

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135161.1 - phase: 0
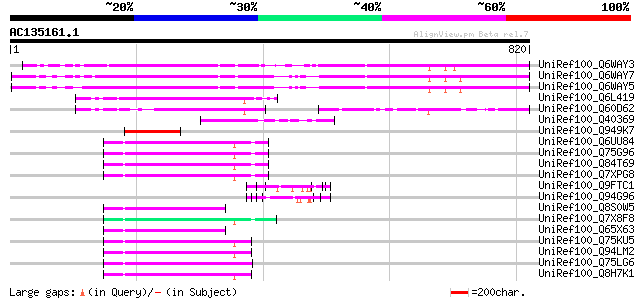
(820 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum] 358 3e-97
UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum] 352 3e-95
UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum] 350 1e-94
UniRef100_Q6L419 Putative gag polyprotein [Solanum demissum] 176 3e-42
UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solan... 130 2e-28
UniRef100_Q40369 RPE15 protein [Medicago sativa] 121 1e-25
UniRef100_Q949K7 Putative gag polyprotein [Cicer arietinum] 111 1e-22
UniRef100_Q6UU84 Putative polyprotein [Oryza sativa] 79 7e-13
UniRef100_Q75G96 Putative polyprotein [Oryza sativa] 78 9e-13
UniRef100_Q84T69 Putative GAG-POL [Oryza sativa] 78 1e-12
UniRef100_Q7XPG8 OSJNBb0003B01.15 protein [Oryza sativa] 77 2e-12
UniRef100_Q9FTC1 Gamma-gliadin [Aegilops speltoides] 77 2e-12
UniRef100_Q94G96 Gamma-gliadin [Triticum aestivum] 77 2e-12
UniRef100_Q8S0W5 Putative polyprotein [Oryza sativa] 77 3e-12
UniRef100_Q7X8F8 OSJNBb0002J11.5 protein [Oryza sativa] 76 4e-12
UniRef100_Q65X63 Putative polyprotein [Oryza sativa] 76 4e-12
UniRef100_Q75KU5 Putative polyprotein [Oryza sativa] 76 5e-12
UniRef100_Q94LM2 Putative polyprotein [Oryza sativa] 76 5e-12
UniRef100_Q75LG6 Putative gag protein [Oryza sativa] 75 6e-12
UniRef100_Q8H7K1 Putative polyprotein [Oryza sativa] 75 6e-12
>UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 358 bits (920), Expect = 3e-97
Identities = 271/843 (32%), Positives = 403/843 (47%), Gaps = 116/843 (13%)
Query: 20 AQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEASSSWTLCADTPRQSAPQR 79
AQ Q +AQ Q + +A+ + Q +A PPP +A DTP + P
Sbjct: 16 AQFMHMMQGVAQGQEELRALVQRQ---EAVTPPPN-----QALPEGNPVHDTPAAAIPVN 67
Query: 80 SAPWFPPFTAGEIFRPITCEAQMPTHQYTAQVPLPAMRVTPATMTYSAPVIHTIPQTEEP 139
+ + GE I + Q P A RV A P++
Sbjct: 68 N------YAVGEELMGIRVDGQ-PIAPDAANA-----RVIHAPARNRIPIVDRQEDLFTM 115
Query: 140 IFHSGNVEAYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDF 199
++ + D K D L ++A+ + G ++ LV +++P+KFK P F
Sbjct: 116 FSEDEDIPGRNDARD--RKVDALAEKIRAMECQNSLGFDVTNMGLVEGLRIPYKFKAPSF 173
Query: 200 EKYKGSSCPEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDL 259
+KY G+SCP H++ Y R++ AY D+++ +Y+FQ+SL+G + WY L ++ +RDL
Sbjct: 174 DKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLDWYMELKSDSIRCWRDL 233
Query: 260 CEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFL 319
EAF+ QY +N+DM P R+ LQ++ Q E+FKEYAQRWR+ AA+V P + E+E+T +F+
Sbjct: 234 GEAFLRQYKHNMDMAPSRTQLQSLCQKSNESFKEYAQRWRELAARVQPPMLERELTDMFI 293
Query: 320 KTLNHFYYKKMVGSTP-KSFAEMVGMGVQLEEGVREGRLVKNTTPASGTKKTGNHFPRKK 378
TL + +M GS P SF+++V G + E ++ G++ +S +KK PR++
Sbjct: 294 GTLQGVFMDRM-GSCPFGSFSDVVICGERTESLIKTGKI--QDVGSSSSKKPFAGAPRRR 350
Query: 379 EQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQ 438
E E V H Q Y+ AA+T IP P P+
Sbjct: 351 EGETNAVQHRRDQNRI-EYRQAAAVT--------------IPA-------------PQPR 382
Query: 439 NVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINPIPVTYAELL 498
Q Q VQQ PQQ QQ+PYQ P+Q P R + +P++YAELL
Sbjct: 383 QQQQQRVQQ----------------PQQQQQQRPYQ--PRQRMPDRR-FDSLPMSYAELL 423
Query: 499 PGLLKKNLVQTRTAPPIPEKLPSWYRLDQTCDFHEGGRGHNIETCYAFKSTVQRLINDGK 558
P LL+ LV+ T P P LP Y + CDFH G GH+ E C A + VQ LI+
Sbjct: 424 PELLRLGLVELCTMAP-PTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDANA 482
Query: 559 ITFTDSAPNVQTNPLPNHGAATVNMIEDCQKTRPILNVQHIRTPLVPLHAKLCKVDLFEH 618
I F PNV NP+P HG VN IE + + NV ++T L+ + ++L ++
Sbjct: 483 INFA-PVPNVVNNPMPQHGGHRVNNIEGKEAEDLVDNVDDVQTSLLVVKSRLLNEGVYSG 541
Query: 619 DHDLCEICLMNSGGCQKVRNDIQGLLDRGELVVERKCDD---VCVIT-----PEGPLEVF 670
+ C C + GC ++R IQG++D G L R D V IT EG +
Sbjct: 542 CDEDCLGCAESENGCDQLRAGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPSEGHGQRV 601
Query: 671 YDSRKSTITPLVICLP------------GPLPYASEKAIPYKY----------------- 701
+ + TP+ I +P G + +A+P+KY
Sbjct: 602 VSAPATNGTPVTIPVPVTISAPTTIVASGRRAVENSRAVPWKYDNAYRSNRRVESQTKPV 661
Query: 702 NATMIEEG--REVPIPHLSSVDNIVEGSRVLRNGRVVPIVFPKKIDAIINKEVRTKDAGI 759
N T + G VP +VDN+ R+GR+ + +A + + K A +
Sbjct: 662 NQTPVTIGLANRVPATVGPAVDNVGGPGGFTRSGRLFAPQPLRDNNAEALAKAKGKQAVV 721
Query: 760 AKEVDQPNGAGTSAEFD--EILKLIKKSEYKVVDQLMQTPSKISIMSLLLNSEAHKDALM 817
+E Q S E D E +K+IKKS+YK+VDQL QTPSKISI+SLLL SEAH++AL+
Sbjct: 722 EEEPVQKEAPEGSFEKDVEEFMKMIKKSDYKIVDQLNQTPSKISILSLLLCSEAHRNALL 781
Query: 818 KVL 820
K+L
Sbjct: 782 KML 784
>UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 352 bits (903), Expect = 3e-95
Identities = 266/862 (30%), Positives = 402/862 (45%), Gaps = 126/862 (14%)
Query: 3 EMAELIKTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEAS 62
E+AE+ MA+ Q Q Q + A Q Q A+ + APP P+
Sbjct: 7 ELAEMKANMAQFMNMMQGVAQGQEELRALVQRQEAAIPPV---NHAPPEGGPVNGN---- 59
Query: 63 SSWTLCADTPRQSAPQRSAPWFPPFTAGEIFRPITCEAQMPTHQYTAQVPLPAMRVTPAT 122
+ A P + + G+ I Q P A R A
Sbjct: 60 ---NVAAAVPINN-----------YAVGDELGGIRINGQ-PIAPDVANA-----RAVRAP 99
Query: 123 MTYSAPVIHTIPQTEEPIFH--SGNVEAYEEVSDLREKYDELRRDMKALREKGKFGKTAY 180
AP++ +E +F S + + V + K D L ++A+ + G
Sbjct: 100 ARNPAPIV----DRQEDMFSLLSEDEDVLGRVDERDRKVDALAEKIRAMECQNSLGFDVT 155
Query: 181 DLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQDDQILIYYFQESLTGP 240
++ LV +++P+KFK P F+KY G+SCP H++ Y R++ AY D+++ +Y+FQ+SL+G
Sbjct: 156 NMGLVEGLRIPYKFKAPSFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGA 215
Query: 241 ASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRD 300
+ WY L + ++ ++DL EAF+ QY +N+DM P R+ LQ++ Q E+FKEYAQRWR+
Sbjct: 216 SLDWYMELKRDSIRCWKDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFKEYAQRWRE 275
Query: 301 TAAQVSPRIEEKEMTKLFLKTLNHFYYKKMVGSTP-KSFAEMVGMGVQLEEGVREGRLVK 359
AA+V P + E+E+T +F+ TL + +M GS P SF+++V G + E ++ G++
Sbjct: 276 LAARVQPPMLERELTDMFIGTLQGVFMDRM-GSCPFVSFSDVVICGERTESLIKTGKI-- 332
Query: 360 NTTPASGTKKTGNHFPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQI 419
+S +KK PR++E E V + Q Q A P P QQ Q
Sbjct: 333 QDAGSSSSKKPFAGAPRRREGETNAVQYRRDQNRSQRCQVAAVTIPAPQPRQQQQQRVQ- 391
Query: 420 PQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQ 479
QPQ QQ QQ+PYQ P Q+ P + F
Sbjct: 392 ---------------------QPQQQQQ---QQRPYQ--PRQRMPDRRF----------- 414
Query: 480 PRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYRLDQTCDFHEGGRGHN 539
+ +P++YAELLP LL+ +V+ RT P P LP Y + CDFH G GH+
Sbjct: 415 --------DSLPMSYAELLPELLRLGMVELRTMAP-PTVLPPGYDANVRCDFHSGAPGHH 465
Query: 540 IETCYAFKSTVQRLINDGKITFTDSAPNVQTNPLPNHGAATVNMIEDCQKTRPILNVQHI 599
E C A + VQ LI+ I F PNV NP+P HG VN IE + ++NV +
Sbjct: 466 TEKCRALQHKVQDLIDAKAINFA-PVPNVVNNPMPQHGGHRVNNIEGKEAEDLVVNVDDV 524
Query: 600 RTPLVPLHAKLCKVDLFEHDHDLCEICLMNSGGCQKVRNDIQGLLDRGELVVERKCDD-- 657
+T L+ + +L ++ + C C + GC ++R IQG++D G L R D
Sbjct: 525 QTSLLVVKGRLLNGGVYSGCDEDCLGCAESENGCDQLRTGIQGMMDEGCLQFSRAVKDRG 584
Query: 658 -VCVIT-----PEGPLEVFYDSRKSTITPLVICLP------------GPLPYASEKAIPY 699
V IT EG + + + TP+ I +P G + +A+P+
Sbjct: 585 TVSTITIYFKPSEGRGQRVVSAPATNGTPVTISVPVTISAPTTIAASGRRAVENSRAVPW 644
Query: 700 KYNATMIEEGR-------------------EVPIPHLSSVDNIVEGSRVLRNGRVVPIVF 740
KY+ R VP +VDN+ R+GR+
Sbjct: 645 KYDNAYRSNRRAESQTRPVNQAPVTIGLPNRVPATVGPAVDNVGGPGGFTRSGRLFAPQP 704
Query: 741 PKKIDAIINKEVRTKDAGIAKEVDQPNGAGTSAEFD--EILKLIKKSEYKVVDQLMQTPS 798
+ +A + + K A + +E Q S E D E +K+IKKS+YK+VDQL QTPS
Sbjct: 705 LRDNNAEALAKAKGKQAVVEEEPVQKEAPEGSFEKDVEEFMKIIKKSDYKIVDQLNQTPS 764
Query: 799 KISIMSLLLNSEAHKDALMKVL 820
KISI+SLLL SEAH++AL+K+L
Sbjct: 765 KISILSLLLCSEAHRNALLKML 786
>UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum]
Length = 1105
Score = 350 bits (898), Expect = 1e-94
Identities = 262/864 (30%), Positives = 403/864 (46%), Gaps = 130/864 (15%)
Query: 3 EMAELIKTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEAS 62
E+AE+ MA+ Q Q Q + A Q Q A+ + APP P+
Sbjct: 7 ELAEMKANMAQFMNMMQGVAQGQEELRAMVQRQEAAIPPV---NHAPPEGGPVNDN---- 59
Query: 63 SSWTLCADTPRQSAPQRSAPWFPPFTAGEIFRPITCEAQMPTHQYTAQVPLPAMRVTPAT 122
+ A P + + G+ I +Q P +
Sbjct: 60 ---NVAAAVPINN-----------YDVGDELGGIRINSQ------------PIVPDAANA 93
Query: 123 MTYSAPVIHTIP--QTEEPIFH--SGNVEAYEEVSDLREKYDELRRDMKALREKGKFGKT 178
T AP + +P +E +F S + + V + K D L ++A+ +
Sbjct: 94 RTIRAPARNPVPIVDRQEDMFTLLSEDEDVLGRVDERDRKVDALAEKIRAMECQNSLSFD 153
Query: 179 AYDLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQDDQILIYYFQESLT 238
++ LV +++P+KFK P F+KY +SCP H++ Y R++ AY D+++ +Y+FQ+SL+
Sbjct: 154 VTNMGLVEGLRIPYKFKAPSFDKYNDTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLS 213
Query: 239 GPASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRW 298
G + WY L + ++ +RDL EAF+ QY +N+DM P R+ LQ++ Q E+FKEYAQRW
Sbjct: 214 GASLDWYMELKRDSIRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFKEYAQRW 273
Query: 299 RDTAAQVSPRIEEKEMTKLFLKTLNHFYYKKMVGSTP-KSFAEMVGMGVQLEEGVREGRL 357
R+ AA+V P + E+E+T +F+ TL + +M GS P SF+++V G + E ++ G++
Sbjct: 274 RELAARVQPPMLERELTDMFIGTLQGVFMDRM-GSCPFGSFSDVVICGERTESLIKTGKI 332
Query: 358 VKNTTPASGTKKTGNHFPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHP 417
+S +KK PR++E E +V H Q Q A P P QQ
Sbjct: 333 --QDVGSSSSKKPFAGAPRRREGETNVVQHRRDQNRIEYRQAAAVTIPAPQPRQQQQQRV 390
Query: 418 QIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRP 477
Q QPQ QQ QQ+PYQ P Q+ P + F
Sbjct: 391 Q----------------------QPQQQQQ---QQRPYQ--PRQRMPDRRF--------- 414
Query: 478 QQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYRLDQTCDFHEGGRG 537
+ +P++YAELLP LL+ +V+ RT P P LP Y + CDFH G G
Sbjct: 415 ----------DSLPMSYAELLPELLRLGMVELRTMAP-PTVLPPGYDANVRCDFHSGAPG 463
Query: 538 HNIETCYAFKSTVQRLINDGKITFTDSAPNVQTNPLPNHGAATVNMIEDCQKTRPILNVQ 597
H+ E C A + VQ LI+ I F PNV NP+P HG VN IE + ++NV
Sbjct: 464 HHTEKCRALQHKVQDLIDAKAINFA-PVPNVVNNPMPQHGGHRVNNIEGKEAEDLVVNVD 522
Query: 598 HIRTPLVPLHAKLCKVDLFEHDHDLCEICLMNSGGCQKVRNDIQGLLDRGELVVERKCDD 657
++T L+ + +L ++ + C C + GC ++R IQG++D G L R D
Sbjct: 523 DVQTSLLVVKGRLLNGGVYSGCDEDCLGCAESENGCDQLRAGIQGMMDEGCLQFSRAVKD 582
Query: 658 ---VCVIT-----PEGPLEVFYDSRKSTITPLVICLP------------GPLPYASEKAI 697
V IT EG + + + TP+ I +P G + + +
Sbjct: 583 RGTVSTITIYFKPTEGRGQRVVSAPATNDTPVTISVPVTISAPTTIVASGRRAVENSRVV 642
Query: 698 PYKYNATMIEEGR-------------------EVPIPHLSSVDNIVEGSRVLRNGRVVPI 738
P+KY+ R VP +VDN+ R+GR+
Sbjct: 643 PWKYDNAYRSNRRAESQTRPVNQAPVTIGLPNRVPATVGPAVDNVGGPGGFTRSGRLFAP 702
Query: 739 VFPKKIDAIINKEVRTKDAGIAKE-VDQPNGAGT-SAEFDEILKLIKKSEYKVVDQLMQT 796
+ +A + + K A + +E V + GT + +E +K+IKKS+YK+VDQL QT
Sbjct: 703 QPLRDNNAEALAKAKGKQAVVEEEPVQKEAPEGTFEKDVEEFMKIIKKSDYKIVDQLNQT 762
Query: 797 PSKISIMSLLLNSEAHKDALMKVL 820
PSKISI+SLL+ SEAH++AL+K+L
Sbjct: 763 PSKISILSLLMCSEAHRNALLKML 786
>UniRef100_Q6L419 Putative gag polyprotein [Solanum demissum]
Length = 421
Score = 176 bits (446), Expect = 3e-42
Identities = 109/322 (33%), Positives = 167/322 (51%), Gaps = 28/322 (8%)
Query: 105 HQYTAQVPLPAMRVTPATMTYSAPVIHTIPQTEEPIFHSGNVEAYEEVSDLREKYDELRR 164
H YT + L P T YS+P + E+ + + E +EE++ + + R
Sbjct: 119 HGYTPEEALKIPSSYPQTHQYSSPF-----KIEKTVKN----EEHEEMTKKMKSLEHSIR 169
Query: 165 DMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQ 224
DM+ L G G + DLC+ P V +P FK P+FEKY G P HLK Y ++
Sbjct: 170 DMQGLG--GHKGISFSDLCMFPHVHLPTGFKTPNFEKYDGHGDPIAHLKRYYNQLRGAEG 227
Query: 225 DDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMT 284
+++L+ YF ESL G ASKW+ + D T+ DL FV+Q+ YN+D+ PDRS L M
Sbjct: 228 KEELLMAYFGESLVGIASKWFIDQDIANWHTWDDLARCFVQQFQYNIDIVPDRSSLANMR 287
Query: 285 QGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNHFYYKKMVGSTPKSFAEMVGM 344
+ E F+EYA RWR+ AA+V P ++E EM +FL+ Y+ ++ + K+F E++ +
Sbjct: 288 KKTTENFREYAVRWREQAARVKPPMKELEMIDVFLQAQEPDYFHYLLSAVGKTFTEVIKV 347
Query: 345 GVQLEEGVREGRLVKNTTPASGTKK----TGNHFPRKKEQEVGMVTHGGPQQTYPAYQHI 400
G +E G++ G++V + T+ +GN +K+ ++V V +TY H
Sbjct: 348 GEMVENGIKSGKIVSQAALKATTQALPNGSGNIGGKKRREDVATVV--SAPRTYAQDNH- 404
Query: 401 AAITPTSHPFQQTNNHPQIPQY 422
T H F PQIPQY
Sbjct: 405 -----TQHYFP-----PQIPQY 416
>UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solanum demissum]
Length = 1784
Score = 130 bits (326), Expect = 2e-28
Identities = 87/303 (28%), Positives = 148/303 (48%), Gaps = 43/303 (14%)
Query: 105 HQYTAQVPLPAMRVTPATMTYSAPVIHTIPQTEEPIFHSGNVEAYEEVSDLREKYDELRR 164
H YT + L P T YS+P + E+ + + E +EE++ + ++ R
Sbjct: 639 HGYTPEEALKIPSSYPQTHQYSSPF-----KIEKTVKN----EEHEEMAKKMKSLEQSIR 689
Query: 165 DMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQ 224
DM+ L G G + DLC+ P V +P G+ EE
Sbjct: 690 DMQGLG--GHKGISFSDLCMFPHVHLP-----------TGAEGKEE-------------- 722
Query: 225 DDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMT 284
+L+ YF ESL G AS+W+ + D T+ +L FV+Q+ YN+D+ PDRS L M
Sbjct: 723 ---LLMAYFGESLVGIASEWFIDQDIANWHTWDNLARCFVQQFQYNIDIVPDRSSLANMR 779
Query: 285 QGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNHFYYKKMVGSTPKSFAEMVGM 344
+ E F+EYA RWR+ AA+V ++E EM +FL+ Y+ ++ + K+F E++ +
Sbjct: 780 KKTTENFREYAVRWREQAARVKLSMKESEMIDVFLQAQEPDYFHYLLSAVGKTFVEVIKV 839
Query: 345 GVQLEEGVREGRLVKNTTPASGTKK----TGNHFPRKKEQEVGMVTHGGPQQTYPAYQHI 400
G +E G++ G++V + T+ +GN +K+ ++V V +G + P +
Sbjct: 840 GEMVENGIKSGKIVSQAALKATTQALQNGSGNIGGKKRREDVATVGNGAKDEFTPIGESY 899
Query: 401 AAI 403
A++
Sbjct: 900 ASL 902
Score = 104 bits (260), Expect = 9e-21
Identities = 91/345 (26%), Positives = 161/345 (46%), Gaps = 52/345 (15%)
Query: 489 PIPVTYAELLPGLLKKNL---VQTRTAPPIPEKLPSWYRLDQTCDFHEGGRGHNIETCYA 545
PI +YA L L N+ ++ + + P P L Q C + GHNIE C+
Sbjct: 894 PIGESYASLFQKLRTLNVLSPIERKMSNPPPRNLD----YSQHCAYCSDAPGHNIERCWY 949
Query: 546 FKSTVQRLINDGKITF-TDSAPNVQTNPLPNHGAATVNMIEDCQKTRPILNVQHIRTPLV 604
K +Q LI+ +I + + PN+ NPLP H NM+E + N + + P
Sbjct: 950 LKKAIQDLIDTRRIIVESPNGPNINQNPLPRH--TETNMLE------MVNNHEEVAVPYK 1001
Query: 605 PLHAKLCKVDL-FEHDHDLCEICLMNSGGCQKVRNDIQGLLDRGELVVERKCDDVC---- 659
P+ KV+ E ++ ++ + G + + L ++ +DV
Sbjct: 1002 PI----LKVETGMESSTNVIDLTKIMPSGAESTSEKLTPS-SAPILTIKGALEDVWASQR 1056
Query: 660 ---VITPEGPLEVFYDSRKSTITPLVICLPGPLPYASEKAIPYKYNATMIE-EGREVPIP 715
++ P GP + + + I P++I LP + KA+P+ Y T++ EG+E+
Sbjct: 1057 EARLVVPRGPDKPILIVQGAYIPPVIIRPVSQLPMTNPKAVPWNYEPTVVTYEGKEIN-- 1114
Query: 716 HLSSVDNIVEGSRVLRNGRVVPIVFPKKIDAIINKEVRTKDAGIAKEVDQPNGAGTSAEF 775
+ + E + R+GR + E+R K+ +V P T E
Sbjct: 1115 -----EEVDEIGGMTRSGRCYALT-----------ELR-KNKNDQMQVKSPV---TEGEA 1154
Query: 776 DEILKLIKKSEYKVVDQLMQTPSKISIMSLLLNSEAHKDALMKVL 820
+E L+ +K S+Y +V+QL +TP++IS++SLL++S+AH+ A++K+L
Sbjct: 1155 EEFLRKMKLSDYSIVEQLRKTPAQISLLSLLIHSDAHRKAVIKIL 1199
>UniRef100_Q40369 RPE15 protein [Medicago sativa]
Length = 219
Score = 121 bits (303), Expect = 1e-25
Identities = 87/216 (40%), Positives = 113/216 (52%), Gaps = 34/216 (15%)
Query: 302 AAQVSPRIEEKEMTKLFLKTLNHFYYKKMVGSTPKSFAEMVGMGVQLEEGVREGRLV--K 359
AA+VSPR++E+E+T+ FLKTL + ++M+ S P +F++MV MG +LEE VREG +V K
Sbjct: 14 AARVSPRLDEEELTQTFLKTLKKEHKERMIVSAPNNFSDMVTMGTRLEEAVREGIIVLEK 73
Query: 360 NTTPASGTKKTGN-HFPRKKEQEVGMVT--HGGPQQTYPAYQHIAAITPTSHPFQQTNNH 416
AS KK GN H RK+ ++VGMV+ H P A P + + Q + H
Sbjct: 74 GECSASAPKKYGNGHHKRKESKKVGMVSGNHDTPVN--------ATQLPLPYQYMQISQH 125
Query: 417 PQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQR 476
P P P Y Q+P P V P N Q Q QP Q QQQ QQ
Sbjct: 126 PFFP-----PFYQQYPLPPGQPPV-PVNAMAQQVQHQPPAQ-----------QQQQQQQG 168
Query: 477 PQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTA 512
P+ P+ PIP+ YAELLP LL K TR A
Sbjct: 169 PR----PKTYFPPIPMRYAELLPTLLAKGHCVTRPA 200
>UniRef100_Q949K7 Putative gag polyprotein [Cicer arietinum]
Length = 88
Score = 111 bits (277), Expect = 1e-22
Identities = 43/88 (48%), Positives = 70/88 (78%)
Query: 182 LCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPA 241
+CLVP V +P KFK+ +FEKYKG++CP+ HL MY R+M ++A +D++LI++FQ+SL+G +
Sbjct: 1 MCLVPDVTIPRKFKVLEFEKYKGATCPKNHLTMYGRKMASHAHNDKLLIHFFQDSLSGAS 60
Query: 242 SKWYTNLDKTRVQTFRDLCEAFVEQYSY 269
WY +L++ R+ +++DL +AF+ QY Y
Sbjct: 61 LSWYMHLERNRIHSWKDLADAFLRQYKY 88
>UniRef100_Q6UU84 Putative polyprotein [Oryza sativa]
Length = 2027
Score = 78.6 bits (192), Expect = 7e-13
Identities = 65/269 (24%), Positives = 113/269 (41%), Gaps = 13/269 (4%)
Query: 148 AYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSC 207
A E S R + R D ++ G G A+ + + +V+ P +F+ EKY GS
Sbjct: 192 ATEGASSSRVSPRQGREDQPSMPPAGGVGCRAF-VASLRNVRWPPRFRPTITEKYDGSVN 250
Query: 208 PEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQY 267
P E L++Y + A DD+++ +F +L G A W NL V ++ DLC+ F +
Sbjct: 251 PTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLMNLPPASVHSWEDLCQQFTMNF 310
Query: 268 SYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNH-FY 326
+ +DL A+ +GD E+ + Y QR+ + P I + F + H
Sbjct: 311 QGTYPRPGEEADLHAVQRGDDESLRSYIQRFCQVCNTI-PCIPAHAVIYAFRGGVRHNRM 369
Query: 327 YKKMVGSTPKSFAEMVGMGVQLEEGVR------EGRLVKNTTPASGTKKTGNHFPRKKEQ 380
+K+ P++ AE+ + ++ G V + +TG R+K++
Sbjct: 370 LEKIASKEPQTTAELFQLADRVARKEEAWTWNPSGSGVAASAAPGSAAQTGRRDRRRKKR 429
Query: 381 EVGMVTHGGPQQTYPAYQHIAAITPTSHP 409
V H G + A + + T P
Sbjct: 430 SV----HSGDEGHVLAVEGVPRATRKGRP 454
>UniRef100_Q75G96 Putative polyprotein [Oryza sativa]
Length = 2027
Score = 78.2 bits (191), Expect = 9e-13
Identities = 65/269 (24%), Positives = 113/269 (41%), Gaps = 13/269 (4%)
Query: 148 AYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSC 207
A E S R + R D ++ G G A+ + + +V+ P +F+ EKY GS
Sbjct: 192 ATEGASSSRVSPRQGREDQPSMPPAGGVGCRAF-VASLRNVRWPPRFRPTITEKYDGSVN 250
Query: 208 PEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQY 267
P E L++Y + A DD+++ +F +L G A W NL V ++ DLC+ F +
Sbjct: 251 PTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLMNLPPASVHSWEDLCQQFTMNF 310
Query: 268 SYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNH-FY 326
+ +DL A+ +GD E+ + Y QR+ + P I + F + H
Sbjct: 311 QGTYPRPGEEADLHAVQRGDDESLRSYIQRFCQVRNTI-PCIPAHAVIYAFRGGVRHNRM 369
Query: 327 YKKMVGSTPKSFAEMVGMGVQLEEGVR------EGRLVKNTTPASGTKKTGNHFPRKKEQ 380
+K+ P++ AE+ + ++ G V + +TG R+K++
Sbjct: 370 LEKIASKEPQTTAELFQLADRVARKEEAWTWNPSGSGVAASAAPGSAAQTGRRDRRRKKR 429
Query: 381 EVGMVTHGGPQQTYPAYQHIAAITPTSHP 409
V H G + A + + T P
Sbjct: 430 SV----HSGDEGHVLAVEGVPRATRKGRP 454
>UniRef100_Q84T69 Putative GAG-POL [Oryza sativa]
Length = 2027
Score = 77.8 bits (190), Expect = 1e-12
Identities = 65/269 (24%), Positives = 113/269 (41%), Gaps = 13/269 (4%)
Query: 148 AYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSC 207
A E S R R+D ++ G G A+ + + +V+ P +F+ EKY GS
Sbjct: 192 ATEGASSSRVSPPHGRKDQPSVPPVGGVGCRAF-VASLRNVRWPPRFRPTITEKYDGSVN 250
Query: 208 PEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQY 267
P E L++Y + A DD+++ +F +L G A W NL V ++ DLC+ F +
Sbjct: 251 PTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLMNLPPASVHSWEDLCQQFTMNF 310
Query: 268 SYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNH-FY 326
+ +DL A+ +GD E+ + Y QR+ + P I + F + H
Sbjct: 311 QGTYPRPGEEADLHAVQRGDDESLRSYIQRFCQVRNTI-PCIPAHAVIYAFRGGVRHNRM 369
Query: 327 YKKMVGSTPKSFAEMVGMGVQLEEGVR------EGRLVKNTTPASGTKKTGNHFPRKKEQ 380
+K+ P++ AE+ + ++ G V + +TG R+K++
Sbjct: 370 LEKIASKEPQTTAELFQLADRVARKEEAWTWNPSGSGVAASAAPGSAAQTGRRDRRRKKR 429
Query: 381 EVGMVTHGGPQQTYPAYQHIAAITPTSHP 409
V H G + A + + T P
Sbjct: 430 SV----HSGDEGHVLAVEGVPRATRKGRP 454
>UniRef100_Q7XPG8 OSJNBb0003B01.15 protein [Oryza sativa]
Length = 2030
Score = 77.4 bits (189), Expect = 2e-12
Identities = 65/269 (24%), Positives = 112/269 (41%), Gaps = 13/269 (4%)
Query: 148 AYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSC 207
A E S R R D ++ G G A+ + + +V+ P +F+ EKY GS
Sbjct: 192 ATEGASSSRVSPPHGREDQPSVPPVGGVGCRAF-VASLRNVRWPPRFRPTITEKYDGSVN 250
Query: 208 PEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQY 267
P E L++Y + A DD+++ +F +L G A W NL V ++ DLC+ F +
Sbjct: 251 PTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLMNLPPASVHSWEDLCQQFTMNF 310
Query: 268 SYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNH-FY 326
+ +DL A+ +GD E+ + Y QR+ + P I + F + H
Sbjct: 311 QGTYPRPGEEADLHAVQRGDDESLRSYIQRFCQVRNTI-PCIPAHAVIYAFRGGVRHNCM 369
Query: 327 YKKMVGSTPKSFAEMVGMGVQLEEGVR------EGRLVKNTTPASGTKKTGNHFPRKKEQ 380
+K+ P++ AE+ + ++ G V + +TG R+K++
Sbjct: 370 LEKIASKEPQTTAELFQLADRVARKEEAWTWNPSGSGVAASAAPGSVAQTGRRDRRRKKR 429
Query: 381 EVGMVTHGGPQQTYPAYQHIAAITPTSHP 409
V H G + A + + T P
Sbjct: 430 SV----HSGDEGHVLAVEGVPRATRKGRP 454
>UniRef100_Q9FTC1 Gamma-gliadin [Aegilops speltoides]
Length = 256
Score = 77.0 bits (188), Expect = 2e-12
Identities = 54/116 (46%), Positives = 59/116 (50%), Gaps = 14/116 (12%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQ-IPQYPQIPQYPQFPQNPSPQNVQPQNVQQQ 448
PQQT+P Q P QQ Q PQ PQ P YPQ PQ P PQ QPQ + Q
Sbjct: 15 PQQTFPHQQQQQQFPQPQQPQQQFLQPQQPFPQQPQQP-YPQQPQQPFPQTQQPQQLFPQ 73
Query: 449 NFQ-QQPYQQYPYQQYPQQNFQ-----QQPYQ-----QRPQQPRPPRMPINPIPVT 493
+ Q QQPY Q P Q YPQQ Q QQP Q Q+PQQP P+ P P P T
Sbjct: 74 SQQPQQPYPQQPQQPYPQQPQQPFPQTQQPQQLFPQSQQPQQPY-PQQPQQPFPQT 128
Score = 73.6 bits (179), Expect = 2e-11
Identities = 50/120 (41%), Positives = 61/120 (50%), Gaps = 8/120 (6%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQ--YPQIPQYPQFPQNPSPQNVQPQNVQ- 446
PQQ +P Q + P S QQ +PQ PQ YPQ PQ P FPQ PQ + PQ+ Q
Sbjct: 57 PQQPFPQTQQPQQLFPQSQQPQQP--YPQQPQQPYPQQPQQP-FPQTQQPQQLFPQSQQP 113
Query: 447 QQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINPIPVTYAELLPGLLKKNL 506
QQ + QQP Q +P Q PQQ F Q YQQ QQ P+ P P + L++ L
Sbjct: 114 QQPYPQQPQQPFPQTQQPQQQFPQ--YQQPQQQFPQPQQPQQSFPQQQPSFVQPSLQQQL 171
Score = 70.5 bits (171), Expect = 2e-10
Identities = 51/133 (38%), Positives = 59/133 (44%), Gaps = 23/133 (17%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQI-PQ-------YPQIPQ--YPQFPQNPSPQN 439
PQQ +P PF QT Q+ PQ YPQ PQ YPQ PQ P PQ
Sbjct: 41 PQQPFPQQPQQPYPQQPQQPFPQTQQPQQLFPQSQQPQQPYPQQPQQPYPQQPQQPFPQT 100
Query: 440 VQPQNV------QQQNFQQQPYQQYPYQQYPQQNF-------QQQPYQQRPQQPRPPRMP 486
QPQ + QQ + QQP Q +P Q PQQ F QQ P Q+PQQ P + P
Sbjct: 101 QQPQQLFPQSQQPQQPYPQQPQQPFPQTQQPQQQFPQYQQPQQQFPQPQQPQQSFPQQQP 160
Query: 487 INPIPVTYAELLP 499
P +L P
Sbjct: 161 SFVQPSLQQQLNP 173
Score = 65.9 bits (159), Expect = 5e-09
Identities = 44/107 (41%), Positives = 52/107 (48%), Gaps = 5/107 (4%)
Query: 374 FPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQ--YPQF 431
FP+ ++ + PQQ YP PF QT Q+ Q PQ YPQ
Sbjct: 61 FPQTQQPQQLFPQSQQPQQPYPQQPQQPYPQQPQQPFPQTQQPQQLFPQSQQPQQPYPQQ 120
Query: 432 PQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNF-QQQPYQQRP 477
PQ P PQ QPQ QQ QQP QQ+P Q PQQ+F QQQP +P
Sbjct: 121 PQQPFPQTQQPQ--QQFPQYQQPQQQFPQPQQPQQSFPQQQPSFVQP 165
Score = 61.2 bits (147), Expect = 1e-07
Identities = 47/103 (45%), Positives = 51/103 (48%), Gaps = 19/103 (18%)
Query: 405 PTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQ-QQNFQQQPYQQYPYQ-- 461
P S QQT PQ +P Q QFPQ PQ Q Q +Q QQ F QQP Q YP Q
Sbjct: 3 PFSQQPQQTVPQPQ-QTFPHQQQQQQFPQ---PQQPQQQFLQPQQPFPQQPQQPYPQQPQ 58
Query: 462 ------QYPQQNF-----QQQPYQQRPQQPRPPRMPINPIPVT 493
Q PQQ F QQPY Q+PQQP P+ P P P T
Sbjct: 59 QPFPQTQQPQQLFPQSQQPQQPYPQQPQQPY-PQQPQQPFPQT 100
Score = 43.5 bits (101), Expect = 0.026
Identities = 27/64 (42%), Positives = 33/64 (51%), Gaps = 12/64 (18%)
Query: 441 QPQNVQQQNFQQQPYQQYPYQ---------QYPQQNF--QQQPYQQRPQQPRPPRMPINP 489
QP + Q Q QP Q +P+Q Q PQQ F QQP+ Q+PQQP P+ P P
Sbjct: 2 QPFSQQPQQTVPQPQQTFPHQQQQQQFPQPQQPQQQFLQPQQPFPQQPQQPY-PQQPQQP 60
Query: 490 IPVT 493
P T
Sbjct: 61 FPQT 64
Score = 42.7 bits (99), Expect = 0.044
Identities = 40/100 (40%), Positives = 45/100 (45%), Gaps = 20/100 (20%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYP---------QIPQYPQFPQNPSPQNV 440
PQQ +P Q + P S QQ +PQ PQ P Q PQY Q PQ PQ
Sbjct: 93 PQQPFPQTQQPQQLFPQSQQPQQP--YPQQPQQPFPQTQQPQQQFPQYQQ-PQQQFPQPQ 149
Query: 441 QPQN---VQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRP 477
QPQ QQ +F Q QQ Q P +NF Q Q RP
Sbjct: 150 QPQQSFPQQQPSFVQPSLQQ---QLNPCKNFLLQ--QCRP 184
>UniRef100_Q94G96 Gamma-gliadin [Triticum aestivum]
Length = 337
Score = 77.0 bits (188), Expect = 2e-12
Identities = 52/110 (47%), Positives = 58/110 (52%), Gaps = 15/110 (13%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQ--FPQNPSPQNVQPQNVQQ 447
PQQT+P A P QQT H Q+PQ PQ PQ FPQ P Q QPQ QQ
Sbjct: 56 PQQTFPHQPQQAFPQP-----QQTFPHQPQQQFPQ-PQQPQQPFPQQPQQQFPQPQQPQQ 109
Query: 448 QNFQQQPYQQYPYQQYPQQNFQQQ-----PYQQRPQQPRP-PRMPINPIP 491
F QQP QQ+P Q PQQ F Q P+ Q+PQQP P P+ P P P
Sbjct: 110 P-FPQQPQQQFPQPQQPQQPFPQPQQPQLPFPQQPQQPFPQPQQPQQPFP 158
Score = 68.2 bits (165), Expect = 1e-09
Identities = 50/126 (39%), Positives = 62/126 (48%), Gaps = 12/126 (9%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQ--FPQNPSPQNVQPQNVQQ 447
PQQT+P +Q P Q PQ Q+PQ PQ PQ FPQ P Q QPQ QQ
Sbjct: 71 PQQTFP-HQPQQQFPQPQQPQQPFPQQPQ-QQFPQ-PQQPQQPFPQQPQQQFPQPQQPQQ 127
Query: 448 QNFQQQ----PYQQYPYQQYPQQNFQQQPYQ--QRPQQPRP-PRMPINPIPVTYAELLPG 500
Q Q P+ Q P Q +PQ QQP+ Q+PQQP P P+ P P P L+
Sbjct: 128 PFPQPQQPQLPFPQQPQQPFPQPQQPQQPFPQLQQPQQPLPQPQQPQQPFPQQQQPLIQP 187
Query: 501 LLKKNL 506
L++ +
Sbjct: 188 YLQQQM 193
Score = 67.4 bits (163), Expect = 2e-09
Identities = 53/120 (44%), Positives = 57/120 (47%), Gaps = 17/120 (14%)
Query: 383 GMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQ--IPQYPQ--IPQYPQ-FPQNPSP 437
G V QQ +P Q P S QQ PQ P PQ PQ Q FP P
Sbjct: 27 GQVQWPQQQQPFPQPQQ-----PFSQQPQQIFPQPQQTFPHQPQQAFPQPQQTFPHQPQQ 81
Query: 438 QNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQ-----QRPQQPRP-PRMPINPIP 491
Q QPQ QQ F QQP QQ+P Q PQQ F QQP Q Q+PQQP P P+ P P P
Sbjct: 82 QFPQPQQPQQP-FPQQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQPQQPQLPFP 140
Score = 56.2 bits (134), Expect = 4e-06
Identities = 43/113 (38%), Positives = 51/113 (45%), Gaps = 16/113 (14%)
Query: 374 FPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQ 433
FP++ +Q+ PQQ +P P Q PQ PQ PQ+P +PQ PQ
Sbjct: 93 FPQQPQQQFPQPQQ--PQQPFPQQPQ----QQFPQPQQPQQPFPQ-PQQPQLP-FPQQPQ 144
Query: 434 NPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQ------QPYQQRPQQP 480
P PQ QPQ Q QQP Q P Q PQQ F Q QPY Q+ P
Sbjct: 145 QPFPQPQQPQQPFPQ--LQQPQQPLPQPQQPQQPFPQQQQPLIQPYLQQQMNP 195
Score = 56.2 bits (134), Expect = 4e-06
Identities = 37/103 (35%), Positives = 48/103 (45%), Gaps = 13/103 (12%)
Query: 400 IAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQY- 458
+A T T++ + Q PQ Q PQ P + PQ + PQ QQ F QP Q +
Sbjct: 12 VALTTTTANIQVDPSGQVQWPQQQQPFPQPQQPFSQQPQQIFPQ--PQQTFPHQPQQAFP 69
Query: 459 ---------PYQQYPQQNFQQQPYQQRPQQPRP-PRMPINPIP 491
P QQ+PQ QQP+ Q+PQQ P P+ P P P
Sbjct: 70 QPQQTFPHQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFP 112
Score = 40.4 bits (93), Expect = 0.22
Identities = 38/100 (38%), Positives = 45/100 (45%), Gaps = 10/100 (10%)
Query: 374 FPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQ 433
FP++ +Q+ PQQ +P Q P QQ PQ PQ P PQ Q PQ
Sbjct: 111 FPQQPQQQFPQPQQ--PQQPFPQPQQPQLPFPQQP--QQPFPQPQQPQQP-FPQLQQ-PQ 164
Query: 434 NPSPQNVQPQN--VQQQNFQQQPYQQYPYQQYPQQNFQQQ 471
P PQ QPQ QQQ QPY Q Q P +N+ Q
Sbjct: 165 QPLPQPQQPQQPFPQQQQPLIQPYLQ--QQMNPCKNYLLQ 202
>UniRef100_Q8S0W5 Putative polyprotein [Oryza sativa]
Length = 2030
Score = 76.6 bits (187), Expect = 3e-12
Identities = 53/195 (27%), Positives = 90/195 (45%), Gaps = 3/195 (1%)
Query: 148 AYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSC 207
A E+ S R R D ++ G G A+ + + +V+ P +F+ EKY GS
Sbjct: 192 ATEDASSSRVSPRHGREDQPSVPPAGGVGCRAF-VASLRNVRWPPRFRPTITEKYDGSVN 250
Query: 208 PEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQY 267
P E L++Y + A DD+++ +F +L G A W NL V ++ DLC+ F +
Sbjct: 251 PTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLMNLPPASVHSWEDLCQQFTMNF 310
Query: 268 SYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNH-FY 326
+ +DL A+ +GD E+ + Y QR+ + P I + F + H
Sbjct: 311 QGTYPRPGEEADLHAVQRGDDESLRSYIQRFCQVRNTI-PCIPAHAVIYAFRGGVRHNRM 369
Query: 327 YKKMVGSTPKSFAEM 341
+K+ P++ AE+
Sbjct: 370 LEKIASKEPQTTAEL 384
>UniRef100_Q7X8F8 OSJNBb0002J11.5 protein [Oryza sativa]
Length = 592
Score = 76.3 bits (186), Expect = 4e-12
Identities = 66/281 (23%), Positives = 113/281 (39%), Gaps = 13/281 (4%)
Query: 148 AYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSC 207
A E S R R D ++ G G A+ + + +V+ P +F+ EKY GS
Sbjct: 192 ATEGASSSRVSPRHGREDQPSVPPAGGVGCRAF-VASLRNVRWPPRFRPTITEKYDGSVN 250
Query: 208 PEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQY 267
P E L++Y + A DD+++ +F +L G A W NL V ++ DLC+ F +
Sbjct: 251 PTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLMNLPPASVHSWEDLCQQFTMNF 310
Query: 268 SYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNH-FY 326
+ +DL A+ +GD E+ + Y QR+ + P I + F + H
Sbjct: 311 QGTYPRPGEEADLHAVQRGDDESLRSYIQRFCQVRNTI-PCIPAHAVIYAFRGGVRHNRM 369
Query: 327 YKKMVGSTPKSFAEMVGMGVQLEEGVR------EGRLVKNTTPASGTKKTGNHFPRKKEQ 380
+K+ P++ AE+ + ++ G V + +TG R+K++
Sbjct: 370 LEKIASKEPQTTAELFQLADRVARKEEAWTWNPSGSGVAASAAPGSAAQTGRRDRRRKKR 429
Query: 381 EVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQ 421
V H G + A + T P P+
Sbjct: 430 SV----HSGDEGHVLAVEGALRATRKGRPASDKRKEAGAPE 466
>UniRef100_Q65X63 Putative polyprotein [Oryza sativa]
Length = 2027
Score = 76.3 bits (186), Expect = 4e-12
Identities = 53/195 (27%), Positives = 90/195 (45%), Gaps = 3/195 (1%)
Query: 148 AYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSC 207
A E S R R D ++ G G A+ + + +V+ P +F+ EKY GS
Sbjct: 192 ATEGASSSRVSPRHGREDQPSVPPVGGVGCRAF-VASLRNVRWPPRFRPTITEKYDGSVN 250
Query: 208 PEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQY 267
P E L++Y + A DD+++ +F +L G A W NL V ++ DLC+ F +
Sbjct: 251 PTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLMNLPPASVHSWEDLCQQFTMNF 310
Query: 268 SYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNH-FY 326
+ +DL A+ +GD E+ + Y QR+ + P I + F + H
Sbjct: 311 QGTYPRPGEEADLHAVQRGDDESLRSYIQRFCQVRNTI-PCIPAHAVIYAFRGGVRHNRM 369
Query: 327 YKKMVGSTPKSFAEM 341
++K+ P++ AE+
Sbjct: 370 FEKIASKEPQTTAEL 384
>UniRef100_Q75KU5 Putative polyprotein [Oryza sativa]
Length = 2026
Score = 75.9 bits (185), Expect = 5e-12
Identities = 60/242 (24%), Positives = 104/242 (42%), Gaps = 9/242 (3%)
Query: 148 AYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSC 207
A E S R R D ++ G G A+ + + +V+ P +F+ EKY GS
Sbjct: 191 ATEGASSSRVSPRHGREDQPSVPPAGGIGCRAF-VASLRNVRWPPRFRPTITEKYDGSVN 249
Query: 208 PEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQY 267
P E L++Y + A DD+++ +F +L G A W NL V ++ DLC+ F +
Sbjct: 250 PTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLMNLPPASVHSWEDLCQQFTMNF 309
Query: 268 SYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNH-FY 326
+ +DL A+ +GD E+ + Y QR+ + P I + F + H
Sbjct: 310 QGTYPRPGEEADLHAVQRGDDESLRSYIQRFCQVRNTI-PCIPAHAVIYAFRGGVRHNRM 368
Query: 327 YKKMVGSTPKSFAEMVGMGVQLEEGVR------EGRLVKNTTPASGTKKTGNHFPRKKEQ 380
+K+ P++ AE+ + ++ G V + +TG R+K++
Sbjct: 369 LEKIASKEPQTTAELFQLADRVARKEEAWTWNPSGSGVAASAAPGSAAQTGRRDRRRKKR 428
Query: 381 EV 382
V
Sbjct: 429 SV 430
>UniRef100_Q94LM2 Putative polyprotein [Oryza sativa]
Length = 2027
Score = 75.9 bits (185), Expect = 5e-12
Identities = 60/242 (24%), Positives = 105/242 (42%), Gaps = 9/242 (3%)
Query: 148 AYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSC 207
A E S R + R D ++ G G A+ + + +V+ P +F+ EKY GS
Sbjct: 192 AAEGASSSRVSPRQGREDQPSVPPVGGVGCRAF-VASLRNVRWPPRFRPTITEKYDGSVN 250
Query: 208 PEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQY 267
P E L++Y + A DD+++ +F +L G A W NL V ++ DLC+ F +
Sbjct: 251 PTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLMNLPPASVHSWEDLCQQFTMNF 310
Query: 268 SYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNH-FY 326
+ +DL A+ +GD E+ + Y QR+ + P I + F + H
Sbjct: 311 QGTYPRPGEEADLHAVQRGDDESLRSYIQRFCQVRNTI-PCIPAHAVIYAFRGGVRHNRM 369
Query: 327 YKKMVGSTPKSFAEMVGMGVQLEEGVR------EGRLVKNTTPASGTKKTGNHFPRKKEQ 380
+K+ P++ AE+ + ++ G V + +TG R+K++
Sbjct: 370 LEKIASKEPQTTAELFQLADRVARKEEAWTWNPSGSGVAASAAPGSAAQTGRRDRRRKKR 429
Query: 381 EV 382
V
Sbjct: 430 SV 431
>UniRef100_Q75LG6 Putative gag protein [Oryza sativa]
Length = 641
Score = 75.5 bits (184), Expect = 6e-12
Identities = 62/243 (25%), Positives = 105/243 (42%), Gaps = 9/243 (3%)
Query: 148 AYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSC 207
A E S R R D ++ G A+ + + +V+ P +F+ EKY GS
Sbjct: 192 ATEGASSSRVSPRHGREDQPSVPPAGGVSCRAF-VASLRNVRWPPRFRPTITEKYDGSVN 250
Query: 208 PEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQY 267
P E L++Y + A DD+++ +F +L G A W NL V ++ DLC+ F +
Sbjct: 251 PTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLVNLPPASVHSWEDLCQQFTMNF 310
Query: 268 SYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNH-FY 326
+ +DL A+ +GD E+ + Y QR+ + P I + F + H
Sbjct: 311 QGTYPRPGEEADLHAVQRGDDESLRSYIQRFCQVRNTI-PCIPAHAVIYAFRGGVRHNRM 369
Query: 327 YKKMVGSTPKSFAEMVGMG---VQLEEG---VREGRLVKNTTPASGTKKTGNHFPRKKEQ 380
+K+ P++ AE+ + + EE G V + +TG R+K++
Sbjct: 370 LEKIASKEPQTTAELFQLADRVARKEEAWTWNSSGSGVAASAAPGSAAQTGRRDRRRKKR 429
Query: 381 EVG 383
E G
Sbjct: 430 EQG 432
>UniRef100_Q8H7K1 Putative polyprotein [Oryza sativa]
Length = 2027
Score = 75.5 bits (184), Expect = 6e-12
Identities = 60/242 (24%), Positives = 104/242 (42%), Gaps = 9/242 (3%)
Query: 148 AYEEVSDLREKYDELRRDMKALREKGKFGKTAYDLCLVPSVQVPHKFKIPDFEKYKGSSC 207
A E S R R D ++ G G A+ + + +V+ P +F+ EKY GS
Sbjct: 192 ATEGASSSRVSPRHGREDQPSVPPAGGVGCRAF-VASLRNVRWPPRFRPTITEKYDGSVN 250
Query: 208 PEEHLKMYVRRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQY 267
P E L++Y + A DD+++ +F +L G A W NL V ++ DLC+ F +
Sbjct: 251 PTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLMNLPPASVHSWEDLCQQFTMNF 310
Query: 268 SYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNH-FY 326
+ +DL A+ +GD E+ + Y QR+ + P I + F + H
Sbjct: 311 QGTYPRPGEEADLHAVQRGDDESLRSYIQRFCQVRNTI-PCIPAHAVIYAFRGGVRHNRM 369
Query: 327 YKKMVGSTPKSFAEMVGMGVQLEEGVR------EGRLVKNTTPASGTKKTGNHFPRKKEQ 380
+K+ P++ AE+ + ++ G V + +TG R+K++
Sbjct: 370 LEKIASKEPQTTAELFQLADRVARKEEAWTWNPSGSGVAASAAPGSAAQTGRRDRRRKKR 429
Query: 381 EV 382
V
Sbjct: 430 SV 431
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,444,446,645
Number of Sequences: 2790947
Number of extensions: 67624347
Number of successful extensions: 501860
Number of sequences better than 10.0: 5174
Number of HSP's better than 10.0 without gapping: 2334
Number of HSP's successfully gapped in prelim test: 3086
Number of HSP's that attempted gapping in prelim test: 346799
Number of HSP's gapped (non-prelim): 49808
length of query: 820
length of database: 848,049,833
effective HSP length: 136
effective length of query: 684
effective length of database: 468,481,041
effective search space: 320441032044
effective search space used: 320441032044
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 79 (35.0 bits)
Medicago: description of AC135161.1


