

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133862.8 + phase: 0
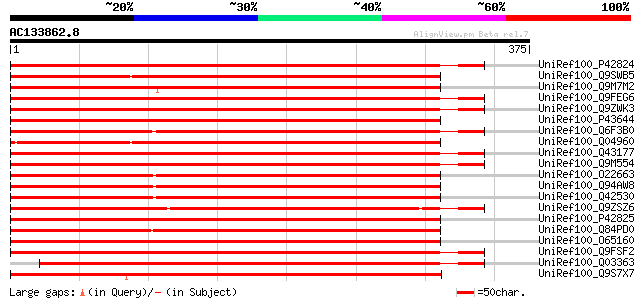
(375 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P42824 DnaJ protein homolog 2 [Allium porrum] 593 e-168
UniRef100_Q9SWB5 Seed maturation protein PM37 [Glycine max] 590 e-167
UniRef100_Q9M7M2 DnaJ-like protein [Lycopersicon esculentum] 587 e-166
UniRef100_Q9FEG6 J2P [Daucus carota] 582 e-165
UniRef100_Q9ZWK3 DnaJ homolog [Salix gilgiana] 578 e-164
UniRef100_P43644 DnaJ protein homolog ANJ1 [Atriplex nummularia] 577 e-163
UniRef100_Q6F3B0 Putative DnaJ like protein [Oryza sativa] 574 e-162
UniRef100_Q04960 DnaJ protein homolog [Cucumis sativus] 574 e-162
UniRef100_Q43177 DnaJ protein [Solanum tuberosum] 573 e-162
UniRef100_Q9M554 DnaJ protein [Euphorbia esula] 567 e-160
UniRef100_O22663 DnaJ protein homolog atj3 [Arabidopsis thaliana] 566 e-160
UniRef100_Q94AW8 AT3g44110/F26G5_60 [Arabidopsis thaliana] 564 e-159
UniRef100_Q42530 DnaJ homolog [Arabidopsis thaliana] 564 e-159
UniRef100_Q9ZSZ6 DnaJ protein [Hevea brasiliensis] 562 e-159
UniRef100_P42825 DnaJ protein homolog ATJ2 [Arabidopsis thaliana] 562 e-159
UniRef100_Q84PD0 DnaJ protein, putative [Oryza sativa] 561 e-158
UniRef100_O65160 DnaJ-related protein ZMDJ1 [Zea mays] 561 e-158
UniRef100_Q9FSF2 Putative DNAJ protein [Nicotiana tabacum] 555 e-156
UniRef100_Q03363 DnaJ protein homolog 1 [Allium porrum] 552 e-156
UniRef100_Q9S7X7 DnaJ homolog protein [Salix gilgiana] 528 e-148
>UniRef100_P42824 DnaJ protein homolog 2 [Allium porrum]
Length = 418
Score = 593 bits (1528), Expect = e-168
Identities = 287/343 (83%), Positives = 305/343 (88%), Gaps = 12/343 (3%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNTKYYE+LGVSKNA+P+DLKKAY+KAAIKNHPDKGGDPEKFKE+ QAYEV
Sbjct: 1 MFGRAPKKSDNTKYYEVLGVSKNATPEDLKKAYRKAAIKNHPDKGGDPEKFKEIGQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
L+DPEKRE+YDQYGE+ LKEGMGGGGG HDPFDIF SFFGGGGF GGGSSRGRRQRRGED
Sbjct: 61 LNDPEKREIYDQYGEEGLKEGMGGGGGVHDPFDIFQSFFGGGGFGGGGSSRGRRQRRGED 120
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
VVHPLKVSLEDLY GTSKKLSLSRNVLC+KC GKGSKSGASM CA CQGSGMK+S+R LG
Sbjct: 121 VVHPLKVSLEDLYNGTSKKLSLSRNVLCTKCKGKGSKSGASMNCASCQGSGMKVSIRQLG 180
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
MIQQMQHPCNECKGTGE ISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG
Sbjct: 181 PGMIQQMQHPCNECKGTGEMISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
EADEAPDTVTGDIVFVLQQKEHPKFKRKG+DLF EH+LSLTEALCGFQF LTHLD+RQLL
Sbjct: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGDDLFYEHSLSLTEALCGFQFVLTHLDNRQLL 300
Query: 301 IKSNPGEVVKPGWMSVRRQHFMMSTLKKRREEGSKLSKRHMMR 343
IKSNPGEV+KP K +EG + +R MR
Sbjct: 301 IKSNPGEVIKP------------DQFKGINDEGMPMYQRPFMR 331
>UniRef100_Q9SWB5 Seed maturation protein PM37 [Glycine max]
Length = 417
Score = 590 bits (1522), Expect = e-167
Identities = 288/312 (92%), Positives = 300/312 (95%), Gaps = 2/312 (0%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNT+YYEILGVSKNAS DDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGRAPKKSDNTRYYEILGVSKNASQDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGG-FPGGGSSRGRRQRRGE 119
LSDPEKRE+YDQYGEDALKEGMGGGGG HDPFDIFSSFFGGG F GGSSRGRRQRRGE
Sbjct: 61 LSDPEKREIYDQYGEDALKEGMGGGGG-HDPFDIFSSFFGGGSPFGSGGSSRGRRQRRGE 119
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVHPLKVSLEDLYLGTSKKLSLSRNV+CSKC+GKGSKSGASM CAGCQG+GMK+S+RHL
Sbjct: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVICSKCSGKGSKSGASMKCAGCQGTGMKVSIRHL 179
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G +MIQQMQH CNECKGTGETI+D+DRCPQCKGEKVVQ+KKVLEV VEKGMQNGQKITFP
Sbjct: 180 GPSMIQQMQHACNECKGTGETINDRDRCPQCKGEKVVQEKKVLEVIVEKGMQNGQKITFP 239
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
GEADEAPDT+TGDIVFVLQQKEHPKFKRK EDLFVEHTLSLTEALCGFQF LTHLDSRQL
Sbjct: 240 GEADEAPDTITGDIVFVLQQKEHPKFKRKAEDLFVEHTLSLTEALCGFQFVLTHLDSRQL 299
Query: 300 LIKSNPGEVVKP 311
LIKSNPGEVVKP
Sbjct: 300 LIKSNPGEVVKP 311
>UniRef100_Q9M7M2 DnaJ-like protein [Lycopersicon esculentum]
Length = 419
Score = 587 bits (1512), Expect = e-166
Identities = 283/313 (90%), Positives = 297/313 (94%), Gaps = 2/313 (0%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNTKYYEILGV K AS +DLKKAY+KAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGRAPKKSDNTKYYEILGVPKAASQEDLKKAYRKAAIKNHPDKGGDPEKFKELAQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFP--GGGSSRGRRQRRG 118
LSDPEKRE+YDQYGEDALKEGMGGGGGGH+PFDIF SFFGGGG P GGGSSR RRQRRG
Sbjct: 61 LSDPEKREIYDQYGEDALKEGMGGGGGGHEPFDIFQSFFGGGGNPFGGGGSSRVRRQRRG 120
Query: 119 EDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRH 178
EDV+HPLKVSLEDLY GTSKKLSLSRNVLCSKC GKGSKSGASM C+GCQGSGMK+S+R
Sbjct: 121 EDVIHPLKVSLEDLYNGTSKKLSLSRNVLCSKCKGKGSKSGASMKCSGCQGSGMKVSIRQ 180
Query: 179 LGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITF 238
LG +MIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQ+KKVLEVHVEKGMQNGQKITF
Sbjct: 181 LGPSMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQEKKVLEVHVEKGMQNGQKITF 240
Query: 239 PGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQ 298
PGEADEAPDT+TGDIVFVLQQKEHPKFKRKG+DLFVEHTLSL E+LCGFQF LTHLD+RQ
Sbjct: 241 PGEADEAPDTITGDIVFVLQQKEHPKFKRKGDDLFVEHTLSLDESLCGFQFVLTHLDNRQ 300
Query: 299 LLIKSNPGEVVKP 311
LLIKS PGEVVKP
Sbjct: 301 LLIKSQPGEVVKP 313
>UniRef100_Q9FEG6 J2P [Daucus carota]
Length = 418
Score = 582 bits (1499), Expect = e-165
Identities = 284/344 (82%), Positives = 304/344 (87%), Gaps = 13/344 (3%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNTKYYEILGV K ASPDDLKKAY+KAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGRAPKKSDNTKYYEILGVPKTASPDDLKKAYRKAAIKNHPDKGGDPEKFKELAQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGG-HDPFDIFSSFFGGGGFPGGGSSRGRRQRRGE 119
LSDPEKRE+YDQYGEDALKEGMGGGGGG HDPFDIF SFFGG F GGGSSRGRRQRRGE
Sbjct: 61 LSDPEKREIYDQYGEDALKEGMGGGGGGGHDPFDIFQSFFGGSPFGGGGSSRGRRQRRGE 120
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DV+HPLKVSLEDL GTSKKLSLSRNV+CSKC GKGSKSGASMTC GCQGSGMK+S+RHL
Sbjct: 121 DVIHPLKVSLEDLCNGTSKKLSLSRNVICSKCKGKGSKSGASMTCPGCQGSGMKVSIRHL 180
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G +MIQQMQHPCN+CKGTGETI+DKDRCPQCKG+KVVQ+KK +EV VEKGMQNGQKITFP
Sbjct: 181 GPSMIQQMQHPCNDCKGTGETINDKDRCPQCKGQKVVQEKKAIEVIVEKGMQNGQKITFP 240
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
GEADEAPDTVTGDIVFVLQQKEHPKFKRKG+DLFVEH+L+L+EALCGFQF LTHLD RQL
Sbjct: 241 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGDDLFVEHSLTLSEALCGFQFTLTHLDGRQL 300
Query: 300 LIKSNPGEVVKPGWMSVRRQHFMMSTLKKRREEGSKLSKRHMMR 343
LIKS PGEV+KP K +EG + +R MR
Sbjct: 301 LIKSQPGEVIKP------------DQFKGINDEGMPMYQRPFMR 332
>UniRef100_Q9ZWK3 DnaJ homolog [Salix gilgiana]
Length = 420
Score = 578 bits (1491), Expect = e-164
Identities = 286/345 (82%), Positives = 303/345 (86%), Gaps = 14/345 (4%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNTKYYE+LGVSK+AS DDLKKAY+KAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGRAPKKSDNTKYYEVLGVSKSASQDDLKKAYRKAAIKNHPDKGGDPEKFKELAQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGG-HDPFDIFSSFFGGGG-FPGGGSSRGRRQRRG 118
LSDPEKRE+YDQYGEDALKEGMG GG G HDPFDIF SFFGGG F GGGSSRGRRQRRG
Sbjct: 61 LSDPEKREIYDQYGEDALKEGMGSGGSGAHDPFDIFQSFFGGGNPFGGGGSSRGRRQRRG 120
Query: 119 EDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRH 178
EDV+HPLKVS EDLY GTSKKLSLSRNV+CSKC GKGSKSGAS CAGCQGSGMK+S+RH
Sbjct: 121 EDVIHPLKVSFEDLYNGTSKKLSLSRNVICSKCKGKGSKSGASSKCAGCQGSGMKVSIRH 180
Query: 179 LGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITF 238
LG +MIQQMQH CNECKGTGETI+DKDRCPQCKGEKVVQ+KKVLEV VEKGMQNGQK+TF
Sbjct: 181 LGPSMIQQMQHACNECKGTGETINDKDRCPQCKGEKVVQEKKVLEVVVEKGMQNGQKVTF 240
Query: 239 PGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQ 298
PGEADEAPDTVTGDIVFVLQQK+HPKFKRKG+DLFVEHTLSLTEALCGFQF LTHLD RQ
Sbjct: 241 PGEADEAPDTVTGDIVFVLQQKDHPKFKRKGDDLFVEHTLSLTEALCGFQFVLTHLDGRQ 300
Query: 299 LLIKSNPGEVVKPGWMSVRRQHFMMSTLKKRREEGSKLSKRHMMR 343
LLIKS PGEVVKP K +EG + +R MR
Sbjct: 301 LLIKSQPGEVVKP------------DQFKAINDEGMPMYQRPFMR 333
>UniRef100_P43644 DnaJ protein homolog ANJ1 [Atriplex nummularia]
Length = 417
Score = 577 bits (1486), Expect = e-163
Identities = 277/311 (89%), Positives = 294/311 (94%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSD+T+YYEILGV K+ASP+DLKKAYKKAAIKNHPDKGGDPEKFKELA AYEV
Sbjct: 1 MFGRAPKKSDSTRYYEILGVPKDASPEDLKKAYKKAAIKNHPDKGGDPEKFKELAHAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSDPEKRE+YDQYGEDALKEGMGGGGG HDPFDIF SFFGG F G GSSRGRRQRRGED
Sbjct: 61 LSDPEKREIYDQYGEDALKEGMGGGGGMHDPFDIFQSFFGGSPFGGVGSSRGRRQRRGED 120
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
VVHPLKVSLEDL+ GT+KKLSLSRNV+CSKC GKGSKSGASM C+GCQG+GMK+S+RHLG
Sbjct: 121 VVHPLKVSLEDLFTGTTKKLSLSRNVICSKCTGKGSKSGASMKCSGCQGTGMKVSIRHLG 180
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
+MIQQMQHPCNECKGTGETI+DKDRCPQCKGEKVVQ+KKVLEV VEKGMQ+GQKITFPG
Sbjct: 181 PSMIQQMQHPCNECKGTGETINDKDRCPQCKGEKVVQEKKVLEVVVEKGMQHGQKITFPG 240
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLF EHTLSLTEALCGF+F LTHLD RQLL
Sbjct: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFYEHTLSLTEALCGFRFVLTHLDGRQLL 300
Query: 301 IKSNPGEVVKP 311
IKSN GEVVKP
Sbjct: 301 IKSNLGEVVKP 311
>UniRef100_Q6F3B0 Putative DnaJ like protein [Oryza sativa]
Length = 417
Score = 574 bits (1480), Expect = e-162
Identities = 283/343 (82%), Positives = 302/343 (87%), Gaps = 14/343 (4%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNT+YYE+LGV K+AS DDLKKAY+KAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGRAPKKSDNTRYYEVLGVPKDASQDDLKKAYRKAAIKNHPDKGGDPEKFKELAQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSDPEKRE+YDQYGEDALKEGMG GGG HDPFDIFSSFFGGG GGGSSRGRRQRRGED
Sbjct: 61 LSDPEKREIYDQYGEDALKEGMGPGGGMHDPFDIFSSFFGGGF--GGGSSRGRRQRRGED 118
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
VVHPLKVSLE+LY GTSKKLSLSRNVLCSKCNGKGSKSGASM C+GCQGSGMK+ +R LG
Sbjct: 119 VVHPLKVSLEELYNGTSKKLSLSRNVLCSKCNGKGSKSGASMKCSGCQGSGMKVQIRQLG 178
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
MIQQMQHPCNECKGTGETISDKDRCP CKGEKV Q+KKVLEV VEKGMQNGQKITFPG
Sbjct: 179 PGMIQQMQHPCNECKGTGETISDKDRCPGCKGEKVAQEKKVLEVVVEKGMQNGQKITFPG 238
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
EADEAPDTVTGDI+FVLQQKEHPKFKRKG+DLF EHTL+LTEALCGFQF LTHLD+RQLL
Sbjct: 239 EADEAPDTVTGDIIFVLQQKEHPKFKRKGDDLFYEHTLNLTEALCGFQFVLTHLDNRQLL 298
Query: 301 IKSNPGEVVKPGWMSVRRQHFMMSTLKKRREEGSKLSKRHMMR 343
IKS PGEVVKP + K +EG + +R M+
Sbjct: 299 IKSKPGEVVKP------------DSFKAVNDEGMPMYQRPFMK 329
>UniRef100_Q04960 DnaJ protein homolog [Cucumis sativus]
Length = 413
Score = 574 bits (1480), Expect = e-162
Identities = 281/311 (90%), Positives = 292/311 (93%), Gaps = 2/311 (0%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGR PKKSDNTKYYEILGVSKNAS DDLKKAY+KAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGR-PKKSDNTKYYEILGVSKNASQDDLKKAYRKAAIKNHPDKGGDPEKFKELAQAYEV 59
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSDPEKRE+YDQYGEDALKEGMGGGGG HDPFDIF SFFGG F GGGSSRGRRQRRGED
Sbjct: 60 LSDPEKREIYDQYGEDALKEGMGGGGG-HDPFDIFQSFFGGSPFGGGGSSRGRRQRRGED 118
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
V+HPLKVSLEDLY GTSKKLSLSRNV+CSKC GKGSKSGASM C GCQGSGMK+S+RHLG
Sbjct: 119 VIHPLKVSLEDLYNGTSKKLSLSRNVICSKCKGKGSKSGASMKCPGCQGSGMKVSIRHLG 178
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
+MIQQMQHPCNECKGTGETI+DKDRC QCKGEKVVQ+KKVLEV VEKGMQN QKITFPG
Sbjct: 179 PSMIQQMQHPCNECKGTGETINDKDRCSQCKGEKVVQEKKVLEVIVEKGMQNAQKITFPG 238
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
EADEAPDTVTGDIVFVLQQKEHPKFKRKG+DLFVEHTLSL E+LCGFQF LTHLD RQLL
Sbjct: 239 EADEAPDTVTGDIVFVLQQKEHPKFKRKGDDLFVEHTLSLVESLCGFQFILTHLDGRQLL 298
Query: 301 IKSNPGEVVKP 311
IKS PGEVVKP
Sbjct: 299 IKSLPGEVVKP 309
>UniRef100_Q43177 DnaJ protein [Solanum tuberosum]
Length = 419
Score = 573 bits (1476), Expect = e-162
Identities = 284/344 (82%), Positives = 302/344 (87%), Gaps = 13/344 (3%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAP+KSDNTKYYEILGV K A+ +DLKKAY+KAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGRAPEKSDNTKYYEILGVPKTAAQEDLKKAYRKAAIKNHPDKGGDPEKFKELAQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGG-SSRGRRQRRGE 119
LSDPEKRE+YDQYGEDALKEGMGGGGGGHDPFDIFSSFFGG F GGG SSRGRRQRRGE
Sbjct: 61 LSDPEKREIYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGSPFGGGGGSSRGRRQRRGE 120
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVHPLKVSLEDLY GTSKKLSLSRNVLCSKC GKGSKSGASM C+GCQGSGMK+++R L
Sbjct: 121 DVVHPLKVSLEDLYNGTSKKLSLSRNVLCSKCKGKGSKSGASMKCSGCQGSGMKVTIRQL 180
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G +MIQQMQHPCNECKGTGE I+DKDRC QCKGEKVVQ+KKVLEV VEKGMQNGQKITFP
Sbjct: 181 GPSMIQQMQHPCNECKGTGEMINDKDRCGQCKGEKVVQEKKVLEVVVEKGMQNGQKITFP 240
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
GEADEAPDTVTGDIVFVLQQKEHPKFKRKG+DLFVEHTLSLTEALCGFQF LTHLD+RQL
Sbjct: 241 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGDDLFVEHTLSLTEALCGFQFILTHLDNRQL 300
Query: 300 LIKSNPGEVVKPGWMSVRRQHFMMSTLKKRREEGSKLSKRHMMR 343
+IK GEVVKP K +EG + +R MR
Sbjct: 301 IIKPQAGEVVKP------------DQFKAINDEGMPMYQRPFMR 332
>UniRef100_Q9M554 DnaJ protein [Euphorbia esula]
Length = 418
Score = 567 bits (1462), Expect = e-160
Identities = 278/344 (80%), Positives = 301/344 (86%), Gaps = 13/344 (3%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDN+KYYEILGVSK+AS DDLKKAY+KAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGRAPKKSDNSKYYEILGVSKSASQDDLKKAYRKAAIKNHPDKGGDPEKFKELAQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSDPEKR++YDQYGEDALKEGMGGGGGGHDPFDIF SFFGG F GGGSSRGRRQRRGED
Sbjct: 61 LSDPEKRDIYDQYGEDALKEGMGGGGGGHDPFDIFQSFFGGSPFGGGGSSRGRRQRRGED 120
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
V HPLKVSLEDLY GTSKKLSLSRNV+CSKC GKGSKSGASM C+GCQGSGMK+S+RHLG
Sbjct: 121 VTHPLKVSLEDLYNGTSKKLSLSRNVICSKCKGKGSKSGASMKCSGCQGSGMKVSIRHLG 180
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEK-VVQQKKVLEVHVEKGMQNGQKITFP 239
+MIQQMQHPCNECKGTGETI+DKDR P +G K + +++KVLEVHVEKGMQNGQKITFP
Sbjct: 181 PSMIQQMQHPCNECKGTGETINDKDRVPPVQGRKGLFKRRKVLEVHVEKGMQNGQKITFP 240
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
GEADEAPDTVTGDIVF+LQQ+EHPKFKR+G+DL VEHTLSLTEALCGFQF LTHLD RQL
Sbjct: 241 GEADEAPDTVTGDIVFILQQREHPKFKRRGDDLVVEHTLSLTEALCGFQFILTHLDGRQL 300
Query: 300 LIKSNPGEVVKPGWMSVRRQHFMMSTLKKRREEGSKLSKRHMMR 343
LIKS PGEVVKP K +EG + +R MR
Sbjct: 301 LIKSQPGEVVKP------------DQFKAINDEGMPMYQRPFMR 332
>UniRef100_O22663 DnaJ protein homolog atj3 [Arabidopsis thaliana]
Length = 420
Score = 566 bits (1458), Expect = e-160
Identities = 274/312 (87%), Positives = 291/312 (92%), Gaps = 2/312 (0%)
Query: 1 MFGRAP-KKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 59
MFGR P KKSDNTK+YEILGV K+ASP+DLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE
Sbjct: 1 MFGRGPSKKSDNTKFYEILGVPKSASPEDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 60
Query: 60 VLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGE 119
VLSDPEKRE+YDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGG F GG +SR RRQRRGE
Sbjct: 61 VLSDPEKREIYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGPF-GGNTSRQRRQRRGE 119
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVHPLKVSLED+YLGT KKLSLSRN LCSKCNGKGSKSGAS+ C GCQGSGMK+S+R L
Sbjct: 120 DVVHPLKVSLEDVYLGTMKKLSLSRNALCSKCNGKGSKSGASLKCGGCQGSGMKVSIRQL 179
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G MIQQMQH CNECKGTGETI+D+DRCPQCKG+KV+ +KKVLEV+VEKGMQ+ QKITF
Sbjct: 180 GPGMIQQMQHACNECKGTGETINDRDRCPQCKGDKVIPEKKVLEVNVEKGMQHSQKITFE 239
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
G+ADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQF LTHLD R L
Sbjct: 240 GQADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFVLTHLDGRSL 299
Query: 300 LIKSNPGEVVKP 311
LIKSNPGEVVKP
Sbjct: 300 LIKSNPGEVVKP 311
>UniRef100_Q94AW8 AT3g44110/F26G5_60 [Arabidopsis thaliana]
Length = 420
Score = 564 bits (1454), Expect = e-159
Identities = 273/312 (87%), Positives = 291/312 (92%), Gaps = 2/312 (0%)
Query: 1 MFGRAP-KKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 59
MFGR P KKSDNTK+YEILGV K+ASP+DLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE
Sbjct: 1 MFGRGPSKKSDNTKFYEILGVPKSASPEDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 60
Query: 60 VLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGE 119
VLSDPEKRE+YDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGG F GG +SR RRQRRGE
Sbjct: 61 VLSDPEKREIYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGPF-GGNTSRQRRQRRGE 119
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVHPLKVSLED+YLGT KKLSLSRN LCSKCNGKGSKSGAS+ C GCQGSGMK+S+R L
Sbjct: 120 DVVHPLKVSLEDVYLGTMKKLSLSRNALCSKCNGKGSKSGASLKCGGCQGSGMKVSIRQL 179
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G MIQQMQH CNECKGTGETI+D+DRCPQCKG+KV+ +KKVLEV+V+KGMQ+ QKITF
Sbjct: 180 GPGMIQQMQHACNECKGTGETINDRDRCPQCKGDKVIPEKKVLEVNVKKGMQHSQKITFE 239
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
G+ADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQF LTHLD R L
Sbjct: 240 GQADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFVLTHLDGRSL 299
Query: 300 LIKSNPGEVVKP 311
LIKSNPGEVVKP
Sbjct: 300 LIKSNPGEVVKP 311
>UniRef100_Q42530 DnaJ homolog [Arabidopsis thaliana]
Length = 420
Score = 564 bits (1454), Expect = e-159
Identities = 273/312 (87%), Positives = 291/312 (92%), Gaps = 2/312 (0%)
Query: 1 MFGRAP-KKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 59
MFGR P KKSDNTK+YEILGV K+ASP+DLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE
Sbjct: 1 MFGRGPSKKSDNTKFYEILGVPKSASPEDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 60
Query: 60 VLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGE 119
VLSDPEKRE+YDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGG F GG +SR RRQRRGE
Sbjct: 61 VLSDPEKREIYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGPF-GGNTSRQRRQRRGE 119
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVHPLKVSLED+YLGT KKLSLSRN LCSKCNGKGSKSGAS+ C GCQGSGMK+S+R L
Sbjct: 120 DVVHPLKVSLEDVYLGTMKKLSLSRNALCSKCNGKGSKSGASLKCGGCQGSGMKVSIRQL 179
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G MIQQMQH CNECKGTGETI+D+DRCPQCKG+KV+ +KKVLEV+VEKGMQ+ QKITF
Sbjct: 180 GPGMIQQMQHACNECKGTGETINDRDRCPQCKGDKVIPEKKVLEVNVEKGMQHSQKITFE 239
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
G+ADEAPDTVTGDIVFVLQQKEHP+FKRKGEDLFVEHTLSLTEALCGFQF LTHLD R L
Sbjct: 240 GQADEAPDTVTGDIVFVLQQKEHPQFKRKGEDLFVEHTLSLTEALCGFQFVLTHLDGRSL 299
Query: 300 LIKSNPGEVVKP 311
LIKSNPGEVVKP
Sbjct: 300 LIKSNPGEVVKP 311
>UniRef100_Q9ZSZ6 DnaJ protein [Hevea brasiliensis]
Length = 415
Score = 562 bits (1449), Expect = e-159
Identities = 279/343 (81%), Positives = 297/343 (86%), Gaps = 14/343 (4%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNTKYYEILGVSKNAS DDLKKAY+KAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGRAPKKSDNTKYYEILGVSKNASQDDLKKAYRKAAIKNHPDKGGDPEKFKELAQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSDPEKRE+YDQYGEDALKEGMG GGG HDPFDIF SFFGG F GGGSSRGRR + GED
Sbjct: 61 LSDPEKREIYDQYGEDALKEGMGSGGGAHDPFDIFQSFFGGNPFGGGGSSRGRR-KEGED 119
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
V+HPLKVSLEDLY GTSKKLSLSRNV+CSKC GKGSKSGASM C+GCQGSGMK+S+R LG
Sbjct: 120 VIHPLKVSLEDLYNGTSKKLSLSRNVICSKCKGKGSKSGASMKCSGCQGSGMKVSIRQLG 179
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
+MIQQMQHPCNECKGTGETI+DKDRCPQCKGEKVVQ+KKVLEV VEKGMQNGQ+ITFPG
Sbjct: 180 PSMIQQMQHPCNECKGTGETINDKDRCPQCKGEKVVQEKKVLEVIVEKGMQNGQRITFPG 239
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
EADEAPDT+TGDIVFVLQQKEHPKFKRKG+DL V+HTLSLTEALC QF LTHLD LL
Sbjct: 240 EADEAPDTITGDIVFVLQQKEHPKFKRKGDDLIVDHTLSLTEALCASQFILTHLDG-DLL 298
Query: 301 IKSNPGEVVKPGWMSVRRQHFMMSTLKKRREEGSKLSKRHMMR 343
IKS PGEVVKP K +EG + +R MR
Sbjct: 299 IKSQPGEVVKP------------DQFKAINDEGMPMYQRPFMR 329
>UniRef100_P42825 DnaJ protein homolog ATJ2 [Arabidopsis thaliana]
Length = 419
Score = 562 bits (1448), Expect = e-159
Identities = 269/312 (86%), Positives = 288/312 (92%), Gaps = 1/312 (0%)
Query: 1 MFGRAP-KKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 59
MFGR P +KSDNTK+YEILGV K A+P+DLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE
Sbjct: 1 MFGRGPSRKSDNTKFYEILGVPKTAAPEDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 60
Query: 60 VLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGE 119
VLSDPEKRE+YDQYGEDALKEGMGGGGGGHDPFDIFSSFFG GG P G SRGRRQRRGE
Sbjct: 61 VLSDPEKREIYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGSGGHPFGSHSRGRRQRRGE 120
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVHPLKVSLED+YLGT+KKLSLSR LCSKCNGKGSKSGASM C GCQGSGMKIS+R
Sbjct: 121 DVVHPLKVSLEDVYLGTTKKLSLSRKALCSKCNGKGSKSGASMKCGGCQGSGMKISIRQF 180
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G M+QQ+QH CN+CKGTGETI+D+DRCPQCKGEKVV +KKVLEV+VEKGMQ+ QKITF
Sbjct: 181 GPGMMQQVQHACNDCKGTGETINDRDRCPQCKGEKVVSEKKVLEVNVEKGMQHNQKITFS 240
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
G+ADEAPDTVTGDIVFV+QQKEHPKFKRKGEDLFVEHT+SLTEALCGFQF LTHLD RQL
Sbjct: 241 GQADEAPDTVTGDIVFVIQQKEHPKFKRKGEDLFVEHTISLTEALCGFQFVLTHLDKRQL 300
Query: 300 LIKSNPGEVVKP 311
LIKS PGEVVKP
Sbjct: 301 LIKSKPGEVVKP 312
>UniRef100_Q84PD0 DnaJ protein, putative [Oryza sativa]
Length = 417
Score = 561 bits (1446), Expect = e-158
Identities = 273/311 (87%), Positives = 289/311 (92%), Gaps = 1/311 (0%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNTKYYEILGV K AS DDLKKAY+KAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGRAPKKSDNTKYYEILGVPKTASQDDLKKAYRKAAIKNHPDKGGDPEKFKELAQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSDPEKRE+YDQYGEDALKEGMGGGG DPFDIFSSFFG F GGGSSRGRRQRRGED
Sbjct: 61 LSDPEKREIYDQYGEDALKEGMGGGGSHVDPFDIFSSFFGPS-FGGGGSSRGRRQRRGED 119
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
V+HPLKVSLEDLY GTSKKLSLSRNVLC+KC GKGSKSGASM C GCQGSGMKI++R LG
Sbjct: 120 VIHPLKVSLEDLYNGTSKKLSLSRNVLCAKCKGKGSKSGASMRCPGCQGSGMKITIRQLG 179
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
+MIQQMQ PCNECKGTGE+I++KDRCP CKGEKV+Q+KKVLEVHVEKGMQ+ QKITFPG
Sbjct: 180 PSMIQQMQQPCNECKGTGESINEKDRCPGCKGEKVIQEKKVLEVHVEKGMQHNQKITFPG 239
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
EADEAPDTVTGDIVFVLQQK+H KFKRKG+DLF EHTLSLTEALCGFQF LTHLD+RQLL
Sbjct: 240 EADEAPDTVTGDIVFVLQQKDHSKFKRKGDDLFYEHTLSLTEALCGFQFVLTHLDNRQLL 299
Query: 301 IKSNPGEVVKP 311
IKSNPGEVVKP
Sbjct: 300 IKSNPGEVVKP 310
>UniRef100_O65160 DnaJ-related protein ZMDJ1 [Zea mays]
Length = 419
Score = 561 bits (1446), Expect = e-158
Identities = 272/311 (87%), Positives = 289/311 (92%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNTKYYEILGV K+AS DDLKKAY+KAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGRAPKKSDNTKYYEILGVPKSASQDDLKKAYRKAAIKNHPDKGGDPEKFKELAQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSDPEKRE+YDQYGEDALKEGMGGGG DPFDIFSSFFG GGGSSRGRRQRRGED
Sbjct: 61 LSDPEKREIYDQYGEDALKEGMGGGGSHVDPFDIFSSFFGPSFGGGGGSSRGRRQRRGED 120
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
VVHPLKVSLEDLY GTSKKLSLSRNV+CSKC GKGSKSGASM C GCQGSGMK+++R LG
Sbjct: 121 VVHPLKVSLEDLYNGTSKKLSLSRNVICSKCKGKGSKSGASMRCPGCQGSGMKVTIRQLG 180
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
+MIQQMQ PCNECKGTGE+I++KDRCP CKGEKV+Q+KKVLEVHVEKGMQ+ QKITFPG
Sbjct: 181 PSMIQQMQQPCNECKGTGESINEKDRCPGCKGEKVIQEKKVLEVHVEKGMQHNQKITFPG 240
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
EADEAPDTVTGDIVFVLQQK+H KFKRKGEDLF EHTLSLTEALCGFQF LTHLD+RQLL
Sbjct: 241 EADEAPDTVTGDIVFVLQQKDHSKFKRKGEDLFYEHTLSLTEALCGFQFVLTHLDNRQLL 300
Query: 301 IKSNPGEVVKP 311
IKS+PGEVVKP
Sbjct: 301 IKSDPGEVVKP 311
>UniRef100_Q9FSF2 Putative DNAJ protein [Nicotiana tabacum]
Length = 418
Score = 555 bits (1429), Expect = e-156
Identities = 270/343 (78%), Positives = 297/343 (85%), Gaps = 12/343 (3%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGR PKKSDNT+YYEILGVSKNAS D++KKAY+KAA+KNHPDKGGDPEKFKELAQAYEV
Sbjct: 1 MFGRGPKKSDNTRYYEILGVSKNASDDEIKKAYRKAAMKNHPDKGGDPEKFKELAQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSD +KRE+YDQYGEDALKEGMGGGGG HDPFDIF SFFGG F GGGSSRGRRQRRGED
Sbjct: 61 LSDSQKREIYDQYGEDALKEGMGGGGGMHDPFDIFESFFGGNPFGGGGSSRGRRQRRGED 120
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
VVHPLKVSLEDLY G +KKLSLSRNV+CSKC+GKGSKSGASM C+GC+GSGMK+S+R LG
Sbjct: 121 VVHPLKVSLEDLYSGITKKLSLSRNVICSKCSGKGSKSGASMKCSGCKGSGMKVSIRQLG 180
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
+MIQQMQH CNECKGTGETI DKDRCP+CKGEKVVQ+KKVLEVHVEKGMQNGQKITFPG
Sbjct: 181 PSMIQQMQHACNECKGTGETIDDKDRCPRCKGEKVVQEKKVLEVHVEKGMQNGQKITFPG 240
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
+ADE PD +TGDIVFVLQQK+ KRKG+DLFV+HTLSLTEALCGFQF +THLD RQLL
Sbjct: 241 KADETPDAITGDIVFVLQQKDTRGSKRKGDDLFVDHTLSLTEALCGFQFIMTHLDGRQLL 300
Query: 301 IKSNPGEVVKPGWMSVRRQHFMMSTLKKRREEGSKLSKRHMMR 343
IKSN GEVVKP K +EG+ + +R MR
Sbjct: 301 IKSNLGEVVKP------------DQFKAINDEGTPMYQRPFMR 331
>UniRef100_Q03363 DnaJ protein homolog 1 [Allium porrum]
Length = 397
Score = 552 bits (1422), Expect = e-156
Identities = 268/322 (83%), Positives = 285/322 (88%), Gaps = 12/322 (3%)
Query: 22 KNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEVLSDPEKREVYDQYGEDALKEG 81
KNASPDDLKKAY+KAAIKNHPDKGGDPEKFKELAQAY+VLSDPEKRE+YDQYGEDALKEG
Sbjct: 1 KNASPDDLKKAYRKAAIKNHPDKGGDPEKFKELAQAYDVLSDPEKREIYDQYGEDALKEG 60
Query: 82 MGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHPLKVSLEDLYLGTSKKLS 141
MGGGGG HDPFDIF SFFGGGGF GGGSSRGRRQRRGEDVVHPLKVSLE+LY GTSKKLS
Sbjct: 61 MGGGGGDHDPFDIFQSFFGGGGFGGGGSSRGRRQRRGEDVVHPLKVSLEELYNGTSKKLS 120
Query: 142 LSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLGANMIQQMQHPCNECKGTGETI 201
LSRNV+CSKCNGKGSKSGASM CA CQGSGMK+S+R LG MIQQMQHPCN+CKGTGE I
Sbjct: 121 LSRNVICSKCNGKGSKSGASMRCASCQGSGMKVSIRQLGPGMIQQMQHPCNDCKGTGEMI 180
Query: 202 SDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDTVTGDIVFVLQQKE 261
+DKDRCP CKGEKVVQ+KKVLEVHVEKGMQNGQ+ITFPGEADEAPDTVTGDIVFVLQQKE
Sbjct: 181 NDKDRCPLCKGEKVVQEKKVLEVHVEKGMQNGQRITFPGEADEAPDTVTGDIVFVLQQKE 240
Query: 262 HPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLIKSNPGEVVKPGWMSVRRQHF 321
HPKF+RKG+DLF +HTLSLTEALCGFQF LTHLD RQLLIKSNPGEVVKP
Sbjct: 241 HPKFQRKGDDLFYKHTLSLTEALCGFQFVLTHLDGRQLLIKSNPGEVVKP---------- 290
Query: 322 MMSTLKKRREEGSKLSKRHMMR 343
K +EG + +R MR
Sbjct: 291 --DQFKAINDEGMPMYQRPFMR 310
>UniRef100_Q9S7X7 DnaJ homolog protein [Salix gilgiana]
Length = 423
Score = 528 bits (1359), Expect = e-148
Identities = 248/314 (78%), Positives = 283/314 (89%), Gaps = 2/314 (0%)
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAP++SDNTKYYE+L VSK AS D+LKKAYKKAAIKNHPDKGGDPEKFKEL+QAYEV
Sbjct: 1 MFGRAPRRSDNTKYYEVLAVSKGASQDELKKAYKKAAIKNHPDKGGDPEKFKELSQAYEV 60
Query: 61 LSDPEKREVYDQYGEDALKEGMG--GGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRG 118
LSDP+KRE+YDQYGEDALKEGMG GGGGGH+PFDIF SFFGGGGF GG SSRGRRQ++G
Sbjct: 61 LSDPDKREIYDQYGEDALKEGMGPGGGGGGHNPFDIFESFFGGGGFGGGSSSRGRRQKQG 120
Query: 119 EDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRH 178
EDV HPLKVSLEDLY GTSKKLSLSRN+LC+KC GKGSKSGA C GCQG+GMK+S+R
Sbjct: 121 EDVAHPLKVSLEDLYNGTSKKLSLSRNILCAKCKGKGSKSGAFGKCRGCQGTGMKVSIRQ 180
Query: 179 LGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITF 238
+G M+QQMQH C EC+G+GE IS+KD+CP C+G KV Q+K+VLEVHVE+GMQ+GQKI F
Sbjct: 181 IGLGMMQQMQHVCPECRGSGELISEKDKCPHCRGNKVTQEKRVLEVHVERGMQHGQKIVF 240
Query: 239 PGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQ 298
G+ADEAPDT+TGD+VFVLQ K+H KF+RK +DLFVEH+LSLTEALCG+QFALTHLD RQ
Sbjct: 241 EGQADEAPDTITGDVVFVLQLKKHSKFERKMDDLFVEHSLSLTEALCGYQFALTHLDGRQ 300
Query: 299 LLIKSNPGEVVKPG 312
LLIKSNP E+VKPG
Sbjct: 301 LLIKSNPYEIVKPG 314
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 677,650,323
Number of Sequences: 2790947
Number of extensions: 31365314
Number of successful extensions: 136977
Number of sequences better than 10.0: 3602
Number of HSP's better than 10.0 without gapping: 2806
Number of HSP's successfully gapped in prelim test: 833
Number of HSP's that attempted gapping in prelim test: 123357
Number of HSP's gapped (non-prelim): 5486
length of query: 375
length of database: 848,049,833
effective HSP length: 129
effective length of query: 246
effective length of database: 488,017,670
effective search space: 120052346820
effective search space used: 120052346820
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC133862.8


