

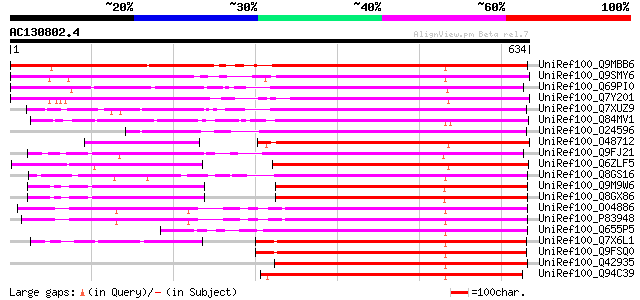
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130802.4 + phase: 0
(634 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9MBB6 Pectin methylesterase [Salix gilgiana] 598 e-169
UniRef100_Q9SMY6 Pectinesterase-like protein [Arabidopsis thaliana] 494 e-138
UniRef100_Q69PI0 Putative pectinesterase [Oryza sativa] 484 e-135
UniRef100_Q7Y201 Putative pectin methylesterase [Arabidopsis tha... 466 e-130
UniRef100_Q7XUZ9 OSJNBa0036B21.20 protein [Oryza sativa] 413 e-114
UniRef100_Q84MV1 Putative pectin methylesterase [Oryza sativa] 395 e-108
UniRef100_O24596 Pectin methylesterase-like protein [Zea mays] 392 e-107
UniRef100_O48712 Putative pectinesterase [Arabidopsis thaliana] 391 e-107
UniRef100_Q9FJ21 Pectin methylesterase [Arabidopsis thaliana] 389 e-106
UniRef100_Q6ZLF5 Putative pectinesterase [Oryza sativa] 385 e-105
UniRef100_Q8GS16 Putative thermostable pectinesterase [Citrus si... 351 4e-95
UniRef100_Q9M9W6 Putative pectinesterase [Arabidopsis thaliana] 348 2e-94
UniRef100_Q8GX86 Putative pectinesterase [Arabidopsis thaliana] 345 3e-93
UniRef100_O04886 Pectinesterase 1 precursor [Citrus sinensis] 337 6e-91
UniRef100_P83948 Pectinesterase 3 precursor [Citrus sinensis] 335 3e-90
UniRef100_Q655P5 Putative pectinesterase [Oryza sativa] 330 6e-89
UniRef100_Q7X6L1 OSJNBb0079B02.7 protein [Oryza sativa] 327 5e-88
UniRef100_Q9FSQ0 Putative pectin methylesterase [Oryza sativa] 327 5e-88
UniRef100_Q42935 Pectin methylesterase precursor [Nicotiana plum... 327 9e-88
UniRef100_Q94C39 At1g11580/T23J18_33 [Arabidopsis thaliana] 326 1e-87
>UniRef100_Q9MBB6 Pectin methylesterase [Salix gilgiana]
Length = 596
Score = 598 bits (1543), Expect = e-169
Identities = 317/641 (49%), Positives = 413/641 (63%), Gaps = 54/641 (8%)
Query: 1 MAFEDFDLVSERRKANAKQHLKKKILIGVTSVVLIACVIAAVTFVIVKR---SGPDHNNN 57
M F+DFD +SERR+ +Q L+KKI+I S + ++ A F +V S P +N
Sbjct: 1 MVFQDFDQLSERRRLERQQKLRKKIIIASVSSIAFFVIVGAGVFSLVSNHDISSPGNNGG 60
Query: 58 DKKPVQNAPPEPERVDKYSRLVTMLCSHSEYKEKCVTTL-KEALKKDPKLKEPKGLLMVF 116
+ + + +R++ +C+ + Y++ C TL K L KDP +PK LL +
Sbjct: 61 SPSAATRPAEKAKPISHVARVIKTVCNATTYQDTCQNTLEKGVLGKDPSSVQPKDLLKIA 120
Query: 117 MLVAKNEINNAFNKTANLKFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLAS 176
+ A EI+ K ++ KF EK A++DC +L EDAKEE+ S+ +G DI KLAS
Sbjct: 121 IKAADEEIDKVIKKASSFKFDKPREKAAFDDCLELIEDAKEELKNSVDCIGN-DIGKLAS 179
Query: 177 KEAELNNWLSAVISYQDTCSDGFPEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVN-- 234
+L+NWLSAV+SYQ TC DGFPEG+LK ME F +R+L SNSLA+VS + +
Sbjct: 180 NAPDLSNWLSAVMSYQQTCIDGFPEGKLKSDMEKTFKATRELTSNSLAMVSSLVSFLKNF 239
Query: 235 AFQGGLSGFKLPWGKSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPG 294
+F G L+ L ++ +P D D V P D+ I +
Sbjct: 240 SFSGTLNRRLLAEEQN-----SPSLDKDGV----------PGWMSHEDRRILK------- 277
Query: 295 AAPIGAPRVDAPPSWAAPAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGR 354
GA + +KP PNV+VAKDGSGDFKTISEALAA+P Y+GR
Sbjct: 278 ----GADK------------------DKPKPNVSVAKDGSGDFKTISEALAAMPAKYEGR 315
Query: 355 YVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGF 414
YV++VK+GVYDETVTVTKKM N+TMYGDG K+I+TGNKNF DGV+TF+TA+F VLGDGF
Sbjct: 316 YVIFVKQGVYDETVTVTKKMANITMYGDGSQKTIVTGNKNFADGVQTFRTATFAVLGDGF 375
Query: 415 VGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTI 474
+ + MGFRNTAG K QAVA RVQAD +IF+NC FEGYQDTLYAQTHRQFYR CVI+GT+
Sbjct: 376 LCKFMGFRNTAGPEKHQAVAIRVQADRAIFLNCRFEGYQDTLYAQTHRQFYRSCVITGTV 435
Query: 475 DFIFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---P 531
DFIFG A++VFQNC + +RKPL+NQ+NI+TA GRID T VLQ C I+ + DL
Sbjct: 436 DFIFGDATSVFQNCLITVRKPLENQQNIVTAQGRIDGHETTGIVLQSCRIEPDKDLVPVK 495
Query: 532 STTKNYIGRPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTD 591
+ ++Y+GRPWKE+SRT+IM+S I I P GWLPW+GDF LKTLYY EY N G GA+T+
Sbjct: 496 NKIRSYLGRPWKEFSRTVIMDSTIGDFIHPGGWLPWQGDFGLKTLYYAEYSNKGGGAQTN 555
Query: 592 ARVKWIGRKDIKRGEALTYTVEPFLDGSWINGTGVPAHLGL 632
AR+KW G IK+ EA+ +T+E F G WI+ +G P HLGL
Sbjct: 556 ARIKWPGYHIIKKEEAMKFTIENFYQGDWISASGSPVHLGL 596
>UniRef100_Q9SMY6 Pectinesterase-like protein [Arabidopsis thaliana]
Length = 609
Score = 494 bits (1271), Expect = e-138
Identities = 271/661 (40%), Positives = 375/661 (55%), Gaps = 80/661 (12%)
Query: 1 MAFEDFDLVSERRKANAKQHLKKKILIGVTSVVLIACVIAAVTFVIV------KRSGPDH 54
MAF+DFD + ER A K+ +K+I++GV SV+++A I F V + G
Sbjct: 1 MAFQDFDKIQERVNAERKRKFRKRIILGVVSVLVVAAAIIGGAFAYVTYENKTQEQGKTT 60
Query: 55 NNNDK-KPVQNAPPEPE---------RVDKYSRLVTMLCSHSEYKEKCVTTLKEALKKDP 104
NN K P ++ P P+ + + +++ LC+ + YK C TLK KKD
Sbjct: 61 NNKSKDSPTKSESPSPKPPSSAAQTVKAGQVDKIIQTLCNSTLYKPTCQNTLKNETKKDT 120
Query: 105 KLKEPKGLLMVFMLVAKNEINNAFNKTANLKFASKEEKGAYEDCKQLFEDAKEEMGFSIT 164
+P+ LL ++ ++++ F + +LK +K++K A CK L ++AKEE+G S+
Sbjct: 121 PQTDPRSLLKSAIVAVNDDLDQVFKRVLSLKTENKDDKDAIAQCKLLVDEAKEELGTSMK 180
Query: 165 EVGQLDISKLASKEAELNNWLSAVISYQDTCSDGFPEGELKKKMEMIFAESRQLLSNSLA 224
+ +++ A +L++WLSAV+SYQ+TC DGF EG+LK ++ F S+ L SNSLA
Sbjct: 181 RINDSEVNNFAKIVPDLDSWLSAVMSYQETCVDGFEEGKLKTEIRKNFNSSQVLTSNSLA 240
Query: 225 VVSQVSQIVNAFQGGLSGFKLPWGKSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQP 284
+ + + G LS P T + +
Sbjct: 241 M-------IKSLDGYLSS-------------VPKVKTRLLLE------------------ 262
Query: 285 IFEAPIGAPGAAPIGAPRVDAPPSWAA-------PAVDLPGSTEKPTPNVTVAKDGSGDF 337
A A D SW + AVD+ PN TVAKDGSG+F
Sbjct: 263 -----------ARSSAKETDHITSWLSNKERRMLKAVDVKALK----PNATVAKDGSGNF 307
Query: 338 KTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVD 397
TI+ AL A+P Y+GRY +Y+K G+YDE+V + KK N+TM GDG K+I+TGNK+
Sbjct: 308 TTINAALKAMPAKYQGRYTIYIKHGIYDESVIIDKKKPNVTMVGDGSQKTIVTGNKSHAK 367
Query: 398 GVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLY 457
+RTF TA+FV G+GF+ + MGFRNTAG QAVA RVQ+D S+F+NC FEGYQDTLY
Sbjct: 368 KIRTFLTATFVAQGEGFMAQSMGFRNTAGPEGHQAVAIRVQSDRSVFLNCRFEGYQDTLY 427
Query: 458 AQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAF 517
A THRQ+YR CVI GT+DFIFG A+A+FQNC + +RK L QKN +TA GR+D T F
Sbjct: 428 AYTHRQYYRSCVIIGTVDFIFGDAAAIFQNCDIFIRKGLPGQKNTVTAQGRVDKFQTTGF 487
Query: 518 VLQKCVIKGEDDL---PSTTKNYIGRPWKEYSRTIIMESDIPALIQPEGWLPW-EGDFAL 573
V+ C + +DL + K+Y+GRPWK +SRT++MES I +I P GWL W E DFA+
Sbjct: 488 VIHNCTVAPNEDLKPVKAQFKSYLGRPWKPHSRTVVMESTIEDVIDPVGWLRWQETDFAI 547
Query: 574 KTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDGSWINGTGVPAHLGLY 633
TL Y EY N G T ARVKW G + + + EA+ +TV PFL G WI G P LGLY
Sbjct: 548 DTLSYAEYKNDGPSGATAARVKWPGFRVLNKEEAMKFTVGPFLQGEWIQAIGSPVKLGLY 607
Query: 634 N 634
+
Sbjct: 608 D 608
>UniRef100_Q69PI0 Putative pectinesterase [Oryza sativa]
Length = 617
Score = 484 bits (1247), Expect = e-135
Identities = 270/648 (41%), Positives = 381/648 (58%), Gaps = 63/648 (9%)
Query: 2 AFEDFDLVSERRKANAKQHLKKKILI--GVTSVVLIACVIAAVTFVIVKRSGPDHNNNDK 59
AF DF ++ERR+A + +++I+I G S+++I V+ A + + K
Sbjct: 4 AFGDFGPLTERRRAEKARQQRRRIMIALGTVSIIIILIVMGAAAITYSGKKSEEDEGGSK 63
Query: 60 KPVQNAPPEPERVDK--------------YSRLVTMLCSHSEYKEKCVTTLKEALKKDPK 105
+ D S+ + M+C+ +++ + C T++ +A +
Sbjct: 64 GSSKGKSKGGGGGDDEDGGGGGGKADLRAVSKSIKMMCAQTDFADSCATSIGKAA--NAS 121
Query: 106 LKEPKGLLMVFMLVAKNEINNAFNKTANLKFASKEEKGAYEDCKQLFEDAKEEMGFSITE 165
+ PK ++ + V ++ AF++ + K A DCK+LF+DAK+++ ++
Sbjct: 122 VSSPKDIIRTAVDVIGGAVDQAFDRADLIMSNDPRVKAAVADCKELFDDAKDDLNCTLKG 181
Query: 166 VGQLDISKLASKEAELNNWLSAVISYQDTCSDGFPEGELKKKMEMIFAESRQLLSNSLAV 225
+ D K + +L WLSAVI+ +TC DGFP+GE + K++ F R+ SN+LA+
Sbjct: 182 IDGKDGLK---QGFQLRVWLSAVIANMETCIDGFPDGEFRDKVKESFNNGREFTSNALAL 238
Query: 226 VSQVSQIVNAFQGGLSGFKLPWGKSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQPI 285
+ + S ++A +G S +L G+ D A D A+A+D P+ PD D+ +
Sbjct: 239 IEKASSFLSALKG--SQRRLLAGEEDNGGGAADPHL-ALAEDG-----IPEWVPDGDRRV 290
Query: 286 FEAPIGAPGAAPIGAPRVDAPPSWAAPAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALA 345
+ G TPNV VAKDGSG FKTI+EALA
Sbjct: 291 LKGG----------------------------GFKNNLTPNVIVAKDGSGKFKTINEALA 322
Query: 346 AIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTA 405
A+P+TY GRYV+YVKEGVY E VT+TKKM ++TMYGDG KSI+TG+KNF DG+ TF+TA
Sbjct: 323 AMPKTYSGRYVIYVKEGVYAEYVTITKKMASVTMYGDGSRKSIVTGSKNFADGLTTFKTA 382
Query: 406 SFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFY 465
+F GDGF+ MGF+NTAGA K QAVA VQ+D S+F+NC +G+QDTLYA + QFY
Sbjct: 383 TFAAQGDGFMAIGMGFQNTAGAAKHQAVALLVQSDKSVFLNCWMDGFQDTLYAHSKAQFY 442
Query: 466 RDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIK 525
R+CVI+GTIDF+FG A+AVFQNC L LR+P+DNQ+NI TA GR D + T FVLQKC
Sbjct: 443 RNCVITGTIDFVFGDAAAVFQNCVLTLRRPMDNQQNIATAQGRADGREATGFVLQKCEFN 502
Query: 526 GEDDLPST----TKNYIGRPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEY 581
E L +NY+GRPW+E+SRT+IMESDIPA+I G++PW G+FALKTLYY EY
Sbjct: 503 AEPALTDAKLPPIRNYLGRPWREFSRTVIMESDIPAIIDKAGYMPWNGEFALKTLYYAEY 562
Query: 582 DNVGAGAKTDARVKWIG-RKDIKRGEALTYTVEPFLDGS-WINGTGVP 627
N G GA T RV W G +K I + +A +TV+ FL WI+ TG P
Sbjct: 563 ANKGPGADTAGRVAWPGYKKVISKADATKFTVDNFLHAKPWIDPTGTP 610
>UniRef100_Q7Y201 Putative pectin methylesterase [Arabidopsis thaliana]
Length = 614
Score = 466 bits (1200), Expect = e-130
Identities = 273/668 (40%), Positives = 369/668 (54%), Gaps = 89/668 (13%)
Query: 1 MAFEDFDLVSERRKANAKQHLKKKILIGVTSVVLIACVIAAVTFVIV------------- 47
MAF+DFD + ER AN K+ +K+I++G S++++ I F V
Sbjct: 1 MAFQDFDKIQERVNANRKRKFRKRIIVGTVSLLVVVAAIVGGAFAYVAYEKRNEQQQQQQ 60
Query: 48 -----KRSGPDHN---NNDKK------PVQNAP---PEPERVDKYSRLVTMLCSHSEYKE 90
+SG +N ++DKK P Q AP + + + +++ LCS + Y +
Sbjct: 61 QAKNHNKSGSGNNVVKDSDKKSPSPPTPSQKAPVSAAQSVKPGQGDKIIQTLCSSTLYMQ 120
Query: 91 KCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKFASKEEKGAYEDCKQ 150
C TLK K L P L + +++ K +LK ++++K A E CK
Sbjct: 121 ICEKTLKNRTDKGFALDNPTTFLKSAIEAVNEDLDLVLEKVLSLKTENQDDKDAIEQCKL 180
Query: 151 LFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDGFPEGELKKKMEM 210
L EDAKEE S+ ++ +++ +L +WLSAV+SYQ+TC DGF EG LK +++
Sbjct: 181 LVEDAKEETVASLNKINVTEVNSFEKVVPDLESWLSAVMSYQETCLDGFEEGNLKSEVKT 240
Query: 211 IFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAHAPAPDADTDAVADDDED 270
S+ L SNSLA++ ++ ++ + L D V++DD
Sbjct: 241 SVNSSQVLTSNSLALIKTFTENLSPVMKVVERHLL------------DGIPSWVSNDDRR 288
Query: 271 LADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAPAVDLPGSTEKPTPNVTVA 330
+ A D V A A A D GS + T N
Sbjct: 289 MLRAVD--------------------------VKALKPNATVAKD--GSGDFTTIN---- 316
Query: 331 KDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIIT 390
+AL A+P+ Y+GRY++YVK+G+YDE VTV KK NLTM GDG K+I+T
Sbjct: 317 -----------DALRAMPEKYEGRYIIYVKQGIYDEYVTVDKKKANLTMVGDGSQKTIVT 365
Query: 391 GNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFE 450
GNK+ +RTF TA+FV G+GF+ + MGFRNTAG+ QAVA RVQ+D SIF+NC FE
Sbjct: 366 GNKSHAKKIRTFLTATFVAQGEGFMAQSMGFRNTAGSEGHQAVAIRVQSDRSIFLNCRFE 425
Query: 451 GYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIITANGRID 510
GYQDTLYA THRQ+YR CVI GTIDFIFG A+A+FQNC + +RK L QKN +TA GR+D
Sbjct: 426 GYQDTLYAYTHRQYYRSCVIVGTIDFIFGDAAAIFQNCNIFIRKGLPGQKNTVTAQGRVD 485
Query: 511 SKSNTAFVLQKCVIKGEDDLPSTT---KNYIGRPWKEYSRTIIMESDIPALIQPEGWLPW 567
T FV+ C I +DL K+Y+GRPWK YSRTIIMES I +I P GWL W
Sbjct: 486 KFQTTGFVVHNCKIAANEDLKPVKEEYKSYLGRPWKNYSRTIIMESKIENVIDPVGWLRW 545
Query: 568 -EGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDGSWINGTGV 626
E DFA+ TLYY EY+N G+ T +RVKW G K I + EAL YTV PFL G WI+ +G
Sbjct: 546 QETDFAIDTLYYAEYNNKGSSGDTTSRVKWPGFKVINKEEALNYTVGPFLQGDWISASGS 605
Query: 627 PAHLGLYN 634
P LGLY+
Sbjct: 606 PVKLGLYD 613
>UniRef100_Q7XUZ9 OSJNBa0036B21.20 protein [Oryza sativa]
Length = 568
Score = 413 bits (1062), Expect = e-114
Identities = 239/621 (38%), Positives = 345/621 (55%), Gaps = 66/621 (10%)
Query: 21 LKKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVT 80
+ +K L+G +L+ V+ V V RSG +N P + + + V
Sbjct: 1 MAQKFLLGGVGAILVVAVVVGVV-ATVTRSGNKAGDNFTVPGE------ANLATSGKSVE 53
Query: 81 MLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKN---EINNAFNKTANL--- 134
LC+ + YKE C TL A KE VF VAK+ I +A K+ +
Sbjct: 54 SLCAPTLYKESCEKTLTTATSGTENPKE------VFSTVAKSALESIKSAVEKSKAIGEA 107
Query: 135 KFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDT 194
K + + A EDCK L ED+ +++ + E+ D+ L S+ +L +WL+ V+++ DT
Sbjct: 108 KTSDSMTESAREDCKALLEDSVDDLR-GMVEMAGGDVKVLFSRSDDLEHWLTGVMTFMDT 166
Query: 195 CSDGFPEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAHA 254
C+DGF + +LK M + + +L SN+LA+ + + I L FK G++ H
Sbjct: 167 CADGFADEKLKADMHSVLRNASELSSNALAITNTLGAIFKKLD--LDMFK---GENPIHR 221
Query: 255 PAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAPAV 314
++ + E + P D+ + +
Sbjct: 222 --------SLIAEQETVGGFPSWMKAPDRKLLAS-------------------------- 247
Query: 315 DLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKM 374
G +P PN VA+DGSG FKTI EA+ ++P+ ++GRYV+YVK G+YDE V V K
Sbjct: 248 ---GDRNRPQPNAVVAQDGSGQFKTIQEAVNSMPKGHQGRYVIYVKAGLYDEIVMVPKDK 304
Query: 375 VNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVA 434
VN+ MYGDG +S +TG K+F DG+ T +TA+F V GF+ ++MGF NTAGA + QAVA
Sbjct: 305 VNIFMYGDGPKRSRVTGRKSFADGITTMKTATFSVEAAGFICKNMGFHNTAGAERHQAVA 364
Query: 435 ARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRK 494
R+ D F NC F+ +QDTLY RQF+R+CVISGTIDFIFG+++AVFQNC ++ R+
Sbjct: 365 LRINGDLGAFYNCRFDAFQDTLYVHARRQFFRNCVISGTIDFIFGNSAAVFQNCLIITRR 424
Query: 495 PLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL-PSTTK--NYIGRPWKEYSRTIIM 551
P+DNQ+N +TA+GR D + V+Q C + + L P K +Y+GRPWKEYSR +IM
Sbjct: 425 PMDNQQNSVTAHGRTDPNMKSGLVIQNCRLVPDQKLFPDRFKIPSYLGRPWKEYSRLVIM 484
Query: 552 ESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYT 611
ES I I+PEG++PW G+FAL TLYY E++N G GA T RV W G + I + EA +T
Sbjct: 485 ESTIADFIKPEGYMPWNGEFALNTLYYAEFNNRGPGAGTSKRVNWKGFRVIGQKEAEQFT 544
Query: 612 VEPFLD-GSWINGTGVPAHLG 631
PF+D G+W+ TG P LG
Sbjct: 545 AGPFVDGGTWLKFTGTPHFLG 565
>UniRef100_Q84MV1 Putative pectin methylesterase [Oryza sativa]
Length = 603
Score = 395 bits (1015), Expect = e-108
Identities = 238/630 (37%), Positives = 353/630 (55%), Gaps = 65/630 (10%)
Query: 26 LIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTMLCSH 85
LI V+L+A V+ V +V+V R+G D + K+ +++ R V + C+
Sbjct: 15 LIIAAVVMLLAIVLGTVAYVVVDRAGDD-DGLSKRGMKST----------MRSVDLFCAP 63
Query: 86 SEYKEKCVTTLKEALKK--DPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKF--ASKEE 141
++Y+ C TL+ L + DP P + + E+ F++++ L+ AS +
Sbjct: 64 TDYRVACKDTLERVLARSSDPA-DHPHAAAAAAITAVERELARGFDRSSVLEAVRASNDS 122
Query: 142 K--GAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDGF 199
+ A DC+ L D + ++ ++T + + ++ +L WLSAVI++Q +C D F
Sbjct: 123 RVAEALRDCRTLLGDCRGDVSRALTSIAWRGVDAVSQ---DLQAWLSAVITFQGSCVDMF 179
Query: 200 PEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAHAPAPDA 259
P+G +K ++ ++R++ SN++A++ Q + AF L D HA A
Sbjct: 180 PQGPIKDQVREAMEKAREISSNAIAIIQQGA----AFAAML----------DLHASESHA 225
Query: 260 DTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAPAVDLPGS 319
D D D+ D + DQ + P P + + + + A
Sbjct: 226 AEGEELDVDHDIQHHVDRHLE-DQSLPPVP---PWLSDEDRRMLTSGEEFVAGL------ 275
Query: 320 TEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTM 379
TPNVTVAKDGSGDF IS AL A+P+ Y G+Y++YVKEGVYDETV VT +M N+TM
Sbjct: 276 ----TPNVTVAKDGSGDFTNISAALDALPEAYAGKYIIYVKEGVYDETVNVTSRMANITM 331
Query: 380 YGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQA 439
YGDG KSI+TG+KN DGVR ++TA+F V GD F +G RNTAG K+QA+A RV+A
Sbjct: 332 YGDGSKKSIVTGSKNIADGVRMWKTATFAVDGDRFTAMRLGIRNTAGEEKQQALALRVKA 391
Query: 440 DCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQ 499
D SIF NC EG QDTL+AQ +RQFYR CVISGT+DFIFG A+A+FQ C ++++ PL +
Sbjct: 392 DKSIFFNCRIEGNQDTLFAQAYRQFYRSCVISGTVDFIFGDAAAMFQRCIILVKPPLPGK 451
Query: 500 KNIITANGRIDSKSNTAFVLQKCVIKGEDDL------PSTTKN---------YIGRPWKE 544
++TA+GR D + T FVL + ++D S T + Y+GRPWKE
Sbjct: 452 PAVVTAHGRRDRQQTTGFVLHHSQVVADEDFAGAGGGSSNTSSSSGAAPRLAYLGRPWKE 511
Query: 545 YSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKR 604
++RTI+MES I + +G++PWEG L +YGEY N G GA + R++ G + R
Sbjct: 512 HARTIVMESVIGGFVHAQGYMPWEGKDNLGEAFYGEYGNSGQGANSTGRMEMRGFHVLDR 571
Query: 605 GEALTYTVEPFLDGS-WINGTGVPAHLGLY 633
+A+ +TV FL G+ WI TG P +GL+
Sbjct: 572 EKAMQFTVGRFLHGADWIPETGTPVTIGLF 601
>UniRef100_O24596 Pectin methylesterase-like protein [Zea mays]
Length = 563
Score = 392 bits (1007), Expect = e-107
Identities = 210/495 (42%), Positives = 295/495 (59%), Gaps = 53/495 (10%)
Query: 142 KGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDGFPE 201
+ A EDCK+L EDA +++ + E+ DI L S+ +L WL+ V+++ DTC DGF +
Sbjct: 114 ESAREDCKKLLEDAADDLR-GMLEMAGGDIKVLFSRSDDLETWLTGVMTFMDTCVDGFVD 172
Query: 202 GELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAHAPAPDADT 261
+LK M + + +L SN+LA+ + + I+ K D + D+
Sbjct: 173 EKLKADMHSVVRNATELSSNALAITNSLGGILK--------------KMDLGMFSKDSRR 218
Query: 262 DAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWA-APAVDLPGST 320
++ + ++ P W +P L S
Sbjct: 219 RLLSSEQDE---------------------------------KGWPVWMRSPERKLLASG 245
Query: 321 EKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMY 380
+P PN VAKDGSG FK+I +A+ A+P+ ++GRYV+YVK G+YDE V V K VN+ MY
Sbjct: 246 NQPKPNAIVAKDGSGQFKSIQQAVDAVPKGHQGRYVIYVKAGLYDEIVMVPKDKVNIFMY 305
Query: 381 GDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQAD 440
GDG +S +TG K+F DG+ T +TA+F V GF+ ++MGF NTAGA + QAVA RVQ D
Sbjct: 306 GDGPKQSRVTGRKSFADGITTMKTATFSVEASGFICKNMGFHNTAGAERHQAVALRVQGD 365
Query: 441 CSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQK 500
+ F NC F+ +QDTLY Q RQF+R+CV+SGTIDFIFG+++AVFQNC ++ R+P+DNQ+
Sbjct: 366 LAAFYNCRFDAFQDTLYVQPRRQFFRNCVVSGTIDFIFGNSAAVFQNCLIITRRPMDNQQ 425
Query: 501 NIITANGRIDSKSNTAFVLQKCVIKGEDDL-PSTTK--NYIGRPWKEYSRTIIMESDIPA 557
N +TA+G D + V+Q C + + L P K +Y+GRPWKE+SR +IMES I
Sbjct: 426 NSVTAHGPTDPNMKSGLVIQNCRLVPDQKLFPDRFKIPSYLGRPWKEFSRLVIMESTIAD 485
Query: 558 LIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLD 617
++PEG++PW GDFALKTLYY EY+N G GA T RV W G I R EA +T PF+D
Sbjct: 486 FVKPEGYMPWNGDFALKTLYYAEYNNRGPGAGTSKRVNWPGFHVIGRKEAEPFTAGPFID 545
Query: 618 GS-WINGTGVPAHLG 631
G+ W+ TG P LG
Sbjct: 546 GAMWLKYTGAPHILG 560
>UniRef100_O48712 Putative pectinesterase [Arabidopsis thaliana]
Length = 496
Score = 391 bits (1005), Expect = e-107
Identities = 197/343 (57%), Positives = 239/343 (69%), Gaps = 15/343 (4%)
Query: 303 VDAPPSWAAP-------AVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRY 355
+D PSW + AVD+ PN TVAKDGSGDF TI++AL A+P+ Y+GRY
Sbjct: 157 LDDIPSWVSNDDRRMLRAVDVKALK----PNATVAKDGSGDFTTINDALRAMPEKYEGRY 212
Query: 356 VVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFV 415
++YVK+G+YDE VTV KK NLTM GDG K+I+TGNK+ +RTF TA+FV G+GF+
Sbjct: 213 IIYVKQGIYDEYVTVDKKKANLTMVGDGSQKTIVTGNKSHAKKIRTFLTATFVAQGEGFM 272
Query: 416 GRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTID 475
+ MGFRNTAG QAVA RVQ+D SIF+NC FEGYQDTLYA THRQ+YR CVI GTID
Sbjct: 273 AQSMGFRNTAGPEGHQAVAIRVQSDRSIFLNCRFEGYQDTLYAYTHRQYYRSCVIVGTID 332
Query: 476 FIFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDLPSTT- 534
FIFG A+A+FQNC + +RK L QKN +TA GR+D T FV+ C I +DL
Sbjct: 333 FIFGDAAAIFQNCNIFIRKGLPGQKNTVTAQGRVDKFQTTGFVVHNCKIAANEDLKPVKE 392
Query: 535 --KNYIGRPWKEYSRTIIMESDIPALIQPEGWLPW-EGDFALKTLYYGEYDNVGAGAKTD 591
K+Y+GRPWK YSRTIIMES I +I P GWL W E DFA+ TLYY EY+N G+ T
Sbjct: 393 EYKSYLGRPWKNYSRTIIMESKIENVIDPVGWLRWQETDFAIDTLYYAEYNNKGSSGDTT 452
Query: 592 ARVKWIGRKDIKRGEALTYTVEPFLDGSWINGTGVPAHLGLYN 634
+RVKW G K I + EAL YTV PFL G WI+ +G P LGLY+
Sbjct: 453 SRVKWPGFKVINKEEALNYTVGPFLQGDWISASGSPVKLGLYD 495
Score = 92.8 bits (229), Expect = 3e-17
Identities = 49/140 (35%), Positives = 76/140 (54%)
Query: 92 CVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKFASKEEKGAYEDCKQL 151
C TLK K L P L + +++ K +LK ++++K A E CK L
Sbjct: 4 CEKTLKNRTDKGFALDNPTTFLKSAIEAVNEDLDLVLEKVLSLKTENQDDKDAIEQCKLL 63
Query: 152 FEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDGFPEGELKKKMEMI 211
EDAKEE S+ ++ +++ +L +WLSAV+SYQ+TC DGF EG LK +++
Sbjct: 64 VEDAKEETVASLNKINVTEVNSFEKVVPDLESWLSAVMSYQETCLDGFEEGNLKSEVKTS 123
Query: 212 FAESRQLLSNSLAVVSQVSQ 231
S+ L SNSLA++ ++
Sbjct: 124 VNSSQVLTSNSLALIKTFTE 143
>UniRef100_Q9FJ21 Pectin methylesterase [Arabidopsis thaliana]
Length = 571
Score = 389 bits (999), Expect = e-106
Identities = 237/618 (38%), Positives = 335/618 (53%), Gaps = 74/618 (11%)
Query: 22 KKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTM 81
KK I+ GV + +L+ V+A V + + ++ +K V P ++ + V
Sbjct: 10 KKCIIAGVITALLVLMVVA------VGITTSRNTSHSEKIV------PVQIKTATTAVEA 57
Query: 82 LCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTA---NLKFAS 138
+C+ ++YKE CV +L +K P +P L+ + V I ++ K + K A+
Sbjct: 58 VCAPTDYKETCVNSL---MKASPDSTQPLDLIKLGFNVTIRSIEDSIKKASVELTAKAAN 114
Query: 139 -KEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSD 197
K+ KGA E C++L DA +++ + I ++ +L WLS I+YQ TC D
Sbjct: 115 DKDTKGALELCEKLMNDATDDLKKCLDNFDGFSIPQIEDFVEDLRVWLSGSIAYQQTCMD 174
Query: 198 GFPE--GELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAHAP 255
F E +L + M+ IF SR+L SN LA+++ +S ++ F ++G GK
Sbjct: 175 TFEETNSKLSQDMQKIFKTSRELTSNGLAMITNISNLLGEFN--VTGVTGDLGKYA---- 228
Query: 256 APDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAPAVD 315
L A DG P SW P
Sbjct: 229 -------------RKLLSAEDGIP----------------------------SWVGPNTR 247
Query: 316 LPGSTEKPTP-NVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKM 374
+T+ NV VA DGSG +KTI+EAL A+P+ + +V+Y+K+GVY+E V VTKKM
Sbjct: 248 RLMATKGGVKANVVVAHDGSGQYKTINEALNAVPKANQKPFVIYIKQGVYNEKVDVTKKM 307
Query: 375 VNLTMYGDGGLKSIITGNKNFVDG-VRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAV 433
++T GDG K+ ITG+ N+ G V+T+ TA+ + GD F +++GF NTAG QAV
Sbjct: 308 THVTFIGDGPTKTKITGSLNYYIGKVKTYLTATVAINGDNFTAKNIGFENTAGPEGHQAV 367
Query: 434 AARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLR 493
A RV AD ++F NC +GYQDTLY +HRQF+RDC +SGT+DFIFG V QNC +V+R
Sbjct: 368 ALRVSADLAVFYNCQIDGYQDTLYVHSHRQFFRDCTVSGTVDFIFGDGIVVLQNCNIVVR 427
Query: 494 KPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGED---DLPSTTKNYIGRPWKEYSRTII 550
KP+ +Q +ITA GR D + +T VLQ C I GE + S K Y+GRPWKE+SRTII
Sbjct: 428 KPMKSQSCMITAQGRSDKRESTGLVLQNCHITGEPAYIPVKSINKAYLGRPWKEFSRTII 487
Query: 551 MESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTY 610
M + I +I P GWLPW GDFAL TLYY EY+N G G+ RVKW G K + +AL +
Sbjct: 488 MGTTIDDVIDPAGWLPWNGDFALNTLYYAEYENNGPGSNQAQRVKWPGIKKLSPKQALRF 547
Query: 611 TVEPFLDGS-WINGTGVP 627
T FL G+ WI VP
Sbjct: 548 TPARFLRGNLWIPPNRVP 565
>UniRef100_Q6ZLF5 Putative pectinesterase [Oryza sativa]
Length = 664
Score = 385 bits (988), Expect = e-105
Identities = 192/318 (60%), Positives = 234/318 (73%), Gaps = 6/318 (1%)
Query: 322 KPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYG 381
K PNV VAKDGSG FKTI++ALAA+P+ Y GRYV+YVKEGVY+E VT+TKKM N+TMYG
Sbjct: 346 KLKPNVVVAKDGSGKFKTINDALAAMPKKYTGRYVIYVKEGVYEEYVTITKKMANVTMYG 405
Query: 382 DGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADC 441
DG K+IITGN+NFVDG+ T++TA+F GDGF+G +GFRNTA A K QAVA VQ+D
Sbjct: 406 DGAKKTIITGNRNFVDGLTTYKTATFNAQGDGFMGVALGFRNTARAAKHQAVALLVQSDK 465
Query: 442 SIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKN 501
SIF+NC EG+QDTLYA + QFYR+CVISGT+DFIFG A+AVFQNC +VLR+PLDNQ+N
Sbjct: 466 SIFLNCRMEGHQDTLYAHSKAQFYRNCVISGTVDFIFGDAAAVFQNCVIVLRRPLDNQQN 525
Query: 502 IITANGRIDSKSNTAFVLQKCVIKGEDDLPSTT----KNYIGRPWKEYSRTIIMESDIPA 557
I TA GR D + T FVLQ E L + ++Y+ RPW+EYSRT+IM SDIPA
Sbjct: 526 IATAQGRADRREATGFVLQHYRFAAESALGDASRPAVRSYLARPWREYSRTLIMNSDIPA 585
Query: 558 LIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIG-RKDIKRGEALTYTVEPFL 616
+ G+LPW GDF LKTL+Y EY N GAGA T RV W G +K I + EA +TV+ FL
Sbjct: 586 FVDKAGYLPWSGDFGLKTLWYAEYGNKGAGAATAGRVSWPGYKKVISKKEATKFTVQNFL 645
Query: 617 DGS-WINGTGVPAHLGLY 633
WI TG P G++
Sbjct: 646 HAEPWIKPTGTPVKYGMW 663
Score = 118 bits (295), Expect = 6e-25
Identities = 64/238 (26%), Positives = 123/238 (50%), Gaps = 6/238 (2%)
Query: 3 FEDFDLVSERRKANAKQHLKKKILI--GVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKK 60
F DFD ++ERR ++ ++++++ G SV+LI V+ G +++
Sbjct: 5 FSDFDPITERRHVERQRQERRRVMVAAGAASVILIIIVMGGAAVAYNASFGDGGSSSSSG 64
Query: 61 PVQNAPPEPERVDKYSRLVTMLCSHSEYKEKCVTTLKEALKK---DPKLKEPKGLLMVFM 117
+P + S+++ +C+ ++YK+ C +L +A PK ++ +
Sbjct: 65 SASGGGAQPS-LHGVSKIIKAMCAQTDYKDTCEKSLAKAAANASASSSSSSPKDVVRASV 123
Query: 118 LVAKNEINNAFNKTANLKFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASK 177
V + I AF+K++ + K A DCK+++E+AK+++ ++ + + L
Sbjct: 124 AVIGDAIEKAFDKSSVIVSDDPRVKAAVADCKEIYENAKDDLDRTLAGIDAGGVDGLTKG 183
Query: 178 EAELNNWLSAVISYQDTCSDGFPEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNA 235
+L WLSAVI++Q+TC DGFP+G+LK KM ++L SN+LA++ + S + A
Sbjct: 184 GYQLRVWLSAVIAHQETCIDGFPDGDLKDKMRDAMESGKELTSNALALIGKASSFLAA 241
>UniRef100_Q8GS16 Putative thermostable pectinesterase [Citrus sinensis]
Length = 631
Score = 351 bits (900), Expect = 4e-95
Identities = 224/625 (35%), Positives = 318/625 (50%), Gaps = 72/625 (11%)
Query: 23 KKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTML 82
+K+ + + +VVL V+ AV +I R N ND+ +A + V + S
Sbjct: 64 RKLFLALFAVVL---VVTAVVTIIATRKNNSSNTNDEHIAAHAHAYAQPVIRSS------ 114
Query: 83 CSHSEYKEKCVTTLKEA-LKKDPKLKEPKGLLMVFMLVAKNEINN---AFNK--TANLKF 136
CS + Y E C + L A K+ PK ++ + + + + + A K T
Sbjct: 115 CSATLYPELCFSALSAAPAAAVSKVNSPKDVIRLSLNLTITAVQHNYFAIKKLITTRKST 174
Query: 137 ASKEEKGAYEDCKQLFEDAKEEMGFSITEV----GQLDISKLASKEAELNNWLSAVISYQ 192
+K EK + DC ++ ++ +E+ + E+ + +A + EL +SA ++ Q
Sbjct: 175 LTKREKTSLHDCLEMVDETLDELYKTEHELQGYPAAANNKSIAEQADELKILVSAAMTNQ 234
Query: 193 DTCSDGFPEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDA 252
+TC DGF KK+ +L+ + V S + + G D
Sbjct: 235 ETCLDGFSHERADKKIR------EELMEGQMHVFHMCSNALAMIKNMTDG--------DI 280
Query: 253 HAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAP 312
D + A DDE P+ D+ + +A
Sbjct: 281 GKDIVDHYSKARRLDDE--TKWPEWLSAGDRRLLQATT---------------------- 316
Query: 313 AVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTK 372
P+VTVA DGSG++ T++ A+AA P+ RY++ +K G Y E V V K
Sbjct: 317 ----------VVPDVTVAADGSGNYLTVAAAVAAAPEGSSRRYIIRIKAGEYRENVEVPK 366
Query: 373 KMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQA 432
K +NL GDG +IITG++N VDG TF +A+ V+GDGF+ RD+ F+NTAG K QA
Sbjct: 367 KKINLMFIGDGRTTTIITGSRNVVDGSTTFNSATVAVVGDGFLARDITFQNTAGPSKHQA 426
Query: 433 VAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVL 492
VA RV +D S F C+ YQDTLY + RQFY C+I+GT+DFIFG+A+AVFQNC +
Sbjct: 427 VALRVGSDLSAFYRCDMLAYQDTLYVHSLRQFYTSCIIAGTVDFIFGNAAAVFQNCDIHA 486
Query: 493 RKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTKNYIGRPWKEYSRTI 549
R+P NQ+N++TA GR D NT V+QKC I DL + + Y+GRPWK YSRT+
Sbjct: 487 RRPNPNQRNMVTAQGRDDPNQNTGIVIQKCRIGATSDLLAVKGSFETYLGRPWKRYSRTV 546
Query: 550 IMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDI-KRGEAL 608
+M+SDI +I P GW W G+FAL TL+Y EY N GAGA T RVKW K I EA
Sbjct: 547 VMQSDISDVINPAGWYEWSGNFALDTLFYAEYQNTGAGADTSNRVKWSTFKVITSAAEAQ 606
Query: 609 TYTVEPFLDGS-WINGTGVPAHLGL 632
TYT F+ GS W+ TG P LGL
Sbjct: 607 TYTAANFIAGSTWLGSTGFPFSLGL 631
>UniRef100_Q9M9W6 Putative pectinesterase [Arabidopsis thaliana]
Length = 669
Score = 348 bits (894), Expect = 2e-94
Identities = 169/312 (54%), Positives = 219/312 (70%), Gaps = 4/312 (1%)
Query: 325 PNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGG 384
P++ VA+DGSG +KTI+EAL +P+ +VV++K G+Y E V V K M +L GDG
Sbjct: 254 PDIVVAQDGSGQYKTINEALQFVPKKRNTTFVVHIKAGLYKEYVQVNKTMSHLVFIGDGP 313
Query: 385 LKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIF 444
K+II+GNKN+ DG+ T++TA+ ++G+ F+ +++GF NTAGAIK QAVA RVQ+D SIF
Sbjct: 314 DKTIISGNKNYKDGITTYRTATVAIVGNYFIAKNIGFENTAGAIKHQAVAVRVQSDESIF 373
Query: 445 VNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIIT 504
NC F+GYQDTLY +HRQF+RDC ISGTIDF+FG A+AVFQNC L++RKPL NQ IT
Sbjct: 374 FNCRFDGYQDTLYTHSHRQFFRDCTISGTIDFLFGDAAAVFQNCTLLVRKPLPNQACPIT 433
Query: 505 ANGRIDSKSNTAFVLQKCVIKGEDD---LPSTTKNYIGRPWKEYSRTIIMESDIPALIQP 561
A+GR D + +T FV Q C I GE D + T+K Y+GRPWKEYSRTIIM + IP +QP
Sbjct: 434 AHGRKDPRESTGFVFQGCTIAGEPDYLAVKETSKAYLGRPWKEYSRTIIMNTFIPDFVQP 493
Query: 562 EGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDG-SW 620
+GW PW GDF LKTL+Y E N G G+ RV W G K + + L +T ++ G W
Sbjct: 494 QGWQPWLGDFGLKTLFYSEVQNTGPGSALANRVTWAGIKTLSEEDILKFTPAQYIQGDDW 553
Query: 621 INGTGVPAHLGL 632
I G GVP GL
Sbjct: 554 IPGKGVPYTTGL 565
Score = 83.2 bits (204), Expect = 2e-14
Identities = 57/220 (25%), Positives = 108/220 (48%), Gaps = 19/220 (8%)
Query: 22 KKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTM 81
++ I+I ++SV+LI+ V+A V + + H+ + K + V+ + V
Sbjct: 13 RRYIVITISSVLLISMVVAVTVGVSLNK----HDGDSKGKAE--------VNASVKAVKD 60
Query: 82 LCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKFASKEE 141
+C+ ++Y++ C TL +K +P L+ V +I +A K+ + K+
Sbjct: 61 VCAPTDYRKTCEDTL---IKNGKNTTDPMELVKTAFNVTMKQITDAAKKSQTIMELQKDS 117
Query: 142 KG--AYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDGF 199
+ A + CK+L + A +E+ S E+G+ + L L WLSA IS+++TC +GF
Sbjct: 118 RTRMALDQCKELMDYALDELSNSFEELGKFEFHLLDEALINLRIWLSAAISHEETCLEGF 177
Query: 200 --PEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQ 237
+G + M+ + +L N LA++S++S V Q
Sbjct: 178 QGTQGNAGETMKKALKTAIELTHNGLAIISEMSNFVGQMQ 217
>UniRef100_Q8GX86 Putative pectinesterase [Arabidopsis thaliana]
Length = 669
Score = 345 bits (884), Expect = 3e-93
Identities = 167/312 (53%), Positives = 218/312 (69%), Gaps = 4/312 (1%)
Query: 325 PNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGG 384
P++ VA+DGSG +KTI+EAL +P+ +VV++K G+Y E V V K M +L GDG
Sbjct: 254 PDIVVAQDGSGQYKTINEALQFVPKKRNTTFVVHIKAGLYKEYVQVNKTMSHLVFIGDGP 313
Query: 385 LKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIF 444
K+II+GNKN+ DG+ ++TA+ ++G+ F+ +++GF NTAGAIK QAVA RVQ+D SIF
Sbjct: 314 DKTIISGNKNYKDGITAYRTATVAIVGNYFIAKNIGFENTAGAIKHQAVAVRVQSDESIF 373
Query: 445 VNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIIT 504
NC F+GYQ+TLY +HRQF+RDC ISGTIDF+FG A+AVFQNC L++RKPL NQ IT
Sbjct: 374 FNCRFDGYQNTLYTHSHRQFFRDCTISGTIDFLFGDAAAVFQNCTLLVRKPLPNQACPIT 433
Query: 505 ANGRIDSKSNTAFVLQKCVIKGEDD---LPSTTKNYIGRPWKEYSRTIIMESDIPALIQP 561
A+GR D + +T FV Q C I GE D + T+K Y+GRPWKEYSRTIIM + IP +QP
Sbjct: 434 AHGRKDPRESTGFVFQGCTIAGEPDYLAVKETSKAYLGRPWKEYSRTIIMNTFIPDFVQP 493
Query: 562 EGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDG-SW 620
+GW PW GDF LKTL+Y E N G G+ RV W G K + + L +T ++ G W
Sbjct: 494 QGWQPWLGDFGLKTLFYSEVQNTGPGSALANRVTWAGIKTLSEEDILKFTPAQYIQGDDW 553
Query: 621 INGTGVPAHLGL 632
I G GVP GL
Sbjct: 554 IPGKGVPYTTGL 565
Score = 83.2 bits (204), Expect = 2e-14
Identities = 57/220 (25%), Positives = 108/220 (48%), Gaps = 19/220 (8%)
Query: 22 KKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTM 81
++ I+I ++SV+LI+ V+A V + + H+ + K + V+ + V
Sbjct: 13 RRYIVITISSVLLISMVVAVTVGVSLNK----HDGDSKGKAE--------VNASVKAVKD 60
Query: 82 LCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKFASKEE 141
+C+ ++Y++ C TL +K +P L+ V +I +A K+ + K+
Sbjct: 61 VCAPTDYRKTCEDTL---IKNGKNTTDPMELVKTAFNVTMKQITDAAKKSQTIMELQKDS 117
Query: 142 KG--AYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDGF 199
+ A + CK+L + A +E+ S E+G+ + L L WLSA IS+++TC +GF
Sbjct: 118 RTRMALDQCKELMDYALDELSNSFEELGKFEFHLLDEALINLRIWLSAAISHEETCLEGF 177
Query: 200 --PEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQ 237
+G + M+ + +L N LA++S++S V Q
Sbjct: 178 QGTQGNAGETMKKALKTAIELTHNGLAIISEMSNFVGQMQ 217
>UniRef100_O04886 Pectinesterase 1 precursor [Citrus sinensis]
Length = 584
Score = 337 bits (864), Expect = 6e-91
Identities = 213/636 (33%), Positives = 322/636 (50%), Gaps = 80/636 (12%)
Query: 10 SERRKANAKQHLKKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEP 69
S + +N + KK L +++++A VI V V +++ D+ N
Sbjct: 16 SNQNISNIPKKKKKLFLALFATLLVVAAVIGIVAGVNSRKNSGDNGN------------- 62
Query: 70 ERVDKYSRLVTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAF- 128
+ + ++ CS + Y + C + + + K+ K ++ + + + + + +
Sbjct: 63 ---EPHHAILKSSCSSTRYPDLCFSAIAAVPEASKKVTSQKDVIEMSLNITTTAVEHNYF 119
Query: 129 --NKTANLKFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAE-LNNWL 185
K +K EK A DC + ++ +E+ ++ ++ + K S+ A+ L +
Sbjct: 120 GIQKLLKRTNLTKREKVALHDCLETIDETLDELHKAVEDLEEYPNKKSLSQHADDLKTLM 179
Query: 186 SAVISYQDTCSDGFPEGELKKKMEMIFAESR----QLLSNSLAVVSQVSQIVNAFQGGLS 241
SA ++ Q TC DGF + K + ++ + ++ SN+LA++ ++
Sbjct: 180 SAAMTNQGTCLDGFSHDDANKHVRDALSDGQVHVEKMCSNALAMIKNMT----------- 228
Query: 242 GFKLPWGKSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAP 301
DTD + + + D G P G
Sbjct: 229 ------------------DTDMMIMRTSNNRKLTEETSTVD--------GWPAWLSPGDR 262
Query: 302 RVDAPPSWAAPAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKE 361
R+ L S+ P V G +FKT++ A+AA PQ RY++ +K
Sbjct: 263 RL------------LQSSSVTPNAVVAADGSG--NFKTVAAAVAAAPQGGTKRYIIRIKA 308
Query: 362 GVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGF 421
GVY E V VTKK N+ GDG ++IITG++N VDG TF++A+ V+G+GF+ RD+ F
Sbjct: 309 GVYRENVEVTKKHKNIMFIGDGRTRTIITGSRNVVDGSTTFKSATAAVVGEGFLARDITF 368
Query: 422 RNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHA 481
+NTAG K QAVA RV AD S F NC+ YQDTLY ++RQF+ +C+I+GT+DFIFG+A
Sbjct: 369 QNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIFGNA 428
Query: 482 SAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTKNYI 538
+AV QNC + RKP QKN++TA GR D NT V+QK I DL + Y+
Sbjct: 429 AAVLQNCDIHARKPNSGQKNMVTAQGRTDPNQNTGIVIQKSRIGATSDLKPVQGSFPTYL 488
Query: 539 GRPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIG 598
GRPWKEYSRT+IM+S I LI P GW W+G+FAL TL+YGE+ N GAGA T RVKW G
Sbjct: 489 GRPWKEYSRTVIMQSSITDLIHPAGWHEWDGNFALNTLFYGEHQNSGAGAGTSGRVKWKG 548
Query: 599 RKDIKRG-EALTYTVEPFLDG-SWINGTGVPAHLGL 632
+ I EA +T F+ G SW+ TG P LGL
Sbjct: 549 FRVITSATEAQAFTPGSFIAGSSWLGSTGFPFSLGL 584
>UniRef100_P83948 Pectinesterase 3 precursor [Citrus sinensis]
Length = 584
Score = 335 bits (858), Expect = 3e-90
Identities = 210/631 (33%), Positives = 320/631 (50%), Gaps = 80/631 (12%)
Query: 15 ANAKQHLKKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDK 74
+N + KK L +++++A VI V V +++ D+ N +
Sbjct: 21 SNIPKKKKKLFLALFATLLVVAAVIGIVAGVNSRKNSGDNGN----------------EP 64
Query: 75 YSRLVTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAF---NKT 131
+ ++ CS + Y + C + + + K+ K ++ + + + + + + K
Sbjct: 65 HHAILKSSCSSTRYPDLCFSAIAAVPEASKKVTSQKDVIEMSLNITTTAVEHNYFGIQKL 124
Query: 132 ANLKFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAE-LNNWLSAVIS 190
+K EK A DC + ++ +E+ ++ ++ + K S+ A+ L +SA ++
Sbjct: 125 LKRTNLTKREKVALHDCLETIDETLDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMT 184
Query: 191 YQDTCSDGFPEGELKKKMEMIFAESR----QLLSNSLAVVSQVSQIVNAFQGGLSGFKLP 246
Q TC DGF + K + ++ + ++ SN+LA++ ++
Sbjct: 185 NQGTCLDGFSHDDANKHVRDALSDGQVHVEKMCSNALAMIKNMT---------------- 228
Query: 247 WGKSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAP 306
DTD + + + D G P G R+
Sbjct: 229 -------------DTDMMIMRTSNNRKLIEETSTVD--------GWPAWLSTGDRRL--- 264
Query: 307 PSWAAPAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDE 366
L S+ P V G +FKT++ ++AA PQ RY++ +K GVY E
Sbjct: 265 ---------LQSSSVTPNVVVAADGSG--NFKTVAASVAAAPQGGTKRYIIRIKAGVYRE 313
Query: 367 TVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAG 426
V VTKK N+ GDG ++IITG++N VDG TF++A+ V+G+GF+ RD+ F+NTAG
Sbjct: 314 NVEVTKKHKNIMFIGDGRTRTIITGSRNVVDGSTTFKSATVAVVGEGFLARDITFQNTAG 373
Query: 427 AIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQ 486
K QAVA RV AD S F NC+ YQDTLY ++RQF+ +C+I+GT+DFIFG+A+AV Q
Sbjct: 374 PSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIFGNAAAVLQ 433
Query: 487 NCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTKNYIGRPWK 543
NC + RKP QKN++TA GR D NT V+QK I DL + Y+GRPWK
Sbjct: 434 NCDIHARKPNSGQKNMVTAQGRADPNQNTGIVIQKSRIGATSDLKPVQGSFPTYLGRPWK 493
Query: 544 EYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIK 603
EYSRT+IM+S I +I P GW W+G+FAL TL+YGE+ N GAGA T RVKW G + I
Sbjct: 494 EYSRTVIMQSSITDVIHPAGWHEWDGNFALNTLFYGEHQNAGAGAGTSGRVKWKGFRVIT 553
Query: 604 RG-EALTYTVEPFLDG-SWINGTGVPAHLGL 632
EA +T F+ G SW+ TG P LGL
Sbjct: 554 SATEAQAFTPGSFIAGSSWLGSTGFPFSLGL 584
>UniRef100_Q655P5 Putative pectinesterase [Oryza sativa]
Length = 426
Score = 330 bits (847), Expect = 6e-89
Identities = 191/458 (41%), Positives = 258/458 (55%), Gaps = 45/458 (9%)
Query: 185 LSAVISYQDTCSDGF--PEGE-LKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLS 241
LSA ++ Q TC DGF +GE ++ ME +++SNSLA+ ++ A GG++
Sbjct: 4 LSAAMTNQYTCLDGFDYKDGERVRHYMESSIHHVSRMVSNSLAM---AKKLPGAGGGGMT 60
Query: 242 GFKLPWGKSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAP 301
+ +PD T + ++ + P G G P
Sbjct: 61 ----------PSSSSPDTATQS----------------ESSETTQRQPFMGYGQMANGFP 94
Query: 302 RVDAPPSWAAPAVD--LPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYV 359
+ W P L TP+ VAKDGSG + T+S A+AA P RYV+++
Sbjct: 95 K------WVRPGDRRLLQAPASSITPDAVVAKDGSGGYTTVSAAVAAAPANSNKRYVIHI 148
Query: 360 KEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDM 419
K G Y E V V K NL GDG K++I ++N VDG TF++A+ V+G+ F+ RD+
Sbjct: 149 KAGAYMENVEVGKSKKNLMFIGDGIGKTVIKASRNVVDGSTTFRSATVAVVGNNFLARDL 208
Query: 420 GFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFG 479
N+AG K QAVA RV AD S F C+F GYQDTLY + RQF+R+C I GTIDFIFG
Sbjct: 209 TIENSAGPSKHQAVALRVGADLSAFYRCSFVGYQDTLYVHSLRQFFRECDIYGTIDFIFG 268
Query: 480 HASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTKN 536
+++ VFQ+C L R+PL NQ N+ TA GR D NT +QKC + DL S+ K
Sbjct: 269 NSAVVFQSCNLYARRPLPNQSNVYTAQGREDPNQNTGISIQKCKVAAASDLLAVQSSFKT 328
Query: 537 YIGRPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKW 596
Y+GRPWK+YSRT+ M+S++ +++ P GWL W G+FAL TLYYGEY N G GA T RVKW
Sbjct: 329 YLGRPWKQYSRTVFMQSELDSVVNPAGWLEWSGNFALDTLYYGEYQNTGPGASTSNRVKW 388
Query: 597 IGRKDI-KRGEALTYTVEPFLDGS-WINGTGVPAHLGL 632
G + I EA T+TV F+DG W+ GT VP +GL
Sbjct: 389 KGYRVITSASEASTFTVGNFIDGDVWLAGTSVPFTVGL 426
>UniRef100_Q7X6L1 OSJNBb0079B02.7 protein [Oryza sativa]
Length = 971
Score = 327 bits (839), Expect = 5e-88
Identities = 168/337 (49%), Positives = 218/337 (63%), Gaps = 8/337 (2%)
Query: 301 PRVDAPPSWAAPAVD--LPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVY 358
P+ PSW + L T+KP + VAKDGSGDFKTI+EA+ A+P+ R+V+Y
Sbjct: 634 PKPGEFPSWVSAHQRRLLQAGTQKP--DKVVAKDGSGDFKTITEAVNAVPKNSPTRFVIY 691
Query: 359 VKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRD 418
VK G Y+E VT+ + N+ MYGDG K+ + GNK+ DGV T T +F G+GFV +
Sbjct: 692 VKAGEYNEYVTIPSSLPNIFMYGDGPTKTRVLGNKSNKDGVATMATRTFSAEGNGFVCKS 751
Query: 419 MGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIF 478
MGF NTAG QAVA VQ D S+F NC FEGYQDTLY +RQF+R+C ++GTID+IF
Sbjct: 752 MGFVNTAGPEGHQAVALHVQGDMSVFFNCKFEGYQDTLYVHANRQFFRNCEVTGTIDYIF 811
Query: 479 GHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTK 535
G+++AVFQ+C + +RKP+DNQ N++TA+GR D T VLQ C I E L
Sbjct: 812 GNSAAVFQSCLMTVRKPMDNQANMVTAHGRTDPNMPTGIVLQDCRIVPEQALFPVRLQIA 871
Query: 536 NYIGRPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVK 595
+Y+GRPWKEY+RT++MES I I+PEGW W GD LKTLYY EY N G GA T RV
Sbjct: 872 SYLGRPWKEYARTVVMESVIGDFIKPEGWSEWMGDVGLKTLYYAEYANTGPGAGTSKRVT 931
Query: 596 WIGRKDIKRGEALTYTVEPFLDG-SWINGTGVPAHLG 631
W G + I + EA +T F+DG +W+ T P +G
Sbjct: 932 WPGYRVIGQAEATQFTAGVFIDGLTWLKNTATPNVMG 968
Score = 82.0 bits (201), Expect = 5e-14
Identities = 58/212 (27%), Positives = 105/212 (49%), Gaps = 29/212 (13%)
Query: 26 LIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTMLCSH 85
++ VV + C IAAVT S D N V+ ++ +CS
Sbjct: 10 IVAAVGVVAVVCAIAAVT-----SSKKDRNGELTANVR---------------LSTVCSV 49
Query: 86 SEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANL--KFASKEEKG 143
+ Y +C +L + +P+ +L + VA E+ +AFN++ ++ +K K
Sbjct: 50 TRYPGRCEQSLGPVVNDTI---DPESVLRAALQVALEEVTSAFNRSMDVGKDDDAKITKS 106
Query: 144 AYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDGFPEGE 203
A E CK+L +DA E++ + L ++ +L WLS+V++Y TC+DGF + E
Sbjct: 107 AIEMCKKLLDDAIEDL----RGMASLKPEEVTKHVNDLRCWLSSVMTYIYTCADGFDKPE 162
Query: 204 LKKKMEMIFAESRQLLSNSLAVVSQVSQIVNA 235
LK+ M+ + S +L SN+LA+++ + +++ A
Sbjct: 163 LKEAMDKLLQNSTELSSNALAIITSLGELMPA 194
>UniRef100_Q9FSQ0 Putative pectin methylesterase [Oryza sativa]
Length = 717
Score = 327 bits (839), Expect = 5e-88
Identities = 168/337 (49%), Positives = 218/337 (63%), Gaps = 8/337 (2%)
Query: 301 PRVDAPPSWAAPAVD--LPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVY 358
P+ PSW + L T+KP + VAKDGSGDFKTI+EA+ A+P+ R+V+Y
Sbjct: 380 PKPGEFPSWVSAHQRRLLQAGTQKP--DKVVAKDGSGDFKTITEAVNAVPKNSPTRFVIY 437
Query: 359 VKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRD 418
VK G Y+E VT+ + N+ MYGDG K+ + GNK+ DGV T T +F G+GFV +
Sbjct: 438 VKAGEYNEYVTIPSSLPNIFMYGDGPTKTRVLGNKSNKDGVATMATRTFSAEGNGFVCKS 497
Query: 419 MGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIF 478
MGF NTAG QAVA VQ D S+F NC FEGYQDTLY +RQF+R+C ++GTID+IF
Sbjct: 498 MGFVNTAGPEGHQAVALHVQGDMSVFFNCKFEGYQDTLYVHANRQFFRNCEVTGTIDYIF 557
Query: 479 GHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTK 535
G+++AVFQ+C + +RKP+DNQ N++TA+GR D T VLQ C I E L
Sbjct: 558 GNSAAVFQSCLMTVRKPMDNQANMVTAHGRTDPNMPTGIVLQDCRIVPEQALFPVRLQIA 617
Query: 536 NYIGRPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVK 595
+Y+GRPWKEY+RT++MES I I+PEGW W GD LKTLYY EY N G GA T RV
Sbjct: 618 SYLGRPWKEYARTVVMESVIGDFIKPEGWSEWMGDVGLKTLYYAEYANTGPGAGTSKRVT 677
Query: 596 WIGRKDIKRGEALTYTVEPFLDG-SWINGTGVPAHLG 631
W G + I + EA +T F+DG +W+ T P +G
Sbjct: 678 WPGYRVIGQAEATQFTAGVFIDGLTWLKNTATPNVMG 714
>UniRef100_Q42935 Pectin methylesterase precursor [Nicotiana plumbaginifolia]
Length = 315
Score = 327 bits (837), Expect = 9e-88
Identities = 163/313 (52%), Positives = 205/313 (65%), Gaps = 4/313 (1%)
Query: 324 TPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDG 383
T NV VAKDGSG +KT+ EA+A++P RYV+YVK+G+Y E V + KK N+ + GDG
Sbjct: 3 TANVIVAKDGSGKYKTVKEAVASVPDNSNSRYVIYVKKGIYKENVEIGKKKKNVMLVGDG 62
Query: 384 GLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSI 443
+IITGN N VDG TF +A+ +GDGF+ +D+ F+NTAGA K QAVA RV AD S+
Sbjct: 63 MDATIITGNLNVVDGATTFNSATVAAVGDGFIAQDVQFQNTAGAAKHQAVALRVGADQSV 122
Query: 444 FVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNII 503
C + +QDTLY + RQFYRDC I+GT+DFIFG+A+ VFQN ++ RKP QKN++
Sbjct: 123 INRCKIDAFQDTLYTHSLRQFYRDCYITGTVDFIFGNAAVVFQNSKIAARKPGSGQKNMV 182
Query: 504 TANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTKNYIGRPWKEYSRTIIMESDIPALIQ 560
TA GR D NT +Q C I DL + K Y+GRPWK YSRT+ M+S+I I
Sbjct: 183 TAQGREDPNQNTGTSIQNCDIIPSSDLAPVKGSVKTYLGRPWKAYSRTVFMQSNIGDHID 242
Query: 561 PEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDGS- 619
PEGW W+GDFALKTLYYGEY N G GA T RVKW G + EA +TV + G
Sbjct: 243 PEGWSVWDGDFALKTLYYGEYMNKGPGAGTSKRVKWPGYHILSAAEATKFTVGQLIQGGV 302
Query: 620 WINGTGVPAHLGL 632
W+ TGV GL
Sbjct: 303 WLKSTGVAYTEGL 315
>UniRef100_Q94C39 At1g11580/T23J18_33 [Arabidopsis thaliana]
Length = 557
Score = 326 bits (836), Expect = 1e-87
Identities = 168/329 (51%), Positives = 212/329 (64%), Gaps = 9/329 (2%)
Query: 307 PSWAAPA----VDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEG 362
PSW ++ T K T NV VAKDG+G FKT++EA+AA P+ RYV+YVK+G
Sbjct: 223 PSWLTALDRKLLESSPKTLKVTANVVVAKDGTGKFKTVNEAVAAAPENSNTRYVIYVKKG 282
Query: 363 VYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFR 422
VY ET+ + KK NL + GDG +IITG+ N +DG TF++A+ GDGF+ +D+ F+
Sbjct: 283 VYKETIDIGKKKKNLMLVGDGKDATIITGSLNVIDGSTTFRSATVAANGDGFMAQDIWFQ 342
Query: 423 NTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHAS 482
NTAG K QAVA RV AD ++ C + YQDTLY T RQFYRD I+GT+DFIFG+++
Sbjct: 343 NTAGPAKHQAVALRVSADQTVINRCRIDAYQDTLYTHTLRQFYRDSYITGTVDFIFGNSA 402
Query: 483 AVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTKNYIG 539
VFQNC +V R P QKN++TA GR D NTA +QKC I DL + K ++G
Sbjct: 403 VVFQNCDIVARNPGAGQKNMLTAQGREDQNQNTAISIQKCKITASSDLAPVKGSVKTFLG 462
Query: 540 RPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGR 599
RPWK YSRT+IM+S I I P GW PW+G+FAL TLYYGEY N G GA T RV W G
Sbjct: 463 RPWKLYSRTVIMQSFIDNHIDPAGWFPWDGEFALSTLYYGEYANTGPGADTSKRVNWKGF 522
Query: 600 KDIKRG-EALTYTVEPFLDGS-WINGTGV 626
K IK EA +TV + G W+ TGV
Sbjct: 523 KVIKDSKEAEQFTVAKLIQGGLWLKPTGV 551
Score = 38.1 bits (87), Expect = 0.79
Identities = 38/193 (19%), Positives = 78/193 (39%), Gaps = 26/193 (13%)
Query: 19 QHLKKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRL 78
++LK K L V S V I + +V F + + NNND + +
Sbjct: 13 KYLKHKNLCLVLSFVAI---LGSVAFFTAQLISVNTNNNDDSLLTTS------------- 56
Query: 79 VTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKFAS 138
+C + ++ C L E LL VF+ + + + + + S
Sbjct: 57 --QICHGAHDQDSCQALLSEFTTLSLSKLNRLDLLHVFLKNSVWRLESTMTMVSEARIRS 114
Query: 139 K--EEKGAYEDCKQLFEDAKEEMGFSITEV--GQLDISKLASKEAELNNWLSAVISYQDT 194
+K + DC+++ + +K+ M S+ E+ G ++ ++ ++ WLS+V++ T
Sbjct: 115 NGVRDKAGFADCEEMMDVSKDRMMSSMEELRGGNYNLESYSN----VHTWLSSVLTNYMT 170
Query: 195 CSDGFPEGELKKK 207
C + + + K
Sbjct: 171 CLESISDVSVNSK 183
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.134 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,103,738,689
Number of Sequences: 2790947
Number of extensions: 50187980
Number of successful extensions: 165650
Number of sequences better than 10.0: 646
Number of HSP's better than 10.0 without gapping: 273
Number of HSP's successfully gapped in prelim test: 416
Number of HSP's that attempted gapping in prelim test: 163123
Number of HSP's gapped (non-prelim): 1649
length of query: 634
length of database: 848,049,833
effective HSP length: 134
effective length of query: 500
effective length of database: 474,062,935
effective search space: 237031467500
effective search space used: 237031467500
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC130802.4


