

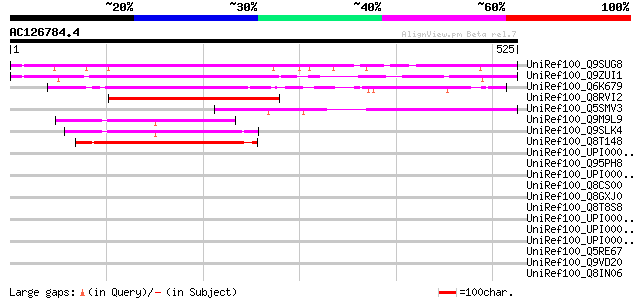
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126784.4 - phase: 0 /pseudo
(525 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SUG8 Hypothetical protein T10C21.130 [Arabidopsis th... 394 e-108
UniRef100_Q9ZUI1 At2g24100/F27D4.1 [Arabidopsis thaliana] 378 e-103
UniRef100_Q6K679 Hypothetical protein P0700F06.23 [Oryza sativa] 301 2e-80
UniRef100_Q8RVI2 Hypothetical protein [Pinus pinaster] 229 1e-58
UniRef100_Q5SMV3 Hypothetical protein P0470C02.5-1 [Oryza sativa] 167 7e-40
UniRef100_Q9M9L9 F10A16.6 protein [Arabidopsis thaliana] 133 1e-29
UniRef100_Q9SLK4 F20D21.12 protein [Arabidopsis thaliana] 132 2e-29
UniRef100_Q8T148 Similar to Dictyostelium discoideum (Slime mold... 132 2e-29
UniRef100_UPI00003C22CB UPI00003C22CB UniRef100 entry 42 0.057
UniRef100_Q95PH8 Histidine kinase DhkH [Dictyostelium discoideum] 38 0.82
UniRef100_UPI00003AB6D7 UPI00003AB6D7 UniRef100 entry 37 1.4
UniRef100_Q8CS00 Hypothetical protein SE1494 [Staphylococcus epi... 37 1.4
UniRef100_Q8GXJ0 Hypothetical protein [Arabidopsis thaliana] 37 1.4
UniRef100_Q8T8S8 AT30755p [Drosophila melanogaster] 37 1.8
UniRef100_UPI000024CF29 UPI000024CF29 UniRef100 entry 36 2.4
UniRef100_UPI00000848BE UPI00000848BE UniRef100 entry 36 2.4
UniRef100_UPI0000499528 UPI0000499528 UniRef100 entry 36 2.4
UniRef100_Q5RE67 Hypothetical protein DKFZp469D1917 [Pongo pygma... 36 2.4
UniRef100_Q9VD20 CG31169-PA, isoform A [Drosophila melanogaster] 36 2.4
UniRef100_Q8IN06 CG31169-PB, isoform B [Drosophila melanogaster] 36 2.4
>UniRef100_Q9SUG8 Hypothetical protein T10C21.130 [Arabidopsis thaliana]
Length = 589
Score = 394 bits (1011), Expect = e-108
Identities = 263/595 (44%), Positives = 334/595 (55%), Gaps = 88/595 (14%)
Query: 2 VRFMGSKNPRSQWESSSATSSISPKFEI-EDSIQDQHAPLNKRHK-------------AT 47
+R G P S+ ++ + K EI ED ++++H PLNKR + A
Sbjct: 4 MRKSGGLRPESE-SAARCRDELPVKLEIAEDDLEEEHGPLNKRSRLWSPGTSSSTMAPAK 62
Query: 48 NDILNEPSPLGLSLRKSPSLLDLIQM--TLCQE----NSVNANTANDNLNSKANKNGRA- 100
+ L+EPSPLGLSL+KSPSLL+LIQM T C + ++ A L ++ A
Sbjct: 63 YNPLDEPSPLGLSLKKSPSLLELIQMKITHCGDPKAAETLKAGALGSGLKRESKTIAAAA 122
Query: 101 ---------SVEKLKASNFPATHLKIGSWEYKSKYEGDLVAKCYFAKQKLVWEVLEGELK 151
S+EKLKASNFPA+ LKIG WEYKS+YEGDLVAKCYFAK KLVWEVLE LK
Sbjct: 123 SVGPTLAPGSIEKLKASNFPASLLKIGQWEYKSRYEGDLVAKCYFAKHKLVWEVLEQGLK 182
Query: 152 SKIEIQWSDISQLKANCPDDGPSTLTLMVARQPLFFRETNPQPRKHTLWQSTTDFTGGQA 211
SKIEIQWSDI LKANCP+DGP TLTL++ARQPLFFRETNPQPRKHTLWQ+T+DFT GQA
Sbjct: 183 SKIEIQWSDIMALKANCPEDGPGTLTLVLARQPLFFRETNPQPRKHTLWQATSDFTDGQA 242
Query: 212 SIHRRHVLQCEQGLLIKHYEKLVQCNDRLKFLSQQPEIMVDSPHFDPRSAAIENPHNLKD 271
S++R+H LQC QG++ KH+EKLVQC+ RL LS+QPEI +DSP+FD R + E+P K
Sbjct: 243 SMNRQHFLQCAQGIMNKHFEKLVQCDHRLFHLSRQPEIAIDSPYFDARQSIFEDPSESKG 302
Query: 272 ---CDLHQGNGSAVSCFQNMGSPHSSLSPS----FTTEHSDPSAI--------------- 309
+L+ G ++S QN+ SP + S S + E PS++
Sbjct: 303 HPFGNLNLSTGPSISGTQNLASPVGAQSSSEHMYLSHEAPSPSSVIDARANEGIGGSEAV 362
Query: 310 ----TLDSVPCEAPS-SSSA*IMKFSSKIC------QMSII*NL**LCKSVIVVLAC*FH 358
D EAP S + F + +C +++ ++ L +S+ V +
Sbjct: 363 NSRNRTDCGQIEAPGIHQSMSLSDFLAVLCDTKNTTDLNLADDVDGLHQSMSVSDFVAY- 421
Query: 359 Q*FNMLGSRN---WDQIKLPGLRPSMSMSDFLGHIEHHISKEMASGDPSFSAERLEYQQM 415
+ SRN DQIK+PGL SMS+SDF+G + + A G E+ E +
Sbjct: 422 ----LSDSRNITDSDQIKVPGLHQSMSVSDFVG-----LLSDSAGGSHPEHMEKFEIMK- 471
Query: 416 MDGITQHLLNDNQVTTDSDEKSLMSRVNSLRCLLQMDPPAVPNSHDNTGFIEGPNDAKVN 475
Q LL+DN DEKSLM RVNSL LL DP NS NT G
Sbjct: 472 -----QQLLSDNIQFEAPDEKSLMPRVNSLFNLLYKDPNVAANSQLNTEMSVGLKSEPKG 526
Query: 476 IDIKATEENSR-----DVYGGNPAPGMSRKDSFGDLLLSLPRIASLPKFLFDISE 525
I N+ D + GM RKDSF DLLL LPRI SLPKFL +ISE
Sbjct: 527 IVSDNNNNNNNNNRVLDTASSSKPQGMLRKDSFSDLLLHLPRITSLPKFLSNISE 581
>UniRef100_Q9ZUI1 At2g24100/F27D4.1 [Arabidopsis thaliana]
Length = 466
Score = 378 bits (970), Expect = e-103
Identities = 240/537 (44%), Positives = 304/537 (55%), Gaps = 85/537 (15%)
Query: 1 MVRFMGSKNPRSQWESSSATSSISPKFEI-EDSIQDQHAPLNKRHKATND--------IL 51
MV M S+N Q S ++ K EI EDS++++HAPLNKR K ++ +L
Sbjct: 1 MVEMMRSENHLRQ-ASKHRNNNFPVKLEIIEDSLEEEHAPLNKRSKLWSNGTSVSKFSLL 59
Query: 52 NEPSPLGLSLRKSPSLLDLIQMTLCQENSVNANTANDNLNSKANKNGRASVEKLKASNFP 111
EPSPLGLSL+KSPS +LI+M L Q + + +N G +VEKLKASNFP
Sbjct: 60 EEPSPLGLSLKKSPSFQELIEMKLSQ----SGDDSNSVKKESFGFGGVGTVEKLKASNFP 115
Query: 112 ATHLKIGSWEYKSKYEGDLVAKCYFAKQKLVWEVLEGELKSKIEIQWSDISQLKANCPDD 171
AT L+IG WEYKS+YEGDLVAKCYFAK KLVWEVLE LKSKIEIQWSDI LKAN P+D
Sbjct: 116 ATILRIGQWEYKSRYEGDLVAKCYFAKHKLVWEVLEQGLKSKIEIQWSDIMALKANLPED 175
Query: 172 GPSTLTLMVARQPLFFRETNPQPRKHTLWQSTTDFTGGQASIHRRHVLQCEQGLLIKHYE 231
P TLT+++AR+PLFFRETNPQPRKHTLWQ+T+DFT GQAS++R+H LQC G++ KH+E
Sbjct: 176 EPGTLTIVLARRPLFFRETNPQPRKHTLWQATSDFTDGQASMNRQHFLQCPPGIMNKHFE 235
Query: 232 KLVQCNDRLKFLSQQPEIMVDSPHFDPRSAAIENPHNLKDCDLHQGNGSAVSCFQNMGSP 291
KLVQC+ RL LS+QPEI + +P FD R + E+P ++ G A S +++
Sbjct: 236 KLVQCDHRLFCLSRQPEINLAAPFFDSRLSIFEDPSVSGSHNIASPVG-AQSSSEHVSLS 294
Query: 292 HSSLSPSFTTEHSDPSAITLDSVPCEAPSSSSA*IMKFSSKICQMSII*NL**LCKSVIV 351
H +LSPS +D+ E S
Sbjct: 295 HDALSPS----------SVMDARAIEGVGGS----------------------------- 315
Query: 352 VLAC*FHQ*FNMLGSRNWDQIKLPGLRPSMSMSDFLGHIEHHISKEMASGDPSFSAERLE 411
+ + W QIK+PGL S+SM+DFL + S + E
Sbjct: 316 ---------IDSRNTNGWSQIKMPGLHQSISMNDFLTFL---------------SDQACE 351
Query: 412 YQQMMDGITQHLLNDNQVTTDSDEKSLMSRVNSLRCLLQMDPPAVPNSHDNTGFIEGPND 471
Q + + Q LL+DN T SDEKS+MS+VNS LLQ + NS N +
Sbjct: 352 NNQEFEEMKQLLLSDNTQTDPSDEKSVMSKVNSFCNLLQ----SAANSQLNIETADTERV 407
Query: 472 AKVNIDIKATEENSRDV---YGGNPAPGMSRKDSFGDLLLSLPRIASLPKFLFDISE 525
V+ + E R V P GMSRKDSF DLL+ LPRI SLPKFLF+ISE
Sbjct: 408 VGVDNNRHMPEGGKRVVDPASSSKPLQGMSRKDSFSDLLVHLPRITSLPKFLFNISE 464
>UniRef100_Q6K679 Hypothetical protein P0700F06.23 [Oryza sativa]
Length = 523
Score = 301 bits (772), Expect = 2e-80
Identities = 205/496 (41%), Positives = 272/496 (54%), Gaps = 81/496 (16%)
Query: 40 LNKRHKATNDILNEPSPLGLSLRKSPSLLDLIQMTLCQENSVNANTANDNLNSKANKNGR 99
L R K IL+ SPLGL LRKSPSLL+LIQM L EN T +++ S++
Sbjct: 83 LGDRLKWDMHILDGSSPLGLRLRKSPSLLELIQMKLAMEN-----TKKEDIKSRS----L 133
Query: 100 ASVEKLKASNFPATHLKIGSWEYKSKYEGDLVAKCYFAKQKLVWEVLEGELKSKIEIQWS 159
+ E++KASNF A LKIG+WE S+YEGDLVAKCYFAK KLVWEVL+ LK KIEIQWS
Sbjct: 134 IASERVKASNFAADFLKIGTWECTSQYEGDLVAKCYFAKHKLVWEVLDAGLKRKIEIQWS 193
Query: 160 DISQLKANCPDDGPSTLTLMVARQPLFFRETNPQPRKHTLWQSTTDFTGGQASIHRRHVL 219
DI LKA CP++G TL L++AR P FF+ET+PQPRKHTLWQ +DFTGGQASI RRH+L
Sbjct: 194 DIIALKATCPENGIGTLDLVLARPPTFFKETDPQPRKHTLWQVASDFTGGQASIKRRHIL 253
Query: 220 QCEQGLLIKHYEKLVQCNDRLKFLSQQPEIMVDSPHFDPRS--AAIENPHNLKDCDLHQG 277
QC+ LL K++EKL+QC+ RL +LS QP M+DSP F P++ + ENP+ K + G
Sbjct: 254 QCQSSLLSKNFEKLIQCDQRLNYLSLQP-YMIDSPVFRPKTEGSIFENPNKSKS---YHG 309
Query: 278 NGSAVSCFQNMGSPHSSLSPSFTTEHSDPSAITLDSVPCEAPSSSSA*IMKFSSKICQMS 337
F + H S + S PC+ P S MK Q S
Sbjct: 310 -------FSYLEGEHESHLSKYIDHVS----------PCDFPLMSKKDGMKDDIANQQQS 352
Query: 338 II*NL**LCKSVIVVLAC*FHQ*FNMLGSRNWD-------QIKLP-----GLRPSMSMSD 385
F + N G+ + D ++K P S+S+ D
Sbjct: 353 -------------------FSRPINW-GASDVDLQVDVSQELKSPHPNSLSQARSLSIDD 392
Query: 386 FLGHIEHHISKEMASG-DPSFSAERLEYQQMMDGITQHLLNDNQVTTDSDEKSLMSRVNS 444
L H++ I ++ +G +PS ++++ ITQ LL+D+ V SDEK +M+RV S
Sbjct: 393 LLSHLDDCIVEQKPAGNNPSLPISEASSNELLEKITQQLLSDSHVAPASDEKRVMARVGS 452
Query: 445 LRCLLQMD------PPAVPNSHDNTGFIEGPNDAKVNIDIKATEENSRDVYGGNPAPGMS 498
L LLQ D P PN G +E + +++ I G NP PG+S
Sbjct: 453 LLSLLQKDAVPANLPKFEPNDSGKIGVVEVGISSALDMGI---------ANGTNP-PGIS 502
Query: 499 RKDSFGDLLLSLPRIA 514
RKDS+ +LL +L I+
Sbjct: 503 RKDSYEELLSNLFNIS 518
>UniRef100_Q8RVI2 Hypothetical protein [Pinus pinaster]
Length = 197
Score = 229 bits (584), Expect = 1e-58
Identities = 111/177 (62%), Positives = 136/177 (76%)
Query: 103 EKLKASNFPATHLKIGSWEYKSKYEGDLVAKCYFAKQKLVWEVLEGELKSKIEIQWSDIS 162
+KLKASNFP ++L+IG+WE S+YEGDLVAKCYFAK KLVWEVL+G LKSKIEIQWSDI+
Sbjct: 15 DKLKASNFPVSNLRIGTWECISRYEGDLVAKCYFAKHKLVWEVLDGGLKSKIEIQWSDIT 74
Query: 163 QLKANCPDDGPSTLTLMVARQPLFFRETNPQPRKHTLWQSTTDFTGGQASIHRRHVLQCE 222
LKA+ +D P TL + V+R PLFFRETNPQPRKHTLWQ+T+DFTGGQA+I RRH LQC
Sbjct: 75 ALKASYLEDEPGTLDIEVSRPPLFFRETNPQPRKHTLWQATSDFTGGQATICRRHFLQCP 134
Query: 223 QGLLIKHYEKLVQCNDRLKFLSQQPEIMVDSPHFDPRSAAIENPHNLKDCDLHQGNG 279
GLL +HYEKL+QC+ RL LS++ + SP FD +SA ++ H+ G
Sbjct: 135 HGLLNRHYEKLIQCDPRLNLLSKKGFLSEASPLFDSKSAVFQDLDEQSSYAKHKKRG 191
>UniRef100_Q5SMV3 Hypothetical protein P0470C02.5-1 [Oryza sativa]
Length = 299
Score = 167 bits (423), Expect = 7e-40
Identities = 114/334 (34%), Positives = 171/334 (51%), Gaps = 60/334 (17%)
Query: 213 IHRRHVLQCEQGLLIKHYEKLVQCNDRLKFLSQQPEIMVDSPHFDPRSAAIENP------ 266
++RRH LQC LL K++EKL+QC+ RL LSQQP+I++DSP F+PR + E+P
Sbjct: 1 MNRRHFLQCPSSLLSKNFEKLLQCDQRLNQLSQQPDIILDSPVFEPRCSIFEDPVESKCQ 60
Query: 267 --HNLKD-CDLHQGNGSAVSCFQNMGSPHSSLSPSFTTE--------HSDPSAITLDSVP 315
NLKD +L +GS C + S ++ S T+ + PSA+ + V
Sbjct: 61 GFTNLKDEHELSGFSGSLSPCAGSSMSAKIEVNDSIATQAGFLAQPGNPGPSAVNVQGVS 120
Query: 316 CEAPSSSSA*IMKFSSKICQMSII*NL**LCKSVIVVLAC*FHQ*FNMLGSRNWDQIKLP 375
+ I + W Q+K+P
Sbjct: 121 RNVNGAPELNIPSW---------------------------------------WSQLKVP 141
Query: 376 GLRPSMSMSDFLGHIEHHISKEMASGDPSFSAERLEYQQMMDGITQHLLNDNQ--VTTDS 433
GLRPSMS+ D + H+ + IS+++ S +P+ + + ++ ++ I Q+LL D Q + S
Sbjct: 142 GLRPSMSVDDLVNHLGNCISEQITSVNPTLPSNEVPTKETLEEIAQYLLGDAQGPPASTS 201
Query: 434 DEKSLMSRVNSLRCLLQMDPPAV--PNSHDNTGFIEGPNDAKVNIDIKATEENSRDVYGG 491
DE+SLM+RV+SL CL+Q D P V P N G + + + + ++ ++ G
Sbjct: 202 DERSLMARVDSLCCLIQKDTPPVAQPKPEPNDSDSIGGDGTEGSDEEFSSAASTVKTTGP 261
Query: 492 NPAPGMSRKDSFGDLLLSLPRIASLPKFLFDISE 525
P MSRKDSFGDLL++LPRIASLP+FLF I E
Sbjct: 262 AQPPAMSRKDSFGDLLMNLPRIASLPQFLFKIPE 295
>UniRef100_Q9M9L9 F10A16.6 protein [Arabidopsis thaliana]
Length = 410
Score = 133 bits (334), Expect = 1e-29
Identities = 85/197 (43%), Positives = 112/197 (56%), Gaps = 14/197 (7%)
Query: 48 NDILNEPSPLGLSLRKSPSLLDLIQMTL-CQENSVNANTANDNLNSKANKNGRASVEKLK 106
N ++E L L L K+P L++ I+ L + T N + S K S EKLK
Sbjct: 17 NRFVDEGPRLNLPLTKTPELINKIESYLKVHYTCPHQQTENSSKTSTLPK----SPEKLK 72
Query: 107 ASNFPATHLKIGSWEYKSKYEGDLVAKCYFAKQKLVWEVLEGE-------LKSKIEIQWS 159
A NFP + +KIG + +K D+VAK YFAK+KL+WE L GE LKSKIEIQW+
Sbjct: 73 AMNFPISTIKIGDCVFVAKNPDDIVAKFYFAKKKLLWEFLFGEPVANMPRLKSKIEIQWN 132
Query: 160 DISQLKANCPD-DGPSTLTLMVARQPLFFRETNPQPRKHTLW-QSTTDFTGGQASIHRRH 217
D+S + + D L + + ++P FF ETNPQ KHT W Q DFTG QAS +RRH
Sbjct: 133 DVSSFEESINSRDETGILKIELKKRPTFFTETNPQAGKHTQWKQLDYDFTGDQASYYRRH 192
Query: 218 VLQCEQGLLIKHYEKLV 234
L G+L K+ EKL+
Sbjct: 193 TLHFPPGVLQKNLEKLL 209
>UniRef100_Q9SLK4 F20D21.12 protein [Arabidopsis thaliana]
Length = 444
Score = 132 bits (333), Expect = 2e-29
Identities = 90/211 (42%), Positives = 119/211 (55%), Gaps = 15/211 (7%)
Query: 57 LGLSLRKSPSLLDLIQMTLCQENSV-NANTANDNLNSKANKNGRASVEKLKASNFPATHL 115
L LSL KSP L++ I+ L + + T N + S K S EKLKA NFP + +
Sbjct: 8 LNLSLTKSPELINKIESYLNGHCTCPHQQTENSSKRSTLPK----SPEKLKAMNFPISTI 63
Query: 116 KIGSWEYKSKYEGDLVAKCYFAKQKLVWEVLEGE-------LKSKIEIQWSDISQLKANC 168
+IG W +K D+VAK YFAK+KL+WE L GE LK KIEIQW+D+S + +
Sbjct: 64 RIGGWVVVAKNPDDIVAKFYFAKKKLIWEFLFGEPETNTLRLKRKIEIQWNDVSSFEESI 123
Query: 169 PD-DGPSTLTLMVARQPLFFRETNPQPRKHTLW-QSTTDFTGGQASIHRRHVLQCEQGLL 226
D L + + ++P FF ETNPQ KHT W Q DFTG AS +RRH L G+L
Sbjct: 124 SSRDETGILKIELKKRPTFFIETNPQAGKHTQWKQLDHDFTGDHASNYRRHTLHFPPGVL 183
Query: 227 IKHYEKLVQCNDRLKFLSQQPEIMVDSPHFD 257
K+ EKLV + K L + P + +S +FD
Sbjct: 184 QKNLEKLVTDSFWSK-LYEVPFPVHESRYFD 213
>UniRef100_Q8T148 Similar to Dictyostelium discoideum (Slime mold).
Homeobox-containing protein [Dictyostelium discoideum]
Length = 1108
Score = 132 bits (333), Expect = 2e-29
Identities = 70/191 (36%), Positives = 116/191 (60%), Gaps = 11/191 (5%)
Query: 69 DLIQMTLCQENSVNANTANDNLNSKA--NKNGRASVEKLKASNFPATHLKIGSWEYKSKY 126
+++ T +S+ NT N +N A + +G KLK SN AT +K+G WE + +
Sbjct: 442 NILSSTSTSSSSIPINT-NPLINDSAVDDDDGLTETSKLKPSNLAATKIKVGEWECSASF 500
Query: 127 EGDLVAKCYFAKQKLVWEVLEGELKSKIEIQWSDISQLKANCPDDGPSTLTLMVARQPLF 186
GD++AK YF K+++VWE+L+ LKSK+ I +S+I+ +K D +TL++ +++ P F
Sbjct: 501 PGDIIAKFYFTKKQIVWELLKLGLKSKMVISFSEITAVKVEDTSDDSATLSIEISKAPKF 560
Query: 187 FRETNPQPRKHTLWQSTTDFTGGQ-ASIHRRHVLQCEQGLLIKHYEKLVQCNDRLKFLSQ 245
++E NPQP+K+T W + DFT Q AS +RRH+L ++ L K+ KL + + +LK L
Sbjct: 561 YKEVNPQPKKNTTWNLSNDFTDNQSASTYRRHILTFKKSSLAKNIVKLHKSDQKLKKL-- 618
Query: 246 QPEIMVDSPHF 256
+++ HF
Sbjct: 619 -----IENQHF 624
>UniRef100_UPI00003C22CB UPI00003C22CB UniRef100 entry
Length = 1084
Score = 41.6 bits (96), Expect = 0.057
Identities = 51/197 (25%), Positives = 76/197 (37%), Gaps = 48/197 (24%)
Query: 78 ENSVNANTANDNLNSKANKNGRASVEKLKAS--NFPATHLKIGSWEYKSKYEGDLVAKCY 135
+ ++ +A DN + GR +L S P + L IG+W S C+
Sbjct: 328 DKDAHSKSAGDNA-----RLGRGDPAELVPSVTAIPTSALCIGTWRRVSPLI------CF 376
Query: 136 FAK--QKLVWEVLEGELKSKIEIQWSDISQLKANCPDDGPST----------------LT 177
F++ Q L W + + K+EI W+ I + DGPS
Sbjct: 377 FSRRLQSLTWYLTSESIGFKLEIPWTSI----RSAYFDGPSNPSIAERAEGVRVPLGHFV 432
Query: 178 LMVARQPLFFRE----------TNPQPRKHTLWQSTTDFT-GGQASIHRRHVLQCEQGLL 226
+ + R P FF E +N +P+ W+ DFT QA RHV+ L
Sbjct: 433 IDLERPPTFFMEVFRSAPLKEGSNEEPK--VSWRQCEDFTEDHQAMTVSRHVINGPYEEL 490
Query: 227 IKHYEKLVQCNDRLKFL 243
+ L +CND LK L
Sbjct: 491 RIAVQALARCNDVLKRL 507
>UniRef100_Q95PH8 Histidine kinase DhkH [Dictyostelium discoideum]
Length = 1147
Score = 37.7 bits (86), Expect = 0.82
Identities = 19/51 (37%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Query: 60 SLRKSPSLLDLIQMTLCQEN-SVNANTANDNLNSKANKNGRASVEKLKASN 109
SL SPS+ L +L N +++ N +N+N+N+ N +G ++ +KLK SN
Sbjct: 847 SLSSSPSIQGLTNSSLSINNINISGNNSNNNINNNNNNSGSSTPKKLKKSN 897
>UniRef100_UPI00003AB6D7 UPI00003AB6D7 UniRef100 entry
Length = 632
Score = 37.0 bits (84), Expect = 1.4
Identities = 39/150 (26%), Positives = 59/150 (39%), Gaps = 16/150 (10%)
Query: 366 SRNWDQIKLPGLRPSMSMSDFLGHIEHHISKEMA-SGDPSFSAERLEYQQMMDGITQHLL 424
S W +I+ L ++ S + +E+A S P ERLE D T HLL
Sbjct: 134 STTWSRIRAHSLN-TVKQSSLAEPVSPSKQQEIAPSSPPGLLEERLE-----DASTVHLL 187
Query: 425 NDNQVTTDSDEKSLMSRVNSLRCLLQMDPPAVPNSHDNTGFIEGPNDAKVNIDIKATEEN 484
VT D +++ SR + L + D + TG P D + + + + N
Sbjct: 188 RTRSVTPDKSDRTHNSRNHVLSAQERDDTSVLCLRKSETGTHSVPADHQESSKLTVSNRN 247
Query: 485 SRDVYGGNPAPGMSRKDSFGDLLLSLPRIA 514
S P + KD G L+ S+ R A
Sbjct: 248 S---------PSSALKDLSGKLMKSIERDA 268
>UniRef100_Q8CS00 Hypothetical protein SE1494 [Staphylococcus epidermidis]
Length = 765
Score = 37.0 bits (84), Expect = 1.4
Identities = 32/109 (29%), Positives = 47/109 (42%), Gaps = 10/109 (9%)
Query: 397 EMASGDPSFSAERLEYQQMMDGITQHLLNDNQVTTDSDEKSLMSRVNSLRCLLQMDPPAV 456
E A A+ + + M+D + DNQ + DEKS ++ N LQ D
Sbjct: 460 ENARSSAKSVADEIHDKHMLDNAGSYNKRDNQ--KNKDEKSFINPDNPN---LQQDKKQG 514
Query: 457 PNSHDNTGFIEGPNDAKVNIDIKATEENSRDVYGGNPAPGMSRKDSFGD 505
NS+ +TG ND+K+N TEE + G + KDS G+
Sbjct: 515 ENSYSDTG-----NDSKLNKSNNQTEEAEDNKNGHDKNIDSKEKDSLGN 558
>UniRef100_Q8GXJ0 Hypothetical protein [Arabidopsis thaliana]
Length = 107
Score = 37.0 bits (84), Expect = 1.4
Identities = 24/62 (38%), Positives = 32/62 (50%), Gaps = 3/62 (4%)
Query: 274 LHQGNGSAVSCFQNMGSPHSSLSPSFTTEHSD---PSAITLDSVPCEAPSSSSA*IMKFS 330
L + + A SC + S S S SF+T S P+ + S C APSSSS+ I + S
Sbjct: 7 LSKKDALAASCSSSSTSSKSKFSRSFSTSASSSKAPAFVRSSSTKCSAPSSSSSSISRSS 66
Query: 331 SK 332
SK
Sbjct: 67 SK 68
>UniRef100_Q8T8S8 AT30755p [Drosophila melanogaster]
Length = 1193
Score = 36.6 bits (83), Expect = 1.8
Identities = 38/166 (22%), Positives = 75/166 (44%), Gaps = 24/166 (14%)
Query: 16 SSSATSSISPKF---EIEDSIQDQHAPLNKRHKATNDILNEPSPLGLSLRKSPSLLDLIQ 72
S +A + IS + + +D+ + H + ++ + ++ P GL+ + SP +LDL
Sbjct: 362 SDAALTVISSRLTAGQEDDNARIGHGHITNTSAESDHMTSKSIPSGLTTKDSPDVLDL-- 419
Query: 73 MTLCQENSVNANTANDNLNSKANKNG---------RASVEKLKASNFPATHLKIGSWEYK 123
C+ ++++ N L A N + ++K +A+NF A I S
Sbjct: 420 --SCESEPLHSSKQNSQLMEDAETNFTERLKVNQLKEQIDKQEAANFKAQENLIKSIRQT 477
Query: 124 SKYEGD------LVAKCYFAKQ--KLVWEVLEGELKSKIEIQWSDI 161
+ GD L+AK + +L +V +LKS++E + S++
Sbjct: 478 IQALGDKEKMENLIAKSKMEEDPFQLKSQVESSQLKSQVEGESSEL 523
>UniRef100_UPI000024CF29 UPI000024CF29 UniRef100 entry
Length = 2430
Score = 36.2 bits (82), Expect = 2.4
Identities = 56/256 (21%), Positives = 105/256 (40%), Gaps = 31/256 (12%)
Query: 83 ANTAN--DNLNSKANKNGRASVEKLKASNFPATHLKIGSWEYK-SKYEGDLVAKCYFAKQ 139
A+TA+ +L SK K+G+ ++ K S+ WE + E V C A++
Sbjct: 2061 AHTASLLSSLTSKGKKDGKQESKRDKESDLEKAKANAALWEARCDMTESARVEYCEAARR 2120
Query: 140 -----KLVWEVLEGELKSKIEIQWSDISQLKANCPDDGPSTLTLMVARQPLFFRET---- 190
K + + KS IEI I+ L+ + L L+ L R +
Sbjct: 2121 LNKTNKQLNDQQHNTEKSSIEI----IAVLRDKDMLNEEKNLDLITQDSDLITRSSDFTT 2176
Query: 191 -NPQPRKHT--LWQSTTDFTGGQASI--HRRHVLQCEQGLLIKHYEKLVQCNDRLKFLSQ 245
N H L ++DFT + + H ++ C L+ ++ + L+ CN F++Q
Sbjct: 2177 QNSDLITHNSDLITHSSDFTTQNSDLITHNSDLITCNSDLITQNSD-LITCNS--DFITQ 2233
Query: 246 QPEIMVDSPHFDPRSAAIENPHNLKDCDLHQGNGSAVSCFQNMGSPHSSL---SPSFTTE 302
+++ + RS + ++ DL N ++C + + +S L + F T+
Sbjct: 2234 NLDLITQNSDLITRS----SDFTTQNSDLITHNSDLITCNSDFITQNSDLITCNSDFITQ 2289
Query: 303 HSDPSAITLDSVPCEA 318
+SD D + C++
Sbjct: 2290 NSDLITCNSDFITCKS 2305
>UniRef100_UPI00000848BE UPI00000848BE UniRef100 entry
Length = 1186
Score = 36.2 bits (82), Expect = 2.4
Identities = 38/166 (22%), Positives = 75/166 (44%), Gaps = 24/166 (14%)
Query: 16 SSSATSSISPKF---EIEDSIQDQHAPLNKRHKATNDILNEPSPLGLSLRKSPSLLDLIQ 72
S +A + IS + + +D+ + H + ++ + ++ P GL+ + SP +LDL
Sbjct: 355 SDAALTVISSRLTAGQEDDNARIGHGHITNTSAESDHMSSKSIPSGLTTKDSPDVLDL-- 412
Query: 73 MTLCQENSVNANTANDNLNSKANKNG---------RASVEKLKASNFPATHLKIGSWEYK 123
C+ ++++ N L A N + ++K +A+NF A I S
Sbjct: 413 --SCESEPLHSSKQNSQLMEDAETNFTERLKVNQLKEQIDKQEAANFKAQENLIKSIRQT 470
Query: 124 SKYEGD------LVAKCYFAKQ--KLVWEVLEGELKSKIEIQWSDI 161
+ GD L+AK + +L +V +LKS++E + S++
Sbjct: 471 IQALGDKEKMENLIAKSKMEEDPFQLKSQVESSQLKSQVEGESSEL 516
>UniRef100_UPI0000499528 UPI0000499528 UniRef100 entry
Length = 1026
Score = 36.2 bits (82), Expect = 2.4
Identities = 29/102 (28%), Positives = 50/102 (48%), Gaps = 13/102 (12%)
Query: 22 SISPKFEIEDSIQDQH---APLNKRHKATNDILNEPSPLGLSLRKS------PSLLDLIQ 72
S +P+ +E SI+ +H +P NK K + +L+ P PL + +++ P+ D+ +
Sbjct: 701 STNPQPTVESSIKQKHFYLSPTNKLKKIDDSLLSIPLPLRVPTKRTIPITIQPNNNDVYK 760
Query: 73 MTLCQENSVNANTANDNLNSKANKNGRASVEKLKASNFPATH 114
+ +SV A T N N+NS S L+ PAT+
Sbjct: 761 ----KSDSVRAITGNRNVNSTQPLTKSKSNVMLRQITMPATN 798
>UniRef100_Q5RE67 Hypothetical protein DKFZp469D1917 [Pongo pygmaeus]
Length = 954
Score = 36.2 bits (82), Expect = 2.4
Identities = 26/95 (27%), Positives = 42/95 (43%), Gaps = 6/95 (6%)
Query: 409 RLEYQQMMDGITQHLLNDNQVTTDSDEKSLMSRVNSLRCLLQMDPPAVPNSHDNTGFIEG 468
R E Q+++D + +HL+ND V S R L C+ +P + P + N I
Sbjct: 502 RSEIQKVLDSLQEHLMNDPDVQAQVQVLSAALRAAQLDCV--NEPESKPTAGLNEVSISH 559
Query: 469 PNDAKVNIDIKATEENSRDVYGG----NPAPGMSR 499
P+ A N A + +++ +P PG SR
Sbjct: 560 PSSASDNQITLAASSSQNELFVARILQSPDPGESR 594
>UniRef100_Q9VD20 CG31169-PA, isoform A [Drosophila melanogaster]
Length = 1469
Score = 36.2 bits (82), Expect = 2.4
Identities = 38/166 (22%), Positives = 75/166 (44%), Gaps = 24/166 (14%)
Query: 16 SSSATSSISPKF---EIEDSIQDQHAPLNKRHKATNDILNEPSPLGLSLRKSPSLLDLIQ 72
S +A + IS + + +D+ + H + ++ + ++ P GL+ + SP +LDL
Sbjct: 638 SDAALTVISSRLTAGQEDDNARIGHGHITNTSAESDHMSSKSIPSGLTTKDSPDVLDL-- 695
Query: 73 MTLCQENSVNANTANDNLNSKANKNG---------RASVEKLKASNFPATHLKIGSWEYK 123
C+ ++++ N L A N + ++K +A+NF A I S
Sbjct: 696 --SCESEPLHSSKQNSQLMEDAETNFTERLKVNQLKEQIDKQEAANFKAQENLIKSIRQT 753
Query: 124 SKYEGD------LVAKCYFAKQ--KLVWEVLEGELKSKIEIQWSDI 161
+ GD L+AK + +L +V +LKS++E + S++
Sbjct: 754 IQALGDKEKMENLIAKSKMEEDPFQLKSQVESSQLKSQVEGESSEL 799
>UniRef100_Q8IN06 CG31169-PB, isoform B [Drosophila melanogaster]
Length = 1304
Score = 36.2 bits (82), Expect = 2.4
Identities = 38/166 (22%), Positives = 75/166 (44%), Gaps = 24/166 (14%)
Query: 16 SSSATSSISPKF---EIEDSIQDQHAPLNKRHKATNDILNEPSPLGLSLRKSPSLLDLIQ 72
S +A + IS + + +D+ + H + ++ + ++ P GL+ + SP +LDL
Sbjct: 473 SDAALTVISSRLTAGQEDDNARIGHGHITNTSAESDHMSSKSIPSGLTTKDSPDVLDL-- 530
Query: 73 MTLCQENSVNANTANDNLNSKANKNG---------RASVEKLKASNFPATHLKIGSWEYK 123
C+ ++++ N L A N + ++K +A+NF A I S
Sbjct: 531 --SCESEPLHSSKQNSQLMEDAETNFTERLKVNQLKEQIDKQEAANFKAQENLIKSIRQT 588
Query: 124 SKYEGD------LVAKCYFAKQ--KLVWEVLEGELKSKIEIQWSDI 161
+ GD L+AK + +L +V +LKS++E + S++
Sbjct: 589 IQALGDKEKMENLIAKSKMEEDPFQLKSQVESSQLKSQVEGESSEL 634
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 885,447,664
Number of Sequences: 2790947
Number of extensions: 37482405
Number of successful extensions: 122701
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 122594
Number of HSP's gapped (non-prelim): 79
length of query: 525
length of database: 848,049,833
effective HSP length: 132
effective length of query: 393
effective length of database: 479,644,829
effective search space: 188500417797
effective search space used: 188500417797
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC126784.4


