

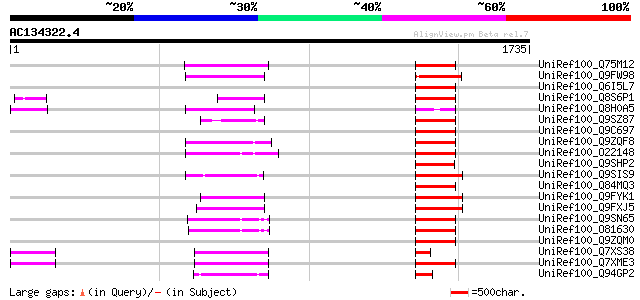
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134322.4 - phase: 0 /pseudo
(1735 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa] 153 4e-35
UniRef100_Q9FW98 Putative non-LTR retroelement reverse transcrip... 147 4e-33
UniRef100_Q6I5L7 Hypothetical protein OSJNBb0088F07.2 [Oryza sat... 144 2e-32
UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa] 142 1e-31
UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa] 139 8e-31
UniRef100_Q9SZ87 RNA-directed DNA polymerase-like protein [Arabi... 139 1e-30
UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [A... 139 1e-30
UniRef100_Q9ZQF8 Putative non-LTR retroelement reverse transcrip... 137 2e-30
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 130 5e-28
UniRef100_Q9SHP2 Putative non-LTR retroelement reverse transcrip... 129 8e-28
UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcrip... 129 1e-27
UniRef100_Q84MQ3 Putative reverse transcriptase [Oryza sativa] 128 1e-27
UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana] 128 2e-27
UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana] 128 2e-27
UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis tha... 127 2e-27
UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana] 127 2e-27
UniRef100_Q9ZQM0 Putative non-LTR retroelement reverse transcrip... 127 4e-27
UniRef100_Q7XS38 OSJNBa0081G05.2 protein [Oryza sativa] 125 1e-26
UniRef100_Q7XME3 OSJNBa0061G20.16 protein [Oryza sativa] 125 1e-26
UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa] 125 1e-26
>UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa]
Length = 1936
Score = 153 bits (387), Expect = 4e-35
Identities = 83/283 (29%), Positives = 140/283 (49%), Gaps = 1/283 (0%)
Query: 583 GLPRPMSIIAWNCRGLGNLRVIPKIKFLVRYYKQDILFLCETMVEVNKIEEFRYLLGYDF 642
G MS +AWNCRGLGN + ++ L++ ++FLCET V K+ R L +
Sbjct: 631 GAAGAMSCLAWNCRGLGNTATVQDLRALIQKAGSQLVFLCETRQSVEKMSRLRRKLAFRG 690
Query: 643 CFAPARNNRSGGIAYFWRNTVNCSIVNYSANHISAKIE-EVSRSAWTFTGFYGYPEIARR 701
+ +SGG+A +W +V+ + + + +I A + W T YG P + R
Sbjct: 691 FVGVSSEGKSGGLALYWDESVSVDVKDINKRYIDAYVRLSPDEPQWHITFVYGEPRVENR 750
Query: 702 RDSWNFIRGLARQINLPWCQMGDFNDILHSDEKKGRATRPNWLIRGFRHSIQDVGLIDVH 761
W+ +R + + LPW +GDFN+ L E + R ++ FR ++ D L D+
Sbjct: 751 HRMWSLLRTIRQSSALPWMVIGDFNETLWQFEHFSKNPRCETQMQNFRDALYDCDLQDLG 810
Query: 762 MEGYLFTWFKSLGTTRVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDHFPILLDRTLM 821
+G T+ R V+ +LDRA+A + W LFP A++ +LV+P SDH PILL+ +
Sbjct: 811 FKGVPHTYDNRRDGWRNVKVRLDRAVADDKWRDLFPEAQVSHLVSPCSDHSPILLEFIVK 870
Query: 822 VRSHRVERSFKFENARRVEEVVNDVVRGSWLGSTVSNVINNMN 864
+ ++ +E E V+ +W+ + V + ++N
Sbjct: 871 DTTRPRQKCLHYEIVWEREPESVQVIEEAWINAGVKTDLGDIN 913
Score = 120 bits (301), Expect = 4e-25
Identities = 58/133 (43%), Positives = 85/133 (63%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+A E H + K + A KLD+SKAYDR+DW +L ++K+GF+ + + WIM+ V
Sbjct: 1191 LLAFECFHSIQKNKKANSAACAYKLDLSKAYDRVDWRFLELALNKLGFAHRWVSWIMLCV 1250
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
TV YSV N + P GLRQG+PLSP+LF+ A+GLS ++ ++ L +++C
Sbjct: 1251 TTVRYSVKFNGTLLRSFAPTRGLRQGEPLSPFLFLFVADGLSLLLKEKVAQNSLTPLKIC 1310
Query: 1476 RIAPRVSHLLFAD 1488
R AP +S+LLFAD
Sbjct: 1311 RQAPGISYLLFAD 1323
Score = 46.2 bits (108), Expect = 0.009
Identities = 27/99 (27%), Positives = 52/99 (52%), Gaps = 1/99 (1%)
Query: 54 PLTLAMDTTEIWAQKNELPFGFMDKKVGALVGSHIGKMVKFDEENNYGL*RKYMRVRVEI 113
P + ++T IW + +LP M+K+ G + GS +GK+ + D + + + R+RV++
Sbjct: 305 PSMVPLETVPIWVRIYDLPLVMMNKERGVIYGSKLGKVREVDVQEDGCNKHDFFRIRVDL 364
Query: 114 ALDAPLQQELIIEWE-EGDNIKLVFKFRETR*VLLCLWC 151
++ PL++ L I+ + +G F R R C +C
Sbjct: 365 PVNRPLKRMLAIKIKVKGAEEIRRFNLRYERIPHFCFFC 403
>UniRef100_Q9FW98 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1382
Score = 147 bits (370), Expect = 4e-33
Identities = 82/270 (30%), Positives = 132/270 (48%), Gaps = 5/270 (1%)
Query: 587 PMSIIAW--NCRGLGNLRVIPKIKFLVRYYKQDILFLCETMVEVNKIEEFRYLLGYDFCF 644
P ++ W NCRGLG+ + ++++LV+ + ++FL ET + + + LG+ F
Sbjct: 3 PNKLVTWGRNCRGLGSAATVGELRWLVKSLRPSLVFLSETKMRDKQARNLMWSLGFSGSF 62
Query: 645 APARNNRSGGIAYFWRNTVNCSIVNYSANHISAKIEEVSRSAWTFTGFYGYPEIARRRDS 704
A + SGG+A FW S+ ++++ I + W + YG P+ R
Sbjct: 63 AVSCEGLSGGLALFWTTAYTVSLRGFNSHFIDVLVSTEELPPWRISFVYGEPKRELRHFF 122
Query: 705 WNFIRGLARQINLPWCQMGDFNDILHSDEKKGRATRPNWLIRGFRHSIQDVGLIDVHMEG 764
WN +R L Q PW GDFN++L DE G R ++ FR + D GLID+ G
Sbjct: 123 WNLLRRLHDQWRGPWLCCGDFNEVLCLDEHLGMRERSEPHMQHFRSCLDDCGLIDLGFVG 182
Query: 765 YLFTWFKSLGTTRVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDHFPILLDRTLMVRS 824
FTW + +LDRA+A + + F + +EN++ SSDH+ I +D +
Sbjct: 183 PKFTWSNKQDANSNSKVRLDRAVANGEFSRYFEDCLVENVITTSSDHYAISIDLSRRNHG 242
Query: 825 HR---VERSFKFENARRVEEVVNDVVRGSW 851
R +++ F+FE A E +VV SW
Sbjct: 243 QRRIPIQQGFRFEAAWLRAEDYREVVENSW 272
Score = 132 bits (333), Expect = 7e-29
Identities = 72/154 (46%), Positives = 98/154 (62%), Gaps = 2/154 (1%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
LVA E +H + K K ALK+D+ KAYDR++WAYL +SK+GFS+ I +M V
Sbjct: 567 LVAYECLH-TIRKQHNKNPFFALKIDMMKAYDRVEWAYLSGCLSKLGFSQDWINTVMRCV 625
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
+V Y+V +N E P++P G+RQGDP+SPYLF+LC EGLS + E G+LQGI+
Sbjct: 626 SSVRYAVKINGELTKPVVPSRGIRQGDPISPYLFLLCTEGLSCLLHKKEVAGELQGIKNG 685
Query: 1476 RIAPRVSHLLFADIDSV*GSIRSSYQSSRVRNLL 1509
R P +SHLLFAD DS+ + S ++N L
Sbjct: 686 RHGPPISHLLFAD-DSIFFAKADSRNVQALKNTL 718
>UniRef100_Q6I5L7 Hypothetical protein OSJNBb0088F07.2 [Oryza sativa]
Length = 1264
Score = 144 bits (364), Expect = 2e-32
Identities = 68/133 (51%), Positives = 96/133 (72%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+A EL HY+ + KGK A+KLD+SKAYDR++W +LR +M ++GF ++ + +M V
Sbjct: 899 LLAYELTHYLNQRKKGKNGVAAIKLDMSKAYDRVEWDFLRHMMLRLGFHDQWVNLVMKCV 958
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
+V Y + +N E I P GLRQGDPLSPYLFI+CAEGLSA ++ A++ G ++GI+VC
Sbjct: 959 TSVTYRIKINGEHSDQIYPQRGLRQGDPLSPYLFIICAEGLSALLQKAQADGKIEGIKVC 1018
Query: 1476 RIAPRVSHLLFAD 1488
R PR++HL FAD
Sbjct: 1019 RDTPRINHLFFAD 1031
Score = 43.9 bits (102), Expect = 0.044
Identities = 25/84 (29%), Positives = 43/84 (50%), Gaps = 1/84 (1%)
Query: 735 KGRATRPNWLIRGFRHSIQDVGLIDVHMEGYLFTWFKSLG-TTRVVEEKLDRALATNSWM 793
+G A R + FR +++D L D+ G FTW + ++E+LDRA+A +W
Sbjct: 487 EGGAPRSESCMAKFRQALEDCQLHDLGFVGDAFTWRNHHHLASNYIKERLDRAVANGAWR 546
Query: 794 QLFPNAKLENLVAPSSDHFPILLD 817
FP ++ N SDH ++++
Sbjct: 547 ARFPLVRVINGDPRHSDHRSVIVE 570
>UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa]
Length = 1509
Score = 142 bits (357), Expect = 1e-31
Identities = 71/133 (53%), Positives = 88/133 (65%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+A E+ HYM K G+ A KLD+SKAYDR++W++L D+M K+GF + IM V
Sbjct: 752 LIAYEMTHYMRNKRSGQVGYAAFKLDMSKAYDRVEWSFLHDMMLKLGFHTDWVNLIMKCV 811
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
TV Y + +N E P GLRQGDPLSPYLF+LCAEG SA + E G L GIR+C
Sbjct: 812 STVTYRIRVNGELSESFSPERGLRQGDPLSPYLFLLCAEGFSALLSKTEEEGRLHGIRIC 871
Query: 1476 RIAPRVSHLLFAD 1488
+ AP VSHLLFAD
Sbjct: 872 QGAPSVSHLLFAD 884
Score = 94.4 bits (233), Expect = 3e-17
Identities = 56/163 (34%), Positives = 80/163 (48%), Gaps = 4/163 (2%)
Query: 693 YGYPEIARRRDSWNFIRGLARQINLPWCQMGDFNDILHSDEKKGRATRPNWLIRGFRHSI 752
YG + +W +RGL PW GDFN+IL S EK+ + + FRH++
Sbjct: 397 YGDAHSETKHRTWTTMRGLIDNPTTPWLMAGDFNEILFSHEKQCGRMKAQSAMDEFRHAL 456
Query: 753 QDVGLIDVHMEGYLFTWFK-SLGTTRVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDH 811
D GL D+ EG FTW S + E LDRA+A W +FP A++ N SDH
Sbjct: 457 TDCGLDDLGFEGDAFTWRNHSHSQEGYIREWLDRAVANPEWRAMFPAARVINGDPRHSDH 516
Query: 812 FPILLD---RTLMVRSHRVERSFKFENARRVEEVVNDVVRGSW 851
P++++ + VR F+FE A EE +VV+ +W
Sbjct: 517 RPVIIELEGKNKGVRGRNGHNDFRFEAAWLEEEKFKEVVKEAW 559
Score = 50.4 bits (119), Expect = 5e-04
Identities = 26/111 (23%), Positives = 53/111 (47%), Gaps = 8/111 (7%)
Query: 15 NKFMVQLYSKADLVRILDRSPWLLDNNMIILKKVAIGEDPLTLAMDT----TEIWAQKNE 70
N F+ + + R L+ PW+ + +++++ I D L D IW + +
Sbjct: 43 NHFLFTFHQASGKRRALEDGPWMFNKDLVVM----IDLDETKLIEDMIFAFVPIWVRAMK 98
Query: 71 LPFGFMDKKVGALVGSHIGKMVKFDEENNYGL*RKYMRVRVEIALDAPLQQ 121
LP G M K+ G +G +G+ + D E + +++R+++ I + PL +
Sbjct: 99 LPLGLMTKETGMAIGREVGEFMTMDLEEDGSAVGQFLRIKIRIDIRKPLMR 149
>UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa]
Length = 1557
Score = 139 bits (350), Expect = 8e-31
Identities = 76/230 (33%), Positives = 119/230 (51%), Gaps = 1/230 (0%)
Query: 588 MSIIAWNCRGLGNLRVIPKIKFLVRYYKQDILFLCETMVEVNKIEEFRYLLGYDFCFAPA 647
M+ ++WNCRG+GN + ++ LV+ +FLCET V+KI+ R LG +
Sbjct: 434 MNCLSWNCRGIGNATTVQELCALVKEASSQFVFLCETRQPVDKIKRLRNRLGLRGFAGVS 493
Query: 648 RNNRSGGIAYFWRNTVNCSIVNYSANHISAKIE-EVSRSAWTFTGFYGYPEIARRRDSWN 706
+ SGG+A FW +V+ I+N + I A I + W T YG P + R W+
Sbjct: 494 SDGMSGGLALFWHESVHVEILNKNERFIDAYIRLSPNDPLWHATFVYGEPRVENRHKMWS 553
Query: 707 FIRGLARQINLPWCQMGDFNDILHSDEKKGRATRPNWLIRGFRHSIQDVGLIDVHMEGYL 766
+ + + NL W +GDFN+ L E R R + ++ FR + L D+ + G
Sbjct: 554 LLNSIKQSSNLSWLVIGDFNEALWQFEHFSRKPRSDAQMQAFRDVLDTCELHDLGISGLP 613
Query: 767 FTWFKSLGTTRVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDHFPILL 816
T+ G V+ +LDRALA + W LF ++++LV+P S+H PI+L
Sbjct: 614 HTYDNRRGGWNNVKVRLDRALADDEWRDLFSQYQVQHLVSPCSNHCPIVL 663
Score = 88.6 bits (218), Expect = 2e-15
Identities = 48/133 (36%), Positives = 67/133 (50%), Gaps = 19/133 (14%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+A E H++ + + A KLD+SKAYDR+DW +L M K GFS + W+M +
Sbjct: 939 LLAFECFHFIQRNKNPRSAACAYKLDLSKAYDRVDWRFLEQSMYKWGFSHCWVSWVMTCI 998
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
TV +G P+LF+ A+GLS + +G + IRVC
Sbjct: 999 STV-------------------CDKGTRCDPFLFLFVADGLSLLLEEKVEQGAISPIRVC 1039
Query: 1476 RIAPRVSHLLFAD 1488
AP +SHLLFAD
Sbjct: 1040 HQAPGISHLLFAD 1052
Score = 50.8 bits (120), Expect = 4e-04
Identities = 29/126 (23%), Positives = 58/126 (46%)
Query: 1 WEPKYQITLILMEGNKFMVQLYSKADLVRILDRSPWLLDNNMIILKKVAIGEDPLTLAMD 60
W + + + N F+V S+ D + D PWL + +++ + P + ++
Sbjct: 193 WRLRADMNYKSLRDNLFIVSFSSEGDYKFVKDGGPWLHRGDALLVAEFDGLTSPSQVPLE 252
Query: 61 TTEIWAQKNELPFGFMDKKVGALVGSHIGKMVKFDEENNYGL*RKYMRVRVEIALDAPLQ 120
IW + +LP M K G L GS +G + + D E + + R+RV++ ++ L+
Sbjct: 253 VVPIWVRIYDLPLVLMTKDRGILYGSKLGHVREVDVEADGRNKHDFFRIRVDLPVNRALK 312
Query: 121 QELIIE 126
+ I+
Sbjct: 313 THIAIK 318
>UniRef100_Q9SZ87 RNA-directed DNA polymerase-like protein [Arabidopsis thaliana]
Length = 1274
Score = 139 bits (349), Expect = 1e-30
Identities = 67/133 (50%), Positives = 93/133 (69%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+ E++H++ K S+A+K D+SKAYDRI W +L++V+ ++GF +K I W+M V
Sbjct: 492 LITHEILHFLRVSGAKKYCSMAIKTDMSKAYDRIKWNFLQEVLMRLGFHDKWIRWVMQCV 551
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
TV YS ++N G ++P GLRQGDPLSPYLFILC E LS R A+ +G + GIRV
Sbjct: 552 CTVSYSFLINGSPQGSVVPSRGLRQGDPLSPYLFILCTEVLSGLCRKAQEKGVMVGIRVA 611
Query: 1476 RIAPRVSHLLFAD 1488
R +P+V+HLLFAD
Sbjct: 612 RGSPQVNHLLFAD 624
Score = 87.4 bits (215), Expect = 3e-15
Identities = 62/216 (28%), Positives = 95/216 (43%), Gaps = 30/216 (13%)
Query: 638 LGYDFCFAPARNNRSGGIAYFWRNTVNCSIVNYSANHISAKIEEVSRSAWTFTGFYGYPE 697
+GY F SGG+A +W+ V I+ + N I +RS +
Sbjct: 17 MGYAHRFTIPPEGLSGGLALYWKENVEVEILEAAPNFID------NRSVF---------- 60
Query: 698 IARRRDSWNFIRGLARQINLPWCQMGDFNDILHSDEKKGRATRPNWLIRGFRHSIQDVGL 757
W+ I L Q + W GDFNDIL + EK+G R FR + GL
Sbjct: 61 -------WDKISSLGAQRSSAWLLTGDFNDILDNSEKQGGPLRWEGFFLAFRSFVSQNGL 113
Query: 758 IDVHMEGYLFTWFKSLGTTRVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDHFPIL-- 815
D++ G +W + + ++ +LDRAL SW +LFP +K E L SDH P++
Sbjct: 114 WDINHTGNSLSW-RGTRYSHFIKSRLDRALGNCSWSELFPMSKCEYLRFEGSDHRPLVTY 172
Query: 816 LDRTLMVRSHRVERSFKFENARRVEEVVNDVVRGSW 851
+ RS + F+F+ R +E + +V+ W
Sbjct: 173 FGAPPLKRS----KPFRFDRRLREKEEIRALVKEVW 204
>UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
Length = 1142
Score = 139 bits (349), Expect = 1e-30
Identities = 70/133 (52%), Positives = 92/133 (68%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+A E+ H + T K+K +A+K D+SKAYD+++W ++ ++ KMGF EK I WIM +
Sbjct: 335 LIAQEMFHGLRTNSSCKDKFMAIKTDMSKAYDQVEWNFIEALLRKMGFCEKWISWIMWCI 394
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
TV Y V++N + G IIP GLRQGDPLSPYLFILC E L A IR AE + + GI+V
Sbjct: 395 TTVQYKVLINGQPKGLIIPERGLRQGDPLSPYLFILCTEVLIANIRKAERQNLITGIKVA 454
Query: 1476 RIAPRVSHLLFAD 1488
+P VSHLLFAD
Sbjct: 455 TPSPAVSHLLFAD 467
>UniRef100_Q9ZQF8 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1225
Score = 137 bits (346), Expect = 2e-30
Identities = 64/133 (48%), Positives = 95/133 (71%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+A E H + T K+K +A+K D+SKAYDR++W++LR +M KMGF++K + WI+ +
Sbjct: 498 LIAQENFHALRTNPACKKKYMAIKTDMSKAYDRVEWSFLRALMLKMGFAQKWVDWIIFCI 557
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
+V Y ++LN G I P G+RQGDP+SP+LFILC E L A++++AE G +QG+++
Sbjct: 558 SSVSYKILLNGSPKGFIKPSRGIRQGDPISPFLFILCTEALVAKLKDAEWHGRIQGLQIS 617
Query: 1476 RIAPRVSHLLFAD 1488
R +P SHLLFAD
Sbjct: 618 RASPSTSHLLFAD 630
Score = 102 bits (254), Expect = 1e-19
Identities = 76/296 (25%), Positives = 130/296 (43%), Gaps = 12/296 (4%)
Query: 588 MSIIAWNCRGLGNLRVIPKIKFLVRYYKQDILFLCETMVEVNKIEEFRYLLGYDFCFAPA 647
M +I+WNC+G+G +++ + R Y LFL ET ++ ++ + LG+D
Sbjct: 1 MRLISWNCQGVGPKTTSRRLEEMCRMYSPGFLFLSETKNDLVYLQNVQVSLGFDCLKTVE 60
Query: 648 RNNRSGGIAYFWRNTVNCSIVNYSANHISAKIEEVSRSAWTFTGF-YGYPEIARRRDSWN 706
SGG+A F+ + I IE + F F YG P + R W
Sbjct: 61 PIGNSGGLALFYSRDYPVKFIYVCDRLID--IETIIDGNRVFITFVYGDPVVQYRELVWK 118
Query: 707 FIRGLARQINLPWCQMGDFNDILHSDEKKGRATRPNWLIRGFRHSIQDVGLIDVHMEGYL 766
+ + + PW +GDFN+I+ + EK+G R F I++ G+ID G L
Sbjct: 119 RLTRIGIVRSEPWFMIGDFNEIIGNHEKRGGKKRSESSFLPFCCMIENCGMIDFPSTGSL 178
Query: 767 FTWF--KSLGTT-----RVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDHFPILLDRT 819
F+W +S G +++ +LDRA+ W ++ + +E L SDH P+L +
Sbjct: 179 FSWVGKRSCGVAGRKRRDLIKCRLDRAMGNEEWHSIYSHTNVEYLQHRGSDHKPLL--AS 236
Query: 820 LMVRSHRVERSFKFENARRVEEVVNDVVRGSWLGSTVSNVINNMNKYINCLCLISL 875
+ + +R + F F+ + + V+ W + + K NC IS+
Sbjct: 237 IQNKPYRPYKHFIFDKRWINKPGFKESVQEGWAFPSRGEGVPFFQKIKNCRQTISI 292
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 130 bits (326), Expect = 5e-28
Identities = 88/311 (28%), Positives = 144/311 (46%), Gaps = 7/311 (2%)
Query: 588 MSIIAWNCRGLGNLRVIPKIKFLVRYYKQDILFLCETMVEVNKIEEFRYLLGYDFCFAPA 647
M I++WNC+G+GN + ++ + Y +++FLCET N +E LG+
Sbjct: 1 MRILSWNCQGVGNTPTVRHLREIRGLYFPEVIFLCETKKRRNYLENVVGHLGFFDLHTVE 60
Query: 648 RNNRSGGIAYFWRNTVNCSIVNYSANHISAKIEEVSRSAWTFTGFYGYPEIARRRDSWNF 707
+SGG+A W+++V ++ I A + + + T YG P A R + W
Sbjct: 61 PIGKSGGLALMWKDSVQIKVLQSDKRLIDALLIWQDKEFY-LTCIYGEPVQAERGELWER 119
Query: 708 IRGLARQINLPWCQMGDFNDILHSDEKKGRATRPNWLIRGFRHSIQDVGLIDVHMEGYLF 767
+ L + PW GDFN+++ EK G R FR + GL +V+ GY F
Sbjct: 120 LTRLGLSRSGPWMLTGDFNELVDPSEKIGGPARKESSCLEFRQMLNSCGLWEVNHSGYQF 179
Query: 768 TWFKSLGTTRVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDHFPILLDRTLMVRSHRV 827
+W+ + +V+ +LDR +A +WM+LFP AK L SDH P++ L+ + R
Sbjct: 180 SWYGN-RNDELVQCRLDRTVANQAWMELFPQAKATYLQKICSDHSPLI--NNLVGDNWRK 236
Query: 828 ERSFKFENARRVEEVVNDVVRGSWLGSTVSNVINNMNKYINCLCLISLS*G*EKIYKFSS 887
FK++ E D++ W + M K +C IS +++ K SS
Sbjct: 237 WAGFKYDKRWVQREGFKDLLCNFWSQQSTKTNALMMEKIASCRREISKW---KRVSKPSS 293
Query: 888 SEGILHISVKL 898
+ I + KL
Sbjct: 294 AVRIQELQFKL 304
Score = 129 bits (325), Expect = 6e-28
Identities = 64/133 (48%), Positives = 90/133 (67%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+A EL+H + + K E+ +A+K DISKAYDR++W +L M +GF++ I IM V
Sbjct: 551 LIAHELLHALSSNNKCSEEFIAIKTDISKAYDRVEWPFLEKAMRGLGFADHWIRLIMECV 610
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
+V Y V++N G IIP GLRQGDPLSPYLF++C E L +++AE + + G++V
Sbjct: 611 KSVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLFVICTEMLVKMLQSAEQKNQITGLKVA 670
Query: 1476 RIAPRVSHLLFAD 1488
R AP +SHLLFAD
Sbjct: 671 RGAPPISHLLFAD 683
>UniRef100_Q9SHP2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 773
Score = 129 bits (324), Expect = 8e-28
Identities = 64/132 (48%), Positives = 90/132 (67%), Gaps = 1/132 (0%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+A EL+H + TK K + VA KLDI+KA+D+I+W ++ +M +MGFSEK WIM +
Sbjct: 7 LIAHELIHSLHTK-KLVQPFVATKLDITKAFDKIEWGFIEAIMKQMGFSEKWCNWIMTCI 65
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
T YS+++N + + IIP G+RQGDP+SPYL++LC EGLSA I+ + L G +
Sbjct: 66 TTTTYSILINGQPVRRIIPKRGIRQGDPISPYLYLLCTEGLSALIQASIKAKQLHGFKAS 125
Query: 1476 RIAPRVSHLLFA 1487
R P +SHLLFA
Sbjct: 126 RNGPAISHLLFA 137
>UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1715
Score = 129 bits (323), Expect = 1e-27
Identities = 69/158 (43%), Positives = 102/158 (63%), Gaps = 2/158 (1%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
LVA EL+H + ++ + + VA+K DISKAYDR++W +L VM ++GF+ + + WIM V
Sbjct: 913 LVAHELLHSLKSRRECQSGYVAVKTDISKAYDRVEWNFLEKVMIQLGFAPRWVKWIMTCV 972
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
+V Y V++N G I P G+RQGDPLSPYLF+ CAE LS +R AE + G+++
Sbjct: 973 TSVSYEVLINGSPYGKIFPSRGIRQGDPLSPYLFLFCAEVLSNMLRKAEVNKQIHGMKIT 1032
Query: 1476 RIAPRVSHLLFADIDSV*GSIRSSYQSSRVRNLLWQNY 1513
+ +SHLLFAD DS+ R+S Q+ L+++ Y
Sbjct: 1033 KDCLAISHLLFAD-DSL-FFCRASNQNIEQLALIFKKY 1068
Score = 116 bits (290), Expect = 7e-24
Identities = 71/265 (26%), Positives = 130/265 (48%), Gaps = 7/265 (2%)
Query: 587 PMSIIAWNCRGLGNLRVIPKIKFLVRYYKQDILFLCETMVEVNKIEEFRYLLGY-DFCFA 645
PM + WNC+GLG + +++ + R Y D+LFL ET + N + +G+ D C
Sbjct: 362 PMRVGFWNCQGLGQPLTVRRLEEVQRVYFLDMLFLIETKQQDNYTRDLGVKMGFEDMCII 421
Query: 646 PARNNRSGGIAYFWRNTVNCSIVNYSANHISAKIEEVSRSAWTFTGFYGYPEIARRRDSW 705
R SGG+ +W+ ++ ++++ + +E + + + + YG+P + R W
Sbjct: 422 SPR-GLSGGLVVYWKKHLSIQVISHDVRLVDLYVEYKNFNFY-LSCIYGHPIPSERHHLW 479
Query: 706 NFIRGLARQINLPWCQMGDFNDILHSDEKKGRATRPNWLIRGFRHSIQDVGLIDVHMEGY 765
++ ++ + PW GDFN+IL+ +EKKG R ++ F + I + D+ +G
Sbjct: 480 EKLQRVSAHRSGPWMMCGDFNEILNLNEKKGGRRRSIGSLQNFTNMINCCNMKDLKSKGN 539
Query: 766 LFTWFKSLGTTRVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDHFPILLDRTLMVRSH 825
++W +E LDR + W FP + E L SDH P+++D V +
Sbjct: 540 PYSWV-GKRQNETIESCLDRVFINSDWQASFPAFETEFLPIAGSDHAPVIIDIAEEVCTK 598
Query: 826 RVERSFKFENAR-RVEEVVNDVVRG 849
R + F+++ + E+ V+ V RG
Sbjct: 599 RGQ--FRYDRRHFQFEDFVDSVQRG 621
Score = 45.4 bits (106), Expect = 0.015
Identities = 31/135 (22%), Positives = 65/135 (47%), Gaps = 6/135 (4%)
Query: 11 LMEGNKFMVQLYSKADLVRILDRSPWLLDNNMIILKKVAIGEDPLTLAMDTTEIWAQKNE 70
+++ +F S+ L +L R PW + M+++++ DPL L + W Q E
Sbjct: 72 IIQNRRFQFIFPSEESLDTVLRRGPWSFADRMLVVERWTPDMDPLVL--NFIPFWIQVRE 129
Query: 71 LPFGFMDKKVGALVGSHIG--KMVKFDEENNYGL*RKYMRVRVEIALDAPLQQELIIEWE 128
+P F++ +V + +G K V FD + +++R++++ ++ PL+ + ++
Sbjct: 130 IPLQFLNLEVIDNIAGSLGERKAVDFDPFTTTRV--EFVRIQIKWDVNHPLRFQRNYQFS 187
Query: 129 EGDNIKLVFKFRETR 143
G N L F + R
Sbjct: 188 LGVNTVLSFYYERLR 202
>UniRef100_Q84MQ3 Putative reverse transcriptase [Oryza sativa]
Length = 818
Score = 128 bits (322), Expect = 1e-27
Identities = 66/133 (49%), Positives = 87/133 (64%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+A E HY+ + + ALKLD+SKA+DR+DW +L V+ ++GF+EK WIM V
Sbjct: 72 LIAFECFHYIQKCNRESQMFCALKLDLSKAFDRVDWDFLDGVLLRLGFAEKWRKWIMSCV 131
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
V YSV N + P P GLRQGDPLSPYLF+ A+GLS + ++ G +Q ++VC
Sbjct: 132 TFVQYSVRFNGNLLEPFKPSRGLRQGDPLSPYLFLFIADGLSNILEKCKTDGLIQPLKVC 191
Query: 1476 RIAPRVSHLLFAD 1488
R AP VSHLLFAD
Sbjct: 192 RGAPGVSHLLFAD 204
>UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana]
Length = 1270
Score = 128 bits (321), Expect = 2e-27
Identities = 72/159 (45%), Positives = 102/159 (63%), Gaps = 2/159 (1%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
LVA ELVH + + + +A+K D+SKAYDR++W+YLR ++ +GF K + WIM+ V
Sbjct: 514 LVAHELVHSLKVHPRISSEFMAVKSDMSKAYDRVEWSYLRSLLLSLGFHLKWVNWIMVCV 573
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
+V YSV++N G II GLRQGDPLSP+LF+LC EGL+ + A+ G L+GI+
Sbjct: 574 SSVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGLTHLLNKAQWEGALEGIQFS 633
Query: 1476 RIAPRVSHLLFADIDSV*GSIRSSYQSSRVRNLLWQNYG 1514
P V HLLFAD DS+ ++S + S V + + YG
Sbjct: 634 ENGPMVHHLLFAD-DSL-FLCKASREQSLVLQKILKVYG 670
Score = 91.7 bits (226), Expect = 2e-16
Identities = 58/214 (27%), Positives = 95/214 (44%), Gaps = 4/214 (1%)
Query: 638 LGYDFCFAPARNNRSGGIAYFWRNTVNCSIVNYSANHISAKIEEVSRSAWTFTGFYGYPE 697
L YD + + GG+A W+++V + N + A+++ + + YG P+
Sbjct: 14 LEYDQVYTVEPVGKCGGLALLWKSSVQVDLKFVDKNLMDAQVQ-FGAVNFCVSCVYGDPD 72
Query: 698 IARRRDSWNFIRGLARQINLPWCQMGDFNDILHSDEKKGRATRPNWLIRGFRHSIQDVGL 757
++R +W I + WC GDFNDILH+ EK G R + + F I+ L
Sbjct: 73 RSKRSQAWERISRIGVGRRDKWCMFGDFNDILHNGEKNGGPRRSDLDCKAFNEMIKGCDL 132
Query: 758 IDVHMEGYLFTWFKSLGTTRVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDHFPILLD 817
+++ G FTW G ++ +LDRA W FP + L SDH P+L+
Sbjct: 133 VEMPAHGNGFTWAGRRG-DHWIQCRLDRAFGNKEWFCFFPVSNQTFLDFRGSDHRPVLI- 190
Query: 818 RTLMVRSHRVERSFKFENARRVEEVVNDVVRGSW 851
LM F+F+ +E V + + +W
Sbjct: 191 -KLMSSQDSYRGQFRFDKRFLFKEDVKEAIIRTW 223
>UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana]
Length = 1254
Score = 128 bits (321), Expect = 2e-27
Identities = 72/159 (45%), Positives = 102/159 (63%), Gaps = 2/159 (1%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
LVA ELVH + + + +A+K D+SKAYDR++W+YLR ++ +GF K + WIM+ V
Sbjct: 517 LVAHELVHSLKVHPRISSEFMAVKSDMSKAYDRVEWSYLRSLLLSLGFHLKWVNWIMVCV 576
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
+V YSV++N G II GLRQGDPLSP+LF+LC EGL+ + A+ G L+GI+
Sbjct: 577 SSVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGLTHLLNKAQWEGALEGIQFS 636
Query: 1476 RIAPRVSHLLFADIDSV*GSIRSSYQSSRVRNLLWQNYG 1514
P V HLLFAD DS+ ++S + S V + + YG
Sbjct: 637 ENGPMVHHLLFAD-DSL-FLCKASREQSLVLQKILKVYG 673
Score = 94.7 bits (234), Expect = 2e-17
Identities = 61/229 (26%), Positives = 102/229 (43%), Gaps = 4/229 (1%)
Query: 623 ETMVEVNKIEEFRYLLGYDFCFAPARNNRSGGIAYFWRNTVNCSIVNYSANHISAKIEEV 682
ETM + + + + L YD + + GG+A W+++V + N + A+++
Sbjct: 2 ETMHSRDDLVDIQSWLEYDQVYTVEPVGKCGGLALLWKSSVQVDLKFVDKNLMDAQVQ-F 60
Query: 683 SRSAWTFTGFYGYPEIARRRDSWNFIRGLARQINLPWCQMGDFNDILHSDEKKGRATRPN 742
+ + YG P+ ++R +W I + WC GDFNDILH+ EK G R +
Sbjct: 61 GAVNFCVSCVYGDPDRSKRSQAWERISRIGVGRRDKWCMFGDFNDILHNGEKNGGPRRSD 120
Query: 743 WLIRGFRHSIQDVGLIDVHMEGYLFTWFKSLGTTRVVEEKLDRALATNSWMQLFPNAKLE 802
+ F I+ L+++ G FTW G ++ +LDRA W FP +
Sbjct: 121 LDCKAFNEMIKGCDLVEMPAHGNGFTWAGRRG-DHWIQCRLDRAFGNKEWFCFFPVSNQT 179
Query: 803 NLVAPSSDHFPILLDRTLMVRSHRVERSFKFENARRVEEVVNDVVRGSW 851
L SDH P+L+ LM F+F+ +E V + + +W
Sbjct: 180 FLDFRGSDHRPVLI--KLMSSQDSYRGQFRFDKRFLFKEDVKEAIIRTW 226
>UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis thaliana]
Length = 1294
Score = 127 bits (320), Expect = 2e-27
Identities = 64/133 (48%), Positives = 91/133 (68%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
++A E++H + T+ + + +A+K D+SKAYDR++W +L M GFSE I WIM V
Sbjct: 913 MIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGFSETWIKWIMGAV 972
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
+V+YSV++N G I P G+RQGDPLSPYLFILCA+ L+ I+N + GD++GIR+
Sbjct: 973 KSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHLIKNRVAEGDIRGIRIG 1032
Query: 1476 RIAPRVSHLLFAD 1488
P V+HL FAD
Sbjct: 1033 NGVPGVTHLQFAD 1045
Score = 118 bits (296), Expect = 1e-24
Identities = 82/274 (29%), Positives = 128/274 (45%), Gaps = 13/274 (4%)
Query: 596 RGLGNLRVIPKIKFLVRYYKQDILFLCETMVEVNKIEEFRYLLGYDFCFAPARNNRSGGI 655
+G+G ++ L + +K D+LFL ET+ + I +LG+ SGG+
Sbjct: 370 KGIGVPLTQSQLSNLCKVFKFDVLFLIETLNKCEVISNLASVLGFPNVITQPPQGHSGGL 429
Query: 656 AYFWRNTVNCSIVNYSANHISAKIEEVSRSAWTFTGFYGYPEIARRRDSWNFIRGLARQI 715
A W+++V S + HI I ++ + + YG+P + R W L++
Sbjct: 430 ALLWKDSVRLSNLYQDDRHIDVHIS-INNINFYLSRVYGHPCQSERHSLWTHFENLSKTR 488
Query: 716 NLPWCQMGDFNDILHSDEKKGRATRPNWLIRGFRHSIQDVGLIDVHMEGYLFTWFKSLGT 775
N PW +GDFN+IL ++EK G R W RGFR+ + L D+ G F+W
Sbjct: 489 NDPWILIGDFNEILSNNEKIGGPQRDEWTFRGFRNMVSTCDLKDIRSIGDRFSWVGE-RH 547
Query: 776 TRVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDHFPILLDRTLMVRSHRVERSFKFEN 835
+ V+ LDRA + LFP A+LE L SDH P+ L +L R R F+F+
Sbjct: 548 SHTVKCCLDRAFINSEGAFLFPFAELEFLEFTGSDHKPLFL--SLEKTETRKMRPFRFD- 604
Query: 836 ARRVEEV--VNDVVRGSWLGSTVSNVINNMNKYI 867
+R+ EV V+ W + IN K++
Sbjct: 605 -KRLLEVPHFKTYVKAGW-----NKAINGQRKHL 632
Score = 39.3 bits (90), Expect = 1.1
Identities = 29/120 (24%), Positives = 55/120 (45%), Gaps = 15/120 (12%)
Query: 30 ILDRSPWLLDNNMIILKK----VAIGEDPLTLAMDTTEIWAQKNELPFGFMDKKVGALVG 85
+L R PW ++ M+ + + + GE M W Q +P ++ + +VG
Sbjct: 91 VLQRGPWSFNDWMLSVHRWFPNITEGE------MKIIPFWVQIQGIPILYLTNAMARVVG 144
Query: 86 SHIGKM--VKFDEENNYGL*RKYMRVRVEIALDAPLQQELIIEWEEGDNIKLVFKFRETR 143
S +G + V FDE N ++RV++ D L+ + +++E +N + F+F R
Sbjct: 145 SRLGYVTDVDFDENANQ---MGFVRVKLAWNFDDHLRFQRNFQFQENENTIIKFRFERLR 201
>UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana]
Length = 1662
Score = 127 bits (320), Expect = 2e-27
Identities = 64/133 (48%), Positives = 91/133 (68%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
++A E++H + T+ + + +A+K D+SKAYDR++W +L M GFSE I WIM V
Sbjct: 933 MIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGFSETWIKWIMGAV 992
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
+V+YSV++N G I P G+RQGDPLSPYLFILCA+ L+ I+N + GD++GIR+
Sbjct: 993 KSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHLIKNRVAEGDIRGIRIG 1052
Query: 1476 RIAPRVSHLLFAD 1488
P V+HL FAD
Sbjct: 1053 NGVPGVTHLQFAD 1065
Score = 117 bits (294), Expect = 2e-24
Identities = 82/273 (30%), Positives = 127/273 (46%), Gaps = 13/273 (4%)
Query: 597 GLGNLRVIPKIKFLVRYYKQDILFLCETMVEVNKIEEFRYLLGYDFCFAPARNNRSGGIA 656
G+G ++ L + +K D+LFL ET+ + I +LG+ SGG+A
Sbjct: 391 GIGVPLTQSQLSNLCKVFKFDVLFLIETLNKCEVISNLASVLGFPNVITQPPQGHSGGLA 450
Query: 657 YFWRNTVNCSIVNYSANHISAKIEEVSRSAWTFTGFYGYPEIARRRDSWNFIRGLARQIN 716
W+++V S + HI I ++ + + YG+P + R W L++ N
Sbjct: 451 LLWKDSVRLSNLYQDDRHIDVHIS-INNINFYLSRVYGHPCQSERHSLWTHFENLSKTRN 509
Query: 717 LPWCQMGDFNDILHSDEKKGRATRPNWLIRGFRHSIQDVGLIDVHMEGYLFTWFKSLGTT 776
PW +GDFN+IL ++EK G R W RGFR+ + L D+ G F+W +
Sbjct: 510 DPWILIGDFNEILSNNEKIGGPQRDEWTFRGFRNMVSTCDLKDIRSIGDRFSWVGE-RHS 568
Query: 777 RVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDHFPILLDRTLMVRSHRVERSFKFENA 836
V+ LDRA + LFP A+LE L SDH P+ L +L R R F+F+
Sbjct: 569 HTVKCCLDRAFINSEGAFLFPFAELEFLEFTGSDHKPLFL--SLEKTETRKMRPFRFD-- 624
Query: 837 RRVEEV--VNDVVRGSWLGSTVSNVINNMNKYI 867
+R+ EV V+ W + IN K++
Sbjct: 625 KRLLEVPHFKTYVKAGW-----NKAINGQRKHL 652
Score = 39.3 bits (90), Expect = 1.1
Identities = 29/120 (24%), Positives = 55/120 (45%), Gaps = 15/120 (12%)
Query: 30 ILDRSPWLLDNNMIILKK----VAIGEDPLTLAMDTTEIWAQKNELPFGFMDKKVGALVG 85
+L R PW ++ M+ + + + GE M W Q +P ++ + +VG
Sbjct: 91 VLQRGPWSFNDWMLSVHRWFPNITEGE------MKIIPFWVQIQGIPILYLTNAMARVVG 144
Query: 86 SHIGKM--VKFDEENNYGL*RKYMRVRVEIALDAPLQQELIIEWEEGDNIKLVFKFRETR 143
S +G + V FDE N ++RV++ D L+ + +++E +N + F+F R
Sbjct: 145 SRLGYVTDVDFDENANQ---MGFVRVKLAWNFDDHLRFQRNFQFQENENTIIKFRFERLR 201
>UniRef100_Q9ZQM0 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1044
Score = 127 bits (318), Expect = 4e-27
Identities = 64/133 (48%), Positives = 89/133 (66%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+ E +HY+ + K +A+K ++SKAYDRI+W +++ VM +MGF + I WI+ +
Sbjct: 377 LITHEALHYLKSLGAEKRCFMAVKTNMSKAYDRIEWDFIKLVMQEMGFHQTWISWILQCI 436
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
TV YS +LN G + P GLRQGDPLSP+LFI+C+E LS R A+ G L G+RV
Sbjct: 437 TTVSYSFLLNGSAQGAVTPERGLRQGDPLSPFLFIICSEVLSGLCRKAQLDGSLLGLRVS 496
Query: 1476 RIAPRVSHLLFAD 1488
+ PRV+HLLFAD
Sbjct: 497 KGNPRVNHLLFAD 509
Score = 38.1 bits (87), Expect = 2.4
Identities = 24/95 (25%), Positives = 46/95 (48%), Gaps = 3/95 (3%)
Query: 757 LIDVHMEGYLFTWFKSLGTTRVVEEKLDRALATNSWMQLFPNAKLENLVAPSSDHFPILL 816
L D+ G +W + VV +LDRAL+ +W + +P ++ L SDH P+L
Sbjct: 6 LYDLRHSGNFLSW-RGKRHDHVVHCRLDRALSNGAWAEDYPASRCIYLCFEGSDHRPLLT 64
Query: 817 DRTLMVRSHRVERSFKFENARRVEEVVNDVVRGSW 851
L + + + F+++ + + V +V+ +W
Sbjct: 65 HFDLSKK--KKKGVFRYDRRLKNNDEVTALVQEAW 97
>UniRef100_Q7XS38 OSJNBa0081G05.2 protein [Oryza sativa]
Length = 1711
Score = 125 bits (314), Expect = 1e-26
Identities = 71/248 (28%), Positives = 121/248 (48%), Gaps = 1/248 (0%)
Query: 618 ILFLCETMVEVNKIEEFRYLLGYDFCFAPARNNRSGGIAYFWRNTVNCSIVNYSANHISA 677
++FLCET V K+ R L + + SGG+A +W +V+ + + + +I A
Sbjct: 578 LVFLCETRQSVEKMSRLRRKLAFRGFVGVSSEGMSGGLALYWDESVSVDVKDINKRYIDA 637
Query: 678 KIEEVS-RSAWTFTGFYGYPEIARRRDSWNFIRGLARQINLPWCQMGDFNDILHSDEKKG 736
+ S W T YG P + R W+ +R + + LPW +GDFN+ L E
Sbjct: 638 YVRLSSDEPQWHITFVYGEPRVENRHRMWSLLRTIRQSSALPWMVIGDFNETLWQFEHFS 697
Query: 737 RATRPNWLIRGFRHSIQDVGLIDVHMEGYLFTWFKSLGTTRVVEEKLDRALATNSWMQLF 796
+ R ++ FR ++ D L D+ +G T+ R V+ +LDRA+A + W LF
Sbjct: 698 KNPRCETQMQNFRDALYDCDLQDLGFKGVPHTYDNRRDGWRNVKVRLDRAVADDKWRDLF 757
Query: 797 PNAKLENLVAPSSDHFPILLDRTLMVRSHRVERSFKFENARRVEEVVNDVVRGSWLGSTV 856
P A++ +LV+P SDH PILL+ + + ++ +E E V+ +W+ + V
Sbjct: 758 PEAQVSHLVSPCSDHSPILLEFIVKDTTRPRQKCLHYEIVWEREPESVQVIEEAWINAGV 817
Query: 857 SNVINNMN 864
+ ++N
Sbjct: 818 KTDLGDIN 825
Score = 58.5 bits (140), Expect = 2e-06
Identities = 36/152 (23%), Positives = 75/152 (48%), Gaps = 1/152 (0%)
Query: 1 WEPKYQITLILMEGNKFMVQLYSKADLVRILDRSPWLLDNNMIILKKVAIGEDPLTLAMD 60
W + +++ ++ N F+V ++ D +L PWL + +++ P + ++
Sbjct: 193 WRLRAEMSYKSLKDNLFIVSFSAEGDHKFVLQGGPWLHRGDALLVADFNGLVSPSMVPLE 252
Query: 61 TTEIWAQKNELPFGFMDKKVGALVGSHIGKMVKFDEENNYGL*RKYMRVRVEIALDAPLQ 120
T IW + +LP M+K+ G + GS +GK+ + D + + + R+RV++ ++ PL+
Sbjct: 253 TMPIWVRIYDLPLVMMNKERGVIYGSKLGKVREVDVQEDGCNKHDFFRIRVDLPVNRPLK 312
Query: 121 QELIIEWE-EGDNIKLVFKFRETR*VLLCLWC 151
+ L I+ + +G F R R C +C
Sbjct: 313 RMLAIKIKVKGAEEIRRFNLRYERIPHFCFFC 344
Score = 50.4 bits (119), Expect = 5e-04
Identities = 21/51 (41%), Positives = 32/51 (62%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK 1406
L+A E H + K + A KLD+SKAYDR+DW +L ++K+GF+ +
Sbjct: 1103 LLAFECFHSIQKNKKANSAACAYKLDLSKAYDRVDWRFLELALNKLGFAHR 1153
>UniRef100_Q7XME3 OSJNBa0061G20.16 protein [Oryza sativa]
Length = 1285
Score = 125 bits (314), Expect = 1e-26
Identities = 71/248 (28%), Positives = 121/248 (48%), Gaps = 1/248 (0%)
Query: 618 ILFLCETMVEVNKIEEFRYLLGYDFCFAPARNNRSGGIAYFWRNTVNCSIVNYSANHISA 677
++FLCET V K+ R L + + SGG+A +W +V+ + + + +I A
Sbjct: 578 LVFLCETRQSVEKMSRLRRKLAFRGFVGVSSEGMSGGLALYWDESVSVDVKDINKRYIDA 637
Query: 678 KIEEVS-RSAWTFTGFYGYPEIARRRDSWNFIRGLARQINLPWCQMGDFNDILHSDEKKG 736
+ S W T YG P + R W+ +R + + LPW +GDFN+ L E
Sbjct: 638 YVRLSSDEPQWHITFVYGEPRVENRHRMWSLLRTIRQSSALPWMVIGDFNETLWQFEHFS 697
Query: 737 RATRPNWLIRGFRHSIQDVGLIDVHMEGYLFTWFKSLGTTRVVEEKLDRALATNSWMQLF 796
+ R ++ FR ++ D L D+ +G T+ R V+ +LDRA+A + W LF
Sbjct: 698 KNPRCETQMQNFRDALYDCDLQDLGFKGVPHTYDNRRDGWRNVKVRLDRAVADDKWRDLF 757
Query: 797 PNAKLENLVAPSSDHFPILLDRTLMVRSHRVERSFKFENARRVEEVVNDVVRGSWLGSTV 856
P A++ +LV+P SDH PILL+ + + ++ +E E V+ +W+ + V
Sbjct: 758 PEAQVSHLVSPCSDHSPILLEFIVKDTTRPRQKCLHYEIVWEREPESVQVIEEAWINAGV 817
Query: 857 SNVINNMN 864
+ ++N
Sbjct: 818 KTDLGDIN 825
Score = 120 bits (300), Expect = 5e-25
Identities = 59/133 (44%), Positives = 83/133 (62%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMMWV 1415
L+A E H + K + A KLD+SKAYDR+DW +L ++K+GF+ + + WIM V
Sbjct: 1103 LLAFECFHSIQKNKKANSAACAYKLDLSKAYDRVDWRFLELALNKLGFAHRWVSWIMSCV 1162
Query: 1416 GTVDYSVILNKEKIGPIIPGFGLRQGDPLSPYLFILCAEGLSAQIRNAESRGDLQGIRVC 1475
TV YSV N + P GLRQGDPLSP+LF+ A+GLS ++ ++ L +++C
Sbjct: 1163 TTVRYSVKFNGTLLRSFAPTRGLRQGDPLSPFLFLFVADGLSLLLKEKVAQNSLTPLKIC 1222
Query: 1476 RIAPRVSHLLFAD 1488
AP +SHLLFAD
Sbjct: 1223 CQAPGISHLLFAD 1235
Score = 58.5 bits (140), Expect = 2e-06
Identities = 36/152 (23%), Positives = 75/152 (48%), Gaps = 1/152 (0%)
Query: 1 WEPKYQITLILMEGNKFMVQLYSKADLVRILDRSPWLLDNNMIILKKVAIGEDPLTLAMD 60
W + +++ ++ N F+V ++ D +L PWL + +++ P + ++
Sbjct: 193 WRLRAEMSYKSLKDNLFIVSFSAEGDHKFVLQGGPWLHRGDALLVADFNGLVSPSMVPLE 252
Query: 61 TTEIWAQKNELPFGFMDKKVGALVGSHIGKMVKFDEENNYGL*RKYMRVRVEIALDAPLQ 120
T IW + +LP M+K+ G + GS +GK+ + D + + + R+RV++ ++ PL+
Sbjct: 253 TMPIWVRIYDLPLVMMNKERGVIYGSKLGKVREVDVQEDGCNKHDFFRIRVDLPVNRPLK 312
Query: 121 QELIIEWE-EGDNIKLVFKFRETR*VLLCLWC 151
+ L I+ + +G F R R C +C
Sbjct: 313 RMLAIKIKVKGAEEIRRFNLRYERIPHFCFFC 344
>UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa]
Length = 1833
Score = 125 bits (313), Expect = 1e-26
Identities = 71/253 (28%), Positives = 123/253 (48%), Gaps = 8/253 (3%)
Query: 614 YKQDILFLCETMVEVNKIEEFRYLLGYDFCFAPARNNRSGGIAYFWRNTVNCSIVNYSAN 673
++ I+FL ET + ++ +G CF + GG+A FW ++N + +Y++
Sbjct: 10 HRPKIVFLAETRQDQYMVQN---RIGLRRCFTVNGVGKGGGLALFWDESMNVVLKSYNSR 66
Query: 674 HISAKIEEVSRSAWTFTGFYGYPEIARRRDSWNFIRGLARQINLPWCQMGDFNDILHSDE 733
HI I E+ W T YG P R W+ +R + PW +GDFN+ + E
Sbjct: 67 HIDVLITELDCCTWRATFVYGEPRAQDRHLMWSLLRRIRSNSGDPWLMIGDFNEAMWQTE 126
Query: 734 KKGRATRPNWLIRGFRHSIQDVGLIDVHMEGYLFTWFKSLGTTRVVEEKLDRALATNSWM 793
K R +R FR + + L D+ +G +T+ R V+ +LDR +A+ +W
Sbjct: 127 HKSHRKRSESQMRDFREVLSECDLHDIGFQGAPWTFCNMQREGRNVKVRLDRGVASPAWS 186
Query: 794 QLFPNAKLENLVAPSSDHFPILLDR--TLMVRSHRVERSFKFENARRVEEVVNDVVRGSW 851
FP A + +L PSSDH P+LL+R T + R ++ R +E E + +V++ +W
Sbjct: 187 SRFPQAVITHLTTPSSDHAPLLLEREETTLARPMKIMR---YEEVWERESSLPEVIQEAW 243
Query: 852 LGSTVSNVINNMN 864
++ + ++N
Sbjct: 244 TMGADASTLGDIN 256
Score = 58.9 bits (141), Expect = 1e-06
Identities = 27/58 (46%), Positives = 37/58 (63%)
Query: 1356 LVAIELVHYMMTKMKGKEKSVALKLDISKAYDRIDWAYLRDVMSKMGFSEK*I*WIMM 1413
L+A E H + + + ALKLD+SKAYDR+DW +L VM ++GF +K WIMM
Sbjct: 536 LIAFECFHSIKNCKRENQNFCALKLDLSKAYDRVDWEFLNGVMERLGFCDKWRRWIMM 593
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.357 0.159 0.586
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,486,671,730
Number of Sequences: 2790947
Number of extensions: 94221227
Number of successful extensions: 395143
Number of sequences better than 10.0: 611
Number of HSP's better than 10.0 without gapping: 340
Number of HSP's successfully gapped in prelim test: 271
Number of HSP's that attempted gapping in prelim test: 391739
Number of HSP's gapped (non-prelim): 2068
length of query: 1735
length of database: 848,049,833
effective HSP length: 142
effective length of query: 1593
effective length of database: 451,735,359
effective search space: 719614426887
effective search space used: 719614426887
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 82 (36.2 bits)
Medicago: description of AC134322.4


