYQ LYQPLYTKR+EIVNG+ EV+GA
Sbjct: 48 SPNVRKRVEALREIQTEHDELEAKFFEERAALEAKYQKLYQPLYTKRFEIVNGVVEVDGA 107
Query: 79 -----------DIDEPEVKGVPYFWLVALQNNDEIADEITERDEDALKYLKDIKYTRTTE 127
D D E KGVP FWL A++NN+ +A+EITERDE+ALK+L+DIK++R +
Sbjct: 108 TTEAAAADKQEDKDAVE-KGVPDFWLTAMKNNEVLAEEITERDEEALKFLRDIKWSRIDD 166
Query: 128 PQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEWLPGKSLTE------- 180
P+GFKL+FFF++NPYF N++LTKTYHM+DEDE I+EKAI TEIEW PGK LT+
Sbjct: 167 PKGFKLDFFFETNPYFKNSVLTKTYHMIDEDEPILEKAIATEIEWYPGKCLTQKILKKKP 226
Query: 181 KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQNEMEHDYDIGSTIRDK 240
KK AK +TE+ +SFFNFF+ ++P+D +DE+A EELQN ME DYDIGSTIRDK
Sbjct: 227 KKGSKNAKPITKTEQCESFFNFFSPPQVPEDEEDIDEDAAEELQNLMEQDYDIGSTIRDK 286
Query: 241 IIPNAVSWYTGEASDGESGDL-DDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNE 299
IIP+AVSW+TGEA++ + +L DD+D+D+DEEDDE+E E E++EED D+DEE+D +
Sbjct: 287 IIPHAVSWFTGEAAEDDYAELEDDEDEDDDEEDDEDEDEEEEDEED-----DEDEEEDED 341
Query: 300 DVNNPKK 306
+ KK
Sbjct: 342 ETKTKKK 348
>UniRef100_Q39892 Nucleosome assembly protein 1 [Glycine max]
Length = 358
Score = 363 bits (931), Expect = 4e-99
Identities = 186/311 (59%), Positives = 236/311 (75%), Gaps = 21/311 (6%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
IQ+L L+ L SP V KR+E L+EIQ +HD+L+A F +ER ALEAKYQ LYQPL
Sbjct: 31 IQSLAGAHSDVLETL--SPNVRKRVESLREIQGKHDELEADFLKEREALEAKYQKLYQPL 88
Query: 62 YTKRYEIVNGLAEVEGA------DIDEPEVKGVPYFWLVALQNNDEIADEITERDEDALK 115
YTKRYEIVNG+ EVEGA + +E + KGVP FWL A++NND +A+EI+ERDE ALK
Sbjct: 89 YTKRYEIVNGVTEVEGAANESTDESEENKEKGVPSFWLNAMENNDVLAEEISERDEGALK 148
Query: 116 YLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEWLPG 175
+LKDIK++R P+GFKL+FFFD+NPYFSNT+LTKTYHM+DEDE I+EKAIGTEIEW PG
Sbjct: 149 FLKDIKWSRIENPKGFKLDFFFDTNPYFSNTVLTKTYHMIDEDEPILEKAIGTEIEWYPG 208
Query: 176 KSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQNEME 228
K LT+ KK AK +TE +SFFNFF E+P+D +DE+ EELQN+ME
Sbjct: 209 KCLTQKVLKKKPKKGSKNAKPITKTESCESFFNFFKPPEVPEDDADIDEDLAEELQNQME 268
Query: 229 HDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNY 288
DYDIGST+RDKIIP+AVSW+TGEA+ G+ + +D +DDEDEE+DE+E E E+++ED+
Sbjct: 269 QDYDIGSTLRDKIIPHAVSWFTGEAAQGD--EFEDLEDDEDEEEDEDEDEDEEDDEDE-- 324
Query: 289 GLDDDEEDDNE 299
DD+EEDD +
Sbjct: 325 --DDEEEDDTK 333
>UniRef100_Q84UX6 Nucleosome/chromatin assembly factor group A [Zea mays]
Length = 380
Score = 362 bits (928), Expect = 9e-99
Identities = 182/324 (56%), Positives = 240/324 (73%), Gaps = 28/324 (8%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q+L Q L+ L SP V KR+E L+EIQ +HD+++AKFFEERAALEAKYQ LY+PL
Sbjct: 40 LQSLAGQHADVLETL--SPNVRKRVEFLREIQSQHDEIEAKFFEERAALEAKYQKLYEPL 97
Query: 62 YTKRYEIVNGLAEVEGA----------DIDEPEVKGVPYFWLVALQNNDEIADEITERDE 111
YTKRY IVNG EV+G + EP+ KGVP FWL A++ N+ +++EI ERDE
Sbjct: 98 YTKRYGIVNGEVEVDGVCDEPASENTVEGKEPDAKGVPDFWLTAMKTNEVLSEEIQERDE 157
Query: 112 DALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIE 171
ALKYLKDIK++R +P+GFKL+FFFD+NP+F N++LTKTYHMVDED+ I+EKAIGTEIE
Sbjct: 158 AALKYLKDIKWSRIEDPKGFKLDFFFDTNPFFKNSVLTKTYHMVDEDDPILEKAIGTEIE 217
Query: 172 WLPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQ 224
W PGK+LT+ KK K +TE +SFFNFFN ++P+D +DE+ +ELQ
Sbjct: 218 WYPGKNLTQKILKKKPKKGSKNTKPITKTEDCESFFNFFNPPQVPEDDDDIDEDTADELQ 277
Query: 225 NEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGES-GDLDDDDDDEDEEDDEEEVEVEKEE 283
++MEHDYDIG+TIRDKIIP+AVSW+TGEA GE G+L DDD+D+D+EDD+E+ E E+E
Sbjct: 278 SQMEHDYDIGTTIRDKIIPHAVSWFTGEAVQGEDFGELGDDDEDDDDEDDDEDDEDEEE- 336
Query: 284 EDDNYGLDDDEEDDNEDVNNPKKR 307
DDDEE+D+E+ + P ++
Sbjct: 337 -------DDDEEEDDEEESKPARK 353
>UniRef100_Q9ZUP3 Putative nucleosome assembly protein [Arabidopsis thaliana]
Length = 379
Score = 362 bits (928), Expect = 9e-99
Identities = 181/321 (56%), Positives = 238/321 (73%), Gaps = 22/321 (6%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q L Q L+ L +P V KR+E L+EIQ ++D+++AKFFEERAALEAKYQ LYQPL
Sbjct: 33 LQNLAGQHSDVLENL--TPPVRKRVEFLREIQNQYDEMEAKFFEERAALEAKYQKLYQPL 90
Query: 62 YTKRYEIVNGLAEVEGA---------DIDEPEVKGVPYFWLVALQNNDEIADEITERDED 112
YTKRYEIVNG+ EVEGA + E KGVP FWL+AL+NN+ A+EITERDE
Sbjct: 91 YTKRYEIVNGVVEVEGAAEEVKSEQGEDKSAEEKGVPDFWLIALKNNEITAEEITERDEG 150
Query: 113 ALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEW 172
ALKYLKDIK++R EP+GFKLEFFFD NPYF NT+LTKTYHM+DEDE I+EKA+GTEIEW
Sbjct: 151 ALKYLKDIKWSRVEEPKGFKLEFFFDQNPYFKNTVLTKTYHMIDEDEPILEKALGTEIEW 210
Query: 173 LPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQN 225
PGK LT+ KK K +TE +SFFNFF+ ++P D +D++ +ELQ
Sbjct: 211 YPGKCLTQKILKKKPKKGSKNTKPITKTEDCESFFNFFSPPQVPDDDEDLDDDMADELQG 270
Query: 226 EMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEED 285
+MEHDYDIGSTI++KII +AVSW+TGEA + + D++DDDD+ DE+DDEE+ E ++++E+
Sbjct: 271 QMEHDYDIGSTIKEKIISHAVSWFTGEAVEADDLDIEDDDDEIDEDDDEEDEEDDEDDEE 330
Query: 286 DNYGLDDDEEDDNEDVNNPKK 306
+ DD+++D+ E+ + KK
Sbjct: 331 E----DDEDDDEEEEADQGKK 347
>UniRef100_Q70Z17 Nucleosome assembly protein 1-like protein 3 [Nicotiana tabacum]
Length = 377
Score = 359 bits (921), Expect = 6e-98
Identities = 187/324 (57%), Positives = 242/324 (73%), Gaps = 25/324 (7%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q LT + L+ L SP V KR+EVL+EIQ +HD+L+AKFFEERAALEAKYQ YQPL
Sbjct: 33 LQGLTGKHSNVLENL--SPNVRKRVEVLREIQTQHDELEAKFFEERAALEAKYQKQYQPL 90
Query: 62 YTKRYEIVNGLAEVEG-----ADIDEPE-----VKGVPYFWLVALQNNDEIADEITERDE 111
Y KR EIVNG+ EV+G A DE E KGVP FW+ A++NN+ +A+EITERDE
Sbjct: 91 YAKRSEIVNGVVEVDGEATQTAAADEEEDKDSVEKGVPDFWVTAMKNNEVLAEEITERDE 150
Query: 112 DALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIE 171
ALK+LKDIK++R P+GFKLEFFF++NPYF+NT+LTKTYHM+DEDE I+EKAIGTEIE
Sbjct: 151 GALKFLKDIKWSRIENPKGFKLEFFFETNPYFTNTVLTKTYHMIDEDEPILEKAIGTEIE 210
Query: 172 WLPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQ 224
W PGK LT+ KK +K + E+ +SFFNFF+ ++P D +DE+A EELQ
Sbjct: 211 WHPGKCLTQKILKKKPKKGSKNSKPITKIEQCESFFNFFSPPQVPDDEEDIDEDAAEELQ 270
Query: 225 NEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESG-DLDDDDDDEDEEDDEEEVEVEKEE 283
N ME DYDIGSTIRDKIIP+AVSW+TGEA+ E DL+DD+D+ED+ED++E+ E E+EE
Sbjct: 271 NLMEQDYDIGSTIRDKIIPHAVSWFTGEAAQDEDYIDLEDDEDEEDDEDEDEDEEDEEEE 330
Query: 284 EDDNYGLDDDEEDDNEDVNNPKKR 307
++ D+D++D++E V KK+
Sbjct: 331 DE-----DEDDDDEDEHVTKTKKK 349
>UniRef100_Q94K07 Nucleosome assembly protein [Arabidopsis thaliana]
Length = 374
Score = 358 bits (919), Expect = 1e-97
Identities = 184/322 (57%), Positives = 233/322 (72%), Gaps = 24/322 (7%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q L Q L+ L +P + +R+EVL+EIQ +HD+++ KF EERAALEAKYQ LYQPL
Sbjct: 33 LQNLAGQHSDVLENL--TPKIRRRVEVLREIQGKHDEIETKFREERAALEAKYQKLYQPL 90
Query: 62 YTKRYEIVNGLAEVEGADIDEP---------EVKGVPYFWLVALQNNDEIADEITERDED 112
Y KRYEIVNG EVEGA D E KGVP FWL AL+NND I++EITERDE
Sbjct: 91 YNKRYEIVNGATEVEGAPEDAKMDQGDEKTAEEKGVPSFWLTALKNNDVISEEITERDEG 150
Query: 113 ALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEW 172
AL YLKDIK+ + EP+GFKLEFFFD NPYF NT+LTK YHM+DEDE ++EKAIGTEI+W
Sbjct: 151 ALIYLKDIKWCKIEEPKGFKLEFFFDQNPYFKNTLLTKAYHMIDEDEPLLEKAIGTEIDW 210
Query: 173 LPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQN 225
PGK LT+ KK AK +TE +SFFNFFN ++P D +DEE EELQN
Sbjct: 211 YPGKCLTQKILKKKPKKGAKNAKPITKTEDCESFFNFFNPPQVPDDDEDIDEERAEELQN 270
Query: 226 EMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEED 285
ME DYDIGSTIR+KIIP+AVSW+TGEA +GE ++D+DD+D+ +ED++E+ E E E+E
Sbjct: 271 LMEQDYDIGSTIREKIIPHAVSWFTGEAIEGEEFEIDNDDEDDIDEDEDEDEEDEDEDE- 329
Query: 286 DNYGLDDDEEDDNEDVNNPKKR 307
++D+ED+ E+V+ KK+
Sbjct: 330 -----EEDDEDEEEEVSKTKKK 346
>UniRef100_Q9LTS5 Nucleosome assembly protein [Arabidopsis thaliana]
Length = 374
Score = 358 bits (919), Expect = 1e-97
Identities = 184/322 (57%), Positives = 233/322 (72%), Gaps = 24/322 (7%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q L Q L+ L +P + +R+EVL+EIQ +HD+++ KF EERAALEAKYQ LYQPL
Sbjct: 33 LQNLAGQHSDVLENL--TPKIRRRVEVLREIQGKHDEIETKFREERAALEAKYQKLYQPL 90
Query: 62 YTKRYEIVNGLAEVEGADIDEP---------EVKGVPYFWLVALQNNDEIADEITERDED 112
Y KRYEIVNG EVEGA D E KGVP FWL AL+NND I++EITERDE
Sbjct: 91 YNKRYEIVNGATEVEGAPEDAKMDQGDEKTAEEKGVPSFWLTALKNNDVISEEITERDEG 150
Query: 113 ALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEW 172
AL YLKDIK+ + EP+GFKLEFFFD NPYF NT+LTK YHM+DEDE ++EKAIGTEI+W
Sbjct: 151 ALIYLKDIKWCKIEEPKGFKLEFFFDQNPYFKNTLLTKAYHMIDEDEPLLEKAIGTEIDW 210
Query: 173 LPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQN 225
PGK LT+ KK AK +TE +SFFNFFN ++P D +DEE EELQN
Sbjct: 211 YPGKCLTQKILKKKPKKGAKNAKPITKTEDCESFFNFFNPPQVPDDDEDIDEERAEELQN 270
Query: 226 EMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEED 285
ME DYDIGSTIR+KIIP+AVSW+TGEA +GE ++D+DD+D+ +ED++E+ E E E+E
Sbjct: 271 LMEQDYDIGSTIREKIIPHAVSWFTGEAIEGEEFEIDNDDEDDIDEDEDEDEEDEDEDE- 329
Query: 286 DNYGLDDDEEDDNEDVNNPKKR 307
++D+ED+ E+V+ KK+
Sbjct: 330 -----EEDDEDEEEEVSKTKKK 346
>UniRef100_Q5VND6 Putative nucleosome assembly protein 1 [Oryza sativa]
Length = 378
Score = 357 bits (917), Expect = 2e-97
Identities = 183/324 (56%), Positives = 238/324 (72%), Gaps = 29/324 (8%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q+L Q L+ L SP V KR+E L+EIQ +HD+++ KFFEERAALEAKYQ LY+PL
Sbjct: 40 LQSLAGQHTDVLEAL--SPNVRKRVEYLREIQGQHDEIELKFFEERAALEAKYQKLYEPL 97
Query: 62 YTKRYEIVNGLAEVEGADIDEP-----------EVKGVPYFWLVALQNNDEIADEITERD 110
YTKRY IVNG+ EV+G + DEP + KGVP FWL A++ N+ +++EI ERD
Sbjct: 98 YTKRYNIVNGVVEVDGGN-DEPASENAAEGKDADAKGVPDFWLTAMKTNEVLSEEIQERD 156
Query: 111 EDALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEI 170
E ALKYLKDIK+ R +P+GFKL+FFFD+NP+F N++LTKTYHMVDEDE I+EKAIGTEI
Sbjct: 157 EPALKYLKDIKWARIDDPKGFKLDFFFDTNPFFKNSVLTKTYHMVDEDEPILEKAIGTEI 216
Query: 171 EWLPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEEL 223
EW PGK+LT+ KK AK +TE +SFFNFF+ ++P D +DE+ +EL
Sbjct: 217 EWYPGKNLTQKILKKKPKKGSKNAKPITKTEVCESFFNFFSPPQVPDDDEDIDEDTADEL 276
Query: 224 QNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEE 283
Q +MEHDYDIG+TIRDKIIP+AVSW+TGEA E D DD ++DEEDDE++ E E+EE
Sbjct: 277 QGQMEHDYDIGTTIRDKIIPHAVSWFTGEAVQAE----DFDDMEDDEEDDEDDDEDEEEE 332
Query: 284 EDDNYGLDDDEEDDNEDVNNPKKR 307
E+D +D++EDD E+ + PKK+
Sbjct: 333 EED----EDEDEDDEEEKSKPKKK 352
>UniRef100_Q70Z21 Nucleosome assembly protein 1-like protein 1 [Oryza sativa]
Length = 377
Score = 356 bits (914), Expect = 4e-97
Identities = 183/324 (56%), Positives = 237/324 (72%), Gaps = 30/324 (9%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q+L Q L+ L SP V KR+E L+EIQ +HD+++ KFFEERAALEAKYQ LY+PL
Sbjct: 40 LQSLAGQHTDVLEAL--SPNVRKRVEYLREIQGQHDEIELKFFEERAALEAKYQKLYEPL 97
Query: 62 YTKRYEIVNGLAEVEGADIDEP-----------EVKGVPYFWLVALQNNDEIADEITERD 110
YTKRY IVNG+ EV+G + DEP + KGVP FWL A++ N+ +++EI ERD
Sbjct: 98 YTKRYNIVNGVVEVDGGN-DEPASENAAEFKDADAKGVPDFWLTAMKTNEVLSEEIQERD 156
Query: 111 EDALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEI 170
E ALKYLKDIK+ R +P+GFKL+FFFD+NP+F N++LTKTYHMVDEDE I+EKAIGTEI
Sbjct: 157 EPALKYLKDIKWARIDDPKGFKLDFFFDTNPFFKNSVLTKTYHMVDEDEPILEKAIGTEI 216
Query: 171 EWLPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEEL 223
EW PGK+LT+ KK AK +TE +SFFNFF+ ++P D +DE+ +EL
Sbjct: 217 EWYPGKNLTQKILKKKPKKGSKNAKPITKTEVCESFFNFFSPPQVPDDDEDIDEDTADEL 276
Query: 224 QNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEE 283
Q +MEHDYDIG+TIRDKIIP+AVSW+TGEA E D DD ++DEEDDE++ E E+EE
Sbjct: 277 QGQMEHDYDIGTTIRDKIIPHAVSWFTGEAVQSE----DFDDMEDDEEDDEDDDEDEEEE 332
Query: 284 EDDNYGLDDDEEDDNEDVNNPKKR 307
ED +D++EDD E+ + PKK+
Sbjct: 333 ED-----EDEDEDDEEEKSKPKKK 351
>UniRef100_Q70Z20 Nucleosome assembly protein 1-like protein 2 [Oryza sativa]
Length = 364
Score = 350 bits (898), Expect = 3e-95
Identities = 177/315 (56%), Positives = 233/315 (73%), Gaps = 29/315 (9%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q L + L+ L P V KR+EVL+EIQ +HDDL+AKFFEERAALEAKYQ +Y+PL
Sbjct: 39 LQGLAARHTDVLESL--EPKVRKRVEVLREIQSQHDDLEAKFFEERAALEAKYQKMYEPL 96
Query: 62 YTKRYEIVNGLAEVEGAD--------IDEPEVKGVPYFWLVALQNNDEIADEITERDEDA 113
Y+KRYEIVNG+ EV+G ++ E KGVP FWL A++N++ +++EI ERDE+A
Sbjct: 97 YSKRYEIVNGVVEVDGVTKEAADETPAEQKEEKGVPEFWLNAMKNHEILSEEIQERDEEA 156
Query: 114 LKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEWL 173
LKYLKDIK+ R +EP+GFKLEF+FD+NP+F N++LTKTYHM+DEDE I+EKAIGTEIEW
Sbjct: 157 LKYLKDIKWYRISEPKGFKLEFYFDTNPFFKNSVLTKTYHMIDEDEPILEKAIGTEIEWF 216
Query: 174 PGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQNE 226
PGK LT+ KK K +TE +SFFNFF+ ++P D +DE+ E+LQN+
Sbjct: 217 PGKCLTQKVLKKKPKKGSKNTKPITKTENCESFFNFFSPPQVPDDDEEIDEDTAEQLQNQ 276
Query: 227 MEHDYDIGSTIRDKIIPNAVSWYTGEASDGE--SGDLDDDDDDEDEEDDEEEVEVEKEEE 284
ME DYDIGSTIRDKIIP+AVSW+TGEA+ E G +DD+DDD++++DD+ E+E
Sbjct: 277 MEQDYDIGSTIRDKIIPHAVSWFTGEAAQDEDFEGIMDDEDDDDEDDDDD-------EDE 329
Query: 285 DDNYGLDDDEEDDNE 299
DD DDDE+D++E
Sbjct: 330 DDE---DDDEDDEDE 341
>UniRef100_Q38809 Arabidopsis thaliana Col-0 nucleosome assembly protein I-like
protein [Arabidopsis thaliana]
Length = 382
Score = 346 bits (887), Expect = 5e-94
Identities = 178/324 (54%), Positives = 229/324 (69%), Gaps = 27/324 (8%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q L QR L+ L +P V KR++ L++IQ +HD+L+AKF EERA LEAKYQ LYQPL
Sbjct: 43 LQNLAGQRSDVLENL--TPNVRKRVDALRDIQSQHDELEAKFREERAILEAKYQTLYQPL 100
Query: 62 YTKRYEIVNGLAEVEGADIDEPEV----------KGVPYFWLVALQNNDEIADEITERDE 111
Y KRYEIVNG EVE A D+ +V KGVP FWL AL+NND I++E+TERDE
Sbjct: 101 YVKRYEIVNGTTEVELAPEDDTKVDQGEEKTAEEKGVPSFWLTALKNNDVISEEVTERDE 160
Query: 112 DALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIE 171
ALKYLKDIK+ + EP+GFKLEFFFD+NPYF NT+LTK+YHM+DEDE ++EKA+GTEI+
Sbjct: 161 GALKYLKDIKWCKIEEPKGFKLEFFFDTNPYFKNTVLTKSYHMIDEDEPLLEKAMGTEID 220
Query: 172 WLPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQ 224
W PGK LT+ KK K + E +SFFNFF+ E+P + +DEE E+LQ
Sbjct: 221 WYPGKCLTQKILKKKPKKGSKNTKPITKLEDCESFFNFFSPPEVPDEDEDIDEERAEDLQ 280
Query: 225 NEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEE 284
N ME DYDIGSTIR+KIIP AVSW+TGEA + E ++DDD++D+ +ED++EE +EE
Sbjct: 281 NLMEQDYDIGSTIREKIIPRAVSWFTGEAMEAEDFEIDDDEEDDIDEDEDEE-----DEE 335
Query: 285 DDNYGLDDDEEDDNEDVNNPKKRV 308
D+ DDD+ED+ E K +
Sbjct: 336 DEE---DDDDEDEEESKTKKKPSI 356
>UniRef100_Q70Z16 Nucleosome assembly protein 1-like protein 4 [Nicotiana tabacum]
Length = 356
Score = 340 bits (872), Expect = 3e-92
Identities = 176/324 (54%), Positives = 240/324 (73%), Gaps = 29/324 (8%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q L Q L+ L +P V KR++VL+E+Q +HD+L++ FFEERAALEAKYQ LY+PL
Sbjct: 41 LQNLAGQHSDILETL--TPQVRKRVDVLRELQSQHDELESHFFEERAALEAKYQKLYEPL 98
Query: 62 YTKRYEIVNGLAEVEGA---------DIDEPEVKGVPYFWLVALQNNDEIADEITERDED 112
YTKRYEIVNG+ EVEG D + KGVP FWL A++ N+ +A+EI+ERDE+
Sbjct: 99 YTKRYEIVNGVVEVEGVNEAPMNQEEDKEAGNEKGVPNFWLTAMKTNEILAEEISERDEE 158
Query: 113 ALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEW 172
ALKYLKDIK+ + + +GFKLEFFFD+NP+F+N++LTKTYHM+D+D+ I+EKAIGT+I+W
Sbjct: 159 ALKYLKDIKWCKIDDRKGFKLEFFFDTNPFFTNSVLTKTYHMIDDDDPILEKAIGTKIDW 218
Query: 173 LPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPK--DGMGVDEEADEEL 223
PGK LT+ KK AK I+TE +SFFNFF ++P+ D +DE+A EEL
Sbjct: 219 CPGKCLTQKILKKKPKKGSKNAKPIIKTETCESFFNFFKPPQVPEDDDDDDIDEDAAEEL 278
Query: 224 QNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDG-ESGDLDDDDDDEDEEDDEEEVEVEKE 282
QN ME DYDIGSTIRDKIIP+AVSW+TGEA++G E D++DDDDD+D++DDE+ +++
Sbjct: 279 QNLMEQDYDIGSTIRDKIIPHAVSWFTGEAAEGDEFEDIEDDDDDDDDDDDED----DED 334
Query: 283 EEDDNYGLDDDEEDDNEDVNNPKK 306
EED+ DD+EE+ ++ ++ K
Sbjct: 335 EEDE----DDEEEEKSKKKSSALK 354
>UniRef100_Q8L8G5 Nucleosome/chromatin assembly factor group A [Zea mays]
Length = 370
Score = 334 bits (856), Expect = 2e-90
Identities = 169/312 (54%), Positives = 224/312 (71%), Gaps = 30/312 (9%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
++ L + L+ L P V KR+E L+EIQ EHD+L+AKFFEERAALEAKYQ LY+PL
Sbjct: 39 LEGLASRHTDVLENL--EPKVRKRVEKLREIQGEHDELEAKFFEERAALEAKYQKLYEPL 96
Query: 62 YTKRYEIVNGLAEVEG------------------ADIDEPEVKGVPYFWLVALQNNDEIA 103
Y+KRYEIVNG+ E+EG ++ E KGVP FWL A++N++ +A
Sbjct: 97 YSKRYEIVNGVVEIEGITKESAAETPEEQKSGDETSAEQKEEKGVPAFWLNAMKNHEILA 156
Query: 104 DEITERDEDALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIME 163
+EI ERDE+ALKYLKDIK+ R +EP+GFKLEF F +N +F N++LTKTYHM+DEDE I+E
Sbjct: 157 EEIQERDEEALKYLKDIKWYRISEPKGFKLEFHFGTNMFFKNSVLTKTYHMIDEDEPILE 216
Query: 164 KAIGTEIEWLPGKSLTEK-------KDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVD 216
KAIGTEIEW PGK LT+K K K +TE +SFFNFF+ ++P D +D
Sbjct: 217 KAIGTEIEWYPGKCLTQKVLKKKPRKGSKNTKPITKTEDCESFFNFFSPPQVPDDDEEID 276
Query: 217 EEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEE 276
E+ E+LQN+ME DYDIGSTIRDKIIP+AVSW+TGEA+ E ++ D +DD+DE+DD+E+
Sbjct: 277 EDTAEQLQNQMEQDYDIGSTIRDKIIPHAVSWFTGEAAQDEDFEVMDGEDDDDEDDDDED 336
Query: 277 VEVEKEEEDDNY 288
+++E+DD+Y
Sbjct: 337 ---DEDEDDDDY 345
>UniRef100_Q9ASD2 Putative nucleosome assembly protein 1 [Oryza sativa]
Length = 301
Score = 320 bits (821), Expect = 2e-86
Identities = 162/282 (57%), Positives = 213/282 (75%), Gaps = 11/282 (3%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+QAL +Q L+ L +P+V KR++VL EIQ +HD+L+AKF EE+AALEA YQ LY PL
Sbjct: 22 LQALAEQHVDVLESL--APVVRKRVDVLIEIQSQHDELEAKFLEEKAALEANYQKLYGPL 79
Query: 62 YTKRYEIVNGLAEVEGADIDEPEVKGVPYFWLVALQNNDEIADEITERDEDALKYLKDIK 121
Y+KR EIV+G+ EVEG + +E E KGVP FWL A++NN+ +A+EI E DE+ALKYLKDIK
Sbjct: 80 YSKRSEIVSGVLEVEG-ETEEREEKGVPDFWLKAMKNNEILAEEIHESDEEALKYLKDIK 138
Query: 122 YTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDED-EFIMEKAIGTEIEWLPGKSLTE 180
+ R +P+GFK EFFFD+NP+F N +LTKTYHM+DED E I+EKAIGTEIEW PG LT+
Sbjct: 139 WCRIDDPKGFKFEFFFDTNPFFKNQVLTKTYHMIDEDDEPILEKAIGTEIEWHPGNCLTQ 198
Query: 181 ----KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQNEMEHDYDIGST 236
K+ + K +TE+++SFFNFF+ ++P+D +DE EELQN+ME DYDI ST
Sbjct: 199 EVLTKESSESTKPITKTEEYESFFNFFSPPQVPEDDAKIDENTAEELQNQMERDYDIAST 258
Query: 237 IRDKIIPNAVSWYTGEASDGE---SGDLDDDDDDEDEEDDEE 275
+RDKIIP+ VSW+TGEA E + +DD++DD+DE DEE
Sbjct: 259 LRDKIIPHVVSWFTGEAVQDEDYGASWVDDEEDDDDEYSDEE 300
>UniRef100_Q9LIC0 Nucleosome assembly protein-like [Arabidopsis thaliana]
Length = 332
Score = 256 bits (654), Expect = 5e-67
Identities = 144/275 (52%), Positives = 179/275 (64%), Gaps = 27/275 (9%)
Query: 19 SPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPLYTK---RYEIVNGLAEV 75
SP V KR+ LK+IQ HD+L+ KF E++ALEA Y LY+PL+ K RYEIVNG+ E
Sbjct: 65 SPKVTKRVLFLKDIQVTHDELEEKFLAEKSALEATYDNLYKPLFAKNLQRYEIVNGVVEA 124
Query: 76 EGADIDEPEVKGVPYFWLVALQNNDEIADEITERDEDALKYLKDIKYTRTTEP-QGFKLE 134
E E +GVP FWL+A++ N+ +A+EITERDE ALKYLKDI+ R + + FKLE
Sbjct: 125 EA------EKEGVPNFWLIAMKTNEMLANEITERDEAALKYLKDIRSCRVEDTSRNFKLE 178
Query: 135 FFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEWLPGKSLTEK-----------KD 183
F FDSN YF N++L+KTYH+ DED ++EK IGT+IEW PGK LT K K
Sbjct: 179 FLFDSNLYFKNSVLSKTYHVNDEDGPVLEKVIGTDIEWFPGKCLTHKVVVKKKTKKGPKK 238
Query: 184 PNTAKQTIETEKFDSFFNFFNSLEIPK-----DGMGVDEEADEELQNEMEHDYDIGSTIR 238
N T +TE +SFFNFF EIP+ D D EELQN M+ DYDI TIR
Sbjct: 239 VNNIPMT-KTENCESFFNFFKPPEIPEIDEVDDYDDFDTIMTEELQNLMDQDYDIAVTIR 297
Query: 239 DKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDD 273
DK+IP+AVSW+TGEA E D+DDDD DE+ D
Sbjct: 298 DKLIPHAVSWFTGEALVDEDDSDDNDDDDNDEKSD 332
>UniRef100_Q69X43 Putative nucleosome assembly protein 1 [Oryza sativa]
Length = 386
Score = 252 bits (643), Expect = 1e-65
Identities = 137/279 (49%), Positives = 180/279 (64%), Gaps = 39/279 (13%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+QAL +Q L+ L +P V KR++VL EIQ +HD+L+ KFFEE+AALEAKYQ LY PL
Sbjct: 141 LQALAEQHVDVLESL--APSVRKRVDVLMEIQSQHDELEVKFFEEKAALEAKYQKLYGPL 198
Query: 62 YTKRYEIVNGLAEVEGADIDEPEVKGVPYFWLVALQNNDEIADEITERDEDALKYLKDIK 121
Y+K KGVP FWL A++NN+ +A+EI E DE+ALKYLKDIK
Sbjct: 199 YSKE--------------------KGVPDFWLNAMKNNEILAEEIHESDEEALKYLKDIK 238
Query: 122 YTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDE-DEFIMEKAIGTEIEWLPGKSLTE 180
+ R +P+GFK EFFF +NP+F N +LTKTYHM+DE DE I+EKAIGTEIEW PG LT+
Sbjct: 239 WCRIDDPKGFKFEFFFYTNPFFKNQVLTKTYHMIDEDDEPILEKAIGTEIEWHPGYCLTQ 298
Query: 181 ----KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQNEMEHDYDIGST 236
K+ + K +TE+ +SFFNFF+ ++P D +DE EELQN+ME DYDI
Sbjct: 299 EVLTKESSESTKPITKTEECESFFNFFSPPQVPDDDAKIDENTAEELQNQMERDYDIAEA 358
Query: 237 IRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEE 275
++D+ SW D +++DD+ DE DEE
Sbjct: 359 VQDE--DYGASWV----------DDEEEDDNNDEYSDEE 385
>UniRef100_Q6RJX6 Induced stolon tip protein NAP1Ps [Capsicum annuum]
Length = 212
Score = 231 bits (590), Expect = 1e-59
Identities = 116/181 (64%), Positives = 143/181 (78%), Gaps = 12/181 (6%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q LT + L+ L SP V KR+EVL+EIQ +HD+L+AKFFEERAALEAKYQ LYQPL
Sbjct: 33 LQDLTGKHTNLLENL--SPNVRKRVEVLREIQTQHDELEAKFFEERAALEAKYQKLYQPL 90
Query: 62 YTKRYEIVNGLAEVEGADIDEPEV----------KGVPYFWLVALQNNDEIADEITERDE 111
YTKR++IVNG+ EV+GA + P V KGVP FW A++NND +A+EI+ERDE
Sbjct: 91 YTKRFDIVNGVVEVDGAVTEVPAVDQEEDKAAVEKGVPDFWFTAMKNNDVLAEEISERDE 150
Query: 112 DALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIE 171
ALK+LKDIK+TR P+GFKLEFFFD+NP+F NT+LTKTYHM+DEDE I+EKAIGTEI+
Sbjct: 151 AALKFLKDIKWTRIDNPKGFKLEFFFDTNPFFKNTVLTKTYHMIDEDEPILEKAIGTEID 210
Query: 172 W 172
W
Sbjct: 211 W 211
>UniRef100_UPI000023CCD5 UPI000023CCD5 UniRef100 entry
Length = 406
Score = 201 bits (512), Expect = 2e-50
Identities = 125/312 (40%), Positives = 180/312 (57%), Gaps = 52/312 (16%)
Query: 22 VEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPLYTKRYEIVNGLAE-----VE 76
V +R+ LK IQK+H L+A+F EE LE KY + PLY KR I+NG +E ++
Sbjct: 86 VRRRVAGLKGIQKDHSKLEAEFQEEVLQLEKKYFAKFSPLYEKRSAIINGKSEPTEDEIK 145
Query: 77 GADIDEPEV-------------------KGVPYFWLVALQNNDEIADEITERDEDALKYL 117
+ D+ EV G+P FWL A++N +A+ IT+RDE ALK+L
Sbjct: 146 AGEEDDEEVDDDDAATKSETKSDVPDNVSGIPEFWLSAMKNQISLAEMITDRDEAALKHL 205
Query: 118 KDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDED----EFIMEKAIGTEIEWL 173
DI+ +P GF+L F F N +FSNT +TKTY+ +E +FI + A G +I+W
Sbjct: 206 TDIRMEYLDKP-GFRLIFEFAPNDFFSNTTITKTYYYQNESGYGGDFIYDHAEGDKIDWS 264
Query: 174 PGKSLT------EKKDPNT-----AKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEE 222
PGK LT ++++ NT K+T+ TE SFFNFF+ + P D DE+A+ +
Sbjct: 265 PGKDLTVRVESKKQRNKNTKQTRIVKKTVPTE---SFFNFFSPPKAPTDDD--DEDAESD 319
Query: 223 LQNEMEHDYDIGSTIRDKIIPNAVSWYTGEA------SDGESGDLD-DDDDDEDEEDDEE 275
++ +E DY +G I++K+IP A+ W+TGEA SD E D DDDDDEDE+D E
Sbjct: 320 IEERLELDYQLGEDIKEKLIPRAIDWFTGEALAFEEISDDELDGADFDDDDDEDEDDLSE 379
Query: 276 EVEVEKEEEDDN 287
E + E+E +DD+
Sbjct: 380 ENDDEEESDDDD 391
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.310 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 581,169,984
Number of Sequences: 2790947
Number of extensions: 29243497
Number of successful extensions: 648187
Number of sequences better than 10.0: 9925
Number of HSP's better than 10.0 without gapping: 6674
Number of HSP's successfully gapped in prelim test: 3577
Number of HSP's that attempted gapping in prelim test: 329359
Number of HSP's gapped (non-prelim): 92497
length of query: 308
length of database: 848,049,833
effective HSP length: 127
effective length of query: 181
effective length of database: 493,599,564
effective search space: 89341521084
effective search space used: 89341521084
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC138526.4


