

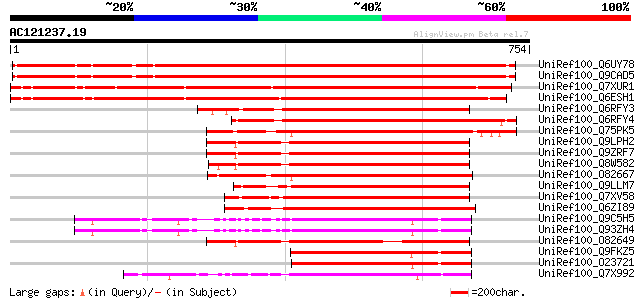
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.19 + phase: 0 /pseudo
(754 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6UY78 YDA [Arabidopsis thaliana] 862 0.0
UniRef100_Q9CAD5 Hypothetical protein F24D7.11 [Arabidopsis thal... 860 0.0
UniRef100_Q7XUR1 OSJNBa0084K11.3 protein [Oryza sativa] 839 0.0
UniRef100_Q6ESH1 Putative MAP3K alpha 1 protein kinase [Oryza sa... 832 0.0
UniRef100_Q6RFY3 MAP3Ka [Lycopersicon esculentum] 428 e-118
UniRef100_Q6RFY4 MAP3Ka [Nicotiana benthamiana] 424 e-117
UniRef100_Q75PK5 Mitogen-activated kinase kinase kinase alpha [L... 418 e-115
UniRef100_Q9LPH2 T3F20.12 protein [Arabidopsis thaliana] 413 e-114
UniRef100_Q9ZRF7 MEK kinase [Arabidopsis thaliana] 413 e-114
UniRef100_Q8W582 At1g53570/F22G10_18 [Arabidopsis thaliana] 412 e-113
UniRef100_O82667 MAP3K alpha 1 protein kinase [Brassica napus] 407 e-112
UniRef100_Q9LLM7 Similar to mitogen-activated protein kinases [O... 400 e-110
UniRef100_Q7XV58 OSJNBa0006B20.13 protein [Oryza sativa] 394 e-108
UniRef100_Q6ZI89 Putative MEK kinase [Oryza sativa] 384 e-105
UniRef100_Q9C5H5 Putative MAP protein kinase [Arabidopsis thaliana] 377 e-103
UniRef100_Q93ZH4 AT5g66850/MUD21_11 [Arabidopsis thaliana] 375 e-102
UniRef100_O82649 MAP3K alpha protein kinase [Arabidopsis thaliana] 364 5e-99
UniRef100_Q9FKZ5 MAP protein kinase [Arabidopsis thaliana] 340 9e-92
UniRef100_O23721 MAP3K gamma protein kinase [Arabidopsis thaliana] 338 3e-91
UniRef100_Q7X992 Putative MAP3K protein kinase [Oryza sativa] 331 6e-89
>UniRef100_Q6UY78 YDA [Arabidopsis thaliana]
Length = 883
Score = 862 bits (2226), Expect = 0.0
Identities = 464/734 (63%), Positives = 542/734 (73%), Gaps = 17/734 (2%)
Query: 4 WWGKSSSKETKKKAGKESIIDTLHRKFKFPSEGKRSTISGGSRRRSNDTISEKGDRSPSE 63
WW KS K+ KKK KESIID +RK F SE + S S SRRR ++ +SE+G S
Sbjct: 3 WWSKS--KDEKKKTNKESIIDAFNRKLGFASEDRSSGRSRKSRRRRDEIVSERGAISRLP 60
Query: 64 SRSPSPS-KVARCQSFAERPHAQPLPLPDLHPSSLGRVDSEISISAKSRLEKPSKPSLFL 122
SRSPSPS +V+RCQSFAER A PLP P + P + DS ++ S + L+ KPS +L
Sbjct: 61 SRSPSPSTRVSRCQSFAERSPAVPLPRPIVRPH-VTSTDSGMNGSQRPGLDANLKPS-WL 118
Query: 123 PLPKPSCIRCGPTPADLDGDMVNASVFSDCSADSDEPADSRNRSPLATDSETGTRTAAGS 182
PLPKP P + D ASV S S D P+DS SPLA+D E G RT
Sbjct: 119 PLPKPHGATSIPDNTGAEPDFATASVSSGSSV-GDIPSDSL-LSPLASDCENGNRT---- 172
Query: 183 PSSLVLKDQSSAVSQPNLREVKKTANILSNHTPSTSPKRKPLRHHVPNLQVPPHG-VFYS 241
P ++ +DQS ++ + K AN N S SP+R+PL HV NLQ+P V S
Sbjct: 173 PVNISSRDQSMHSNKNSAEMFKPVAN--KNRILSASPRRRPLGTHVKNLQIPQRDLVLCS 230
Query: 242 GPDSSLSSPSRSPLRAFGTDQVLNSAFWAGKPYPEINFVGSGHCSSPGSGHNSGHNSMGG 301
PDS LSSPSRSP+R+F DQV N KPY +++ +GSG CSSPGSG+NSG+NS+GG
Sbjct: 231 APDSLLSSPSRSPMRSFIPDQVSNHGLLISKPYSDVSLLGSGQCSSPGSGYNSGNNSIGG 290
Query: 302 DMSGPLFWQPSRGSPEYSPIPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADD 361
DM+ LFW SR SPE SP+PSPRMTSPGPSSRIQSGAVTP+HPRAGG+ T S T R DD
Sbjct: 291 DMATQLFWPQSRCSPECSPVPSPRMTSPGPSSRIQSGAVTPLHPRAGGSTTGSPTRRLDD 350
Query: 362 GKQQSHRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGT 421
+QQSHRLPLPPL ++NT PFS + SAATSPS+PRSPARA++ +S GSRWKKG+LLG G+
Sbjct: 351 NRQQSHRLPLPPLLISNTCPFSPTYSAATSPSVPRSPARAEATVSPGSRWKKGRLLGMGS 410
Query: 422 FGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSET 481
FGHVY+GFNS+SGEMCAMKEVTL SDD KS ESA+QL QE+ +LSRLRH NIVQYYGSET
Sbjct: 411 FGHVYLGFNSESGEMCAMKEVTLCSDDPKSRESAQQLGQEISVLSRLRHQNIVQYYGSET 470
Query: 482 VDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGAN 541
VDDKLYIYLEYVSGGSI+KLLQEYGQFGE AIR+YTQQILSGLAYLHAKNT+HRDIKGAN
Sbjct: 471 VDDKLYIYLEYVSGGSIYKLLQEYGQFGENAIRNYTQQILSGLAYLHAKNTVHRDIKGAN 530
Query: 542 ILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTV 601
ILVDP+GRVKVADFGMAKHIT Q PLSFKGSPYWMAPEVIKNS +L VDIWSLGCTV
Sbjct: 531 ILVDPHGRVKVADFGMAKHITAQSGPLSFKGSPYWMAPEVIKNSNGSNLAVDIWSLGCTV 590
Query: 602 LEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASEL 661
LEMATTKPPWSQYEGV AMFKIGNSKELP IPDHLS EGKDFVRKCLQRNP +RP+A++L
Sbjct: 591 LEMATTKPPWSQYEGVPAMFKIGNSKELPDIPDHLSEEGKDFVRKCLQRNPANRPTAAQL 650
Query: 662 LDHPFVKGAAPLERPIMVPEASDPITGITHGTKALGIGQGRNLSALDSDKLLVHSSRVLK 721
LDH FV+ P+ERPI+ E ++ + + ++L IG R+L LDS+ + + LK
Sbjct: 651 LDHAFVRNVMPMERPIVSGEPAEAMNVASSTMRSLDIGHARSLPCLDSEDATNYQQKGLK 710
Query: 722 NNPHESLKSFHQFP 735
H S S Q P
Sbjct: 711 ---HGSGFSISQSP 721
>UniRef100_Q9CAD5 Hypothetical protein F24D7.11 [Arabidopsis thaliana]
Length = 883
Score = 860 bits (2221), Expect = 0.0
Identities = 463/734 (63%), Positives = 541/734 (73%), Gaps = 17/734 (2%)
Query: 4 WWGKSSSKETKKKAGKESIIDTLHRKFKFPSEGKRSTISGGSRRRSNDTISEKGDRSPSE 63
WW KS K+ KKK KESIID +RK F SE + S S SRRR ++ +SE+G S
Sbjct: 3 WWSKS--KDEKKKTNKESIIDAFNRKLGFASEDRSSGRSRKSRRRRDEIVSERGAISRLP 60
Query: 64 SRSPSPS-KVARCQSFAERPHAQPLPLPDLHPSSLGRVDSEISISAKSRLEKPSKPSLFL 122
SRSPSPS +V+RCQSFAER A PLP P + P + DS ++ S + L+ KPS +L
Sbjct: 61 SRSPSPSTRVSRCQSFAERSPAVPLPRPIVRPH-VTSTDSGMNGSQRPGLDANLKPS-WL 118
Query: 123 PLPKPSCIRCGPTPADLDGDMVNASVFSDCSADSDEPADSRNRSPLATDSETGTRTAAGS 182
PLPKP P + D ASV S S D P+DS SPLA+D E G RT
Sbjct: 119 PLPKPHGATSIPDNTGAEPDFATASVSSGSSV-GDIPSDSL-LSPLASDCENGNRT---- 172
Query: 183 PSSLVLKDQSSAVSQPNLREVKKTANILSNHTPSTSPKRKPLRHHVPNLQVPPHG-VFYS 241
P ++ +DQS ++ + K N N S SP+R+PL HV NLQ+P V S
Sbjct: 173 PVNISSRDQSMHSNKNSAEMFKPVPN--KNRILSASPRRRPLGTHVKNLQIPQRDLVLCS 230
Query: 242 GPDSSLSSPSRSPLRAFGTDQVLNSAFWAGKPYPEINFVGSGHCSSPGSGHNSGHNSMGG 301
PDS LSSPSRSP+R+F DQV N KPY +++ +GSG CSSPGSG+NSG+NS+GG
Sbjct: 231 APDSLLSSPSRSPMRSFIPDQVSNHGLLISKPYSDVSLLGSGQCSSPGSGYNSGNNSIGG 290
Query: 302 DMSGPLFWQPSRGSPEYSPIPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADD 361
DM+ LFW SR SPE SP+PSPRMTSPGPSSRIQSGAVTP+HPRAGG+ T S T R DD
Sbjct: 291 DMATQLFWPQSRCSPECSPVPSPRMTSPGPSSRIQSGAVTPLHPRAGGSTTGSPTRRLDD 350
Query: 362 GKQQSHRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGT 421
+QQSHRLPLPPL ++NT PFS + SAATSPS+PRSPARA++ +S GSRWKKG+LLG G+
Sbjct: 351 NRQQSHRLPLPPLLISNTCPFSPTYSAATSPSVPRSPARAEATVSPGSRWKKGRLLGMGS 410
Query: 422 FGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSET 481
FGHVY+GFNS+SGEMCAMKEVTL SDD KS ESA+QL QE+ +LSRLRH NIVQYYGSET
Sbjct: 411 FGHVYLGFNSESGEMCAMKEVTLCSDDPKSRESAQQLGQEISVLSRLRHQNIVQYYGSET 470
Query: 482 VDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGAN 541
VDDKLYIYLEYVSGGSI+KLLQEYGQFGE AIR+YTQQILSGLAYLHAKNT+HRDIKGAN
Sbjct: 471 VDDKLYIYLEYVSGGSIYKLLQEYGQFGENAIRNYTQQILSGLAYLHAKNTVHRDIKGAN 530
Query: 542 ILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTV 601
ILVDP+GRVKVADFGMAKHIT Q PLSFKGSPYWMAPEVIKNS +L VDIWSLGCTV
Sbjct: 531 ILVDPHGRVKVADFGMAKHITAQSGPLSFKGSPYWMAPEVIKNSNGSNLAVDIWSLGCTV 590
Query: 602 LEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASEL 661
LEMATTKPPWSQYEGV AMFKIGNSKELP IPDHLS EGKDFVRKCLQRNP +RP+A++L
Sbjct: 591 LEMATTKPPWSQYEGVPAMFKIGNSKELPDIPDHLSEEGKDFVRKCLQRNPANRPTAAQL 650
Query: 662 LDHPFVKGAAPLERPIMVPEASDPITGITHGTKALGIGQGRNLSALDSDKLLVHSSRVLK 721
LDH FV+ P+ERPI+ E ++ + + ++L IG R+L LDS+ + + LK
Sbjct: 651 LDHAFVRNVMPMERPIVSGEPAEAMNVASSTMRSLDIGHARSLPCLDSEDATNYQQKGLK 710
Query: 722 NNPHESLKSFHQFP 735
H S S Q P
Sbjct: 711 ---HGSGFSISQSP 721
>UniRef100_Q7XUR1 OSJNBa0084K11.3 protein [Oryza sativa]
Length = 894
Score = 839 bits (2168), Expect = 0.0
Identities = 449/735 (61%), Positives = 541/735 (73%), Gaps = 14/735 (1%)
Query: 1 MPTWWGKSSSKETKKKAGKESIIDTLHRKFKFPSEGKRSTISGGSRRRSNDTISEKGDRS 60
MP WWGKS SK+ KK KE++IDT HR P++ K ST S S RR ND+ EK RS
Sbjct: 1 MPPWWGKSFSKDAKKTT-KENLIDTFHRLIS-PNDQKGSTKSKRSCRRGNDSSVEKSCRS 58
Query: 61 PSESRSPSPSK-VARCQSF-AERPHAQPLPLPDLHPSSLGRVDSEISISAKSRLEKPSKP 118
+ SR SPSK V+RCQSF A+RPHA PLP+P + P + R S+I+ S K LEK KP
Sbjct: 59 TTVSRPTSPSKEVSRCQSFSADRPHAHPLPIPGVRPP-VTRTVSDITES-KPILEKRGKP 116
Query: 119 SLFLPLPKPSCIRCGPTPADLDGDMVNASVFSDCSADSDEPADSRNRSPLATDSETGTRT 178
L LPLPKP+ +++ ++V AS S+CS DSD+ DS+ +SP+ D+E T
Sbjct: 117 PLLLPLPKPNRPPRRHGNSEVVSEIVVASPSSNCS-DSDDHGDSQLQSPVGNDAENATLV 175
Query: 179 AAGSPSSLVLKDQSSAVSQPNLREVKKTAN-ILSNHTPSTSPKRKPLRHHVPNLQVPPHG 237
+ SS K+ ++ N++E+ + AN + +H STSP+ + NLQ P
Sbjct: 176 TLKNKSSNARKECPGPITAKNMKEIHRPANQVHGSHILSTSPRGVAADSYQSNLQNPRPL 235
Query: 238 VFYSGPDSSLSSPSRSPLRAFGTDQVLNSAFWAGKPYPEINFVGSGHCSSPGSGHNSGHN 297
V S P+S +SSPSRSP R D + SAFWA KP+ ++ FVGSG CSSPGSG SGHN
Sbjct: 236 VLDSAPNSLMSSPSRSP-RRICPDHIPTSAFWAVKPHTDVTFVGSGQCSSPGSGQTSGHN 294
Query: 298 SMGGDMSGPLFWQPSRGSPEYSPIPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTG 357
S+GGDM LFWQPSR SPE SPIPSPRMTSPGPSSR+ SG+V+P+HPR+GG ES T
Sbjct: 295 SVGGDMLAQLFWQPSRSSPECSPIPSPRMTSPGPSSRVHSGSVSPLHPRSGGMAPESPTN 354
Query: 358 RADDGKQ-QSHRLPLPPLTVTNTSPFSHSNSAATSP-SMPRSPARADSPMSSGSRWKKGK 415
R DDGK+ Q+H+LPLPPL+++++S F +NS TSP S+PRSP R ++P S GSRWKKGK
Sbjct: 355 RHDDGKKKQTHKLPLPPLSISHSS-FHPNNSTPTSPISVPRSPGRTENPPSPGSRWKKGK 413
Query: 416 LLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQ 475
L+GRGTFGHVY+GFNS SGEMCAMKEVTLF DD KS ESAKQL QE+ LLSRL+HPNIVQ
Sbjct: 414 LIGRGTFGHVYVGFNSDSGEMCAMKEVTLFLDDPKSKESAKQLGQEISLLSRLQHPNIVQ 473
Query: 476 YYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHR 535
YYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQ GE AIRSYTQQILSGLAYLHAKNT+HR
Sbjct: 474 YYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQLGEQAIRSYTQQILSGLAYLHAKNTVHR 533
Query: 536 DIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIW 595
DIKGANILVDP+GRVK+ADFGMAKHI GQ CP SFKGSPYWMAPEVIKNS C+L VDIW
Sbjct: 534 DIKGANILVDPSGRVKLADFGMAKHINGQQCPFSFKGSPYWMAPEVIKNSNGCNLAVDIW 593
Query: 596 SLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDR 655
SLGCTVLEMAT+KPPWSQYEG+AAMFKIGNSKELP IPDHLS GKDF+RKCLQR+P R
Sbjct: 594 SLGCTVLEMATSKPPWSQYEGIAAMFKIGNSKELPPIPDHLSEPGKDFIRKCLQRDPSQR 653
Query: 656 PSASELLDHPFVKGAAPLERPIMVPEASDPITGITHGTKALGIGQGRNLSALDSDKLLVH 715
P+A ELL HPFV+ A LE+ ++ E + + I+ + A RN+S+L + ++
Sbjct: 654 PTAMELLQHPFVQKAVSLEKSVL-SEPLEHLAVISCRSSAKMAAHTRNISSLGLEGQTIY 712
Query: 716 SSRVLK-NNPHESLK 729
R K ++ H ++
Sbjct: 713 QRRGAKFSSKHSDIR 727
>UniRef100_Q6ESH1 Putative MAP3K alpha 1 protein kinase [Oryza sativa]
Length = 894
Score = 832 bits (2148), Expect = 0.0
Identities = 446/726 (61%), Positives = 529/726 (72%), Gaps = 14/726 (1%)
Query: 1 MPTWWGKSSSKETKKKAGKESIIDTLHRKFKFPSEGKRSTISGGSRRRSNDTISEKGDRS 60
MP WWGKSSSKE KK A KE++IDT HR P+E K T S G+RR S D +EKG S
Sbjct: 1 MPPWWGKSSSKEVKKTA-KENLIDTFHRLLS-PNEQKGRTKSRGNRRHSKDPTAEKGCWS 58
Query: 61 PSESRSPSPSK-VARCQSFAE-RPHAQPLPLPDLHPSSLGRVDSEISISAKSRLEKPSKP 118
++SRS SPSK V+RCQSFA R HAQPLPLP + + R S+I+ S K LEK K
Sbjct: 59 TAQSRSASPSKEVSRCQSFAAARAHAQPLPLPRSR-AMVARTASDITES-KVVLEKRGKG 116
Query: 119 SLFLPLPKPSCIRCGPTPADLDGDMVNASVFSDCSADSDEPADSRNRSPLATDSETGTRT 178
LPLP + ++ P + ++ AS+ S S DSD+P D R + P+A D++ +
Sbjct: 117 QQ-LPLPTTNWVKERPETTEPVAELSTASISSHGSIDSDDPGDLRLQGPVANDTDNVAKV 175
Query: 179 AAGSPSSLVLKDQSSAVSQPNLREVKKTAN-ILSNHTPSTSPKRKPLRH-HVPNLQVPPH 236
A SS+V K+ SSA+++ +EV N LSN STSP+ + + NLQ
Sbjct: 176 ATTGNSSVVHKECSSAITRKGTKEVTMPTNAFLSNQILSTSPRGTVVADSYQSNLQNSRK 235
Query: 237 GVFYSGPDSSLSSPSRSPLRAFGTDQVLNSAFWAGKPYPEINFVGSGHCSSPGSGHNSGH 296
V S P+S +SSPSRSP R DQ+ +SAFWA KP+ ++ FVGS CSSPGSG SGH
Sbjct: 236 VVLDSAPNSVMSSPSRSP-RILCPDQIPSSAFWAVKPHTDVTFVGSAQCSSPGSGQTSGH 294
Query: 297 NSMGGDMSGPLFWQPSRGSPEYSPIPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQT 356
NS+GGDM LFWQPSRGSPE SPIPSPRMTSPGPSSR+ SG+V+P+HPRAGG ES T
Sbjct: 295 NSVGGDMLAQLFWQPSRGSPECSPIPSPRMTSPGPSSRVHSGSVSPLHPRAGGMAPESPT 354
Query: 357 GRADDGKQ-QSHRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGK 415
R D+GK+ Q+HRLPLPPL++ N S F +NS TSP + SP R ++P S GSRWKKGK
Sbjct: 355 RRLDEGKRKQTHRLPLPPLSICNNSTFLPNNSTPTSP-ISHSPGRVENPTSPGSRWKKGK 413
Query: 416 LLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQ 475
L+GRGTFGHVYIGFNS GEMCAMKEVTLFSDD KS ESAKQL QE+ LL+RL+HPNIV+
Sbjct: 414 LVGRGTFGHVYIGFNSDKGEMCAMKEVTLFSDDPKSKESAKQLCQEILLLNRLQHPNIVR 473
Query: 476 YYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHR 535
YYGSE VDDKLYIYLEYVSGGSIHKLLQEYGQFGE AIRSYT+QIL GLAYLHAKNT+HR
Sbjct: 474 YYGSEMVDDKLYIYLEYVSGGSIHKLLQEYGQFGEPAIRSYTKQILLGLAYLHAKNTVHR 533
Query: 536 DIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIW 595
DIKGANILVDPNGRVK+ADFGMAKHI GQ C SFKGSPYWMAPEVIKNS C+L VDIW
Sbjct: 534 DIKGANILVDPNGRVKLADFGMAKHINGQQCAFSFKGSPYWMAPEVIKNSNGCNLAVDIW 593
Query: 596 SLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDR 655
SLGCTVLEMAT+KPPWSQYEG+AA+FKIGNSKELP IPDHLS EG+DF+R+CLQRNP R
Sbjct: 594 SLGCTVLEMATSKPPWSQYEGIAAVFKIGNSKELPPIPDHLSEEGRDFIRQCLQRNPSSR 653
Query: 656 PSASELLDHPFVKGAAPLERPIMVPEASDPITGITHGTKALGIGQGRNLSALDSDKLLVH 715
P+A +LL H F++ A+PLE+ + P T K +G RN+S+L + ++
Sbjct: 654 PTAVDLLQHSFIRNASPLEKSLSDPLLQLSTTSCKPDLKV--VGHARNMSSLGLEGQSIY 711
Query: 716 SSRVLK 721
R K
Sbjct: 712 QRRAAK 717
>UniRef100_Q6RFY3 MAP3Ka [Lycopersicon esculentum]
Length = 614
Score = 428 bits (1101), Expect = e-118
Identities = 220/406 (54%), Positives = 280/406 (68%), Gaps = 25/406 (6%)
Query: 273 PYPEINFVGSGHCSSPGSGH---NSGHNSMGGDMSGPLFWQPSR-------GSPEYSPIP 322
P P ++ +G+ H G G +S +S D GP+ ++ G SP+
Sbjct: 69 PMPSVSSLGNDHGVVLGCGSVSVSSTSSSGSSDGGGPVNTDQAQLDTFRGLGDNRLSPLS 128
Query: 323 SPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRLPLPPLTVTNTSPF 382
+ S G ++ +P+HPR +S TG+ DD + + H+LPLPP + + S
Sbjct: 129 RSPVRSRGTTT-----TSSPLHPRFSSLNLDSSTGKLDDVRSECHQLPLPPGSPPSPSAL 183
Query: 383 SHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEV 442
+ PR A+ + S+WKKG+LLGRGTFGHVY+GFN ++G+MCA+KEV
Sbjct: 184 PN----------PRPCVVAEGANVNMSKWKKGRLLGRGTFGHVYLGFNRENGQMCAIKEV 233
Query: 443 TLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLL 502
+ SDD S E KQL QE+ LLS L HPNIV+Y+GSE ++ L +YLEYVSGGSIHKLL
Sbjct: 234 KVVSDDQTSKECLKQLNQEIILLSNLTHPNIVRYHGSELDEETLSVYLEYVSGGSIHKLL 293
Query: 503 QEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHIT 562
QEYG F E I++YT+QILSGL++LHA+NT+HRDIKGANILVDPNG +K+ADFGMAKHIT
Sbjct: 294 QEYGPFREPVIQNYTRQILSGLSFLHARNTVHRDIKGANILVDPNGEIKLADFGMAKHIT 353
Query: 563 GQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFK 622
LSFKGSPYWMAPEV+ N+ L VDIWSLGCT+LEMAT+KPPWSQYEGVAA+FK
Sbjct: 354 SCASVLSFKGSPYWMAPEVVMNTSGYGLAVDIWSLGCTILEMATSKPPWSQYEGVAAIFK 413
Query: 623 IGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
IGNSK+ P IP+HLSN+ K F+R CLQR P RP+AS+LL+HPFVK
Sbjct: 414 IGNSKDFPEIPEHLSNDAKSFIRSCLQREPSLRPTASKLLEHPFVK 459
>UniRef100_Q6RFY4 MAP3Ka [Nicotiana benthamiana]
Length = 611
Score = 424 bits (1090), Expect = e-117
Identities = 227/419 (54%), Positives = 289/419 (68%), Gaps = 17/419 (4%)
Query: 323 SPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRAD-DGKQQSHRLPLPPLTVTNTSP 381
SP SP SR + +P+HPR +S TG+ D D +SH+LPLPP + + S
Sbjct: 129 SPLSRSP-VRSRGTTTTSSPLHPRFSSMNLDSPTGKLDNDVSSESHQLPLPPGSPPSPSA 187
Query: 382 FSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKE 441
+ PR+ A+ + S+WKKGKLLGRGTFGHVY+GFN ++G+MCA+KE
Sbjct: 188 LPN----------PRTCGVAEGSNVNMSKWKKGKLLGRGTFGHVYLGFNRENGQMCAIKE 237
Query: 442 VTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKL 501
V + SDD S E KQL QE+ LLS L HPNIV+YYGSE D+ L +YLEYVSGGSIHKL
Sbjct: 238 VRVVSDDQTSKECLKQLNQEIILLSNLSHPNIVRYYGSELDDETLSVYLEYVSGGSIHKL 297
Query: 502 LQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHI 561
LQEYG F E I++YT+QILSGL++LHA+NT+HRDIKGANILVDPNG +K+ADFGMAKHI
Sbjct: 298 LQEYGAFREPVIQNYTRQILSGLSFLHARNTVHRDIKGANILVDPNGEIKLADFGMAKHI 357
Query: 562 TGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMF 621
T LSFKGSPYWMAPEV+ N+ L VDIWSLGC +LEMA++KPPWSQYEGVAA+F
Sbjct: 358 TSSSLVLSFKGSPYWMAPEVVMNTSGYGLPVDIWSLGCAILEMASSKPPWSQYEGVAAIF 417
Query: 622 KIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGAAPLE-RPIMVP 680
KIGNSK+ P IPDHLSN+ K+F++ CLQR P RP+AS+LL+HPFVK + + + V
Sbjct: 418 KIGNSKDFPEIPDHLSNDAKNFIKLCLQREPSARPTASQLLEHPFVKNQSTTKVTHVGVT 477
Query: 681 EASDPITGITHGTKALGIGQGRNLSALDSDKL---LVHSSRVLKNNPHESLKSFHQFPV 736
+ + P + + T + GRN+S + ++ SR L + P E +K+ PV
Sbjct: 478 KEAYPRSFDGNRTPPVLDSGGRNISPTKGNYASHPVITISRPL-SCPREIVKTITSLPV 535
>UniRef100_Q75PK5 Mitogen-activated kinase kinase kinase alpha [Lotus japonicus]
Length = 627
Score = 418 bits (1074), Expect = e-115
Identities = 230/466 (49%), Positives = 307/466 (65%), Gaps = 38/466 (8%)
Query: 286 SSPGSGHNSGHNSMGGDMSGPLFWQPSRGSPEYSPIPSPRMTSPGPSSRIQSGAVTPIHP 345
S GS +S ++ D S + +RG E P+ P SR + +P+H
Sbjct: 109 SISGSSLSSSASASFDDHSISPHFNANRGQEEVKFNVRPK----SPCSRGPTSPTSPLHQ 164
Query: 346 RAGGTPTESQTG-RADDGKQQSHRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSP 404
R +S TG + D+G Q H LPLPP + TSPS P + RA+
Sbjct: 165 RLHALSLDSPTGGKQDEGTSQCHPLPLPP-------------GSPTSPSAPCN-TRANGV 210
Query: 405 MSSG----SRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQ 460
+ + S+WKKGKLLGRGTFGHVY+GFNS++G+MCA+KEV +FSDD S E KQL Q
Sbjct: 211 LENNTCNLSKWKKGKLLGRGTFGHVYLGFNSENGQMCAIKEVKVFSDDKTSKECLKQLNQ 270
Query: 461 EVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQI 520
E++LL++ HPNIVQYYGSE ++ L +YLEYVSGGSIHKLLQEYG F E I++YT+QI
Sbjct: 271 EINLLNQFSHPNIVQYYGSELGEESLSVYLEYVSGGSIHKLLQEYGAFKEPVIQNYTRQI 330
Query: 521 LSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPE 580
+SGLAYLH++NT+HRDIKGANILVDPNG +K+ADFGM+KHI LSFKGSPYWMAPE
Sbjct: 331 VSGLAYLHSRNTVHRDIKGANILVDPNGEIKLADFGMSKHINSAASMLSFKGSPYWMAPE 390
Query: 581 VIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEG 640
V+ N+ L VDI SLGCT+LEMAT+KPPWSQ+EGVAA+FKIGNSK++P IP+HLS++
Sbjct: 391 VVMNTNGYGLPVDISSLGCTILEMATSKPPWSQFEGVAAIFKIGNSKDMPEIPEHLSDDA 450
Query: 641 KDFVRKCLQRNPRDRPSASELLDHPFVKGAAPLERPIMVPEAS---DPITGITHGTKALG 697
K+F+++CLQR+P RP+A LL+HPF++ + + V AS D ++ G++
Sbjct: 451 KNFIKQCLQRDPLARPTAQSLLNHPFIRDQSATK----VANASITRDAFPYMSDGSRTPP 506
Query: 698 I----GQGRNLSALDSD---KLLVHSSRVLKNNPHESLKSFHQFPV 736
+ +++ LD D K + + R L+ NP +S ++ PV
Sbjct: 507 VLEPHSNRSSITTLDVDYATKPALAAVRTLR-NPRDSTRTITSLPV 551
>UniRef100_Q9LPH2 T3F20.12 protein [Arabidopsis thaliana]
Length = 608
Score = 413 bits (1062), Expect = e-114
Identities = 217/390 (55%), Positives = 271/390 (68%), Gaps = 18/390 (4%)
Query: 286 SSPGSGHNSGHNSMGG-DMSGPLFWQPSRGSPEYSPIPSPRM---TSPGPSSRIQSGAVT 341
S+ GS S +S G D L RG +++ +PR SP ++ I + +
Sbjct: 93 STSGSTSVSSVSSSGSADDQSQLVASRGRGDVKFNVAAAPRSPERVSP-KAATITTRPTS 151
Query: 342 PIHPRAGGTPT-ESQTGRADDGKQQS--HRLPLPPLTVTNTSPFSHSNSAATSPSMPRSP 398
P H R G + ES TGR DDG+ S H LP PP + T+ S S + P
Sbjct: 152 PRHQRLSGVVSLESSTGRNDDGRSSSECHPLPRPPTSPTSPSAVHGSRIGGGYETSP--- 208
Query: 399 ARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQL 458
S S WKKGK LG GTFG VY+GFNS+ G+MCA+KEV + SDD S E KQL
Sbjct: 209 -------SGFSTWKKGKFLGSGTFGQVYLGFNSEKGKMCAIKEVKVISDDQTSKECLKQL 261
Query: 459 MQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQ 518
QE++LL++L HPNIVQYYGSE ++ L +YLEYVSGGSIHKLL++YG F E I++YT+
Sbjct: 262 NQEINLLNQLCHPNIVQYYGSELSEETLSVYLEYVSGGSIHKLLKDYGSFTEPVIQNYTR 321
Query: 519 QILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMA 578
QIL+GLAYLH +NT+HRDIKGANILVDPNG +K+ADFGMAKH+T LSFKGSPYWMA
Sbjct: 322 QILAGLAYLHGRNTVHRDIKGANILVDPNGEIKLADFGMAKHVTAFSTMLSFKGSPYWMA 381
Query: 579 PEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSN 638
PEV+ + + VDIWSLGCT+LEMAT+KPPWSQ+EGVAA+FKIGNSK+ P IPDHLSN
Sbjct: 382 PEVVMSQNGYTHAVDIWSLGCTILEMATSKPPWSQFEGVAAIFKIGNSKDTPEIPDHLSN 441
Query: 639 EGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
+ K+F+R CLQRNP RP+AS+LL+HPF++
Sbjct: 442 DAKNFIRLCLQRNPTVRPTASQLLEHPFLR 471
>UniRef100_Q9ZRF7 MEK kinase [Arabidopsis thaliana]
Length = 608
Score = 413 bits (1062), Expect = e-114
Identities = 217/390 (55%), Positives = 271/390 (68%), Gaps = 18/390 (4%)
Query: 286 SSPGSGHNSGHNSMGG-DMSGPLFWQPSRGSPEYSPIPSPRM---TSPGPSSRIQSGAVT 341
S+ GS S +S G D L RG +++ +PR SP ++ I + +
Sbjct: 92 STSGSTSVSSVSSSGSADDQSQLVASRGRGDVKFNVAAAPRSPERVSP-KAATITTRPTS 150
Query: 342 PIHPRAGGTPT-ESQTGRADDGKQQS--HRLPLPPLTVTNTSPFSHSNSAATSPSMPRSP 398
P H R G + ES TGR DDG+ S H LP PP + T+ S S + P
Sbjct: 151 PRHQRLSGVVSLESSTGRNDDGRSSSECHPLPRPPTSPTSPSAVHGSRIGGGYETSP--- 207
Query: 399 ARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQL 458
S S WKKGK LG GTFG VY+GFNS+ G+MCA+KEV + SDD S E KQL
Sbjct: 208 -------SGFSTWKKGKFLGSGTFGQVYLGFNSEKGKMCAIKEVKVISDDQTSKECLKQL 260
Query: 459 MQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQ 518
QE++LL++L HPNIVQYYGSE ++ L +YLEYVSGGSIHKLL++YG F E I++YT+
Sbjct: 261 NQEINLLNQLCHPNIVQYYGSELSEETLSVYLEYVSGGSIHKLLKDYGSFTEPVIQNYTR 320
Query: 519 QILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMA 578
QIL+GLAYLH +NT+HRDIKGANILVDPNG +K+ADFGMAKH+T LSFKGSPYWMA
Sbjct: 321 QILAGLAYLHGRNTVHRDIKGANILVDPNGEIKLADFGMAKHVTAFSTMLSFKGSPYWMA 380
Query: 579 PEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSN 638
PEV+ + + VDIWSLGCT+LEMAT+KPPWSQ+EGVAA+FKIGNSK+ P IPDHLSN
Sbjct: 381 PEVVMSQNGYTHAVDIWSLGCTILEMATSKPPWSQFEGVAAIFKIGNSKDTPEIPDHLSN 440
Query: 639 EGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
+ K+F+R CLQRNP RP+AS+LL+HPF++
Sbjct: 441 DAKNFIRLCLQRNPTVRPTASQLLEHPFLR 470
>UniRef100_Q8W582 At1g53570/F22G10_18 [Arabidopsis thaliana]
Length = 609
Score = 412 bits (1060), Expect = e-113
Identities = 216/392 (55%), Positives = 269/392 (68%), Gaps = 24/392 (6%)
Query: 290 SGHNSGHNSMGG-------DMSGPLFWQPSRGSPEYSPIPSPRM---TSPGPSSRIQSGA 339
SG SG S+ D L RG +++ +PR SP ++ I +
Sbjct: 91 SGSTSGSTSVSSVSPSGSADDQSQLVASRGRGDVKFNVAAAPRSPERVSP-KAATITTRP 149
Query: 340 VTPIHPRAGGTPT-ESQTGRADDGKQQS--HRLPLPPLTVTNTSPFSHSNSAATSPSMPR 396
+P H R G + ES TGR DDG+ S H LP PP + T+ S S + P
Sbjct: 150 TSPRHQRLSGVVSLESSTGRNDDGRSSSECHPLPRPPTSPTSPSAVHGSRIGGGYETSP- 208
Query: 397 SPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAK 456
S S WKKGK LG GTFG VY+GFNS+ G+MCA+KEV + SDD S E K
Sbjct: 209 ---------SGFSTWKKGKFLGSGTFGQVYLGFNSEKGKMCAIKEVKVISDDQTSKECLK 259
Query: 457 QLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSY 516
QL QE++LL++L HPNIVQYYGSE ++ L +YLEYVSGGSIHKLL++YG F E I++Y
Sbjct: 260 QLNQEINLLNQLCHPNIVQYYGSELSEETLSVYLEYVSGGSIHKLLKDYGSFTEPVIQNY 319
Query: 517 TQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYW 576
T+QIL+GLAYLH +NT+HRDIKGANILVDPNG +K+ADFGMAKH+T LSFKGSPYW
Sbjct: 320 TRQILAGLAYLHGRNTVHRDIKGANILVDPNGEIKLADFGMAKHVTAFSTMLSFKGSPYW 379
Query: 577 MAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHL 636
MAPEV+ + + VDIWSLGCT+LEMAT+KPPWSQ+EGVAA+FKIGNSK+ P IPDHL
Sbjct: 380 MAPEVVMSQNGYTHAVDIWSLGCTILEMATSKPPWSQFEGVAAIFKIGNSKDTPEIPDHL 439
Query: 637 SNEGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
SN+ K+F+R CLQRNP RP+AS+LL+HPF++
Sbjct: 440 SNDAKNFIRLCLQRNPTVRPTASQLLEHPFLR 471
>UniRef100_O82667 MAP3K alpha 1 protein kinase [Brassica napus]
Length = 591
Score = 407 bits (1046), Expect = e-112
Identities = 212/393 (53%), Positives = 272/393 (68%), Gaps = 15/393 (3%)
Query: 288 PGSGHNSGHNSMGGDMSGPLFWQPSRGSPEYSPIPSPRMTSPGPSSRIQSGA--VTPIHP 345
P ++ + S+ G SG + S GS E P+ S ++ ++GA +P+H
Sbjct: 77 PSLSNDQVNGSVSG--SGSVSSVSSSGSGEDQSQPTAPRKSNAAAASPKAGARPTSPLHN 134
Query: 346 RAGGTPTESQ-TGRADDGKQQS-HRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADS 403
R G ES TGR DDG+ H LPLPP SP S S SP+ P S +
Sbjct: 135 RFSGMTLESSSTGRNDDGRSSEYHPLPLPP-----GSPTSPSVVLPCSPTSPSSGVQGSW 189
Query: 404 PMSSG----SRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLM 459
+ S+WKKG+ +G GTFG VY GFNS+ G +CA+KEV + SDD S E KQL
Sbjct: 190 VVGGSEKEISKWKKGRFIGSGTFGKVYQGFNSEEGRICAIKEVKVISDDKNSKECLKQLN 249
Query: 460 QEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQ 519
QE+++LS+L HPNIVQYYGSE ++ L +YLE+VSGGSI+KLL EYG F E I++YT+Q
Sbjct: 250 QEINVLSQLCHPNIVQYYGSELSEETLSVYLEFVSGGSIYKLLTEYGAFTEPVIQNYTRQ 309
Query: 520 ILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAP 579
IL GLAYLH +NT+HRDIKGANILVDPNG +K+ADFGMAKH+T LSF GSPYWMAP
Sbjct: 310 ILYGLAYLHGRNTVHRDIKGANILVDPNGEIKLADFGMAKHVTAYSTMLSFTGSPYWMAP 369
Query: 580 EVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNE 639
EV+ + +L VD+WS+GCT+LEMAT KPPWSQ+EGVAA+FKIGNSK++P IPDHLSN+
Sbjct: 370 EVVMHKNGYTLAVDVWSVGCTILEMATAKPPWSQFEGVAAIFKIGNSKDMPEIPDHLSND 429
Query: 640 GKDFVRKCLQRNPRDRPSASELLDHPFVKGAAP 672
K+F+R CLQRNP RP+A++LL+HPF++ +P
Sbjct: 430 AKNFIRLCLQRNPTVRPTAAQLLEHPFLRVHSP 462
Score = 35.0 bits (79), Expect = 8.3
Identities = 29/111 (26%), Positives = 43/111 (38%), Gaps = 14/111 (12%)
Query: 1 MPTWWGKSSSKETKKKAGKESIIDTLHRKFKFPSEGKRSTISGGSRRRSNDTISEKGDRS 60
MPTWW + SSK L + + S+ IS + + S D + +
Sbjct: 1 MPTWWVRKSSKNKDDS-------HLLQTQTRSVSDKSIRRISADNSKSSPDPV------T 47
Query: 61 PSESRSPSPSKVARCQSFAERPHAQPLPLPDLHPSSL-GRVDSEISISAKS 110
PS + A F++ PLPLP L + G V S+S+ S
Sbjct: 48 PSRCTPRCSREFAGASGFSDEKKCHPLPLPSLSNDQVNGSVSGSGSVSSVS 98
>UniRef100_Q9LLM7 Similar to mitogen-activated protein kinases [Oryza sativa]
Length = 653
Score = 400 bits (1029), Expect = e-110
Identities = 203/343 (59%), Positives = 245/343 (71%), Gaps = 23/343 (6%)
Query: 326 MTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRLPLPPLTVTNTSPFSHS 385
++ PG +I +P+ RA G S T R DD + S PLP
Sbjct: 185 ISPPGSKGQIPC-PTSPVQSRAFGQCPGSPTARQDDSRSSSSPHPLP------------- 230
Query: 386 NSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLF 445
P P S +R S S+WKKGKLLG GTFG VY GFNS+ G+MCA+KEV +
Sbjct: 231 ----RPPGSPCSSSRVVS-----SQWKKGKLLGSGTFGQVYQGFNSEGGQMCAIKEVKVI 281
Query: 446 SDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEY 505
SDD+ S E +QL QE+ LLS+L HPNIVQYYGS+ + L +YLEYVSGGSIHKLLQEY
Sbjct: 282 SDDSNSKECLRQLHQEIVLLSQLSHPNIVQYYGSDLSSETLSVYLEYVSGGSIHKLLQEY 341
Query: 506 GQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQY 565
G FGE +R+YT QILSGLAYLH +NT+HRDIKGANILVDPNG +K+ADFGMAKHI+
Sbjct: 342 GAFGEAVLRNYTAQILSGLAYLHGRNTVHRDIKGANILVDPNGDIKLADFGMAKHISAHT 401
Query: 566 CPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGN 625
SFKGSPYWMAPEVI N+ SL VDIWSLGCT++EMAT +PPW QYEGVAA+FKIGN
Sbjct: 402 SIKSFKGSPYWMAPEVIMNTNGYSLSVDIWSLGCTIIEMATARPPWIQYEGVAAIFKIGN 461
Query: 626 SKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
SK++P IPDHLS E K+F++ CLQR+P RP+A++L++HPFVK
Sbjct: 462 SKDIPDIPDHLSFEAKNFLKLCLQRDPAARPTAAQLMEHPFVK 504
>UniRef100_Q7XV58 OSJNBa0006B20.13 protein [Oryza sativa]
Length = 709
Score = 394 bits (1011), Expect = e-108
Identities = 209/359 (58%), Positives = 248/359 (68%), Gaps = 10/359 (2%)
Query: 312 SRGS--PEYSPIPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRL 369
SRG PE P R SPGP + AV ++ R G S D K H L
Sbjct: 211 SRGRMLPEDFLAPRTRSLSPGPKGH--TFAVNNVNSREFGFSPRSPVKMMDGLKSPPHPL 268
Query: 370 PLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGF 429
PLPP T SP S +A + P + + S S+WKKGKLLG GTFG VY+GF
Sbjct: 269 PLPPGPAT-CSPLPPSPTAYSP-----HPLGPTTCLQSESQWKKGKLLGSGTFGQVYLGF 322
Query: 430 NSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIY 489
NS++G+ CA+KEV + SDD S E KQL QE+ +L +L HPNIVQYYGSE DD L IY
Sbjct: 323 NSENGQFCAIKEVQVISDDPHSKERLKQLNQEIDMLRQLSHPNIVQYYGSEMTDDALSIY 382
Query: 490 LEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGR 549
LE+VSGGSIHKLL+EYG F E IR+YT QILSGLAYLH +NT+HRDIKGANILV PNG
Sbjct: 383 LEFVSGGSIHKLLREYGPFKEPVIRNYTGQILSGLAYLHGRNTVHRDIKGANILVGPNGE 442
Query: 550 VKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKP 609
VK+ADFGMAKHI+ SFKGSPYWMAPEVI N + L VDIWSLGCT++EMAT KP
Sbjct: 443 VKLADFGMAKHISSFAEIRSFKGSPYWMAPEVIMNGRGYHLPVDIWSLGCTIIEMATAKP 502
Query: 610 PWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
PW +YEGVAA+FKI NSKE+P IPD S EGK F++ CL+R+P R +A++L+DHPFV+
Sbjct: 503 PWHKYEGVAAIFKIANSKEIPEIPDSFSEEGKSFLQMCLKRDPASRFTATQLMDHPFVQ 561
>UniRef100_Q6ZI89 Putative MEK kinase [Oryza sativa]
Length = 735
Score = 384 bits (987), Expect = e-105
Identities = 202/367 (55%), Positives = 245/367 (66%), Gaps = 22/367 (5%)
Query: 312 SRGS--PEYSPIPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRL 369
SRG PE + PR SPGP S R G S R DD + S L
Sbjct: 199 SRGRMLPEDTFAVRPRSHSPGPRGHAYSACCA----RDFGFTPRSPVKRMDDPRSPSQPL 254
Query: 370 PLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGF 429
PLPP+ V ++S P+ + + S+WK+GKLLG GTFG VY+GF
Sbjct: 255 PLPPVPVASSS----------------IPSSSITSSQFQSQWKRGKLLGSGTFGQVYLGF 298
Query: 430 NSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIY 489
NS++G+ CA+KEV +F DD+ S E +QL QE+ +L +L H NIVQYYGSE D+ L IY
Sbjct: 299 NSENGQFCAIKEVQVFLDDSHSKERLRQLNQEIDMLKQLSHQNIVQYYGSELADEALSIY 358
Query: 490 LEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGR 549
LEYVSGGSIHKLL+EYG F E IR+YT+QILSGLAYLH +NT+HRDIKGANILV PNG
Sbjct: 359 LEYVSGGSIHKLLREYGPFKEPVIRNYTRQILSGLAYLHGRNTVHRDIKGANILVGPNGE 418
Query: 550 VKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKP 609
VK+ADFGMAKH+T SF+GSPYWMAPEV+ N+K +L VDIWSLGCT++EMAT K
Sbjct: 419 VKLADFGMAKHVTSFAEIRSFRGSPYWMAPEVVMNNKGYNLAVDIWSLGCTIIEMATAKH 478
Query: 610 PWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKG 669
PW YE VAA+FKI NSK++P IPD S EGKDF+ CL+R+P RPSA+ LL HPFV
Sbjct: 479 PWYPYEDVAAIFKIANSKDIPEIPDCFSKEGKDFLSLCLKRDPVQRPSAASLLGHPFVHD 538
Query: 670 AAPLERP 676
+ P
Sbjct: 539 HQAVRAP 545
>UniRef100_Q9C5H5 Putative MAP protein kinase [Arabidopsis thaliana]
Length = 716
Score = 377 bits (967), Expect = e-103
Identities = 255/596 (42%), Positives = 342/596 (56%), Gaps = 81/596 (13%)
Query: 95 SSLGRVDSEISISAKSRLEKPSKPS-----LFLPLPKPSCIRCGPTPADLDG-DMVNASV 148
+S DS ++ S + P PS L LPLP+ + IR LD D +
Sbjct: 76 TSSSTFDSGLTRSPSAFTAVPRSPSAVPLPLPLPLPEVAGIRNAANARGLDDRDRDPERL 135
Query: 149 FSDCSADSDEPADSRNRSPLATDSETGTRTAAGSPSSLVLKDQSSAVSQPNLREVKKTAN 208
SD ++ S P S N A DS T ++ S ++ V+ P + +A
Sbjct: 136 ISDRTS-SGPPLTSVNGG-FARDSRKATENSSYQDFSPRNRN-GYWVNIPTM-----SAP 187
Query: 209 ILSNHTPSTSPKRKPLRHHVPNLQVPPHG-VFYSGPD-----SSLSSPSRSPLRAFGTDQ 262
+P SP+RK H +P +PP +S PD S L P+ + AF TD
Sbjct: 188 TSPYMSPVPSPQRKSTGHDLPFFYLPPKSNQAWSAPDMPLDTSGLPPPAFYDITAFSTD- 246
Query: 263 VLNSAFWAGKPYPEINFVGSGHCSSPGSGHNSGHNSMGGDMSGPLFWQPSRGSPEYSPIP 322
NS + +P S + P QPSR S SP+
Sbjct: 247 --NSPIHSPQP-----------------------RSPRKQIRSP---QPSRPS---SPLH 275
Query: 323 SPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRLPLPPLTVTNTSPF 382
S ++P P + S P+HPR T+ GR D H LPLPP ++S
Sbjct: 276 SVDSSAP-PRDSVSS----PLHPRLS---TDVTNGRRDCCNV--HPLPLPPGATCSSS-- 323
Query: 383 SHSNSAATSPSMPRSPARADS-PMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKE 441
SAA+ PS P++P + DS PM+S +WKKGKL+GRGTFG VY+ NS++G +CAMKE
Sbjct: 324 ----SAASVPS-PQAPLKLDSFPMNS--QWKKGKLIGRGTFGSVYVASNSETGALCAMKE 376
Query: 442 VTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKL 501
V LF DD KS E KQL QE+ LLS L+HPNIVQY+GSETV+D+ +IYLEYV GSI+K
Sbjct: 377 VELFPDDPKSAECIKQLEQEIKLLSNLQHPNIVQYFGSETVEDRFFIYLEYVHPGSINKY 436
Query: 502 LQEY-GQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKH 560
++++ G E +R++T+ ILSGLAYLH K T+HRDIKGAN+LVD +G VK+ADFGMAKH
Sbjct: 437 IRDHCGTMTESVVRNFTRHILSGLAYLHNKKTVHRDIKGANLLVDASGVVKLADFGMAKH 496
Query: 561 ITGQYCPLSFKGSPYWMAPEVIK------NSKECSLGVDIWSLGCTVLEMATTKPPWSQY 614
+TGQ LS KGSPYWMAPE+++ ++ + + VDIWSLGCT++EM T KPPWS++
Sbjct: 497 LTGQRADLSLKGSPYWMAPELMQAVMQKDSNPDLAFAVDIWSLGCTIIEMFTGKPPWSEF 556
Query: 615 EGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGA 670
EG AAMFK+ ++ P IP+ +S EGKDF+R C QRNP +RP+AS LL+H F+K +
Sbjct: 557 EGAAAMFKV--MRDSPPIPESMSPEGKDFLRLCFQRNPAERPTASMLLEHRFLKNS 610
>UniRef100_Q93ZH4 AT5g66850/MUD21_11 [Arabidopsis thaliana]
Length = 716
Score = 375 bits (963), Expect = e-102
Identities = 254/596 (42%), Positives = 342/596 (56%), Gaps = 81/596 (13%)
Query: 95 SSLGRVDSEISISAKSRLEKPSKPS-----LFLPLPKPSCIRCGPTPADLDG-DMVNASV 148
+S DS ++ S + P PS L LPLP+ + IR LD D +
Sbjct: 76 TSSSTFDSGLTRSPSAFTAVPRSPSAVPLPLPLPLPEVAGIRNAANARGLDDRDRDPERL 135
Query: 149 FSDCSADSDEPADSRNRSPLATDSETGTRTAAGSPSSLVLKDQSSAVSQPNLREVKKTAN 208
SD ++ S P S N A DS T ++ S ++ V+ P + +A
Sbjct: 136 ISDRTS-SGPPLTSVNGG-FARDSRKATENSSYQDFSPRNRN-GYWVNIPTM-----SAP 187
Query: 209 ILSNHTPSTSPKRKPLRHHVPNLQVPPHG-VFYSGPD-----SSLSSPSRSPLRAFGTDQ 262
+P SP+RK H +P +PP +S PD S L P+ + AF TD
Sbjct: 188 TSPYMSPVPSPQRKSTGHDLPFFYLPPKSNQAWSAPDMPLDTSGLPPPAFYDITAFSTD- 246
Query: 263 VLNSAFWAGKPYPEINFVGSGHCSSPGSGHNSGHNSMGGDMSGPLFWQPSRGSPEYSPIP 322
NS + +P S + P QPSR S SP+
Sbjct: 247 --NSPIHSPQP-----------------------RSPRKQIRSP---QPSRPS---SPLH 275
Query: 323 SPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRLPLPPLTVTNTSPF 382
S ++P P + S P+HPR T+ GR D H LPLPP ++S
Sbjct: 276 SVDSSAP-PRDSVSS----PLHPRLS---TDVTNGRRDCCNV--HPLPLPPGATCSSS-- 323
Query: 383 SHSNSAATSPSMPRSPARADS-PMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKE 441
SAA+ PS P++P + DS PM+S +W+KGKL+GRGTFG VY+ NS++G +CAMKE
Sbjct: 324 ----SAASVPS-PQAPLKLDSFPMNS--QWEKGKLIGRGTFGSVYVASNSETGALCAMKE 376
Query: 442 VTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKL 501
V LF DD KS E KQL QE+ LLS L+HPNIVQY+GSETV+D+ +IYLEYV GSI+K
Sbjct: 377 VELFPDDPKSAECIKQLEQEIKLLSNLQHPNIVQYFGSETVEDRFFIYLEYVHPGSINKY 436
Query: 502 LQEY-GQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKH 560
++++ G E +R++T+ ILSGLAYLH K T+HRDIKGAN+LVD +G VK+ADFGMAKH
Sbjct: 437 IRDHCGTMTESVVRNFTRHILSGLAYLHNKKTVHRDIKGANLLVDASGVVKLADFGMAKH 496
Query: 561 ITGQYCPLSFKGSPYWMAPEVIK------NSKECSLGVDIWSLGCTVLEMATTKPPWSQY 614
+TGQ LS KGSPYWMAPE+++ ++ + + VDIWSLGCT++EM T KPPWS++
Sbjct: 497 LTGQRADLSLKGSPYWMAPELMQAVMQKDSNPDLAFAVDIWSLGCTIIEMFTGKPPWSEF 556
Query: 615 EGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGA 670
EG AAMFK+ ++ P IP+ +S EGKDF+R C QRNP +RP+AS LL+H F+K +
Sbjct: 557 EGAAAMFKV--MRDSPPIPESMSPEGKDFLRLCFQRNPAERPTASMLLEHRFLKNS 610
>UniRef100_O82649 MAP3K alpha protein kinase [Arabidopsis thaliana]
Length = 582
Score = 364 bits (935), Expect = 5e-99
Identities = 200/390 (51%), Positives = 251/390 (64%), Gaps = 44/390 (11%)
Query: 286 SSPGSGHNSGHNSMGG-DMSGPLFWQPSRGSPEYSPIPSPRM---TSPGPSSRIQSGAVT 341
S+ GS S +S G D L RG +++ +PR SP ++ I + +
Sbjct: 92 STSGSTSVSSVSSSGSADDQSQLVASRGRGDVKFNVAAAPRSPERVSP-KAATITTRPTS 150
Query: 342 PIHPRAGGTPT-ESQTGRADDGKQQS--HRLPLPPLTVTNTSPFSHSNSAATSPSMPRSP 398
P H R G + ES TGR DDG+ S H LP PP + T+ S S + P
Sbjct: 151 PRHQRLSGVVSLESSTGRNDDGRSSSECHPLPRPPTSPTSPSAVHGSRIGGGYETSP--- 207
Query: 399 ARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQL 458
S S WKKGK LG GTFG VY+GFNS+ G+MCA+KEV + SDD S E KQL
Sbjct: 208 -------SGFSTWKKGKFLGSGTFGQVYLGFNSEKGKMCAIKEVKVISDDQTSKECLKQL 260
Query: 459 MQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQ 518
QE++LL++L HPNIVQYYGSE ++ L +YLEYVSGGSIHKLL++YG F E I++YT+
Sbjct: 261 NQEINLLNQLCHPNIVQYYGSELSEETLSVYLEYVSGGSIHKLLKDYGSFTEPVIQNYTR 320
Query: 519 QILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMA 578
QIL+GLAYLH +NT+HRDIKGANIL FKGSPYWMA
Sbjct: 321 QILAGLAYLHGRNTVHRDIKGANIL--------------------------FKGSPYWMA 354
Query: 579 PEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSN 638
PEV+ + + VDIWSLGCT+LEMAT+KPPWSQ+EGVAA+FKIGNSK+ P IPDHLSN
Sbjct: 355 PEVVMSQNGYTHAVDIWSLGCTILEMATSKPPWSQFEGVAAIFKIGNSKDTPEIPDHLSN 414
Query: 639 EGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
+ K+F+R CLQRNP RP+AS+LL+HPF++
Sbjct: 415 DAKNFIRLCLQRNPTVRPTASQLLEHPFLR 444
>UniRef100_Q9FKZ5 MAP protein kinase [Arabidopsis thaliana]
Length = 376
Score = 340 bits (872), Expect = 9e-92
Identities = 164/270 (60%), Positives = 210/270 (77%), Gaps = 10/270 (3%)
Query: 409 SRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRL 468
S+WKKGKL+GRGTFG VY+ NS++G +CAMKEV LF DD KS E KQL QE+ LLS L
Sbjct: 3 SQWKKGKLIGRGTFGSVYVASNSETGALCAMKEVELFPDDPKSAECIKQLEQEIKLLSNL 62
Query: 469 RHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEY-GQFGELAIRSYTQQILSGLAYL 527
+HPNIVQY+GSETV+D+ +IYLEYV GSI+K ++++ G E +R++T+ ILSGLAYL
Sbjct: 63 QHPNIVQYFGSETVEDRFFIYLEYVHPGSINKYIRDHCGTMTESVVRNFTRHILSGLAYL 122
Query: 528 HAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVI----- 582
H K T+HRDIKGAN+LVD +G VK+ADFGMAKH+TGQ LS KGSPYWMAPEV+
Sbjct: 123 HNKKTVHRDIKGANLLVDASGVVKLADFGMAKHLTGQRADLSLKGSPYWMAPEVLMQAVM 182
Query: 583 --KNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEG 640
++ + + VDIWSLGCT++EM T KPPWS++EG AAMFK+ ++ P IP+ +S EG
Sbjct: 183 QKDSNPDLAFAVDIWSLGCTIIEMFTGKPPWSEFEGAAAMFKV--MRDSPPIPESMSPEG 240
Query: 641 KDFVRKCLQRNPRDRPSASELLDHPFVKGA 670
KDF+R C QRNP +RP+AS LL+H F+K +
Sbjct: 241 KDFLRLCFQRNPAERPTASMLLEHRFLKNS 270
>UniRef100_O23721 MAP3K gamma protein kinase [Arabidopsis thaliana]
Length = 372
Score = 338 bits (867), Expect = 3e-91
Identities = 162/268 (60%), Positives = 210/268 (77%), Gaps = 9/268 (3%)
Query: 410 RWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLR 469
+WKKGKL+GRGTFG VY+ NS++G +CAMKEV LF DD KS E KQL QE+ LLS L+
Sbjct: 1 QWKKGKLIGRGTFGSVYVASNSETGALCAMKEVELFPDDPKSAECIKQLEQEIKLLSNLQ 60
Query: 470 HPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEY-GQFGELAIRSYTQQILSGLAYLH 528
HPNIVQY+GSETV+D+ +IYLEYV GSI+K ++++ G E +R++T+ ILSGLAYLH
Sbjct: 61 HPNIVQYFGSETVEDRFFIYLEYVHPGSINKYIRDHCGTMTESVVRNFTRHILSGLAYLH 120
Query: 529 AKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIK----- 583
K T+HRDIKGAN+LVD +G VK+ADFGMAKH+TGQ LS KGSPYWMAPE+++
Sbjct: 121 NKKTVHRDIKGANLLVDASGVVKLADFGMAKHLTGQRADLSLKGSPYWMAPELMQAVMQK 180
Query: 584 -NSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKD 642
++ + + VDIWSLGCT++EM T KPPWS++EG AAMFK+ ++ P IP+ +S EGKD
Sbjct: 181 DSNPDLAFAVDIWSLGCTIIEMFTGKPPWSEFEGAAAMFKV--MRDSPPIPESMSPEGKD 238
Query: 643 FVRKCLQRNPRDRPSASELLDHPFVKGA 670
F+R C QRNP +RP+AS LL+H F+K +
Sbjct: 239 FLRLCFQRNPAERPTASMLLEHRFLKNS 266
>UniRef100_Q7X992 Putative MAP3K protein kinase [Oryza sativa]
Length = 736
Score = 331 bits (848), Expect = 6e-89
Identities = 207/521 (39%), Positives = 285/521 (53%), Gaps = 55/521 (10%)
Query: 166 SPLATDSETGTRTAAGSPSSLVLKDQSSAVSQPNLREVKKTANILSNHTPSTSPKRKPLR 225
SP +S+T G ++ V +S + Q + S T ++S RK R
Sbjct: 142 SPKPVESDTSEPDVGGERATRV----TSQIVQNFPDNNNNLPDNSSKRTTTSSHHRKVFR 197
Query: 226 HHVPN----------LQVPPHGVFYSGPDSSLSSPSRSPLRAFGTDQVLNSAFWAGKPYP 275
+ L +P SG S + SP R + T W+ P
Sbjct: 198 EKFQDKSSTETANFRLNIPAKSAPSSGFSSPVCSPRRFSNAEYTTPTAQGPQAWSA---P 254
Query: 276 EINFVGSGHCSSPGSGHNSGHNSMGGDMSGPLFWQPSRGSPEYSPIPSPRMTSPGPSSRI 335
+ V S SSP +S ++ G E S S + SP S+
Sbjct: 255 SVRSVDSMATSSPR-------------ISPEIY----TGVTEQSTF-SNSLRSPILMSKN 296
Query: 336 QSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRLPLPPLTVTNTSPFSHSNSAATSPSMP 395
S +P+HP+ P + + +G H LP PP + N+ S N +A MP
Sbjct: 297 SSAPPSPLHPKL--FPENNMSRIEGNGNVSFHPLPRPPGAI-NSMQTSIVNQSAPKVEMP 353
Query: 396 RSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESA 455
S +W+KG+LLG GTFG VY N Q+G +CAMKEV + DDAKS ES
Sbjct: 354 ----------SVAGQWQKGRLLGSGTFGCVYEATNRQTGALCAMKEVNIIPDDAKSAESL 403
Query: 456 KQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLL-QEYGQFGELAIR 514
KQL QE+ LS+ +H NIVQYYGS+T +D+ YIYLEYV GSI+K + Q YG E +R
Sbjct: 404 KQLEQEIKFLSQFKHENIVQYYGSDTFEDRFYIYLEYVHPGSINKYVKQHYGAMTESVVR 463
Query: 515 SYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSP 574
++T+ IL GLA+LH + +HRDIKGAN+LVD +G VK+ADFGMAKH++ LS KG+P
Sbjct: 464 NFTRHILRGLAFLHGQKIMHRDIKGANLLVDVSGVVKLADFGMAKHLSTAAPNLSLKGTP 523
Query: 575 YWMAPEVIKNSKECSLG----VDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELP 630
YWMAPE+++ + +G VDIWSLGCT++EM KPPWS EG AAMF++ + P
Sbjct: 524 YWMAPEMVQATLNKDVGYDLAVDIWSLGCTIIEMFNGKPPWSDLEGPAAMFRVLHKD--P 581
Query: 631 TIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGAA 671
IPD+LS+EGKDF++ C +RNP +RP+ASELL+HPF++ ++
Sbjct: 582 PIPDNLSHEGKDFLQFCFKRNPAERPTASELLEHPFIRNSS 622
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,365,687,911
Number of Sequences: 2790947
Number of extensions: 65167005
Number of successful extensions: 242445
Number of sequences better than 10.0: 20809
Number of HSP's better than 10.0 without gapping: 14095
Number of HSP's successfully gapped in prelim test: 6774
Number of HSP's that attempted gapping in prelim test: 184063
Number of HSP's gapped (non-prelim): 30303
length of query: 754
length of database: 848,049,833
effective HSP length: 135
effective length of query: 619
effective length of database: 471,271,988
effective search space: 291717360572
effective search space used: 291717360572
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 79 (35.0 bits)
Medicago: description of AC121237.19


