

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135796.2 - phase: 0
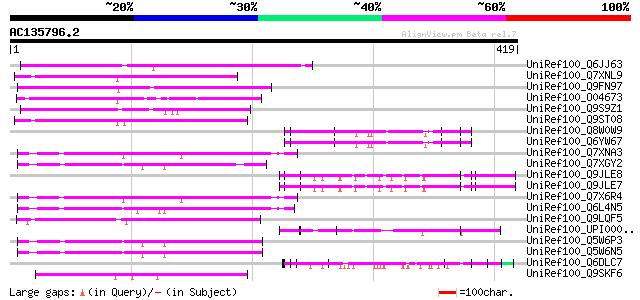
(419 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6JJ63 Putative tnp2 transposase [Ipomoea trifida] 129 2e-28
UniRef100_Q7XNL9 OSJNBb0068N06.11 protein [Oryza sativa] 110 6e-23
UniRef100_Q9FN97 Transposon protein-like [Arabidopsis thaliana] 107 5e-22
UniRef100_O04673 Carrot DNA for transposon Tdc1 [Daucus carota] 100 1e-19
UniRef100_Q9S9Z1 F21H2.10 protein [Arabidopsis thaliana] 99 2e-19
UniRef100_Q9ST08 Transposon-like ORF protein [Brassica campestris] 94 6e-18
UniRef100_Q8W0W9 Repressor protein [Oryza sativa] 90 1e-16
UniRef100_Q6YW67 Repressor protein [Oryza sativa] 90 1e-16
UniRef100_Q7XNA3 OSJNBa0011E07.15 protein [Oryza sativa] 84 7e-15
UniRef100_Q7XGY2 Transposase [Oryza sativa] 84 1e-14
UniRef100_Q9JLE8 GABA-A receptor epsilon-like subunit [Mus muscu... 83 1e-14
UniRef100_Q9JLE7 GABA-A receptor epsilon-like subunit [Mus muscu... 83 1e-14
UniRef100_Q7X6R4 OSJNBa0095H06.16 protein [Oryza sativa] 83 1e-14
UniRef100_Q6L4N5 Putative TNP2 transposable element [Oryza sativa] 82 2e-14
UniRef100_Q9LQF5 F15O4.30 [Arabidopsis thaliana] 82 3e-14
UniRef100_UPI00002A9902 UPI00002A9902 UniRef100 entry 82 4e-14
UniRef100_Q5W6P3 Putative polyprotein [Oryza sativa] 81 5e-14
UniRef100_Q5W6N5 Putative polyprotein [Oryza sativa] 81 5e-14
UniRef100_Q6DLC7 Omega gliadin [Aegilops tauschii] 81 5e-14
UniRef100_Q9SKF6 Putative TNP2-like transposon protein [Arabidop... 81 6e-14
>UniRef100_Q6JJ63 Putative tnp2 transposase [Ipomoea trifida]
Length = 651
Score = 129 bits (323), Expect = 2e-28
Identities = 73/245 (29%), Positives = 123/245 (49%), Gaps = 9/245 (3%)
Query: 10 MYNRRITGRKGFTAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNERFLTPKGVNVH 69
MYNR G++G+T EFL G++EF+++AC P Y I CPC CKN L+ V VH
Sbjct: 1 MYNRLSPGKRGYTDEFLAGVEEFVNYACTLPVYESTNTIRCPCSKCKNRVHLSADEVRVH 60
Query: 70 IRQKGFTPGYWYWTSHGEEAPQINLEVDLHSDAMPYSSQQDTGFGDAG---LDDQNFVQD 126
+ +KGF P YW WT HGE P +N + + S+Q +T D G N + +
Sbjct: 61 LYKKGFEPYYWVWTCHGEPIPTVN---EQFQETRNESNQYETMVFDVGGSSFIPFNVLNN 117
Query: 127 EEVPTNVETTEFCDMLNSTQQTLRPGDKNTSKMSAAIAMLSLKSKHNMSQDCFNDVIKFM 186
+E + +F D+L++ +Q L P +++S A+ M+++K+ N+ Q + + ++
Sbjct: 118 QEEDPHTSAKDFYDLLSTARQPLSPSCVEETELSYALKMMNIKATFNIPQLAMDQIFEYN 177
Query: 187 RESNHTENVTPSNSRGTKRTMQPTKISRKK-KCAQTHAPLIIPSDTVQTHANQATQPPPR 245
+ ++N PS +++ + ++ +K C L SD + PR
Sbjct: 178 KHIMGSDNRVPSRYYDSEKLISKLGLAHEKIDCCINSCMLYYKSDINDRQCKFCGE--PR 235
Query: 246 TKPRT 250
KPR+
Sbjct: 236 FKPRS 240
>UniRef100_Q7XNL9 OSJNBb0068N06.11 protein [Oryza sativa]
Length = 1188
Score = 110 bits (276), Expect = 6e-23
Identities = 61/190 (32%), Positives = 97/190 (50%), Gaps = 8/190 (4%)
Query: 5 EHRTWMYNRRITGRKGFTAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNERFLTPK 64
E R+WMY R + G + E+L+G+K F+ A + Q + + CPC CKN + +
Sbjct: 3 EDRSWMYKRVVNGY--ISDEYLKGVKRFMSHAVSKLQGQAEKRVRCPCTKCKNGQLHDKR 60
Query: 65 GVNVHIRQKGFTPGYWYWTSHGE------EAPQINLEVDLHSDAMPYSSQQDTGFGDAGL 118
V +H+ KGFT Y W+ HGE E P+ +++ +++ Y
Sbjct: 61 IVQMHLCNKGFTENYHVWSRHGESGADTSETPEGSIDNEMNKGGNAYGESDHIDEMVLDA 120
Query: 119 DDQNFVQDEEVPTNVETTEFCDMLNSTQQTLRPGDKNTSKMSAAIAMLSLKSKHNMSQDC 178
NF Q+ E P N E F DML++ + L S++SA +L++KS HNMS C
Sbjct: 121 AGPNFCQNIEEPPNAEAASFFDMLDAADKPLWEKCGKHSQLSAVSRLLTIKSDHNMSVAC 180
Query: 179 FNDVIKFMRE 188
F++++K M+E
Sbjct: 181 FDELVKVMKE 190
>UniRef100_Q9FN97 Transposon protein-like [Arabidopsis thaliana]
Length = 1089
Score = 107 bits (268), Expect = 5e-22
Identities = 69/221 (31%), Positives = 108/221 (48%), Gaps = 14/221 (6%)
Query: 7 RTWMYNRRITGRKGFTAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKN-ERFLTPKG 65
R WMY R + F +GL FL FA QP +LE+ + CPC+ C N R
Sbjct: 11 RHWMYERIDPTTNRVSQAFYEGLDNFLKFAKSQPLFLENKKLFCPCIKCGNGGRQQEEHI 70
Query: 66 VNVHIRQKGFTPGYWYWTSHGEE--------APQINLEVDLHSDAM-PYSSQQDTGFGDA 116
V H+ KGF YW WTSHGE+ + + + + H++ + PY FG
Sbjct: 71 VATHLFNKGFMLNYWVWTSHGEDYNMLINNNSEEDTVRFESHTEPVDPYVEMVSDAFGST 130
Query: 117 GLDDQNFVQDEEVPTNVETTEFCDMLNSTQQTLRPGDK-NTSKMSAAIAMLSLKSKHNMS 175
+ F Q E N E +F D+L++ +Q + G K SK+S A ++SLK+ +N+S
Sbjct: 131 ---ESEFDQMREEDPNFEAKKFYDILDAAKQPIYDGCKEGLSKLSLAARLMSLKTDNNLS 187
Query: 176 QDCFNDVIKFMRESNHTENVTPSNSRGTKRTMQPTKISRKK 216
Q+C + + + M+E N +P + K+ M+ + +K
Sbjct: 188 QNCMDSIAQIMQEYLPEGNNSPKSYYEIKKLMRSLGLPYQK 228
>UniRef100_O04673 Carrot DNA for transposon Tdc1 [Daucus carota]
Length = 520
Score = 99.8 bits (247), Expect = 1e-19
Identities = 66/215 (30%), Positives = 106/215 (48%), Gaps = 21/215 (9%)
Query: 6 HRTWMYNRRITGRKGF-TAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNERFLTPK 64
+R+WMY R T GF + F+ G++EF++ QP + I CPC CKN +F
Sbjct: 4 NRSWMYER--TDESGFLNSLFISGVEEFMNHVISQPTSMNGTSIQCPCTKCKNRKFWNSD 61
Query: 65 GVNVHIRQKGFTPGYWYWTSHGE-----------EAPQINLEVDLHSDAMPYSSQQDTGF 113
V +H+ + GF Y+ W+ HGE A N+ + + Y+ D
Sbjct: 62 IVKLHLLKNGFVRDYYIWSRHGESYIFNGNEDQSSANYSNVARGTDGNNLMYNMVIDA-- 119
Query: 114 GDAGLDDQNFVQDEEVPTNVETTEFCDMLNSTQQTLRPGDKNTSKMSAAIAMLSLKSKHN 173
G D + EE+P N E +MLNS+++ L G + TS+++A MLSLKS H+
Sbjct: 120 GGPSFDPH---RSEEMP-NAEAQNIYNMLNSSERELYDGCE-TSQLAAMAQMLSLKSDHH 174
Query: 174 MSQDCFNDVIKFMRESNHTENVTPSNSRGTKRTMQ 208
S+ C++ +F++ +N + GTK+ M+
Sbjct: 175 WSEACYDQTSQFIKGILPKDNTFLDSFYGTKKHME 209
>UniRef100_Q9S9Z1 F21H2.10 protein [Arabidopsis thaliana]
Length = 504
Score = 99.0 bits (245), Expect = 2e-19
Identities = 63/209 (30%), Positives = 107/209 (51%), Gaps = 24/209 (11%)
Query: 10 MYNRRITGRKGFTAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNERFLTPKGVNVH 69
MYN G T EF +GL +F+ FA Q LE G + CPC C NE+FL + H
Sbjct: 1 MYNYIDEGTGELTKEFKEGLVDFMRFAANQEIPLECGKMYCPCSTCGNEKFLPVDSIWKH 60
Query: 70 IRQKGFTPGYWYWTSHGEEAPQINLEVDLHSDAMPYSSQQDTGFGDAGLDDQNFVQDE-- 127
+ +GF PGY+ W SHG++ L D + +D + +++N ++D
Sbjct: 61 LYSRGFMPGYYIWFSHGKDFE--TLANSNEDDEIHGLEPEDNNSNE---EEENIIRDPTG 115
Query: 128 --EVPTNV--ETTE------------FCDMLNSTQQTLRPGDK-NTSKMSAAIAMLSLKS 170
++ TN ETTE F DML++ ++ + G K S++SAA ++++K+
Sbjct: 116 VIQMVTNAFRETTESGIEEPNLEAKKFFDMLDAAKKPIYDGCKEGHSRLSAATRLMNIKT 175
Query: 171 KHNMSQDCFNDVIKFMRESNHTENVTPSN 199
++N+S+DC + + F+++ N+ P +
Sbjct: 176 EYNLSEDCVDAIADFVKDLLPEPNLAPGS 204
>UniRef100_Q9ST08 Transposon-like ORF protein [Brassica campestris]
Length = 703
Score = 94.4 bits (233), Expect = 6e-18
Identities = 64/209 (30%), Positives = 97/209 (45%), Gaps = 19/209 (9%)
Query: 5 EHRTWMY-NRRITGRKGFTAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNERFLTP 63
E R WMY +R GR T E+L GL+ F+ A P E G + CPC C N +
Sbjct: 9 ELRNWMYMHRDANGR--VTKEYLAGLETFMHQADSTPLAQESGKMFCPCRKCNNSKLANR 66
Query: 64 KGVNVHIRQKGFTPGYWYWTSHGE-------EAPQIN-------LEVDLHSDAMPYSSQQ 109
+ V H+ +GFTP Y+ W HGE EA N + LH++ + +
Sbjct: 67 ENVWKHLINRGFTPNYYIWFQHGEGYNYDQNEASSSNSNFQEEPVNHHLHNEHSYHQEEM 126
Query: 110 -DTGFGDAGLDDQNFVQDEEVPTNVETTEFCDMLNSTQQTLRPG-DKNTSKMSAAIAMLS 167
D + D DE+ N++ +F MLN+ Q L G + SK+S A M++
Sbjct: 127 VDYDRVHDMVADAFVAHDEDEEPNIDAKKFYGMLNAANQPLYSGCREGLSKLSLAARMMN 186
Query: 168 LKSKHNMSQDCFNDVIKFMRESNHTENVT 196
+K+ HN+ + C N+ +E +NV+
Sbjct: 187 IKTDHNLPESCMNEWADLFKEYLPEDNVS 215
>UniRef100_Q8W0W9 Repressor protein [Oryza sativa]
Length = 296
Score = 90.1 bits (222), Expect = 1e-16
Identities = 62/147 (42%), Positives = 77/147 (52%), Gaps = 13/147 (8%)
Query: 233 QTHANQATQPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPTQ---PQRR 289
+ A Q TQ PP+ + + +P P+ Q + Q Q PQ L PQ Q PQ +
Sbjct: 147 EPEAQQQTQQPPQPQLHPQPQQPLQPQLQLHPQPQQQPSQLHPQQLLHPQSQQTPQPQPQ 206
Query: 290 LQPQPTQP---QAPPQLRRKPQPTQGRSQRQLDLQPQPTQP-QLQPQLRMRPQPTQPRPQ 345
+ PQP QP Q PQL ++PQ Q + Q Q L PQP QP QLQ Q ++ PQP QP
Sbjct: 207 VHPQPQQPPQLQPQPQLLQQPQLPQ-QLQPQSQLPPQPQQPPQLQLQSQLHPQPQQP--- 262
Query: 346 PQIRLQPQPTQHTSPPPPPPPQSRIQP 372
PQ LQPQP H P P QS+ QP
Sbjct: 263 PQ--LQPQPQLHQQPQPQAELQSQSQP 287
Score = 63.5 bits (153), Expect = 1e-08
Identities = 40/123 (32%), Positives = 56/123 (45%), Gaps = 20/123 (16%)
Query: 269 QSQQSQPQLFLQPQPTQPQRRLQPQPTQPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQPQ 328
Q+ Q ++F + + +P+ +P+A Q ++ PQP L PQP QP
Sbjct: 120 QAVAEQQRMFAEARARMNNGAAKPKEPEPEAQQQTQQPPQP---------QLHPQPQQP- 169
Query: 329 LQPQLRMRPQPTQP----------RPQPQIRLQPQPTQHTSPPPPPPPQSRIQPIMSQTI 378
LQPQL++ PQP Q PQ Q QPQP H P PP Q + Q + +
Sbjct: 170 LQPQLQLHPQPQQQPSQLHPQQLLHPQSQQTPQPQPQVHPQPQQPPQLQPQPQLLQQPQL 229
Query: 379 PTQ 381
P Q
Sbjct: 230 PQQ 232
Score = 63.2 bits (152), Expect = 1e-08
Identities = 52/134 (38%), Positives = 64/134 (46%), Gaps = 35/134 (26%)
Query: 228 PSDTVQTHANQATQPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPTQPQ 287
P + + Q QP P+ P+ P+ P L Q QPQL QPQ Q
Sbjct: 189 PQQLLHPQSQQTPQPQPQVHPQ-----PQQPPQL----------QPQPQLLQQPQLPQ-- 231
Query: 288 RRLQPQPT---QPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQ-PQLQPQLRMRPQPTQPR 343
+LQPQ QPQ PPQL Q Q L PQP Q PQLQPQ ++ QP QP+
Sbjct: 232 -QLQPQSQLPPQPQQPPQL-----------QLQSQLHPQPQQPPQLQPQPQLHQQP-QPQ 278
Query: 344 PQPQIRLQPQPTQH 357
+ Q + QPQ T+H
Sbjct: 279 AELQSQSQPQ-TEH 291
>UniRef100_Q6YW67 Repressor protein [Oryza sativa]
Length = 181
Score = 90.1 bits (222), Expect = 1e-16
Identities = 62/147 (42%), Positives = 77/147 (52%), Gaps = 13/147 (8%)
Query: 233 QTHANQATQPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPTQ---PQRR 289
+ A Q TQ PP+ + + +P P+ Q + Q Q PQ L PQ Q PQ +
Sbjct: 32 EPEAQQQTQQPPQPQLHPQPQQPLQPQLQLHPQPQQQPSQLHPQQLLHPQSQQTPQPQPQ 91
Query: 290 LQPQPTQP---QAPPQLRRKPQPTQGRSQRQLDLQPQPTQP-QLQPQLRMRPQPTQPRPQ 345
+ PQP QP Q PQL ++PQ Q + Q Q L PQP QP QLQ Q ++ PQP QP
Sbjct: 92 VHPQPQQPPQLQPQPQLLQQPQLPQ-QLQPQSQLPPQPQQPPQLQLQSQLHPQPQQP--- 147
Query: 346 PQIRLQPQPTQHTSPPPPPPPQSRIQP 372
PQ LQPQP H P P QS+ QP
Sbjct: 148 PQ--LQPQPQLHQQPQPQAELQSQSQP 172
Score = 63.5 bits (153), Expect = 1e-08
Identities = 40/123 (32%), Positives = 56/123 (45%), Gaps = 20/123 (16%)
Query: 269 QSQQSQPQLFLQPQPTQPQRRLQPQPTQPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQPQ 328
Q+ Q ++F + + +P+ +P+A Q ++ PQP L PQP QP
Sbjct: 5 QAVAEQQRMFAEARARMNNGAAKPKEPEPEAQQQTQQPPQP---------QLHPQPQQP- 54
Query: 329 LQPQLRMRPQPTQP----------RPQPQIRLQPQPTQHTSPPPPPPPQSRIQPIMSQTI 378
LQPQL++ PQP Q PQ Q QPQP H P PP Q + Q + +
Sbjct: 55 LQPQLQLHPQPQQQPSQLHPQQLLHPQSQQTPQPQPQVHPQPQQPPQLQPQPQLLQQPQL 114
Query: 379 PTQ 381
P Q
Sbjct: 115 PQQ 117
Score = 63.2 bits (152), Expect = 1e-08
Identities = 52/134 (38%), Positives = 64/134 (46%), Gaps = 35/134 (26%)
Query: 228 PSDTVQTHANQATQPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPTQPQ 287
P + + Q QP P+ P+ P+ P L Q QPQL QPQ Q
Sbjct: 74 PQQLLHPQSQQTPQPQPQVHPQ-----PQQPPQL----------QPQPQLLQQPQLPQ-- 116
Query: 288 RRLQPQPT---QPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQ-PQLQPQLRMRPQPTQPR 343
+LQPQ QPQ PPQL Q Q L PQP Q PQLQPQ ++ QP QP+
Sbjct: 117 -QLQPQSQLPPQPQQPPQL-----------QLQSQLHPQPQQPPQLQPQPQLHQQP-QPQ 163
Query: 344 PQPQIRLQPQPTQH 357
+ Q + QPQ T+H
Sbjct: 164 AELQSQSQPQ-TEH 176
>UniRef100_Q7XNA3 OSJNBa0011E07.15 protein [Oryza sativa]
Length = 906
Score = 84.0 bits (206), Expect = 7e-15
Identities = 66/248 (26%), Positives = 109/248 (43%), Gaps = 32/248 (12%)
Query: 7 RTWMYNRRITGRKGFTAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNER-FLTPKG 65
R WMY + + EF+ G+ FL A Q G I CPC CKN++ + +
Sbjct: 3 RQWMYADQRS------KEFIDGVYYFLRVAEANNQ---KGFICCPCNKCKNQKEYSASRT 53
Query: 66 VNVHIRQKGFTPGYWYWTSHGEEAPQI---NLEVDLHSDAMPYSSQQDTGFGDAGLD-DQ 121
++ H+ + GF P Y YWTSHGE+ ++ +E D D Y+ + G+ +D D
Sbjct: 54 IHFHLFESGFMPSYNYWTSHGEQGVEMEEDEVEDDKIPDFAQYAGFEGNQTGEEDIDADY 113
Query: 122 NFVQDEEVPTNVETTEFCD----------MLNSTQQTLRPG-DKNTSKMSAAIAMLSLKS 170
N V D+ + E C+ ML + +L PG ++ K+ + L K+
Sbjct: 114 NDVADDHGQMLQDAKEDCESEKGAHKLDKMLEDHRTSLYPGCEQGHKKLDTTLEFLQWKA 173
Query: 171 KHNMSQDCFNDVIKFMRESNHTENVTPSNSRGTKRTMQPTKISRKKKCAQTHAPLIIPSD 230
K+ +S F D++K ++ N P K+ + P + +K HA P+D
Sbjct: 174 KNGVSDKAFGDLLKLVKNILPEGNKLPETMYEAKKIVCPLGLEVQK----IHA---CPND 226
Query: 231 TVQTHANQ 238
+ H +
Sbjct: 227 CILYHGEE 234
>UniRef100_Q7XGY2 Transposase [Oryza sativa]
Length = 2535
Score = 83.6 bits (205), Expect = 1e-14
Identities = 60/223 (26%), Positives = 102/223 (44%), Gaps = 33/223 (14%)
Query: 7 RTWMYNRRITGRKGFTAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNER-FLTPKG 65
R WMY R + EF+ G+ FL A Q G I CPC CKN++ + +
Sbjct: 3 RQWMYADRRS------KEFIDGVHYFLKVAEASRQR---GFICCPCNKCKNQKEYSASRT 53
Query: 66 VNVHIRQKGFTPGYWYWTSHGEEAPQINLEVDLHSDAMPYSSQ------QDTGFGDAGLD 119
++ H+ + GF P Y YWTSHGE+ ++ E ++ D +P +Q TG + D
Sbjct: 54 IHFHLFESGFIPSYNYWTSHGEQGVEME-EEEVEDDNIPDFAQYVGFEGNQTGEEEIAAD 112
Query: 120 DQNFVQD---------EEVPTNVETTEFCDMLNSTQQTLRPG-DKNTSKMSAAIAMLSLK 169
+ D E+ + E + ML + +L PG ++ K+ + +L K
Sbjct: 113 GNDVADDLGQMLQDAREDCESEKEAHKLDKMLEDHRTSLYPGCEQGHKKLDTTLELLQWK 172
Query: 170 SKHNMSQDCFNDVIKFMRESNHTENVTPSNSRGTKRTMQPTKI 212
+K+ +S F D++K ++ N+ P ++ + T + KI
Sbjct: 173 AKNGVSDKAFGDLLKLVK------NILPGGNKLPETTYEAKKI 209
Score = 42.7 bits (99), Expect = 0.019
Identities = 28/87 (32%), Positives = 36/87 (41%), Gaps = 17/87 (19%)
Query: 283 PTQPQRRLQPQPTQPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQPQLQPQLRMRPQPTQP 342
P P P P+ P APP P P Q + L P PT P+ P PT P
Sbjct: 1666 PAPPSPPQDPAPSPPHAPPA----PSPPQAPASTP-PLDPAPTPPRA-------PTPTPP 1713
Query: 343 RPQPQIRLQPQPTQHTSPPPPPPPQSR 369
+ P P++ +PP PPP +R
Sbjct: 1714 QAP-----LPAPSKSRAPPAPPPAHTR 1735
>UniRef100_Q9JLE8 GABA-A receptor epsilon-like subunit [Mus musculus]
Length = 910
Score = 83.2 bits (204), Expect = 1e-14
Identities = 54/150 (36%), Positives = 80/150 (53%), Gaps = 6/150 (4%)
Query: 224 PLIIPSDTVQTHANQATQPPPRTKPRTKFNRPRPPKALTRLQLRNQSQ-QSQPQLFLQPQ 282
PL +P Q QP P+ +P+ + P+ T Q++ Q + +PQ QPQ
Sbjct: 184 PLPLPLPEPQPEPQPDPQPEPQPEPQPEPQPEPQPEPQTPGQVQAGRQPRPRPQPEPQPQ 243
Query: 283 PTQPQRRLQPQPTQPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQPQLQPQLRMRPQPTQP 342
P +PQ QP+P QPQ PQ +PQP + Q + + QP+P QP+ QP+ + P+P QP
Sbjct: 244 P-EPQPEPQPEP-QPQPEPQPEPEPQP-EPEPQPEPEPQPEP-QPEPQPEPKPEPEP-QP 298
Query: 343 RPQPQIRLQPQPTQHTSPPPPPPPQSRIQP 372
P+PQ QP+P P P P P+ + +P
Sbjct: 299 EPEPQPEPQPEPQPEPKPEPEPQPEPKPEP 328
Score = 78.2 bits (191), Expect = 4e-13
Identities = 62/205 (30%), Positives = 93/205 (45%), Gaps = 23/205 (11%)
Query: 228 PSDTVQTHANQATQPPPRTKPRT-------KFNRPRP---PKALTRLQLRNQSQ---QSQ 274
P Q QP P+ +P+T + RPRP P+ Q Q + Q +
Sbjct: 200 PQPEPQPEPQPEPQPEPQPEPQTPGQVQAGRQPRPRPQPEPQPQPEPQPEPQPEPQPQPE 259
Query: 275 PQLFLQPQPT-QPQRRLQPQPTQPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQPQLQPQL 333
PQ +PQP +PQ +PQP +PQ PQ KP+P Q + + QP+P QP+ QP+
Sbjct: 260 PQPEPEPQPEPEPQPEPEPQP-EPQPEPQPEPKPEP---EPQPEPEPQPEP-QPEPQPEP 314
Query: 334 RMRPQPTQPRPQPQIRLQPQPTQHTSPPPPPPPQSRIQPIMSQTIPTQDLEWQDFAMINE 393
+ P+P QP P+P+ QP+P P P P P+ + +P Q P + E +
Sbjct: 315 KPEPEP-QPEPKPEPEPQPEPEPEPEPEPQPEPEPKPEP---QPEPEPEPEPEPEPEPEP 370
Query: 394 TPSSTTSHSSEESSGNRIPILPEAD 418
P E S + + + PE +
Sbjct: 371 EPEPEPEPEPEPESESELDLEPEPE 395
Score = 75.5 bits (184), Expect = 3e-12
Identities = 54/148 (36%), Positives = 74/148 (49%), Gaps = 18/148 (12%)
Query: 241 QPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPT---QPQRRLQPQPTQP 297
QP P+ +P +P P L L Q +PQ QP+P QP+ + +PQP +P
Sbjct: 163 QPEPQPQPLAG-RQPLPQPLPWPLPLPLPEPQPEPQPDPQPEPQPEPQPEPQPEPQP-EP 220
Query: 298 QAPPQLR--RKPQPT-QGRSQRQLDLQPQPT---QPQLQPQLRMRPQPT-------QPRP 344
Q P Q++ R+P+P Q Q Q + QP+P QPQ +PQ PQP +P+P
Sbjct: 221 QTPGQVQAGRQPRPRPQPEPQPQPEPQPEPQPEPQPQPEPQPEPEPQPEPEPQPEPEPQP 280
Query: 345 QPQIRLQPQPTQHTSPPPPPPPQSRIQP 372
+PQ QP+P P P P PQ QP
Sbjct: 281 EPQPEPQPEPKPEPEPQPEPEPQPEPQP 308
Score = 69.3 bits (168), Expect = 2e-10
Identities = 50/154 (32%), Positives = 70/154 (44%), Gaps = 20/154 (12%)
Query: 241 QPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQ---------SQPQLFLQPQPTQPQRRLQ 291
QP P +P T P + Q + Q++ SQPQ QPQP Q
Sbjct: 41 QPQPVPQPLTPPESPAGDTVVFGPQPQPQAESPGHDDVVFSSQPQPMPQPQPMPEP---Q 97
Query: 292 PQPTQPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQPQLQPQLRMRPQPTQPRPQPQIRLQ 351
PQP +P+ PQ + +PQP GR PQP P PQ + P+P +P P+P+ +
Sbjct: 98 PQP-EPEPEPQPQPQPQPLAGRQPL-----PQPL-PWPMPQPQPEPEP-EPEPEPEPEPE 149
Query: 352 PQPTQHTSPPPPPPPQSRIQPIMSQTIPTQDLEW 385
P+P P P P P+ + QP+ + Q L W
Sbjct: 150 PEPEPEPEPEPEPQPEPQPQPLAGRQPLPQPLPW 183
Score = 67.8 bits (164), Expect = 6e-10
Identities = 39/136 (28%), Positives = 72/136 (52%), Gaps = 6/136 (4%)
Query: 241 QPPPRTKPRTKFNRPRPPKALTRLQLRNQSQ-QSQPQLFLQPQPT-QPQRRLQPQPT-QP 297
QP P+ +P+ + PK Q + + + +PQ +P+P QP+ +P+P +P
Sbjct: 307 QPEPQPEPKPEPEPQPEPKPEPEPQPEPEPEPEPEPQPEPEPKPEPQPEPEPEPEPEPEP 366
Query: 298 QAPPQLRRKPQPT-QGRSQRQLDLQPQPTQPQLQPQLRMRPQPTQPRPQPQIRLQPQPTQ 356
+ P+ +P+P + S+ +LDL+P+P +P P P+P +P P+P+ +P+P
Sbjct: 367 EPEPEPEPEPEPEPEPESESELDLEPEP-EPHPHPHPHPHPEP-EPEPEPEPEPEPEPEP 424
Query: 357 HTSPPPPPPPQSRIQP 372
P P P P+ + +P
Sbjct: 425 EPEPEPEPEPEMKSEP 440
Score = 65.1 bits (157), Expect = 4e-09
Identities = 49/181 (27%), Positives = 74/181 (40%), Gaps = 40/181 (22%)
Query: 228 PSDTVQTHANQATQPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPTQPQ 287
P Q +P P+ +P+ R P+ L + + +P+ +P+P +P+
Sbjct: 92 PMPEPQPQPEPEPEPQPQPQPQPLAGRQPLPQPLP-WPMPQPQPEPEPEPEPEPEP-EPE 149
Query: 288 RRLQPQPTQPQAPPQLRRKPQPTQGRS-------------------QRQLDLQPQPTQPQ 328
+P+P +P+ PQ +PQP GR + Q D QP+P QP+
Sbjct: 150 PEPEPEP-EPEPEPQPEPQPQPLAGRQPLPQPLPWPLPLPLPEPQPEPQPDPQPEP-QPE 207
Query: 329 LQPQLRMRPQPT-----------------QPRPQPQIRLQPQPTQHTSPPPPPPPQSRIQ 371
QP+ + PQP QP PQPQ QP+P P P P P+ Q
Sbjct: 208 PQPEPQPEPQPEPQTPGQVQAGRQPRPRPQPEPQPQPEPQPEPQPEPQPQPEPQPEPEPQ 267
Query: 372 P 372
P
Sbjct: 268 P 268
Score = 57.8 bits (138), Expect = 6e-07
Identities = 42/146 (28%), Positives = 66/146 (44%), Gaps = 16/146 (10%)
Query: 241 QPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPTQPQRRLQPQPTQPQAP 300
QP P +P+ + P P L + QP + PQP QP+ +P+P +P+
Sbjct: 91 QPMPEPQPQPE-PEPEPQPQPQPQPLAGRQPLPQPLPWPMPQP-QPEPEPEPEP-EPEPE 147
Query: 301 PQLRRKPQPTQGRSQRQLDLQPQPTQPQLQPQLRMRPQPTQ-----PRPQPQIRLQPQPT 355
P+ +P+P + + +PQP +PQ QP +P P P P P+ + +PQP
Sbjct: 148 PEPEPEPEP-------EPEPEPQP-EPQPQPLAGRQPLPQPLPWPLPLPLPEPQPEPQPD 199
Query: 356 QHTSPPPPPPPQSRIQPIMSQTIPTQ 381
P P P P+ + +P P Q
Sbjct: 200 PQPEPQPEPQPEPQPEPQPEPQTPGQ 225
Score = 38.1 bits (87), Expect = 0.47
Identities = 26/108 (24%), Positives = 53/108 (49%), Gaps = 9/108 (8%)
Query: 241 QPPPRTKPRTKFNRPRPPKALTRLQLRNQSQ---QSQPQLFLQPQPTQPQRRLQPQPTQP 297
+P P +P + P++ + L L + + P +P+P +P+ +P+P +P
Sbjct: 365 EPEPEPEPEPEPEPEPEPESESELDLEPEPEPHPHPHPHPHPEPEP-EPEPEPEPEP-EP 422
Query: 298 QAPPQLRRKPQPTQGRSQRQLDLQPQPT---QPQLQPQLRMRPQPTQP 342
+ P+ +P+P +S+ Q L+PQP+ P +P+L + +P
Sbjct: 423 EPEPEPEPEPEPEM-KSEPQSQLEPQPSGKKLPAKEPELTVDCMTERP 469
>UniRef100_Q9JLE7 GABA-A receptor epsilon-like subunit [Mus musculus]
Length = 912
Score = 83.2 bits (204), Expect = 1e-14
Identities = 54/150 (36%), Positives = 80/150 (53%), Gaps = 6/150 (4%)
Query: 224 PLIIPSDTVQTHANQATQPPPRTKPRTKFNRPRPPKALTRLQLRNQSQ-QSQPQLFLQPQ 282
PL +P Q QP P+ +P+ + P+ T Q++ Q + +PQ QPQ
Sbjct: 184 PLPLPLPEPQPEPQPDPQPEPQPEPQPEPQPEPQPEPQTPGQVQAGRQPRPRPQPEPQPQ 243
Query: 283 PTQPQRRLQPQPTQPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQPQLQPQLRMRPQPTQP 342
P +PQ QP+P QPQ PQ +PQP + Q + + QP+P QP+ QP+ + P+P QP
Sbjct: 244 P-EPQPEPQPEP-QPQPEPQPEPEPQP-EPEPQPEPEPQPEP-QPEPQPEPKPEPEP-QP 298
Query: 343 RPQPQIRLQPQPTQHTSPPPPPPPQSRIQP 372
P+PQ QP+P P P P P+ + +P
Sbjct: 299 EPEPQPEPQPEPQPEPKPEPEPQPEPKPEP 328
Score = 78.2 bits (191), Expect = 4e-13
Identities = 62/205 (30%), Positives = 93/205 (45%), Gaps = 23/205 (11%)
Query: 228 PSDTVQTHANQATQPPPRTKPRT-------KFNRPRP---PKALTRLQLRNQSQ---QSQ 274
P Q QP P+ +P+T + RPRP P+ Q Q + Q +
Sbjct: 200 PQPEPQPEPQPEPQPEPQPEPQTPGQVQAGRQPRPRPQPEPQPQPEPQPEPQPEPQPQPE 259
Query: 275 PQLFLQPQPT-QPQRRLQPQPTQPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQPQLQPQL 333
PQ +PQP +PQ +PQP +PQ PQ KP+P Q + + QP+P QP+ QP+
Sbjct: 260 PQPEPEPQPEPEPQPEPEPQP-EPQPEPQPEPKPEP---EPQPEPEPQPEP-QPEPQPEP 314
Query: 334 RMRPQPTQPRPQPQIRLQPQPTQHTSPPPPPPPQSRIQPIMSQTIPTQDLEWQDFAMINE 393
+ P+P QP P+P+ QP+P P P P P+ + +P Q P + E +
Sbjct: 315 KPEPEP-QPEPKPEPEPQPEPEPEPEPEPQPEPEPKPEP---QPEPEPEPEPEPEPEPEP 370
Query: 394 TPSSTTSHSSEESSGNRIPILPEAD 418
P E S + + + PE +
Sbjct: 371 EPEPEPEPEPEPESESELDLEPEPE 395
Score = 75.5 bits (184), Expect = 3e-12
Identities = 54/148 (36%), Positives = 74/148 (49%), Gaps = 18/148 (12%)
Query: 241 QPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPT---QPQRRLQPQPTQP 297
QP P+ +P +P P L L Q +PQ QP+P QP+ + +PQP +P
Sbjct: 163 QPEPQPQPLAG-RQPLPQPLPWPLPLPLPEPQPEPQPDPQPEPQPEPQPEPQPEPQP-EP 220
Query: 298 QAPPQLR--RKPQPT-QGRSQRQLDLQPQPT---QPQLQPQLRMRPQPT-------QPRP 344
Q P Q++ R+P+P Q Q Q + QP+P QPQ +PQ PQP +P+P
Sbjct: 221 QTPGQVQAGRQPRPRPQPEPQPQPEPQPEPQPEPQPQPEPQPEPEPQPEPEPQPEPEPQP 280
Query: 345 QPQIRLQPQPTQHTSPPPPPPPQSRIQP 372
+PQ QP+P P P P PQ QP
Sbjct: 281 EPQPEPQPEPKPEPEPQPEPEPQPEPQP 308
Score = 69.3 bits (168), Expect = 2e-10
Identities = 50/154 (32%), Positives = 70/154 (44%), Gaps = 20/154 (12%)
Query: 241 QPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQ---------SQPQLFLQPQPTQPQRRLQ 291
QP P +P T P + Q + Q++ SQPQ QPQP Q
Sbjct: 41 QPQPVPQPLTPPESPAGDTVVFGPQPQPQAESPGHDDVVFSSQPQPMPQPQPMPEP---Q 97
Query: 292 PQPTQPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQPQLQPQLRMRPQPTQPRPQPQIRLQ 351
PQP +P+ PQ + +PQP GR PQP P PQ + P+P +P P+P+ +
Sbjct: 98 PQP-EPEPEPQPQPQPQPLAGRQPL-----PQPL-PWPMPQPQPEPEP-EPEPEPEPEPE 149
Query: 352 PQPTQHTSPPPPPPPQSRIQPIMSQTIPTQDLEW 385
P+P P P P P+ + QP+ + Q L W
Sbjct: 150 PEPEPEPEPEPEPQPEPQPQPLAGRQPLPQPLPW 183
Score = 66.6 bits (161), Expect = 1e-09
Identities = 39/137 (28%), Positives = 71/137 (51%), Gaps = 6/137 (4%)
Query: 241 QPPPRTKPRTKFNRPRPPKALTRLQLRNQSQ-QSQPQLFLQPQPT-QPQRRLQPQPT-QP 297
QP P+ +P+ + PK Q + + + +PQ +P+P QP+ +P+P +P
Sbjct: 307 QPEPQPEPKPEPEPQPEPKPEPEPQPEPEPEPEPEPQPEPEPKPEPQPEPEPEPEPEPEP 366
Query: 298 QAPPQLRRKPQPT-QGRSQRQLDLQPQPT-QPQLQPQLRMRPQPTQPRPQPQIRLQPQPT 355
+ P+ +P+P + S+ +LDL+P+P P P P+P +P P+P+ +P+P
Sbjct: 367 EPEPEPEPEPEPEPEPESESELDLEPEPEPHPHPHPHPHPHPEP-EPEPEPEPEPEPEPE 425
Query: 356 QHTSPPPPPPPQSRIQP 372
P P P P+ + +P
Sbjct: 426 PEPEPEPEPEPEMKSEP 442
Score = 65.1 bits (157), Expect = 4e-09
Identities = 49/181 (27%), Positives = 74/181 (40%), Gaps = 40/181 (22%)
Query: 228 PSDTVQTHANQATQPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPTQPQ 287
P Q +P P+ +P+ R P+ L + + +P+ +P+P +P+
Sbjct: 92 PMPEPQPQPEPEPEPQPQPQPQPLAGRQPLPQPLP-WPMPQPQPEPEPEPEPEPEP-EPE 149
Query: 288 RRLQPQPTQPQAPPQLRRKPQPTQGRS-------------------QRQLDLQPQPTQPQ 328
+P+P +P+ PQ +PQP GR + Q D QP+P QP+
Sbjct: 150 PEPEPEP-EPEPEPQPEPQPQPLAGRQPLPQPLPWPLPLPLPEPQPEPQPDPQPEP-QPE 207
Query: 329 LQPQLRMRPQPT-----------------QPRPQPQIRLQPQPTQHTSPPPPPPPQSRIQ 371
QP+ + PQP QP PQPQ QP+P P P P P+ Q
Sbjct: 208 PQPEPQPEPQPEPQTPGQVQAGRQPRPRPQPEPQPQPEPQPEPQPEPQPQPEPQPEPEPQ 267
Query: 372 P 372
P
Sbjct: 268 P 268
Score = 57.8 bits (138), Expect = 6e-07
Identities = 42/146 (28%), Positives = 66/146 (44%), Gaps = 16/146 (10%)
Query: 241 QPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPTQPQRRLQPQPTQPQAP 300
QP P +P+ + P P L + QP + PQP QP+ +P+P +P+
Sbjct: 91 QPMPEPQPQPE-PEPEPQPQPQPQPLAGRQPLPQPLPWPMPQP-QPEPEPEPEP-EPEPE 147
Query: 301 PQLRRKPQPTQGRSQRQLDLQPQPTQPQLQPQLRMRPQPTQ-----PRPQPQIRLQPQPT 355
P+ +P+P + + +PQP +PQ QP +P P P P P+ + +PQP
Sbjct: 148 PEPEPEPEP-------EPEPEPQP-EPQPQPLAGRQPLPQPLPWPLPLPLPEPQPEPQPD 199
Query: 356 QHTSPPPPPPPQSRIQPIMSQTIPTQ 381
P P P P+ + +P P Q
Sbjct: 200 PQPEPQPEPQPEPQPEPQPEPQTPGQ 225
Score = 37.4 bits (85), Expect = 0.80
Identities = 26/110 (23%), Positives = 53/110 (47%), Gaps = 11/110 (10%)
Query: 241 QPPPRTKPRTKFNRPRPPKALTRLQLRNQSQ-----QSQPQLFLQPQPTQPQRRLQPQPT 295
+P P +P + P++ + L L + + P +P+P +P+ +P+P
Sbjct: 365 EPEPEPEPEPEPEPEPEPESESELDLEPEPEPHPHPHPHPHPHPEPEP-EPEPEPEPEP- 422
Query: 296 QPQAPPQLRRKPQPTQGRSQRQLDLQPQPT---QPQLQPQLRMRPQPTQP 342
+P+ P+ +P+P +S+ Q L+PQP+ P +P+L + +P
Sbjct: 423 EPEPEPEPEPEPEPEM-KSEPQSQLEPQPSGKKLPAKEPELTVDCMTERP 471
>UniRef100_Q7X6R4 OSJNBa0095H06.16 protein [Oryza sativa]
Length = 837
Score = 83.2 bits (204), Expect = 1e-14
Identities = 66/248 (26%), Positives = 109/248 (43%), Gaps = 32/248 (12%)
Query: 7 RTWMYNRRITGRKGFTAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNER-FLTPKG 65
R WMY R + EF+ G+ FL A Q G I CPC CKN++ + +
Sbjct: 3 RQWMYTDRRS------KEFINGVHYFLRVAEANKQ---KGFICCPCNKCKNQKEYSASRT 53
Query: 66 VNVHIRQKGFTPGYWYWTSHGEEAPQI---NLEVDLHSDAMPYSSQQDTGFGDAGLD-DQ 121
++ H+ + GF P Y WTSHGE+ ++ +E D D Y+ + G+ +D D
Sbjct: 54 IHFHLFESGFMPSYNCWTSHGEQGVEMKEDEVEDDNIPDFAQYAEFEGNQTGEEDIDADY 113
Query: 122 NFVQDEEVPTNVETTEFCD----------MLNSTQQTLRPG-DKNTSKMSAAIAMLSLKS 170
N V D+ + E C+ ML + +L PG ++ K+ + L K+
Sbjct: 114 NDVADDLSQMLQDAKEDCESEKGAHKLDKMLEDHRTSLYPGCEQRHKKLDTTLEFLQWKA 173
Query: 171 KHNMSQDCFNDVIKFMRESNHTENVTPSNSRGTKRTMQPTKISRKKKCAQTHAPLIIPSD 230
K+ +S F D++K ++ N P + K+ + P + +K HA P+D
Sbjct: 174 KNGVSDKAFGDLLKLVKNILPEGNKLPETTYEAKKIVCPLGLEVQK----IHA---CPND 226
Query: 231 TVQTHANQ 238
+ H +
Sbjct: 227 CILYHGEE 234
>UniRef100_Q6L4N5 Putative TNP2 transposable element [Oryza sativa]
Length = 1087
Score = 82.4 bits (202), Expect = 2e-14
Identities = 68/246 (27%), Positives = 113/246 (45%), Gaps = 34/246 (13%)
Query: 7 RTWMYNRRITGRKGFTAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNER-FLTPKG 65
R WMY R + EF+ G+ FL A + +G ISCPC CKN++ + T +
Sbjct: 3 RQWMYADRRS------KEFIDGMHYFLGVANDNKR---NGFISCPCDKCKNQKEYSTTRT 53
Query: 66 VNVHIRQKGFTPGYWYWTSHGEEAPQINLEVDLHSDAMP----YSSQQDTGFGDAGLDDQ 121
++ H+ Q GF Y WTSHGE + E ++ + +P Y ++ G+A LD +
Sbjct: 54 IHAHLFQWGFMRSYNCWTSHGEVGVAME-EDEVEDENIPDWAQYGGFEENTMGEAELDVE 112
Query: 122 N---------FVQD--EEVPTNVETTEFCDMLNSTQQTLRPG-DKNTSKMSAAIAMLSLK 169
+QD E+ + E + ML + L PG +K K+S + L K
Sbjct: 113 GNHGADELGQILQDLQEDCKSEKEMQKLERMLADHRTMLYPGCEKGYKKLSTILEFLQWK 172
Query: 170 SKHNMSQDCFNDVIKFMRESNHTENVTPSNSRGTKRTMQPTKISRKKKCAQTHAPLIIPS 229
+K+ +S F +++K ++ N P ++ K+T+ P + +K HA P+
Sbjct: 173 AKNGVSDKAFGELLKLVKNILPEGNELPESTYEAKKTVYPLGLEVQK----IHA---CPN 225
Query: 230 DTVQTH 235
D + H
Sbjct: 226 DCILYH 231
>UniRef100_Q9LQF5 F15O4.30 [Arabidopsis thaliana]
Length = 1180
Score = 82.0 bits (201), Expect = 3e-14
Identities = 62/234 (26%), Positives = 106/234 (44%), Gaps = 37/234 (15%)
Query: 6 HRTWMYNR--RITGRKGFTAEFLQGLKEFLDFACQQPQYLE-DGVISCPCMLCKNER-FL 61
+R WMY R +TG +AE++ G++EF+ FA QP G CPC +CKNE+ +
Sbjct: 14 YRDWMYKRFDEVTGN--LSAEYVAGVEEFMTFANSQPIVQSCRGKFHCPCSVCKNEKHII 71
Query: 62 TPKGVNVHIRQKGFTPGYWYWTSHGEEAPQINLE-------------------------- 95
+ + V+ H+ +GF P Y+ W HGEE +N++
Sbjct: 72 SGRRVSSHLFSQGFMPDYYVWYKHGEE---LNMDIGTSYTDRTYFSENHEEVGNVVEDPY 128
Query: 96 VDLHSDAMPYSSQQDTGFG-DAGLDDQNFVQDEEVPTNVETTEFCDMLNSTQQTLRPG-D 153
VD+ +DA ++ D G D Q+ E P + +F D+L L G
Sbjct: 129 VDMVNDAFNFNVGYDDNVGHDDCYHHDGSYQNVEEPVRNHSNKFYDLLEGANNLLYDGCR 188
Query: 154 KNTSKMSAAIAMLSLKSKHNMSQDCFNDVIKFMRESNHTENVTPSNSRGTKRTM 207
+ S++S A ++ K+++NMS+ + V + + N ++ T++ M
Sbjct: 189 EGQSQLSLASRLMHNKAEYNMSEKLVDSVCEMFTDFLPEGNQATTSHYQTEKLM 242
>UniRef100_UPI00002A9902 UPI00002A9902 UniRef100 entry
Length = 285
Score = 81.6 bits (200), Expect = 4e-14
Identities = 53/140 (37%), Positives = 78/140 (54%), Gaps = 10/140 (7%)
Query: 240 TQPPPRTKPRTKFNRPRP-PKALTRLQLRNQSQ-QSQPQLFLQPQPT-QPQRRLQPQPT- 295
+QP P +P + P+P P+ L+ + + Q + +PQL L+PQP QP+ QP+P
Sbjct: 8 SQPEPEPQPEPE---PQPEPEPEPELEPQPEPQLEPEPQLELEPQPEPQPEPEPQPEPEP 64
Query: 296 QPQAPPQLRRKPQPT-QGRSQRQLDLQPQPT-QPQLQPQLRMRPQPT-QPRPQPQIRLQP 352
QP+ PQ +P+P Q + Q + QP+P QP+ +PQ +P+P QP P+PQ QP
Sbjct: 65 QPEPEPQPEPQPEPEPQPEPEPQPEPQPEPEPQPEPEPQPEPQPEPEPQPEPEPQPEPQP 124
Query: 353 QPTQHTSPPPPPPPQSRIQP 372
+P P P P PQ QP
Sbjct: 125 EPEPQPEPQPEPEPQPEPQP 144
Score = 77.8 bits (190), Expect = 5e-13
Identities = 58/196 (29%), Positives = 94/196 (47%), Gaps = 21/196 (10%)
Query: 224 PLIIPSDTVQTHANQATQPPPRTKPRTKFNRPRP-PKALTRLQLRNQSQ-QSQPQLFLQP 281
P + P ++ QP P +P + P+P P+ Q + Q + +PQ QP
Sbjct: 36 PQLEPEPQLELEPQPEPQPEPEPQPEPE---PQPEPEPQPEPQPEPEPQPEPEPQPEPQP 92
Query: 282 QPTQPQRRLQPQPTQPQAPPQLRRKPQPTQGRSQRQLDLQPQPTQPQLQPQLRMRPQPT- 340
+P +PQ +PQP +PQ P+ + +P+P Q Q + + QP+P QP+ +PQ +P+P
Sbjct: 93 EP-EPQPEPEPQP-EPQPEPEPQPEPEP-QPEPQPEPEPQPEP-QPEPEPQPEPQPEPEP 148
Query: 341 QPRPQPQIRLQPQPTQHTSPPPPPPPQSRIQP-----------IMSQTIPTQDLEWQDFA 389
QP PQP+ QP+P P P P PQ +P + T P L + ++
Sbjct: 149 QPEPQPEPEPQPEPEPQPEPQPEPEPQPEPEPQPEPEPEIARLSVQITGPGTTLYFNEYG 208
Query: 390 MINETPSSTTSHSSEE 405
+++ S T S E+
Sbjct: 209 LLSINESFTVSSGEEK 224
Score = 76.6 bits (187), Expect = 1e-12
Identities = 52/139 (37%), Positives = 74/139 (52%), Gaps = 8/139 (5%)
Query: 241 QPPPRTKPRTKFN-RPRP-PKALTRLQLRNQSQ-QSQPQLFLQPQPT-QPQRRLQPQPTQ 296
QP P+ +P + P+P P+ Q + Q + +PQ QP+P QP+ QP+P Q
Sbjct: 33 QPEPQLEPEPQLELEPQPEPQPEPEPQPEPEPQPEPEPQPEPQPEPEPQPEPEPQPEP-Q 91
Query: 297 PQAPPQLRRKPQPT-QGRSQRQLDLQPQPT-QPQLQPQLRMRPQPT-QPRPQPQIRLQPQ 353
P+ PQ +PQP Q + Q + +PQP QP+ +PQ +P+P QP PQP+ QP+
Sbjct: 92 PEPEPQPEPEPQPEPQPEPEPQPEPEPQPEPQPEPEPQPEPQPEPEPQPEPQPEPEPQPE 151
Query: 354 PTQHTSPPPPPPPQSRIQP 372
P P P P PQ QP
Sbjct: 152 PQPEPEPQPEPEPQPEPQP 170
Score = 72.4 bits (176), Expect = 2e-11
Identities = 43/107 (40%), Positives = 59/107 (54%), Gaps = 15/107 (14%)
Query: 271 QQSQPQLFLQPQPTQPQRRLQPQPT-QPQAPPQLRRKPQPTQGRSQRQLDLQPQP-TQPQ 328
++SQP+ QP+P +PQ +P+P +PQ PQL +P QL+L+PQP QP+
Sbjct: 6 RESQPEPEPQPEP-EPQPEPEPEPELEPQPEPQLEPEP---------QLELEPQPEPQPE 55
Query: 329 LQPQLRMRPQP---TQPRPQPQIRLQPQPTQHTSPPPPPPPQSRIQP 372
+PQ PQP QP PQP+ QP+P P P P PQ +P
Sbjct: 56 PEPQPEPEPQPEPEPQPEPQPEPEPQPEPEPQPEPQPEPEPQPEPEP 102
>UniRef100_Q5W6P3 Putative polyprotein [Oryza sativa]
Length = 939
Score = 81.3 bits (199), Expect = 5e-14
Identities = 59/220 (26%), Positives = 99/220 (44%), Gaps = 27/220 (12%)
Query: 7 RTWMYNRRITGRKGFTAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNER-FLTPKG 65
R WMY R + EF+ G+ FL A Q G I CPC CKN++ + +
Sbjct: 3 RQWMYADRRS------KEFIDGVHYFLRVAEANRQR---GFICCPCNKCKNQKEYSASRT 53
Query: 66 VNVHIRQKGFTPGYWYWTSHGEEAPQINLEVDLHSDAMPYSSQ------QDTGFGDAGLD 119
++ H+ + GF P Y WTSHGE+ ++ E ++ D +P +Q TG + D
Sbjct: 54 IHFHLFESGFMPSYNCWTSHGEQGVEME-EDEVEDDNIPDFAQYVGFEGNQTGEEEIAAD 112
Query: 120 DQNFVQD---------EEVPTNVETTEFCDMLNSTQQTLRPG-DKNTSKMSAAIAMLSLK 169
+ D E+ + E + ML + +L PG ++ K+ + +L K
Sbjct: 113 GNDVADDLGQMLQDAREDCESEKEAHKLDKMLEDHRTSLYPGCEQGHKKLDTTLELLQWK 172
Query: 170 SKHNMSQDCFNDVIKFMRESNHTENVTPSNSRGTKRTMQP 209
+K+ +S F D++K ++ EN P + K+ + P
Sbjct: 173 AKNGVSDKAFGDLLKLIKNILPGENKLPETTYEAKKIVCP 212
>UniRef100_Q5W6N5 Putative polyprotein [Oryza sativa]
Length = 1882
Score = 81.3 bits (199), Expect = 5e-14
Identities = 59/220 (26%), Positives = 99/220 (44%), Gaps = 27/220 (12%)
Query: 7 RTWMYNRRITGRKGFTAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNER-FLTPKG 65
R WMY R + EF+ G+ FL A Q G I CPC CKN++ + +
Sbjct: 3 RQWMYADRRS------KEFIDGVHYFLRVAEANRQR---GFICCPCNKCKNQKEYSASRT 53
Query: 66 VNVHIRQKGFTPGYWYWTSHGEEAPQINLEVDLHSDAMPYSSQ------QDTGFGDAGLD 119
++ H+ + GF P Y WTSHGE+ ++ E ++ D +P +Q TG + D
Sbjct: 54 IHFHLFESGFMPSYNCWTSHGEQGVEME-EDEVEDDNIPDFAQYVGFEGNQTGEEEIAAD 112
Query: 120 DQNFVQD---------EEVPTNVETTEFCDMLNSTQQTLRPG-DKNTSKMSAAIAMLSLK 169
+ D E+ + E + ML + +L PG ++ K+ + +L K
Sbjct: 113 GNDVADDLGQMLQDAREDCESEKEAHKLDKMLEDHRTSLYPGCEQGHKKLDTTLELLQWK 172
Query: 170 SKHNMSQDCFNDVIKFMRESNHTENVTPSNSRGTKRTMQP 209
+K+ +S F D++K ++ EN P + K+ + P
Sbjct: 173 AKNGVSDKAFGDLLKLIKNILPGENKLPETTYEAKKIVCP 212
>UniRef100_Q6DLC7 Omega gliadin [Aegilops tauschii]
Length = 392
Score = 81.3 bits (199), Expect = 5e-14
Identities = 64/161 (39%), Positives = 75/161 (45%), Gaps = 11/161 (6%)
Query: 227 IPSDTVQTHANQATQPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPTQP 286
IP Q Q QP P+ + +P+ Q Q QQS PQ QPQ P
Sbjct: 224 IPQQPQQPFPLQPQQPFPQQPQQPFPQQPQQSFPQQPQQPYPQQQQSFPQ---QPQQPFP 280
Query: 287 QRRLQPQPTQPQAPPQLR-RKPQPTQGRSQRQLDLQPQPTQPQ----LQPQLRMRPQPTQ 341
+P P QPQ P LR ++P P Q + +Q LQPQP QPQ LQPQ + QP Q
Sbjct: 281 PTTTKPFPQQPQQPFPLRPQQPFPQQPQQSQQSFLQPQPQQPQQPSILQPQQPLPQQPQQ 340
Query: 342 PRPQPQIRLQPQPTQHTSPPPPPP-PQSRIQPIMSQTIPTQ 381
P QPQ +L QP Q S P P PQ QP Q P Q
Sbjct: 341 PFQQPQQQLSQQPEQTISQQPQQPFPQQPHQP--QQPYPQQ 379
Score = 69.7 bits (169), Expect = 1e-10
Identities = 75/210 (35%), Positives = 90/210 (42%), Gaps = 47/210 (22%)
Query: 226 IIPSDTVQTHANQATQPPPRTKPRTKFNRPRPPKALTRLQ-LRNQSQQSQP------QLF 278
+IP Q Q QP P+ +P+ F +P+ P + Q QSQQSQ QLF
Sbjct: 159 VIPQQPQQPFPLQPQQPFPQ-QPQQPFPQPQQPIPVQPQQSFPQQSQQSQQPFAQPQQLF 217
Query: 279 ----------------LQPQ---PTQPQRRLQPQPTQPQAP-PQLRRKPQPTQGRSQRQL 318
LQPQ P QPQ QP P QPQ PQ ++P P Q +S Q
Sbjct: 218 PELQQPIPQQPQQPFPLQPQQPFPQQPQ---QPFPQQPQQSFPQQPQQPYPQQQQSFPQQ 274
Query: 319 DLQPQP---TQP---QLQPQLRMRPQ---PTQPRPQPQIRLQPQPTQHTSP----PPPPP 365
QP P T+P Q Q +RPQ P QP+ Q LQPQP Q P P P
Sbjct: 275 PQQPFPPTTTKPFPQQPQQPFPLRPQQPFPQQPQQSQQSFLQPQPQQPQQPSILQPQQPL 334
Query: 366 PQSRIQPIMSQTIPTQDLEWQDFAMINETP 395
PQ QP P Q L Q I++ P
Sbjct: 335 PQQPQQPFQQ---PQQQLSQQPEQTISQQP 361
Score = 68.9 bits (167), Expect = 2e-10
Identities = 67/201 (33%), Positives = 85/201 (41%), Gaps = 36/201 (17%)
Query: 238 QATQPPPRTK------PRTKF-NRPRPPKALTRLQLRNQSQQSQPQLFLQPQPTQPQRRL 290
Q QP P+T+ P+ F +P+ P +L QS+Q PQ QP P QPQ+
Sbjct: 117 QPQQPFPQTQQSFPLQPQQPFPQQPQQPFPQPQLPFPQQSEQVIPQQPQQPFPLQPQQPF 176
Query: 291 QPQPTQPQAPPQ--LRRKPQ---PTQGRSQRQLDLQPQ----------PTQPQ----LQP 331
QP QP PQ + +PQ P Q + +Q QPQ P QPQ LQP
Sbjct: 177 PQQPQQPFPQPQQPIPVQPQQSFPQQSQQSQQPFAQPQQLFPELQQPIPQQPQQPFPLQP 236
Query: 332 QLRMRPQPTQPRPQ------PQIRLQPQPTQHTSPPPPPPPQSRIQPIMSQTIPTQDLEW 385
Q QP QP PQ PQ QP P Q S P PQ P ++ P Q
Sbjct: 237 QQPFPQQPQQPFPQQPQQSFPQQPQQPYPQQQQS--FPQQPQQPFPPTTTKPFPQQ--PQ 292
Query: 386 QDFAMINETPSSTTSHSSEES 406
Q F + + P S++S
Sbjct: 293 QPFPLRPQQPFPQQPQQSQQS 313
Score = 68.6 bits (166), Expect = 3e-10
Identities = 62/167 (37%), Positives = 78/167 (46%), Gaps = 25/167 (14%)
Query: 233 QTHANQATQPPPRTKPRTKFNRPRP-----PKALTRLQLRNQSQQ--SQPQLFLQPQPTQ 285
Q+ ++Q P + P+ + +P P + Q QSQQ +QPQ QP P Q
Sbjct: 35 QSFSHQQQPFPQQPYPQQPYPSQQPYPSQQPFPTPQQQFPQQSQQPFTQPQ---QPTPLQ 91
Query: 286 PQRRLQPQPTQPQAP-PQLRR----KPQPTQGRSQRQLDLQPQPTQPQLQPQLRMRPQPT 340
PQ+ QP QPQ P PQ ++ +PQ ++Q+ LQPQ PQ QPQ PQP
Sbjct: 92 PQQPFPQQPQQPQQPFPQPQQPFPWQPQQPFPQTQQSFPLQPQQPFPQ-QPQQPF-PQPQ 149
Query: 341 QPRPQ------PQIRLQPQPTQHTSPPPPPPPQSRIQPIMSQTIPTQ 381
P PQ PQ QP P Q P P P Q QP Q IP Q
Sbjct: 150 LPFPQQSEQVIPQQPQQPFPLQPQQPFPQQPQQPFPQP--QQPIPVQ 194
Score = 63.2 bits (152), Expect = 1e-08
Identities = 63/183 (34%), Positives = 76/183 (41%), Gaps = 37/183 (20%)
Query: 228 PSDTVQTHANQATQPPPRTK----PRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQP 283
P + Q + +Q P P+ + + F +P+ P L Q QQ PQ QPQ
Sbjct: 53 PYPSQQPYPSQQPFPTPQQQFPQQSQQPFTQPQQPTPL-------QPQQPFPQQPQQPQQ 105
Query: 284 TQPQRRLQPQPTQPQAP-----------PQLRRKPQPTQGRSQRQLDLQPQ-----PTQP 327
PQ + QP P QPQ P PQ QP Q Q QL Q P QP
Sbjct: 106 PFPQPQ-QPFPWQPQQPFPQTQQSFPLQPQQPFPQQPQQPFPQPQLPFPQQSEQVIPQQP 164
Query: 328 Q----LQPQLRMRPQPTQPRPQPQ--IRLQPQ---PTQHTSPPPPPPPQSRIQPIMSQTI 378
Q LQPQ QP QP PQPQ I +QPQ P Q P ++ P + Q I
Sbjct: 165 QQPFPLQPQQPFPQQPQQPFPQPQQPIPVQPQQSFPQQSQQSQQPFAQPQQLFPELQQPI 224
Query: 379 PTQ 381
P Q
Sbjct: 225 PQQ 227
Score = 62.8 bits (151), Expect = 2e-08
Identities = 52/129 (40%), Positives = 59/129 (45%), Gaps = 11/129 (8%)
Query: 233 QTHANQATQPPPRTKPRTKFNRPRPPKALTRLQLRNQSQQSQPQLFLQPQPTQPQRRLQP 292
Q+ Q QP P T + +P+ P L Q Q Q Q FLQPQP QPQ QP
Sbjct: 269 QSFPQQPQQPFPPTTTKPFPQQPQQPFPLRPQQPFPQQPQQSQQSFLQPQPQQPQ---QP 325
Query: 293 QPTQPQAP-PQLRRKPQPTQGRSQRQLDLQPQPTQP-QLQPQLRMRPQPTQPRPQPQIRL 350
QPQ P PQ QP Q Q Q L QP Q QPQ QP QP+ QP +
Sbjct: 326 SILQPQQPLPQ-----QPQQPFQQPQQQLSQQPEQTISQQPQQPFPQQPHQPQ-QPYPQQ 379
Query: 351 QPQPTQHTS 359
QP + TS
Sbjct: 380 QPYGSSLTS 388
Score = 60.8 bits (146), Expect = 7e-08
Identities = 59/169 (34%), Positives = 68/169 (39%), Gaps = 18/169 (10%)
Query: 264 LQLRNQSQQSQPQLF-LQPQPTQPQRRLQPQPTQPQAP------PQLRRKP-----QPTQ 311
LQ QS Q Q F QP P QP QP P+Q P PQ ++P QPT
Sbjct: 30 LQSPQQSFSHQQQPFPQQPYPQQPYPSQQPYPSQQPFPTPQQQFPQQSQQPFTQPQQPTP 89
Query: 312 GRSQRQLDLQP-QPTQPQLQPQLRMRPQPTQPRPQPQ--IRLQPQPTQHTSPPPP-PPPQ 367
+ Q+ QP QP QP QPQ QP QP PQ Q LQPQ P P P PQ
Sbjct: 90 LQPQQPFPQQPQQPQQPFPQPQQPFPWQPQQPFPQTQQSFPLQPQQPFPQQPQQPFPQPQ 149
Query: 368 SRIQPIMSQTIPTQDLEWQDFAMINETPSSTTSHSSEESSGNRIPILPE 416
Q IP Q Q F + + P IP+ P+
Sbjct: 150 LPFPQQSEQVIPQQ--PQQPFPLQPQQPFPQQPQQPFPQPQQPIPVQPQ 196
>UniRef100_Q9SKF6 Putative TNP2-like transposon protein [Arabidopsis thaliana]
Length = 1040
Score = 80.9 bits (198), Expect = 6e-14
Identities = 52/198 (26%), Positives = 93/198 (46%), Gaps = 23/198 (11%)
Query: 22 TAEFLQGLKEFLDFACQQPQYLEDGVISCPCMLCKNERFLTPKGVNVHIRQKGFTPGYWY 81
T E+ L+ F+ +P E+G I CPC C NE++L V H+ +GF P Y+
Sbjct: 185 TIEYYDELRGFMYQTTNEPFARENGTILCPCRKCLNEKYLEFNVVKKHLYNRGFMPNYYV 244
Query: 82 WTSHG---EEAPQINLEVDLHS--------------DAMPYSSQQDTGFGDAGLDD---- 120
W HG E A ++ +++ + S D Y Q+ F D +
Sbjct: 245 WLRHGDVVEIAHELVIQIMVSSNLVILIMMRIMQVPDQFGYGYNQENRFHDMVTNAFHET 304
Query: 121 -QNFVQDEEVPTNVETTEFCDMLNSTQQTLRPG-DKNTSKMSAAIAMLSLKSKHNMSQDC 178
+F ++ NV+ F DML++ Q + G + SK+S A M+++K+ +N+S+ C
Sbjct: 305 IASFPENISEEPNVDAQHFYDMLDAANQPIYEGCREGNSKLSLASRMMTIKADNNLSEKC 364
Query: 179 FNDVIKFMRESNHTENVT 196
+ + ++E +N++
Sbjct: 365 MDSWAELIKEYLPPDNIS 382
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.130 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 796,322,318
Number of Sequences: 2790947
Number of extensions: 40748163
Number of successful extensions: 482668
Number of sequences better than 10.0: 12491
Number of HSP's better than 10.0 without gapping: 2981
Number of HSP's successfully gapped in prelim test: 10140
Number of HSP's that attempted gapping in prelim test: 239483
Number of HSP's gapped (non-prelim): 73286
length of query: 419
length of database: 848,049,833
effective HSP length: 130
effective length of query: 289
effective length of database: 485,226,723
effective search space: 140230522947
effective search space used: 140230522947
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 76 (33.9 bits)
Medicago: description of AC135796.2


