

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135161.4 + phase: 0
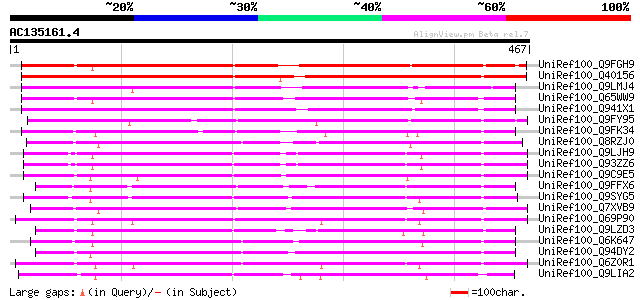
(467 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FGH9 Leucine zipper protein [Arabidopsis thaliana] 357 3e-97
UniRef100_Q40156 L.esculentum protein with leucine zipper [Lycop... 339 9e-92
UniRef100_Q9LMJ4 F10K1.28 protein [Arabidopsis thaliana] 286 1e-75
UniRef100_Q65WW9 Putative leucine zipper protein [Oryza sativa] 256 7e-67
UniRef100_Q941X1 Leucine zipper protein-like [Oryza sativa] 248 2e-64
UniRef100_Q9FY95 Hypothetical protein T19L5_110 [Arabidopsis tha... 236 1e-60
UniRef100_Q9FK34 Gb|AAD25781.1 [Arabidopsis thaliana] 227 5e-58
UniRef100_Q8RZJ0 Putative leucine zipper protein [Oryza sativa] 227 6e-58
UniRef100_Q9LJH9 Gb|AAD25781.1 [Arabidopsis thaliana] 220 8e-56
UniRef100_Q93ZZ6 Hypothetical protein At3g14090 [Arabidopsis tha... 220 8e-56
UniRef100_Q9C9E5 Hypothetical protein T10D10.6 [Arabidopsis thal... 218 4e-55
UniRef100_Q9FFX6 Putative leucine zipper protein [Arabidopsis th... 217 5e-55
UniRef100_Q9SYG5 F15I1.17 protein [Arabidopsis thaliana] 214 4e-54
UniRef100_Q7XVB9 OSJNBa0072D21.9 protein [Oryza sativa] 213 9e-54
UniRef100_Q69P90 Putative leucine zipper-containing protein [Ory... 207 4e-52
UniRef100_Q9LZD3 Hypothetical protein F12E4_320 [Arabidopsis tha... 204 6e-51
UniRef100_Q6K647 Putative leucine zipper-containing protein [Ory... 200 8e-50
UniRef100_Q94DY2 P0403C05.2 protein [Oryza sativa] 198 3e-49
UniRef100_Q6Z0R1 Putative leucine zipper-containing protein [Ory... 197 5e-49
UniRef100_Q9LIA2 Gb|AAF27143.1 [Arabidopsis thaliana] 197 7e-49
>UniRef100_Q9FGH9 Leucine zipper protein [Arabidopsis thaliana]
Length = 624
Score = 357 bits (917), Expect = 3e-97
Identities = 203/458 (44%), Positives = 285/458 (61%), Gaps = 30/458 (6%)
Query: 11 LMLEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKD 70
L+++AL T+N+L E K ML +GF K C VYSSCRRE LEE + + L L+I++
Sbjct: 194 LIIDALPSATINDLHEMAKRMLGAGFGKACSHVYSSCRREFLEESMSR--LGLQKLSIEE 251
Query: 71 VNM---EDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIR 127
V+ ++L I RWIKA VA +ILFP+ER+LCD VFF FS+ +D+SF +VCR TI+
Sbjct: 252 VHKMPWQELEDEIDRWIKAANVALRILFPSERRLCDRVFFGFSSAADLSFMEVCRGSTIQ 311
Query: 128 LLNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGE 187
LLNF + IA + LF++LD++ET+ DL+P FES+F DQ+ LRNE T+ K+LGE
Sbjct: 312 LLNFADAIAIGSRSPERLFKVLDVFETMRDLMPEFESVFSDQFCSVLRNEAVTIWKRLGE 371
Query: 188 TIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVL 247
I G E EN IR + P A GG LHP+ R+VMN+L C R+ LEQVFE+
Sbjct: 372 AIRGIFMELENLIR-RDPAKAAVPGGGLHPITRYVMNYLRAACRSRQTLEQVFEE----- 425
Query: 248 LEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNS 307
S+ S S+ ++QM IME+LES LE ++ DP L YV+LMN+
Sbjct: 426 -------------SNGVPSKDSTLLTVQMSWIMELLESNLEVKSKVYKDPALCYVFLMNN 472
Query: 308 SRYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNM 367
RYI+ K + +LG LLGD +++H+ K++ Y RSSW K+L L++DN N
Sbjct: 473 GRYIVQKVKDGDLGLLLGDDWIRKHNVKVKQYHMNYQRSSWNKMLGLLKVDNT-AAGMNG 531
Query: 368 VGKSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVL 427
+GK+MK++LK FN F+EICK S W + DE LKEE+ + L L+PAY +FI + +
Sbjct: 532 LGKTMKEKLKQFNIQFDEICKVHSTWVVFDEQLKEELKISLARLLVPAYGSFIGRFQNL- 590
Query: 428 KLEVKKPPDGYIEYETKDIKAILNNMFKIYRPSSCGRK 465
++ K D YI+Y +DI+A +N +FK ++ GRK
Sbjct: 591 -GDIGKNADKYIKYGVEDIEARINELFK---GTTTGRK 624
>UniRef100_Q40156 L.esculentum protein with leucine zipper [Lycopersicon esculentum]
Length = 631
Score = 339 bits (870), Expect = 9e-92
Identities = 190/459 (41%), Positives = 280/459 (60%), Gaps = 16/459 (3%)
Query: 11 LMLEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLT-IK 69
+++EAL +++L E K M+ +G++KEC YS RRE LEE L + L ++ ++
Sbjct: 185 ILIEALPAGIISDLHEIAKRMVAAGYDKECSHAYSVSRREFLEESLSRLGLQKLSMDQVQ 244
Query: 70 DVNMEDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIRLL 129
+ +L I++W+KA VA +ILFP+ER+LCD VFF F+++SD+SF +V R TI+LL
Sbjct: 245 KMQWNELEDEIEKWVKAVNVALRILFPSERRLCDRVFFGFNSVSDLSFMEVSRGSTIQLL 304
Query: 130 NFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETI 189
NF + +A LF++LD+YE L DL+P FE +F DQY V LRNE T+ ++LGE I
Sbjct: 305 NFADAVAISSRAPERLFKVLDVYEALRDLMPKFEFMFSDQYCVLLRNEALTIWRRLGEAI 364
Query: 190 VGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFED---HGHV 246
G E EN IR + P P GG LHP+ R+VMN++ C R LEQVF++
Sbjct: 365 RGIFMELENLIR-RDPAKTPVPGGGLHPITRYVMNYIRAACRSRITLEQVFKEIIVPSAS 423
Query: 247 LLEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMN 306
++Y + DD + SSSS ++QM IME+LES LE I+ D L V++MN
Sbjct: 424 AVDYREGDDR---------ALSSSSLAVQMAWIMELLESNLETKSKIYKDSALLAVFMMN 474
Query: 307 SSRYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPN 366
+ RYI+ K ++ELG LLGD +++H+AK++ Y RSSW KV L++DNN + P
Sbjct: 475 NERYIVQKVKDSELGLLLGDDWVRKHAAKVKQYHVNYHRSSWSKVSGVLKIDNNAMSSPT 534
Query: 367 MVGKSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIV 426
+S+K++LK FN F EICK QS W I DE LKEE+ + + L PAY NFI +L
Sbjct: 535 GASRSLKEKLKLFNSYFEEICKTQSTWIIFDEQLKEELRISVAGALSPAYRNFIGRLQ-- 592
Query: 427 LKLEVKKPPDGYIEYETKDIKAILNNMFKIYRPSSCGRK 465
+ + + +I++ +D++A ++ +F+ S GRK
Sbjct: 593 SNNDSSRHTERHIKFSVEDLEARISELFQGSSGSGGGRK 631
>UniRef100_Q9LMJ4 F10K1.28 protein [Arabidopsis thaliana]
Length = 599
Score = 286 bits (731), Expect = 1e-75
Identities = 165/448 (36%), Positives = 256/448 (56%), Gaps = 31/448 (6%)
Query: 11 LMLEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLT-IK 69
+++EAL + +L M+ GF KEC VYSS RRE LEE L + L ++ ++
Sbjct: 176 IVIEALQSSVIGDLNAIAVRMVAGGFAKECSRVYSSRRREFLEESLSRLHLRGLSMEEVQ 235
Query: 70 DVNMEDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEF--SAISDISFTDVCREFTIR 127
+ +DL I RWIKA + F + FP+ER LCD VF + S+++D+SF +VCR T +
Sbjct: 236 ESPWQDLEDEIDRWIKAVTLIFHVFFPSERLLCDRVFSDLPVSSVTDLSFMEVCRGTTTQ 295
Query: 128 LLNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGE 187
LLNF + IA LF+++D+YE + DLIP E+LF D+Y LR+E + K+LGE
Sbjct: 296 LLNFADAIALGSRLPERLFKVVDLYEAMQDLIPKMETLFSDRYCSPLRHEALAIHKRLGE 355
Query: 188 TIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVL 247
I G E EN IR + P F GG +HP+ R+VMN+L C R+ LEQ+ + G
Sbjct: 356 AIRGIFMELENLIR-RDPPKTAFPGGGIHPITRYVMNYLRAACKSRQSLEQILDQTG--- 411
Query: 248 LEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNS 307
+ + S + S+Q+ ++E+LES LE + DP+L ++++MN+
Sbjct: 412 ---------------NETGSDTRPLSVQIIWVLELLESNLEGKKRTYRDPSLCFLFMMNN 456
Query: 308 SRYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNM 367
+YI+ K +NELG +LG+ + +H+AKLR Y RSSW +V+ LR D +P +
Sbjct: 457 DKYILDKAKDNELGLVLGEDWIVKHAAKLRQYHSNYRRSSWNQVVGLLRTDG---PYPKL 513
Query: 368 VGKSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVL 427
V + L+ F F+E+CK QS W + D L+EE+ + + PAY+NFIR+L
Sbjct: 514 V-----ENLRLFKSQFDEVCKVQSQWVVSDGQLREELRSSVAGIVSPAYSNFIRRLKESP 568
Query: 428 KLEVKKPPDGYIEYETKDIKAILNNMFK 455
++ ++ + +I Y +D++ I+ +FK
Sbjct: 569 EINGRR-GEPFIPYTVEDVEFIIKRLFK 595
>UniRef100_Q65WW9 Putative leucine zipper protein [Oryza sativa]
Length = 589
Score = 256 bits (655), Expect = 7e-67
Identities = 154/452 (34%), Positives = 255/452 (56%), Gaps = 29/452 (6%)
Query: 11 LMLEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKD 70
++++ALS +V N+ + + M+++GF +EC VY++ RR ++E + + L T ++
Sbjct: 156 VVIDALSPGSVANVHQIARRMVDAGFGRECAEVYAAARRGFVDESVAR--LGVRPRTAEE 213
Query: 71 VNM---EDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIR 127
V+ E+L I RWI AF + F+IL P+ER+LCD VF + D++F R ++
Sbjct: 214 VHASSWEELEFDIARWIPAFNMVFRILIPSERRLCDRVFDGLAPFGDLAFVAAVRTQALQ 273
Query: 128 LLNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGE 187
L++F + I++ LFR++DMYE + DL+P+ + +F D YS +LR E+ V LG
Sbjct: 274 LISFGDAISSSSRAPERLFRVVDMYEAVRDLLPDLDPVFADPYSAALRAEVTAVCNTLGS 333
Query: 188 TIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFE-DHGHV 246
+I G E EN IR + P GG +HP+ R+VMN+L C R+ LE+V E D G V
Sbjct: 334 SIKGIFMELENLIR-RDPARVAAQGGGIHPITRYVMNYLRAACGSRQTLEEVMEGDFGAV 392
Query: 247 LLEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMN 306
++++ +SS ++ + IM+VL L+ I+ DP+L V+LMN
Sbjct: 393 ----------GGAAAAVDPDRPTSSLAVHIAWIMDVLHKNLDIKSKIYRDPSLACVFLMN 442
Query: 307 SSRYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPN 366
+ +YII K ++ELG LLGD +++ + ++R +Y R +WGKV L+ P
Sbjct: 443 NGKYIIQKVNDSELGVLLGDEWIKQMTNRVRRWSMDYQRVTWGKVTTVLQTGG-----PG 497
Query: 367 MVG---KSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKL 423
+ G +MK++L+ FN F EI + QS W I DE L+ ++ + E ++P YT I +
Sbjct: 498 VGGLPATAMKQKLRMFNTYFQEIYEVQSEWVIADEQLRVDVRAAVAEAVMPVYTALISR- 556
Query: 424 HIVLKLEVKKPPDGYIEYETKDIKAILNNMFK 455
LK + D YI+Y +D++A + ++F+
Sbjct: 557 ---LKSSPEARHDLYIKYTPEDVEACIQHLFE 585
>UniRef100_Q941X1 Leucine zipper protein-like [Oryza sativa]
Length = 652
Score = 248 bits (634), Expect = 2e-64
Identities = 144/447 (32%), Positives = 250/447 (55%), Gaps = 19/447 (4%)
Query: 11 LMLEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLT-IK 69
++++AL +V+++ + + M+++GF +EC Y++ RR ++E + + + + + +
Sbjct: 219 VVIDALPPGSVSDVHQIARRMVDAGFGRECAEAYAAARRGFIDESVARLGIRARTIDEVH 278
Query: 70 DVNMEDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIRLL 129
+ E+L I RWI AFK+ F+IL P+ER+LCD VF + D++F R ++L+
Sbjct: 279 SLPWEELEFDIARWIPAFKMVFRILIPSERRLCDRVFDGLAPYGDLAFVAAVRTQVLQLI 338
Query: 130 NFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETI 189
+F + ++ LFR++DMYE + DL+P+ + +F D YS +LR E++ V LG +I
Sbjct: 339 SFGDAVSAASRAPERLFRVIDMYEAVRDLLPDLDPVFADPYSAALRAEVSAVCNTLGSSI 398
Query: 190 VGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFE-DHGHVLL 248
G E EN IR + P GG +HP+ R+VMN+L C R+ LE+V E D G V
Sbjct: 399 KGIFMELENLIR-RDPARVSVPGGGIHPITRYVMNYLRAACGSRQTLEEVMEGDLGAVGG 457
Query: 249 EYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSS 308
D P +SS ++ + IM+VL LE I+ DP L ++LMN+
Sbjct: 458 AAIAVDPDRP----------TSSLAVHIAWIMDVLHKNLETKSKIYRDPPLASIFLMNNG 507
Query: 309 RYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMV 368
+YII K ++ELG LLGD +++ +++R EY R +W KV+ L+ + ++
Sbjct: 508 KYIIHKVNDSELGVLLGDEWMKQMMSRVRRWSLEYQRGAWAKVMSVLQTGGPGI--GSLP 565
Query: 369 GKSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVLK 428
K++ ++L+ FN EIC QS W I DE L+E++ + +++ AY I + LK
Sbjct: 566 AKALLQKLRMFNGYLEEICAIQSEWVIADEQLREDVRAAITDSVKSAYMGLISR----LK 621
Query: 429 LEVKKPPDGYIEYETKDIKAILNNMFK 455
+ D +I++ +D++A + ++F+
Sbjct: 622 SSPEAAQDLFIKHSPEDVEARIQHLFE 648
>UniRef100_Q9FY95 Hypothetical protein T19L5_110 [Arabidopsis thaliana]
Length = 653
Score = 236 bits (602), Expect = 1e-60
Identities = 144/456 (31%), Positives = 240/456 (52%), Gaps = 16/456 (3%)
Query: 17 SLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLT-IKDVNMED 75
S E+++ L++ M+++G+ EC + Y RR +E L + N+ ++ + E
Sbjct: 208 SPESISTLKKIAGAMISAGYEAECCMSYEMSRRHAFKEELTEVGFEGINVEDVQRIGWES 267
Query: 76 LGLRIKRWIKAFKVAFKILFPTERQLCDIVF--FEFSAISDISFTDVCREFTIRLLNFPN 133
L I WI + +LFP E LC+ VF + S++ FT + TIR L+F
Sbjct: 268 LEGEIASWISIVRRCSTVLFPGELSLCNAVFPDQDHSSVRKRLFTGLVSAVTIRFLDFSG 327
Query: 134 VIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTL 193
+ + ++ LF+ LDMYETL DLIP E Q L E+ +LGE V
Sbjct: 328 AVVLTKRSSEKLFKFLDMYETLRDLIPAVE-----QSDSDLIQEIKLAQTRLGEAAVTIF 382
Query: 194 REFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKH 253
E E +I+S G P G +HPL R+ MN+L + C+Y+E L+QVF+ + +
Sbjct: 383 GELEKSIKSDN-GRTPVPSGAVHPLTRYTMNYLKYACEYKETLDQVFQHYEANQTDNKPE 441
Query: 254 DDTVPSSSSSSSSSSSSSSSL--QMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYI 311
+T P S+ QM R+ME+L++ LE ++ DP+L +++LMN+ RYI
Sbjct: 442 PETKPRQQQREDDEEYKVSAFARQMIRVMELLDANLEIKSRLYRDPSLRFIFLMNNGRYI 501
Query: 312 IIKTMEN-ELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMVGK 370
+ K + E+ L+G ++ S +LR + Y R +WGKVL+ + + L V+ +
Sbjct: 502 LQKIKGSIEIRDLMGQSWTRKRSTELRQYHKSYQRETWGKVLQCMNQEG-LQVNGKVSKP 560
Query: 371 SMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVLKLE 430
+K++ K FN +F+EI K QS W + DE ++ E+ V + ++PAY +F + L+
Sbjct: 561 VLKERFKIFNAMFDEIHKTQSTWIVSDEQMQSELRVSISSLVIPAYRSFFGRYK--QHLD 618
Query: 431 VKKPPDGYIEYETKDIKAILNNMFKIYRPSSCGRKR 466
K D Y++Y+ +DI++ ++++F P+S RKR
Sbjct: 619 SGKQTDKYVKYQPEDIESFIDDLFD-GNPTSMARKR 653
>UniRef100_Q9FK34 Gb|AAD25781.1 [Arabidopsis thaliana]
Length = 637
Score = 227 bits (579), Expect = 5e-58
Identities = 137/461 (29%), Positives = 249/461 (53%), Gaps = 39/461 (8%)
Query: 11 LMLEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKD 70
L ++ ++ V +L+E + M+ +G+ KEC+ VYSS RR+ L++CL+ L + L+I++
Sbjct: 187 LWVDLINPTAVEDLKEIAERMIRAGYEKECVQVYSSVRRDALDDCLM--ILGVEKLSIEE 244
Query: 71 VNMED---LGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIR 127
V D + ++K+WI+A K+ ++L E+++CD +F + ++ F + + ++
Sbjct: 245 VQKIDWKSMDEKMKKWIQAVKITVRVLLVGEKKICDEIFSSSESSKEVCFNETTKSCVMQ 304
Query: 128 LLNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGE 187
LLNF +A + ++ LFR+LDMY+ L +++ E + D + + NE VL+ LG+
Sbjct: 305 LLNFGEAVAIGRRSSEKLFRILDMYDALANVLQTLEVMVTDCF---VCNETKGVLEALGD 361
Query: 188 TIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVL 247
GT EFEN +R++ P G++HP++R+VMN++ I DY L + E +
Sbjct: 362 AARGTFVEFENNVRNE-TSKRPTTNGEVHPMIRYVMNYMKLIVDYAVTLNSLLESN---- 416
Query: 248 LEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEV---LESKLEAMFNIFNDPTLGYVYL 304
SS S S+ S +RI+ + LES LE ++ D L +V++
Sbjct: 417 -----------ESSGVSGDDSTEEMSPLAKRILGLITSLESNLEDKSKLYEDGGLQHVFM 465
Query: 305 MNSSRYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLR---LDNNL 361
MN+ YI+ K ++ELG LLGD +++ ++R Y+R+SW +VL LR + +
Sbjct: 466 MNNIYYIVQKVKDSELGKLLGDDWVRKRRGQIRQYATGYLRASWSRVLSALRDESMGGSS 525
Query: 362 LVHP-------NMVGKSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLP 414
P N ++K++ + FN F E+ + Q+ W + D L+EE+ + + E ++P
Sbjct: 526 SGSPSYGQRSNNSSKMALKERFRGFNASFEELYRLQTAWKVPDPQLREELRISISEKVIP 585
Query: 415 AYTNFIRKLHIVLKLEVKKPPDGYIEYETKDIKAILNNMFK 455
AY F + +LE + YI+Y D+++ L ++F+
Sbjct: 586 AYRAFFGRNR--SQLEGGRHAGKYIKYTPDDLESYLPDLFE 624
>UniRef100_Q8RZJ0 Putative leucine zipper protein [Oryza sativa]
Length = 646
Score = 227 bits (578), Expect = 6e-58
Identities = 136/451 (30%), Positives = 247/451 (54%), Gaps = 23/451 (5%)
Query: 16 LSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKDVNMED 75
+S +TV L+ +ML +G+ E VY RR+ L ECL L D +++++V +
Sbjct: 214 ISPDTVGALRGIADVMLRAGYGPELCQVYGEMRRDTLMECLA--VLGVDKMSLEEVQRVE 271
Query: 76 LGL---RIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIRLLNFP 132
G+ ++K+WI+A KV + L ER++C+ +F + + FT+ + ++LLNF
Sbjct: 272 WGVLDGKMKKWIQALKVVVRGLVAEERRICNQIFAADAEAEEDCFTEAAKGCVLQLLNFG 331
Query: 133 NVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGT 192
+ IA + ++ LFR+L MYE L +++P E LF ++ E +L +LG+ + GT
Sbjct: 332 DAIAIGKRSSEKLFRILGMYEALDEVLPELEGLFSGDARDFIKEEAVGILMRLGDAVRGT 391
Query: 193 LREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTK 252
+ EF N I+ + A GG++HPL R+VMN++ + DY L Q+ +D
Sbjct: 392 VAEFANAIQGETSRRA-LPGGEIHPLTRYVMNYVRLLADYSRSLNQLLKDW--------- 441
Query: 253 HDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYII 312
DT + + + + + + ++ L++K+E ++ D L ++LMN+ YI+
Sbjct: 442 --DTELENGGDNVNMTPLGQCVLI--LITHLQAKIEEKSKLYEDEALQNIFLMNNLLYIV 497
Query: 313 IKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDN--NLLVHPNMVGK 370
K ++EL TLLGD +++ ++R Y+RSSW +VL LR D + + +
Sbjct: 498 QKVKDSELKTLLGDNWIRQRRGQIRRYSTGYLRSSWTRVLACLRDDGLPQTMGSSSALKA 557
Query: 371 SMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVLKLE 430
S+K++ K+FN F E+ K Q+ W ++D L+EE+ + + E +LPAY +F+ + +LE
Sbjct: 558 SLKERFKNFNLAFEELYKTQTTWKVVDPQLREELKISISEKVLPAYRSFVGRFR--GQLE 615
Query: 431 VKKPPDGYIEYETKDIKAILNNMFKIYRPSS 461
+ YI+Y +D++ +++ F+ RP++
Sbjct: 616 GGRNSARYIKYNPEDLENQVSDFFEGRRPNA 646
>UniRef100_Q9LJH9 Gb|AAD25781.1 [Arabidopsis thaliana]
Length = 623
Score = 220 bits (560), Expect = 8e-56
Identities = 135/463 (29%), Positives = 254/463 (54%), Gaps = 21/463 (4%)
Query: 13 LEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKDVN 72
++ +S E V++L+ V+ M+ +G+++EC+ VY + R+ +E + KQ L ++I DV
Sbjct: 173 MDLISPEAVSDLRSIVQRMVAAGYSRECIQVYGTVRKSAME-AIFKQ-LGIVKISIGDVQ 230
Query: 73 M---EDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEF-SAISDISFTDVCREFTIRL 128
E + +I++WI+A KV +++F +E++LC+ +F +A+ + F + + +RL
Sbjct: 231 RLEWEVVEGKIRKWIRAAKVCIRVVFSSEKRLCEQLFDGICTAMDETCFMETVKASALRL 290
Query: 129 LNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGET 188
FP I+ + + LF++LD+++ L D++P+ E++F S ++R + + +L E
Sbjct: 291 FTFPEAISISRRSPEKLFKILDLHDALTDMLPDIEAIFDSDSSDAIRAQAVEIQSRLAEA 350
Query: 189 IVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLL 248
G L EFEN + + P P GG +HPL R+VMN++ I DY++ L+ + +
Sbjct: 351 ARGILSEFENAV-LREPSIVPVPGGTIHPLTRYVMNYIVMISDYKQTLDDLIMSN----- 404
Query: 249 EYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSS 308
T D P + S S L + ++ VL LE + D +L ++++MN+
Sbjct: 405 PSTGSDPNTP-DMDFTELDSKSPLDLHLIWLIVVLHFNLEEKSKHYRDTSLAHIFIMNNI 463
Query: 309 RYIIIKTMEN-ELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNM 367
YI+ K + EL ++GD L++ + R+ Y R++W +VL LR D L V +
Sbjct: 464 HYIVQKVKRSPELREMIGDHYLRKLTGIFRHAATNYQRATWVRVLNSLR-DEGLHVSGSF 522
Query: 368 VG----KSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKL 423
+++++ K+FN +F E+ + QS W + D L+EE+ + L E+L+PAY +F+ +
Sbjct: 523 SSGVSRSALRERFKAFNTMFEEVHRTQSTWSVPDAQLREELRISLSEHLIPAYRSFLGRF 582
Query: 424 HIVLKLEVKKPPDGYIEYETKDIKAILNNMFKIYRPSSCGRKR 466
+E + P+ Y++Y +DI+ I+ + F+ Y R+R
Sbjct: 583 R--GHIESGRHPENYLKYSVEDIETIVLDFFEGYTTPPHLRRR 623
>UniRef100_Q93ZZ6 Hypothetical protein At3g14090 [Arabidopsis thaliana]
Length = 503
Score = 220 bits (560), Expect = 8e-56
Identities = 135/463 (29%), Positives = 254/463 (54%), Gaps = 21/463 (4%)
Query: 13 LEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKDVN 72
++ +S E V++L+ V+ M+ +G+++EC+ VY + R+ +E + KQ L ++I DV
Sbjct: 53 MDLISPEAVSDLRSIVQRMVAAGYSRECIQVYGTVRKSAME-AIFKQ-LGIVKISIGDVQ 110
Query: 73 M---EDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEF-SAISDISFTDVCREFTIRL 128
E + +I++WI+A KV +++F +E++LC+ +F +A+ + F + + +RL
Sbjct: 111 RLEWEVVEGKIRKWIRAAKVCIRVVFSSEKRLCEQLFDGICTAMDETCFMETVKASALRL 170
Query: 129 LNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGET 188
FP I+ + + LF++LD+++ L D++P+ E++F S ++R + + +L E
Sbjct: 171 FTFPEAISISRRSPEKLFKILDLHDALTDMLPDIEAIFDSDSSDAIRAQAVEIQSRLAEA 230
Query: 189 IVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLL 248
G L EFEN + + P P GG +HPL R+VMN++ I DY++ L+ + +
Sbjct: 231 ARGILSEFENAV-LREPSIVPVPGGTIHPLTRYVMNYIVMISDYKQTLDDLIMSN----- 284
Query: 249 EYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSS 308
T D P + S S L + ++ VL LE + D +L ++++MN+
Sbjct: 285 PSTGSDPNTP-DMDFTELDSKSPLDLHLIWLIVVLHFNLEEKSKHYRDTSLAHIFIMNNI 343
Query: 309 RYIIIKTMEN-ELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNM 367
YI+ K + EL ++GD L++ + R+ Y R++W +VL LR D L V +
Sbjct: 344 HYIVQKVKRSPELREMIGDHYLRKLTGIFRHAATNYQRATWVRVLNSLR-DEGLHVSGSF 402
Query: 368 VG----KSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKL 423
+++++ K+FN +F E+ + QS W + D L+EE+ + L E+L+PAY +F+ +
Sbjct: 403 SSGVSRSALRERFKAFNTMFEEVHRTQSTWSVPDAQLREELRISLSEHLIPAYRSFLGRF 462
Query: 424 HIVLKLEVKKPPDGYIEYETKDIKAILNNMFKIYRPSSCGRKR 466
+E + P+ Y++Y +DI+ I+ + F+ Y R+R
Sbjct: 463 R--GHIESGRHPENYLKYSVEDIETIVLDFFEGYTTPPHLRRR 503
>UniRef100_Q9C9E5 Hypothetical protein T10D10.6 [Arabidopsis thaliana]
Length = 633
Score = 218 bits (554), Expect = 4e-55
Identities = 136/463 (29%), Positives = 248/463 (53%), Gaps = 18/463 (3%)
Query: 13 LEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKDV- 71
+E + +E+V +L + M+++G+ +EC+ VY S R+ ++ + L + L+I DV
Sbjct: 178 IELIPIESVIHLSWIARRMVSAGYLRECIQVYGSVRKSAVDSSFRR--LGIEKLSIGDVQ 235
Query: 72 --NMEDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAIS--DISFTDVCREFTIR 127
N E L +I+RWI+A K+ +++F +E+ LC+ VF A++ + F + + I+
Sbjct: 236 RLNWEALEQKIRRWIRAAKICVRVVFASEKLLCEHVFESVGAVNIHEACFMETVKGPAIQ 295
Query: 128 LLNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGE 187
L NF I+ + + LF++LD+++ L +L+P+ ES+F + S S+R + +L +L E
Sbjct: 296 LFNFAEAISISRRSPEKLFKILDLHDALIELLPDIESVFDLKSSDSIRVQAAEILSRLAE 355
Query: 188 TIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVL 247
G L EFEN + + P P GG +HPL R+VMN+++ I +YR L +
Sbjct: 356 AARGILSEFENAV-LREPSRVPVPGGTIHPLTRYVMNYISLISEYRPTLIDLIMSKP--- 411
Query: 248 LEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNS 307
D P S ++ +L + I+ +L+ LE + + L ++++MN+
Sbjct: 412 -SRNATDSNTPDFDFSELENNKGPLALHLIWIIVILQFNLEGKSKYYKNAALSHLFIMNN 470
Query: 308 SRYIIIKTMEN-ELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLR---LDNNLLV 363
+ YI+ K + EL ++GD L++ + K R Y R++W KVL LR L
Sbjct: 471 AHYIVQKIKGSPELREMIGDLYLRKLTGKFRQAATYYQRAAWVKVLYCLRDEGLHTKGSF 530
Query: 364 HPNMVGKSMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKL 423
+ +++++ KSFN LF E+ + QS W + D L+EE+ + + E L PAY +F+ +
Sbjct: 531 SSGVSRSALRERFKSFNALFEEVHRVQSQWLVPDSQLREELKISILEKLSPAYRSFLGRF 590
Query: 424 HIVLKLEVKKPPDGYIEYETKDIKAILNNMFKIYRPSSCGRKR 466
+E K P+ YI+ +D++ + ++F+ Y + R+R
Sbjct: 591 R--SHIESGKHPENYIKISVEDLETEVLDLFEGYSATQHLRRR 631
>UniRef100_Q9FFX6 Putative leucine zipper protein [Arabidopsis thaliana]
Length = 695
Score = 217 bits (553), Expect = 5e-55
Identities = 138/436 (31%), Positives = 233/436 (52%), Gaps = 23/436 (5%)
Query: 24 LQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKDV---NMEDLGLRI 80
L++ + M G+ EC VY RR L L KQ + ++I +V + + L I
Sbjct: 264 LRKIAEKMKAGGYGWECREVYLVGRRNILMRTL-KQDCEFEKVSIDEVQKMSWDTLEREI 322
Query: 81 KRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIRLLNFPNVIANDQS 140
W K FK + FP E +L + +F F V I+ L F +A +
Sbjct: 323 PIWNKTFKDCSSLFFPGELKLAERIF---PGDEGNLFCIVTHGLAIQFLGFAEAVAMTRR 379
Query: 141 NTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTLREFENTI 200
+T LF++LD+YETL D P E LF ++ LRNE+ + +LGET + + E++I
Sbjct: 380 STEKLFKILDIYETLRDSFPAMEELFPEELRSELRNEVTSARSRLGETAIHIFCDLEHSI 439
Query: 201 RSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKHDDTVPSS 260
+S P GG +HPL R+ MN+L + C+Y++ LEQVF+ H + + ++ P
Sbjct: 440 KSDS-SKTPVPGGAVHPLTRYTMNYLKYSCEYKDTLEQVFKSHSKM-----EREEEEPVE 493
Query: 261 SSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKTMEN-E 319
S +S+ +S Q+ RIME+L+ LE + D L +++MN+ RYI+ K + E
Sbjct: 494 SGNSAFAS------QLMRIMELLDGNLETKSKQYKDIPLSCIFMMNNGRYIVQKIKGSAE 547
Query: 320 LGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMVGKSMKKQLKSF 379
+ ++GD +R S++LR + Y R +WGK+L FL L+ + +V ++K++ KSF
Sbjct: 548 IHEVMGDTWCRRRSSELRNYHKNYQRETWGKLLGFLG-HEGLMHNGKIVKPNLKERFKSF 606
Query: 380 NKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVLKLEVKKPPDGYI 439
N F+EI K Q+ W + DE L+ E+ V + ++PAY F+ + L+ + + Y+
Sbjct: 607 NATFDEIHKTQTTWVVNDEQLQSELRVSITAVMIPAYRAFMARFG--QYLDPGRQTEKYV 664
Query: 440 EYETKDIKAILNNMFK 455
+Y+ +DI+ +++ +F+
Sbjct: 665 KYQPEDIEDLIDQLFE 680
>UniRef100_Q9SYG5 F15I1.17 protein [Arabidopsis thaliana]
Length = 622
Score = 214 bits (545), Expect = 4e-54
Identities = 135/453 (29%), Positives = 249/453 (54%), Gaps = 18/453 (3%)
Query: 13 LEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKDV- 71
++ ++ E V++L+ + M+ +G+++EC+ VY + R+ +E ++ + L L I DV
Sbjct: 170 IDLVTPEAVSDLRSIAQRMIGAGYSRECVQVYGNVRKSAME--MIFKQLGIVKLGIGDVQ 227
Query: 72 --NMEDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIRLL 129
+ E + ++I+RWI+A KV +++F +E++LC+ +F + + F ++ + ++L
Sbjct: 228 RLDWEAVEVKIRRWIRAAKVCVRVVFASEKRLCEQIFE--GTMEETCFMEIVKTSALQLF 285
Query: 130 NFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETI 189
NFP I+ + + LF++LD+++ + DL+P+ E +F S S+ + + +L E
Sbjct: 286 NFPEAISISRRSPEKLFKILDLHDAITDLLPDMEEIFDSSSSESILVQATEIQSRLAEAA 345
Query: 190 VGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLE 249
G L EFEN + + P P GG +HPL R+VMN+L I DYRE L + L+
Sbjct: 346 RGILTEFENAV-FREPSVVPVPGGTIHPLTRYVMNYLNLISDYRETLIDLVMTKPCRGLK 404
Query: 250 YTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSR 309
T +D S S +L M M +L+ LE + D L ++++MN+
Sbjct: 405 CT--NDRNDPDMDISELEGISPLALHMIWTMVMLQFNLEEKSLHYRDEPLSHIFVMNNVH 462
Query: 310 YIIIKTMEN-ELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNM- 367
YI+ K + EL L+GD L++ + R +Y R++W +VL LR D L V +
Sbjct: 463 YIVQKVKSSPELMELIGDKYLRKLTGIFRQAATKYQRATWVRVLNSLR-DEGLHVSGSFS 521
Query: 368 --VGKS-MKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLH 424
V KS ++++ K+FN +F E+ + QS W + D L+EE+ + L E+L+PAY +F+ +
Sbjct: 522 SGVSKSALRERFKAFNTMFEEVHRIQSTWSVPDTQLREELRISLSEHLIPAYRSFLGRFR 581
Query: 425 IVLKLEVKKPPDGYIEYETKDIKAILNNMFKIY 457
+E + P+ Y++Y ++++ + + F+ Y
Sbjct: 582 --GHIESGRHPENYLKYSVENLETAVLDFFEGY 612
>UniRef100_Q7XVB9 OSJNBa0072D21.9 protein [Oryza sativa]
Length = 688
Score = 213 bits (542), Expect = 9e-54
Identities = 134/461 (29%), Positives = 240/461 (51%), Gaps = 25/461 (5%)
Query: 19 ETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKDVNMEDLGL 78
E +++L+ + M +G+ E VY RR+ L+ECL L + L+I +V + L
Sbjct: 240 EAIDDLRSIAQRMDRAGYASELEQVYCGVRRDLLDECLA--VLGVERLSIDEVQRMEWKL 297
Query: 79 ---RIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIRLLNFPNVI 135
++K+W+ K + L ER++CD V + D F + + +++LNF + +
Sbjct: 298 LNDKMKKWVHGVKTVVRSLLTGERRICDQVLAVSDELRDECFVESTKGCIMQILNFGDAV 357
Query: 136 ANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTLRE 195
A + L R+LDMYE L ++IP + LF + +L VL++LG+ + GTL E
Sbjct: 358 AVCSRSPEKLSRILDMYEALAEVIPELKELFFGNSGNDVICDLEGVLERLGDAVKGTLLE 417
Query: 196 FENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKHDD 255
F ++ + P G++HP+ R+VMN+L + Y + L+++ D ++ H D
Sbjct: 418 FGKVLQQES-SRRPMMAGEIHPMTRYVMNYLRLLVVYSDTLDKLLGDDSAGDVD---HSD 473
Query: 256 TVPSSSSSSS-SSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIK 314
T S S + +++ LE+ LE ++ D L ++ MN+ YI+ K
Sbjct: 474 THRGGDDEEEYLESLSPLGRHLVKLISYLEANLEEKSKLYEDGALQCIFSMNNILYIVQK 533
Query: 315 TMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMVGKS--- 371
++ELG +LGD ++R K+R N + Y+R SW KVL FL+ D H G
Sbjct: 534 VKDSELGRILGDHWIRRRRGKIRQNSKNYLRISWTKVLSFLKDD----AHGGRSGSGSGS 589
Query: 372 -----MKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIV 426
+K++ K+FN F+EI ++Q+LW + D L+EE+ + + EN++PAY F+ + +
Sbjct: 590 GNSSRIKEKFKNFNLAFDEIYRSQTLWKVPDPQLREELKISISENVIPAYRAFLGRYGSL 649
Query: 427 LKLEVKKPPDGYIEYETKDIKAILNNMFK-IYRPSSCGRKR 466
++ + YI+Y +D++ L+++F+ P++ R+R
Sbjct: 650 --VDSGRNSGRYIKYTPEDLENQLSDLFEGSLGPANHSRRR 688
>UniRef100_Q69P90 Putative leucine zipper-containing protein [Oryza sativa]
Length = 638
Score = 207 bits (528), Expect = 4e-52
Identities = 138/481 (28%), Positives = 246/481 (50%), Gaps = 26/481 (5%)
Query: 6 SLEGTLMLEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDN 65
SL ++ L + +++L M +G+ +EC+ VY+S R+ ++ L + L +
Sbjct: 161 SLRSIREIDLLPADAISDLHAIASRMAVAGYGRECVQVYASVRKPAVDSALRR--LGVEK 218
Query: 66 LTIKDVNM---EDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEF--------SAISD 114
L+I DV E L +I+RWI+A + A + +F +ER+LC ++F + +A D
Sbjct: 219 LSIGDVQRLEWEVLEAKIRRWIRAARAAVRGVFASERRLCFLIFHDLPLSSSTITTATHD 278
Query: 115 ISFTDVCREFTIRLLNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFC-DQYSVS 173
F + + ++L F I+ + + LF+++D+++ + DL+P+ +F + S
Sbjct: 279 APFAEAVKGAALQLFGFAEAISIGRRSPEKLFKIIDLHDAIADLLPDVSDIFAASKAGES 338
Query: 174 LRNELNTVLKKLGETIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYR 233
+ + + +L + + G L EFEN + + P P GG +HPL R+VMN+ + I DY+
Sbjct: 339 IYVQAAEIRSRLADAVRGILSEFENAVL-RDPSKTPVPGGTIHPLTRYVMNYSSLISDYK 397
Query: 234 EILEQVFEDHGHVLLEYTKH-DDTVPSSSSSSSSSSSSSSSLQMERI--MEVLESKLEAM 290
L ++ ++ PS + S L I + VLE LE+
Sbjct: 398 TTLSELIVSRPLACSRIAPEGNENAPSFPDLDLADPDSQLPLAAHLIWIIVVLEHNLESK 457
Query: 291 FNIFNDPTLGYVYLMNSSRYIIIKTMEN-ELGTLLGDGMLQRHSAKLRYNFEEYIRSSWG 349
+++ D L ++++MN+ YI K ++ EL L+GD L++ + K R Y R++W
Sbjct: 458 ASLYKDAALSHLFVMNNVHYIAHKIKDSPELRGLIGDEYLKQLTGKFRLAATRYQRTAWL 517
Query: 350 KVLEFLRLDNNLLVHPNM---VGKS-MKKQLKSFNKLFNEICKAQSLWFIMDETLKEEII 405
K+L LR D L V V KS ++++ KSFN F E + QS W++ D L+EE+
Sbjct: 518 KILNCLR-DEGLHVSGGFSSGVSKSALRERFKSFNAAFEEAHRVQSAWYVPDTQLREELR 576
Query: 406 VYLGENLLPAYTNFIRKLHIVLKLEVKKPPDGYIEYETKDIKAILNNMFKIYRPSSCGRK 465
+ + E LLPAY +F+ + +E + P+ YI+Y +D++ + N F+ PS R+
Sbjct: 577 ISIAEKLLPAYRSFLGRFR--HHIENGRHPELYIKYSVEDLETSVTNFFEGCPPSLHNRR 634
Query: 466 R 466
R
Sbjct: 635 R 635
>UniRef100_Q9LZD3 Hypothetical protein F12E4_320 [Arabidopsis thaliana]
Length = 638
Score = 204 bits (518), Expect = 6e-51
Identities = 134/444 (30%), Positives = 229/444 (51%), Gaps = 36/444 (8%)
Query: 24 LQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKDVNM---EDLGLRI 80
L + + M+ +G ++ L +Y R LEE L K L + L+ +DV E L +I
Sbjct: 210 LHDLAQQMVQAGHQQQLLQIYRDTRSFVLEESLKK--LGVEKLSKEDVQRMQWEVLEAKI 267
Query: 81 KRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIRLLNFPNVIANDQS 140
WI ++A K+LF ERQ+CD +F F ++SD F +V LL+F + IA +
Sbjct: 268 GNWIHFMRIAVKLLFAGERQVCDQIFRGFDSLSDQCFAEVTVSSVSMLLSFGDAIARSKR 327
Query: 141 NTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTLREFENTI 200
+ LF +LDMYE + +L E++F + + +R+ + K+L +T T +FE +
Sbjct: 328 SPEKLFVLLDMYEIMRELHTEIETIFKGKACLEIRDSATGLTKRLAQTAQETFGDFEEAV 387
Query: 201 RSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKHDDTVPSS 260
K G +HPL +V+N++ ++ DY+ L+Q+F LE+ DD
Sbjct: 388 -EKDATKTAVLDGTVHPLTSYVINYVKFLFDYQTTLKQLF-------LEFGNGDD----- 434
Query: 261 SSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKTMENEL 320
S+S +S+ M RIM+ L++ L+ + DP L +++LMN+ Y++ +E
Sbjct: 435 ------SNSQLASVTM-RIMQALQNNLDGKSKQYKDPALTHLFLMNNIHYMVRSVRRSEA 487
Query: 321 GTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLE------FLRLDNNLLVHPNMVGKS--- 371
LLGD +QRH ++ + +Y R +W K+L+ L N G S
Sbjct: 488 KDLLGDDWVQRHRRIVQQHANQYKRVAWTKILQSSSAQGLTSSGGGSLEGGNSSGVSRGL 547
Query: 372 MKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVLKLEV 431
+K++ K FN F+E+ + QS W + D L+E + + + E LLPAY +F+++ + +E
Sbjct: 548 LKERFKMFNMQFDELHQRQSQWTVPDTELRESLRLAVAEVLLPAYRSFLKRFGPL--VES 605
Query: 432 KKPPDGYIEYETKDIKAILNNMFK 455
K P YI+Y +D++ +L +F+
Sbjct: 606 GKNPQKYIKYTAEDLERLLGELFE 629
>UniRef100_Q6K647 Putative leucine zipper-containing protein [Oryza sativa]
Length = 689
Score = 200 bits (508), Expect = 8e-50
Identities = 123/449 (27%), Positives = 236/449 (52%), Gaps = 22/449 (4%)
Query: 19 ETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKDVNM---ED 75
E V++L+ M +G+++E Y RR+ L+E L L + L+I +V +
Sbjct: 238 EAVDDLRAIADRMARAGYSRELADAYCGIRRDLLDEYL--SALGVERLSIDEVQRIEWKH 295
Query: 76 LGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIRLLNFPNVI 135
L ++K+W++A K ++L ER+LCD V + + F + + +++L+F + +
Sbjct: 296 LNDKMKKWVQAVKTVVRVLLAGERRLCDQVLSVSDELREECFIESTKGCIMQILSFGDAV 355
Query: 136 ANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTLRE 195
A + L R+LDMYE L ++IP + L + +++ L +LG+ I GTL E
Sbjct: 356 AVCPRSPEKLSRILDMYEALAEVIPEMKDLCLGSSGDGVISDVQANLDRLGDAIRGTLFE 415
Query: 196 FENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKHDD 255
F ++ + A G++HP+ R+VMN+L + Y + L+ + +D+ ++ + +D
Sbjct: 416 FGKVLQLESSRRA-MTAGEIHPMTRYVMNYLRLLVVYSDTLDALLDDNADDQIDLARAED 474
Query: 256 TVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKT 315
S + ++ +++ LE+ LE ++ D L ++ MN+ YI+ K
Sbjct: 475 -----QDQEHLESMTPLGKRLLKLISYLEANLEEKSKLYEDSALECIFSMNNLLYIVQKV 529
Query: 316 MENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMVGK----- 370
++ELG +LGD ++R + K+R + Y+R SW KVL FL+ D + + G
Sbjct: 530 RDSELGKILGDHWVKRRNGKIRQYSKSYLRISWMKVLSFLKDDGHGSGSGSSSGSGSGHS 589
Query: 371 ----SMKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIV 426
S+K++ K+FN F EI + Q+ W + D L+EE+ + + EN++PAY F+ +
Sbjct: 590 SSRMSIKEKFKNFNLAFEEIYRNQTTWKVPDPQLREELKISISENVIPAYRAFLGRYG-- 647
Query: 427 LKLEVKKPPDGYIEYETKDIKAILNNMFK 455
+++ + YI+Y +D+++ L+++F+
Sbjct: 648 SQVDGGRNSGKYIKYTPEDLESQLSDLFE 676
>UniRef100_Q94DY2 P0403C05.2 protein [Oryza sativa]
Length = 601
Score = 198 bits (503), Expect = 3e-49
Identities = 124/436 (28%), Positives = 228/436 (51%), Gaps = 14/436 (3%)
Query: 24 LQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTIKDV---NMEDLGLRI 80
++ M S ++KEC Y S R+ ++E L L D L+++++ N L I
Sbjct: 170 VKSIANFMFLSEYDKECSQAYISTRQSAVDENLGS--LRIDKLSMEELLSTNWTKLSSLI 227
Query: 81 KRWIKAFKVAFKILFPTERQLCDIVFFEFS-AISDISFTDVCREFTIRLLNFPNVIANDQ 139
KRW +A KV ++ +E++L + VF E S + +D+ F ++ ++LL F +A
Sbjct: 228 KRWNRAMKVFVQVYLTSEKRLSNHVFGELSESTADLCFYEISLSSVMQLLTFYESVAIGP 287
Query: 140 SNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTLREFENT 199
LFR+LDMYE L+DL+P E LF + + E N VL +LGE++ T+ EF+
Sbjct: 288 PKPEKLFRLLDMYEVLNDLLPEVEFLFQEGCDDIVLTEYNEVLLQLGESVRKTITEFKYA 347
Query: 200 IRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYTKHDDTVPS 259
++S NA G++HPL ++VMN++ + Y + L+ + +D ++
Sbjct: 348 VQSYTSSNA-MARGEVHPLTKYVMNYIKALTAYSKTLDSLLKDTDRRCQHFS-----TDI 401
Query: 260 SSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKTMENE 319
S ++ + S+L ++ + +LE LEA ++ D L +++MN+ Y++ K +E
Sbjct: 402 QSMANQCPHFTVSALHLQSVTAILEENLEAGSRLYRDDRLRNIFMMNNIYYMVQKVKNSE 461
Query: 320 LGTLLGDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLRLDNNLLVHPNMVGKSMKKQLKSF 379
L LGD ++ H+ K + Y R+SW VL FL D K +K++ K+F
Sbjct: 462 LKIFLGDDWIRVHNRKFQQQAMSYERASWSHVLSFLSDDGLCAAGDGASRKIIKEKFKNF 521
Query: 380 NKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVLKLEVKKPPDGYI 439
N F + + Q+ W I D+ L+E++ + + ++ AY F + + +L+ + + YI
Sbjct: 522 NLSFEDAYRTQTGWSIPDDQLREDVRISISLKIIQAYRTFTGRYY--SRLDGTRHLERYI 579
Query: 440 EYETKDIKAILNNMFK 455
+Y+ +D++ +L ++F+
Sbjct: 580 KYKPEDLEKLLLDLFE 595
>UniRef100_Q6Z0R1 Putative leucine zipper-containing protein [Oryza sativa]
Length = 632
Score = 197 bits (501), Expect = 5e-49
Identities = 137/484 (28%), Positives = 245/484 (50%), Gaps = 29/484 (5%)
Query: 6 SLEGTLMLEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDN 65
SL ++ L + V++L+ M +G+ +EC VY+S R+ ++ L + L +
Sbjct: 152 SLPSIREIDLLPDDAVSDLRAIASRMAAAGYGRECAQVYASVRKPAVDASLRR--LGVER 209
Query: 66 LTIKDVNMED---LGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFS-----------A 111
L+I DV + L +I+RWI+A + A + +F +ER+LC ++F + A
Sbjct: 210 LSIGDVQRLEWKALEAKIRRWIRAARAAVRGVFASERRLCFLIFHDLPISNITVTAAAPA 269
Query: 112 ISDISFTDVCREFTIRLLNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFC-DQY 170
D F + + ++L F I+ + + LF+++D+++ L DL+P+ +F +
Sbjct: 270 THDTPFAEAVKGAALQLFGFAEAISIGRRSPEKLFKIIDLHDALSDLLPDVSDIFAASKV 329
Query: 171 SVSLRNELNTVLKKLGETIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWIC 230
+ S+ + + +L + + G L EFEN + P A GG +HPL R+VMN+ + I
Sbjct: 330 AESIYVQAAEIRSRLADAVRGILSEFENAVLRDPPKTA-VPGGTVHPLTRYVMNYSSLIS 388
Query: 231 DYREILEQVFEDHGHVLLEYTKH-DDTVPSSSSSSSSSSSSSSSLQMERI--MEVLESKL 287
DY+ L ++ ++ PS + + + L I + VLE L
Sbjct: 389 DYKVTLSELIVSRPSASARLAAEGNELAPSLAELDLPEPDNQTPLAAHIIWIIVVLEHNL 448
Query: 288 EAMFNIFNDPTLGYVYLMNSSRYIIIKTMEN-ELGTLLGDGMLQRHSAKLRYNFEEYIRS 346
E +++ D L +++LMN+ YI+ K ++ +L L+GD L+R + K Y RS
Sbjct: 449 EGKASLYRDTALSHLFLMNNVYYIVHKVKDSPDLWNLIGDDYLKRLTGKFTMAATNYQRS 508
Query: 347 SWGKVLEFLRLDNNLLVHPNM---VGKS-MKKQLKSFNKLFNEICKAQSLWFIMDETLKE 402
+W K+L LR D L V + KS ++++ +SFN F E + QS W + D L+E
Sbjct: 509 AWLKILNCLR-DEGLHVSGGFSSGISKSALRERFRSFNAAFEEAHRVQSGWCVPDTQLRE 567
Query: 403 EIIVYLGENLLPAYTNFIRKLHIVLKLEVKKPPDGYIEYETKDIKAILNNMFKIYRPSSC 462
E+ + + E L+PAY +F+ + +E K P+ YI+Y +D++ +N+ F+ PS
Sbjct: 568 ELRISISEKLVPAYRSFLGRFR--HHIENGKHPELYIKYSAEDLEIAVNDFFEGVPPSPH 625
Query: 463 GRKR 466
R+R
Sbjct: 626 IRRR 629
>UniRef100_Q9LIA2 Gb|AAF27143.1 [Arabidopsis thaliana]
Length = 658
Score = 197 bits (500), Expect = 7e-49
Identities = 129/469 (27%), Positives = 237/469 (50%), Gaps = 39/469 (8%)
Query: 9 GTLMLEALSLETVNNLQETVKLMLNSGFNKECLIVYSSCRRECLEECLVKQFLNSDNLTI 68
G ++++ ++ + + +L+ M+ SG+++EC+ V + R++ L+E L + + L+I
Sbjct: 193 GDVVVDLVNPDVILDLKNIANTMIASGYDRECIQVCTMVRKDALDEFLYNHEV--EKLSI 250
Query: 69 KDVNMED---LGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFT 125
+DV D L IK+W++ + ++ +E+ L + +F + + I F D +
Sbjct: 251 EDVLRMDWATLNTNIKKWVRVMRDIVQVYLLSEKSLDNQIFGDLNEIGLTCFVDTVKAPM 310
Query: 126 IRLLNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKL 185
++LLNF ++ L R+L+MYE +L+P ++LF D S+R E V+++L
Sbjct: 311 MQLLNFGEAVSLGPRQPEKLLRILEMYELASELLPEIDALFLDHPGSSVRTEYREVMRRL 370
Query: 186 GETIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGH 245
G+ T EF++ I + + PF GG +HPL +VMN+L + D++ L+ + +H
Sbjct: 371 GDCARTTFLEFKSAI-AADVSSHPFPGGAVHPLTNYVMNYLMALTDFKHTLDSLLMEHDD 429
Query: 246 VLLEYTKHDDTVPSS-----------SSSSSSSSSSSSSLQMER----IMEVLESKLEAM 290
D T+P S S+ +SSS L M R I VLE+ L+
Sbjct: 430 A------EDLTIPPSPDIINPVMVEEESTYENSSSPEKFLAMTRHFYSITSVLEANLQEK 483
Query: 291 FNIFNDPTLGYVYLMNSSRYIIIKTMENELGTLLGDGMLQRHSAKLRYNFEEYIRSSWGK 350
++ D +L +++L+N+ Y+ K +++EL + GD ++H+ K + EY R++W
Sbjct: 484 SKLYKDVSLQHIFLLNNIHYMTRKVLKSELRHIFGDKWNRKHTWKFQQQATEYERATWLP 543
Query: 351 VLEFLRLDNNLLVHPNMVGKSMK-----KQLKSFNKLFNEICKAQSLWFIMDETLKEEII 405
VL FL+ D + + G K ++ + FN F E+ KAQ+ W I DE L+E++
Sbjct: 544 VLSFLKDDGSGSGPGSGSGSGSKNLRPRERFQGFNTAFEEVYKAQTGWLISDEGLREDVR 603
Query: 406 VYLGENLLPAYTNFIRKLHIVLKLEVKKPPDGYIEYETKDIKAILNNMF 454
++ AY F + + YI+Y T DI+ +L ++F
Sbjct: 604 TKASMWVIQAYWTFYSR-------HKNSVSERYIKYTTDDIERLLLDLF 645
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 762,012,526
Number of Sequences: 2790947
Number of extensions: 32333031
Number of successful extensions: 200908
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 74
Number of HSP's successfully gapped in prelim test: 67
Number of HSP's that attempted gapping in prelim test: 199937
Number of HSP's gapped (non-prelim): 309
length of query: 467
length of database: 848,049,833
effective HSP length: 131
effective length of query: 336
effective length of database: 482,435,776
effective search space: 162098420736
effective search space used: 162098420736
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC135161.4


