

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121233.14 - phase: 0
(275 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
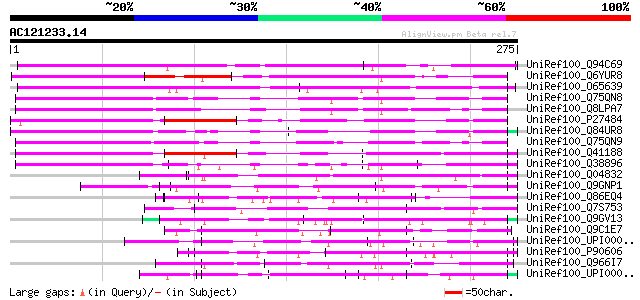
UniRef100_Q94C69 Putative glycine-rich, zinc-finger DNA-binding ... 270 4e-71
UniRef100_Q6YUR8 Putative Glycine-rich protein 2 [Oryza sativa] 264 2e-69
UniRef100_O65639 Glycine-rich protein [Arabidopsis thaliana] 258 1e-67
UniRef100_Q75QN8 Cold shock domain protein 3 [Triticum aestivum] 228 1e-58
UniRef100_Q8LPA7 Cold shock protein-1 [Triticum aestivum] 226 5e-58
UniRef100_P27484 Glycine-rich protein 2 [Nicotiana sylvestris] 213 3e-54
UniRef100_Q84UR8 Putative cold shock protein-1 [Oryza sativa] 207 2e-52
UniRef100_Q75QN9 Cold shock domain protein 2 [Triticum aestivum] 203 3e-51
UniRef100_Q41188 Glycine-rich protein 2 [Arabidopsis thaliana] 197 2e-49
UniRef100_Q38896 Glycine-rich protein 2b [Arabidopsis thaliana] 185 1e-45
UniRef100_Q04832 DNA-binding protein HEXBP [Leishmania major] 147 3e-34
UniRef100_Q9GNP1 Vasa homolog [Ciona savignyi] 138 1e-31
UniRef100_Q86EQ4 Clone ZZD1536 mRNA sequence [Schistosoma japoni... 138 2e-31
UniRef100_Q7S753 Hypothetical protein [Neurospora crassa] 137 4e-31
UniRef100_Q9GV13 Vasa-related protein CnVAS1 [Hydra magnipapillata] 136 7e-31
UniRef100_Q9C1E7 Putative DNA binding protein [Schizophyllum com... 122 8e-27
UniRef100_UPI00004301EE UPI00004301EE UniRef100 entry 122 1e-26
UniRef100_P90606 Nucleic acid binding protein [Trypanosoma equip... 121 2e-26
UniRef100_Q966I7 Hypothetical protein K08D12.3 [Caenorhabditis e... 120 4e-26
UniRef100_UPI00003C130E UPI00003C130E UniRef100 entry 119 9e-26
>UniRef100_Q94C69 Putative glycine-rich, zinc-finger DNA-binding protein [Arabidopsis
thaliana]
Length = 301
Score = 270 bits (689), Expect = 4e-71
Identities = 146/298 (48%), Positives = 172/298 (56%), Gaps = 34/298 (11%)
Query: 5 RISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGKT 64
R G V WF++ KG+GFI PDD E+LFVHQS I S+GFRSL+ G+ VE+ IA G +GKT
Sbjct: 10 RSIGKVSWFSDGKGYGFITPDDGGEELFVHQSSIVSDGFRSLTLGESVEYEIALGSDGKT 69
Query: 65 KAVDVTGPKGEPLQVRQDNH--------------------GGGGGGRGF-RGGERRNGG- 102
KA++VT P G L ++++ GG GG+ F GG RR+GG
Sbjct: 70 KAIEVTAPGGGSLNKKENSSRGSGGNCFNCGEVGHMAKDCDGGSGGKSFGGGGGRRSGGE 129
Query: 103 GGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRG-GGN 161
G CY CGD GH ARDC R SGGGGGG R CY+CG H A+DC G GGN
Sbjct: 130 GECYMCGDVGHFARDC----RQSGGGNSGGGGGG--GRPCYSCGEVGHLAKDCRGGSGGN 183
Query: 162 NNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSG--GGGGGACYKCGEVGHIARDCSNEG 219
GGGG G GG CY CGGVGH ARDC G GGGG CY CG VGHIA+ C+++
Sbjct: 184 RYGGGGGRGSGGDGCYMCGGVGHFARDCRQNGGGNVGGGGSTCYTCGGVGHIAKVCTSKI 243
Query: 220 GRFDGGNGR--YD-DGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDCVEAS 274
GG GR Y+ G G R GSG G GG CF CGK GHFAR+C +
Sbjct: 244 PSGGGGGGRACYECGGTGHLARDCDRRGSGSSGGGGGSNKCFICGKEGHFARECTSVA 301
Score = 80.1 bits (196), Expect = 6e-14
Identities = 37/83 (44%), Positives = 47/83 (56%), Gaps = 19/83 (22%)
Query: 193 SSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDG 252
+S G GG C+ CGEVGH+A+DC DGG+G + FG GGG G
Sbjct: 87 NSSRGSGGNCFNCGEVGHMAKDC-------DGGSG------------GKSFGGGGGRRSG 127
Query: 253 GKGTCFNCGKAGHFARDCVEASG 275
G+G C+ CG GHFARDC ++ G
Sbjct: 128 GEGECYMCGDVGHFARDCRQSGG 150
>UniRef100_Q6YUR8 Putative Glycine-rich protein 2 [Oryza sativa]
Length = 241
Score = 264 bits (675), Expect = 2e-69
Identities = 141/276 (51%), Positives = 165/276 (59%), Gaps = 46/276 (16%)
Query: 2 AEERISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDN 61
A R G V+WFN+TKGFGFI PDD SEDLFVHQS I+++GFRSL+EG++VEF I++ ++
Sbjct: 3 AAARHRGTVKWFNDTKGFGFISPDDGSEDLFVHQSSIKADGFRSLAEGEQVEFAISESED 62
Query: 62 GKTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRS 121
G+TKAVDVTGP G ++ GGGGGG G RGG GGG Y G
Sbjct: 63 GRTKAVDVTGPDGSFVKGGAGGGGGGGGGFGSRGGGGSGGGGRSYGGSWGG--------- 113
Query: 122 DRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGG-GTSCYRCG 180
RSGGGG G C+ CG H ARDC GGG GGGG GGG G C++CG
Sbjct: 114 -----GRRSGGGGPGG---GCFKCGESGHMARDCFNGGGVGVGGGGGGGGGAGGGCFKCG 165
Query: 181 GVGHIARDCATPSSGGGGGG------ACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNG 234
+GH+ARDC GGGGGG ACY CGE GH+ARDC N GG GG G G
Sbjct: 166 EMGHMARDCFNSGGGGGGGGGGGGGGACYNCGETGHLARDCYNGGG---GG------GGG 216
Query: 235 RFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
RFGG GG +C+NCG+AGH ARDC
Sbjct: 217 RFGG-------------GGDRSCYNCGEAGHIARDC 239
Score = 50.8 bits (120), Expect = 4e-05
Identities = 25/49 (51%), Positives = 30/49 (61%), Gaps = 6/49 (12%)
Query: 74 GEPLQVRQDNHGGGGGGRGFRGGERRNGGG--GCYTCGDTGHIARDCDR 120
GE + +D + GGGGG GG R GGG CY CG+ GHIARDC +
Sbjct: 197 GETGHLARDCYNGGGGG----GGGRFGGGGDRSCYNCGEAGHIARDCHK 241
>UniRef100_O65639 Glycine-rich protein [Arabidopsis thaliana]
Length = 299
Score = 258 bits (659), Expect = 1e-67
Identities = 141/297 (47%), Positives = 165/297 (55%), Gaps = 42/297 (14%)
Query: 5 RISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGKT 64
R +G V WFN +KG+GFI PDD S +LFVHQS I SEG+RSL+ GD VEF I G +GKT
Sbjct: 10 RSTGKVNWFNASKGYGFITPDDGSVELFVHQSSIVSEGYRSLTVGDAVEFAITQGSDGKT 69
Query: 65 KAVDVTGPKGEPLQVRQDNHG-----GGGG------------------GRGFRGGERRN- 100
KAV+VT P G L+ ++ G GGGG G G GGERR+
Sbjct: 70 KAVNVTAPGGGSLKKENNSRGNGARRGGGGSGCYNCGELGHISKDCGIGGGGGGGERRSR 129
Query: 101 GGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGG 160
GG GCY CGDTGH ARDC + D+ + GG G CYTCG H ARDC +
Sbjct: 130 GGEGCYNCGDTGHFARDCTSAGNGDQRGATKGGNDG-----CYTCGDVGHVARDCTQKSV 184
Query: 161 NNNNGGGGYGGGGTSCYRCGGVGHIARDC------ATPSSGGGGGGACYKCGEVGHIARD 214
N + G GG CY CG VGH ARDC SGGGG G CY CG VGHIARD
Sbjct: 185 GNGDQRGAVKGGNDGCYTCGDVGHFARDCTQKVAAGNVRSGGGGSGTCYSCGGVGHIARD 244
Query: 215 CSNEGGRFDGGNGRYD-DGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
C+ + G Y G+G + GSGGGG+D C+ CGK GHFAR+C
Sbjct: 245 CAT---KRQPSRGCYQCGGSGHLARDCDQRGSGGGGND---NACYKCGKEGHFAREC 295
Score = 98.2 bits (243), Expect = 2e-19
Identities = 53/129 (41%), Positives = 67/129 (51%), Gaps = 35/129 (27%)
Query: 158 GGG-----NNNNGGGGY-GGGGTSCYRCGGVGHIARDCATPSSGGGG------GGACYKC 205
GGG NN+ G G GGGG+ CY CG +GHI++DC GGGG G CY C
Sbjct: 78 GGGSLKKENNSRGNGARRGGGGSGCYNCGELGHISKDCGIGGGGGGGERRSRGGEGCYNC 137
Query: 206 GEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGH 265
G+ GH ARDC++ G G++R G+ GG+DG C+ CG GH
Sbjct: 138 GDTGHFARDCTSAGN------------------GDQR-GATKGGNDG----CYTCGDVGH 174
Query: 266 FARDCVEAS 274
ARDC + S
Sbjct: 175 VARDCTQKS 183
>UniRef100_Q75QN8 Cold shock domain protein 3 [Triticum aestivum]
Length = 231
Score = 228 bits (582), Expect = 1e-58
Identities = 134/284 (47%), Positives = 151/284 (52%), Gaps = 77/284 (27%)
Query: 4 ERISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGK 63
ER+ G V+WFN TKGFGFI PDD EDLFVHQS I+S+G+RSL+E D VEF I GD+G+
Sbjct: 3 ERVKGTVKWFNVTKGFGFISPDDGGEDLFVHQSAIKSDGYRSLNENDAVEFEIITGDDGR 62
Query: 64 TKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDR 123
TKA DVT P G L G GGG RG RGG GGGG Y G G+
Sbjct: 63 TKASDVTAPGGGALS-GGSRPGEGGGDRGGRGGY--GGGGGGYGGGGGGY---------- 109
Query: 124 NDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGG---TSCYRCG 180
GGGGGG GGG GGGGYGGGG CY+CG
Sbjct: 110 -------GGGGGG--------------------YGGGGGGYGGGGYGGGGGGGRGCYKCG 142
Query: 181 GVGHIARDCATPSSGGGGGG--------------ACYKCGEVGHIARDCSNEGGRFDGGN 226
GHI+RDC P GGGGGG CYKCGE GHI+RDC GG
Sbjct: 143 EDGHISRDC--PQGGGGGGGYGGGGYGGGGGGGRECYKCGEEGHISRDCPQGGG------ 194
Query: 227 GRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
G G GGG R G GGGG CF+CG++GHF+R+C
Sbjct: 195 -----GGGYGGGGGRGGGGGGGG-------CFSCGESGHFSREC 226
>UniRef100_Q8LPA7 Cold shock protein-1 [Triticum aestivum]
Length = 229
Score = 226 bits (576), Expect = 5e-58
Identities = 132/283 (46%), Positives = 151/283 (52%), Gaps = 77/283 (27%)
Query: 4 ERISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGK 63
ER+ G V+WFN TKGFGFI P+D SEDLFVHQS I+S+G+RSL+E D VEF + GD+G+
Sbjct: 3 ERVKGTVKWFNVTKGFGFISPEDGSEDLFVHQSAIKSDGYRSLNENDTVEFEVITGDDGR 62
Query: 64 TKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDR 123
TKA DVT P G L G GGG RG RGG G GG
Sbjct: 63 TKASDVTAPGGGALS-GGSRPGDGGGDRGGRGGYGGGGYGG------------------- 102
Query: 124 NDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGG--TSCYRCGG 181
GGGGGG GGG GGGGYGGGG CY+CG
Sbjct: 103 -------GGGGGGYGG------------------GGGGYGGGGGGYGGGGGGRGCYKCGE 137
Query: 182 VGHIARDCATPSSGGGGGG--------------ACYKCGEVGHIARDCSNEGGRFDGGNG 227
GHI+RDC P GGGGGG CYKCGE GHI+RDC +GG GG G
Sbjct: 138 EGHISRDC--PQGGGGGGGYGGGGYGGGGGGGRECYKCGEEGHISRDCP-QGGGGGGGGG 194
Query: 228 RYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
Y G GR GGG GGGG CF+CG++GHF+R+C
Sbjct: 195 GYGGGGGRGGGG------GGGG-------CFSCGESGHFSREC 224
>UniRef100_P27484 Glycine-rich protein 2 [Nicotiana sylvestris]
Length = 214
Score = 213 bits (543), Expect = 3e-54
Identities = 129/273 (47%), Positives = 143/273 (52%), Gaps = 67/273 (24%)
Query: 1 MAEE---RISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIA 57
MAEE R G V+WF++ KGFGFI PDD EDLFVHQS IRSEGFRSL+EG+ VEF +
Sbjct: 1 MAEESGQRAKGTVKWFSDQKGFGFITPDDGGEDLFVHQSGIRSEGFRSLAEGETVEFEVE 60
Query: 58 DGDNGKTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARD 117
G +G+TKAVDVTGP G +Q GGGGGG G RGG GG G Y G G
Sbjct: 61 SGGDGRTKAVDVTGPDGAAVQ---GGRGGGGGGGG-RGGGGYGGGSGGYGGGGRGG---- 112
Query: 118 CDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCY 177
S G GGGD G + GGG GG YGGGG
Sbjct: 113 ------------SRGYGGGD--------GGY---------GGGGGYGGGSRYGGGGGGYG 143
Query: 178 RCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFG 237
GG G G GGG C+KCGE GH ARDCS GG G
Sbjct: 144 GGGGYG---------GGGSGGGSGCFKCGESGHFARDCSQSGG----------------G 178
Query: 238 GGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
GG RFG GGGG GG G C+ CG+ GHFAR+C
Sbjct: 179 GGGGRFGGGGGG--GGGGGCYKCGEDGHFAREC 209
Score = 55.8 bits (133), Expect = 1e-06
Identities = 23/39 (58%), Positives = 25/39 (63%)
Query: 85 GGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDR 123
GGGGGG F GG GGGGCY CG+ GH AR+C R
Sbjct: 176 GGGGGGGRFGGGGGGGGGGGCYKCGEDGHFARECTSGGR 214
>UniRef100_Q84UR8 Putative cold shock protein-1 [Oryza sativa]
Length = 197
Score = 207 bits (528), Expect = 2e-52
Identities = 120/270 (44%), Positives = 143/270 (52%), Gaps = 78/270 (28%)
Query: 1 MAEERISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGD 60
MA ER+ G V+WF+ TKGFGFI PDD EDLFVHQS ++S+G+RSL++GD VEF++ G+
Sbjct: 1 MASERVKGTVKWFDATKGFGFITPDDGGEDLFVHQSSLKSDGYRSLNDGDVVEFSVGSGN 60
Query: 61 NGKTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDR 120
+G+TKAVDVT P G L + GGG RG+ GG GGGG Y GD G+
Sbjct: 61 DGRTKAVDVTAPGGGALT--GGSRPSGGGDRGYGGG----GGGGRYG-GDRGY------- 106
Query: 121 SDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCG 180
GGGGGG GG GGGGYGGGG
Sbjct: 107 ----------GGGGGG-------------------YGGGDRGYGGGGGYGGGG------- 130
Query: 181 GVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGN 240
GGG ACYKCGE GH+ARDCS GG G G +GGG
Sbjct: 131 ---------------GGGSRACYKCGEEGHMARDCSQGGG-----------GGGGYGGGG 164
Query: 241 RRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
+ GGGG GG G C+NCG+ GH AR+C
Sbjct: 165 GGYRGGGGG--GGGGGCYNCGETGHIAREC 192
Score = 67.4 bits (163), Expect = 4e-10
Identities = 44/127 (34%), Positives = 51/127 (39%), Gaps = 40/127 (31%)
Query: 152 ARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHI 211
A D GG GG GGG Y GGGGG
Sbjct: 66 AVDVTAPGGGALTGGSRPSGGGDRGY------------------GGGGG----------- 96
Query: 212 ARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGG---GHDGGKGTCFNCGKAGHFAR 268
GGR+ G G Y G G +GGG+R +G GGG G GG C+ CG+ GH AR
Sbjct: 97 -------GGRYGGDRG-YGGGGGGYGGGDRGYGGGGGYGGGGGGGSRACYKCGEEGHMAR 148
Query: 269 DCVEASG 275
DC + G
Sbjct: 149 DCSQGGG 155
>UniRef100_Q75QN9 Cold shock domain protein 2 [Triticum aestivum]
Length = 205
Score = 203 bits (517), Expect = 3e-51
Identities = 123/267 (46%), Positives = 142/267 (53%), Gaps = 69/267 (25%)
Query: 4 ERISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGK 63
ER+ G V+WFN TKGFGFI P+D SEDLFVHQS I+++G+RSL+E D VEF + GD+G+
Sbjct: 3 ERLKGTVKWFNVTKGFGFISPEDGSEDLFVHQSAIKADGYRSLNENDVVEFEVITGDDGR 62
Query: 64 TKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDR 123
TKA DVT P G L GGG RG RGG GGG Y G G+
Sbjct: 63 TKATDVTAPGGGAL-AGGSRPSEGGGDRGGRGGGGYGGGG--YGGGGGGY---------- 109
Query: 124 NDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVG 183
GGGGGG GGG + GGGGYGGGG R GG
Sbjct: 110 -------GGGGGG-------------------YGGGGGSYGGGGGYGGGGGGG-RYGG-- 140
Query: 184 HIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRF 243
G GGG CYKCGE GHI+RDC GG GG G Y G GR GGG
Sbjct: 141 -----------GSGGGRECYKCGEEGHISRDCPQGGG---GGGGGYGGGGGRGGGG---- 182
Query: 244 GSGGGGHDGGKGTCFNCGKAGHFARDC 270
GGGG CF+CG++GHF+R+C
Sbjct: 183 --GGGG-------CFSCGESGHFSREC 200
>UniRef100_Q41188 Glycine-rich protein 2 [Arabidopsis thaliana]
Length = 203
Score = 197 bits (501), Expect = 2e-49
Identities = 112/267 (41%), Positives = 132/267 (48%), Gaps = 77/267 (28%)
Query: 4 ERISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGK 63
ER G V+WF+ KGFGFI PDD +DLFVHQS IRSEGFRSL+ + VEF + +N +
Sbjct: 9 ERRKGSVKWFDTQKGFGFITPDDGGDDLFVHQSSIRSEGFRSLAAEEAVEFEVEIDNNNR 68
Query: 64 TKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDR 123
KA+DV+GP G P+Q GGG G RGG
Sbjct: 69 PKAIDVSGPDGAPVQ-----GNSGGGSSGGRGG--------------------------- 96
Query: 124 NDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVG 183
GGG GG R GG GGGGYGG G
Sbjct: 97 ------FGGGRGGGRG------------------SGGGYGGGGGGYGGRG---------- 122
Query: 184 HIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRF 243
GG GG CYKCGE GH+ARDCS GG + GG G Y G G +GGG +
Sbjct: 123 ----------GGGRGGSDCYKCGEPGHMARDCSEGGGGYGGGGGGYGGGGG-YGGGGGGY 171
Query: 244 GSGGGGHDGGKGTCFNCGKAGHFARDC 270
G GG G GG G+C++CG++GHFARDC
Sbjct: 172 GGGGRGGGGGGGSCYSCGESGHFARDC 198
Score = 58.2 bits (139), Expect = 2e-07
Identities = 32/84 (38%), Positives = 37/84 (43%), Gaps = 15/84 (17%)
Query: 192 PSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHD 251
P G GGG+ S G F GG G G +GGG +G GGG
Sbjct: 81 PVQGNSGGGS--------------SGGRGGFGGGRGGGRGSGGGYGGGGGGYGGRGGGGR 126
Query: 252 GGKGTCFNCGKAGHFARDCVEASG 275
GG C+ CG+ GH ARDC E G
Sbjct: 127 GGSD-CYKCGEPGHMARDCSEGGG 149
Score = 55.1 bits (131), Expect = 2e-06
Identities = 24/41 (58%), Positives = 28/41 (67%), Gaps = 2/41 (4%)
Query: 85 GGGGGGRGFRGGERRNGGGG--CYTCGDTGHIARDCDRSDR 123
G GGGG G+ GG R GGGG CY+CG++GH ARDC R
Sbjct: 163 GYGGGGGGYGGGGRGGGGGGGSCYSCGESGHFARDCTSGGR 203
>UniRef100_Q38896 Glycine-rich protein 2b [Arabidopsis thaliana]
Length = 201
Score = 185 bits (469), Expect = 1e-45
Identities = 112/268 (41%), Positives = 140/268 (51%), Gaps = 86/268 (32%)
Query: 4 ERISGVVQWFNNTKGFGFIKPDDDSEDLFVHQSDIRSEGFRSLSEGDRVEFTIADGDNGK 63
+R G V+WF+ KGFGFI P D +DLFVHQS IRSEGFRSL+ + VEF + ++G+
Sbjct: 13 DRRKGTVKWFDTQKGFGFITPSDGGDDLFVHQSSIRSEGFRSLAAEESVEFDVEVDNSGR 72
Query: 64 TKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDR 123
KA++V+GP G P+Q + GGGG G RGG GGG
Sbjct: 73 PKAIEVSGPDGAPVQ----GNSGGGGSSGGRGGF----GGG------------------- 105
Query: 124 NDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVG 183
GG GGG RGGG+ GGGYGG G+ GG G
Sbjct: 106 -------GGRGGG--------------------RGGGSY---GGGYGGRGS-----GGRG 130
Query: 184 HIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRF 243
GGGG +C+KCGE GH+AR+CS GG + G GGG R+
Sbjct: 131 -----------GGGGDNSCFKCGEPGHMARECSQGGGGYSG------------GGGGGRY 167
Query: 244 GSGGGGHDGGKG-TCFNCGKAGHFARDC 270
GSGGGG GG G +C++CG++GHFARDC
Sbjct: 168 GSGGGGGGGGGGLSCYSCGESGHFARDC 195
Score = 78.2 bits (191), Expect = 2e-13
Identities = 64/199 (32%), Positives = 76/199 (38%), Gaps = 64/199 (32%)
Query: 87 GGGGRGFRGGERRNG---------GGGCYTCGDTG------------------------H 113
GGG GG+RR G G G T D G
Sbjct: 3 GGGDVNMSGGDRRKGTVKWFDTQKGFGFITPSDGGDDLFVHQSSIRSEGFRSLAAEESVE 62
Query: 114 IARDCDRSDRNDRNDRSG----------GGGGGDRDRACYTCGSFEHFAR-DCMRGGGNN 162
+ D S R + SG GGGG R + G R GGG
Sbjct: 63 FDVEVDNSGRPKAIEVSGPDGAPVQGNSGGGGSSGGRGGFGGGGGRGGGRGGGSYGGGYG 122
Query: 163 NNGGGGYGGGG--TSCYRCGGVGHIARDCATPS---SGGGGGG---------------AC 202
G GG GGGG SC++CG GH+AR+C+ SGGGGGG +C
Sbjct: 123 GRGSGGRGGGGGDNSCFKCGEPGHMARECSQGGGGYSGGGGGGRYGSGGGGGGGGGGLSC 182
Query: 203 YKCGEVGHIARDCSNEGGR 221
Y CGE GH ARDC++ G R
Sbjct: 183 YSCGESGHFARDCTSGGAR 201
Score = 61.2 bits (147), Expect = 3e-08
Identities = 34/85 (40%), Positives = 39/85 (45%), Gaps = 14/85 (16%)
Query: 192 PSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDD-GNGRFGGGNRRFGSGGGGH 250
P G GGG S G F GG GR G G +GGG GSGG G
Sbjct: 85 PVQGNSGGGGS-------------SGGRGGFGGGGGRGGGRGGGSYGGGYGGRGSGGRGG 131
Query: 251 DGGKGTCFNCGKAGHFARDCVEASG 275
GG +CF CG+ GH AR+C + G
Sbjct: 132 GGGDNSCFKCGEPGHMARECSQGGG 156
>UniRef100_Q04832 DNA-binding protein HEXBP [Leishmania major]
Length = 271
Score = 147 bits (371), Expect = 3e-34
Identities = 79/206 (38%), Positives = 97/206 (46%), Gaps = 46/206 (22%)
Query: 71 GPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGG-----GCYTCGDTGHIARDCDRSDRND 125
G +G + + GG GG G + G GG CY CGD GHI+RDC
Sbjct: 103 GQEGHLSRDCPSSQGGSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDCPNGQ--- 159
Query: 126 RNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGT-SCYRCGGVGH 184
GG G DR CY CG H +RDC NG GGY G G CY+CG GH
Sbjct: 160 ------GGYSGAGDRTCYKCGDAGHISRDCP-------NGQGGYSGAGDRKCYKCGESGH 206
Query: 185 IARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFG 244
++R+C + S G G ACYKCG+ GHI+R+C GG +
Sbjct: 207 MSRECPSAGSTGSGDRACYKCGKPGHISRECPEAGGSY---------------------- 244
Query: 245 SGGGGHDGGKGTCFNCGKAGHFARDC 270
GG GG TC+ CG+AGH +RDC
Sbjct: 245 --GGSRGGGDRTCYKCGEAGHISRDC 268
Score = 136 bits (342), Expect = 7e-31
Identities = 74/194 (38%), Positives = 93/194 (47%), Gaps = 27/194 (13%)
Query: 97 ERRNGGGG---CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFAR 153
E R+G G C+ CG+ GH++RDC S + G + CY CG H +R
Sbjct: 61 EARSGAAGAMTCFRCGEAGHMSRDCPNSAKP----------GAAKGFECYKCGQEGHLSR 110
Query: 154 DCMRGGGNNNNG----------GGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGG--A 201
DC G + G GGY G T CY+CG GHI+RDC G G G
Sbjct: 111 DCPSSQGGSRGGYGQKRGRSGAQGGYSGDRT-CYKCGDAGHISRDCPNGQGGYSGAGDRT 169
Query: 202 CYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCG 261
CYKCG+ GHI+RDC N G + G R G G +R S G G + C+ CG
Sbjct: 170 CYKCGDAGHISRDCPNGQGGYSGAGDRKCYKCGESGHMSRECPSAGSTGSGDRA-CYKCG 228
Query: 262 KAGHFARDCVEASG 275
K GH +R+C EA G
Sbjct: 229 KPGHISRECPEAGG 242
Score = 119 bits (298), Expect = 9e-26
Identities = 65/195 (33%), Positives = 87/195 (44%), Gaps = 34/195 (17%)
Query: 98 RRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMR 157
R C CG GH AR+C +D G +R C+ CG H +R+C
Sbjct: 11 RTESSTSCRNCGKEGHYARECPEADSK----------GDERSTTCFRCGEEGHMSRECP- 59
Query: 158 GGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGG-ACYKCGEVGHIARDCS 216
N G +C+RCG GH++RDC + G G CYKCG+ GH++RDC
Sbjct: 60 ------NEARSGAAGAMTCFRCGEAGHMSRDCPNSAKPGAAKGFECYKCGQEGHLSRDCP 113
Query: 217 NEGGRFDGGNGRYDDGNGRFGGGN---------------RRFGSGGGGHDG-GKGTCFNC 260
+ G GG G+ +G GG + R +G GG+ G G TC+ C
Sbjct: 114 SSQGGSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDCPNGQGGYSGAGDRTCYKC 173
Query: 261 GKAGHFARDCVEASG 275
G AGH +RDC G
Sbjct: 174 GDAGHISRDCPNGQG 188
Score = 41.2 bits (95), Expect = 0.030
Identities = 20/48 (41%), Positives = 26/48 (53%), Gaps = 4/48 (8%)
Query: 74 GEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRS 121
G+P + ++ GG G RGG R CY CG+ GHI+RDC S
Sbjct: 228 GKPGHISRECPEAGGSYGGSRGGGDRT----CYKCGEAGHISRDCPSS 271
>UniRef100_Q9GNP1 Vasa homolog [Ciona savignyi]
Length = 770
Score = 138 bits (348), Expect = 1e-31
Identities = 75/199 (37%), Positives = 104/199 (51%), Gaps = 35/199 (17%)
Query: 82 DNHGGGGG-----GRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGG 136
D+ G GGG G GF + GC+ CG+ GH++R+C + GGGG
Sbjct: 80 DDTGFGGGFGSSSGGGFGDTRGSSRSKGCFKCGEEGHMSRECPQ------------GGGG 127
Query: 137 DRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGG 196
R + C+ CG H +R+C + GGGG GGGG C++CG GH++R+C G
Sbjct: 128 SRGKGCFKCGEEGHMSRECPK-------GGGGGGGGGRGCFKCGEEGHMSRECPKGGDSG 180
Query: 197 GGGGA----CYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDG 252
G + C+KCGE GH++R+C GG GG G G G +R GGG G
Sbjct: 181 FEGRSRSKGCFKCGEEGHMSRECPQGGG---GGRGSGCFKCGEEGHMSRECPQGGG---G 234
Query: 253 GKGT-CFNCGKAGHFARDC 270
G+G+ CF CG+ GH +R+C
Sbjct: 235 GRGSGCFKCGEEGHMSREC 253
Score = 118 bits (295), Expect = 2e-25
Identities = 71/196 (36%), Positives = 94/196 (47%), Gaps = 22/196 (11%)
Query: 88 GGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGG----GGGDRDRACY 143
G G GF G+ +GG G G D D SGGG G R + C+
Sbjct: 52 GFGSGF--GKSDDGGFGSKPNSGFGKSNFDDDTGFGGGFGSSSGGGFGDTRGSSRSKGCF 109
Query: 144 TCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGG-AC 202
CG H +R+C +GGG G G C++CG GH++R+C GGGGGG C
Sbjct: 110 KCGEEGHMSRECPQGGG---------GSRGKGCFKCGEEGHMSRECPKGGGGGGGGGRGC 160
Query: 203 YKCGEVGHIARDCSNEGGRFDGGNGRYDD--GNGRFGGGNRRFGSGGGGHDGGKGT-CFN 259
+KCGE GH++R+C G G R G G +R GGGG G+G+ CF
Sbjct: 161 FKCGEEGHMSRECPKGGDSGFEGRSRSKGCFKCGEEGHMSRECPQGGGG---GRGSGCFK 217
Query: 260 CGKAGHFARDCVEASG 275
CG+ GH +R+C + G
Sbjct: 218 CGEEGHMSRECPQGGG 233
Score = 94.4 bits (233), Expect = 3e-18
Identities = 56/177 (31%), Positives = 79/177 (43%), Gaps = 35/177 (19%)
Query: 39 RSEG-FRSLSEGDRVEFTIADGDNGKTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGE 97
RS+G F+ EG G + K G +G R+ GGGGGG
Sbjct: 104 RSKGCFKCGEEGHMSRECPQGGGGSRGKGCFKCGEEGH--MSRECPKGGGGGG------- 154
Query: 98 RRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMR 157
GG GC+ CG+ GH++R+C + G G R + C+ CG H +R+C +
Sbjct: 155 --GGGRGCFKCGEEGHMSRECPKGG-------DSGFEGRSRSKGCFKCGEEGHMSRECPQ 205
Query: 158 GGGNNNNGG----------------GGYGGGGTSCYRCGGVGHIARDCATPSSGGGG 198
GGG G GG GG G+ C++CG GH++R+C +SG GG
Sbjct: 206 GGGGGRGSGCFKCGEEGHMSRECPQGGGGGRGSGCFKCGEEGHMSRECPRNTSGEGG 262
Score = 45.4 bits (106), Expect = 0.002
Identities = 25/72 (34%), Positives = 39/72 (53%), Gaps = 15/72 (20%)
Query: 74 GEPLQVRQDNHGGGGGGRG---FRGGER--------RNGGGG----CYTCGDTGHIARDC 118
GE + ++ GGGGGRG F+ GE + GGGG C+ CG+ GH++R+C
Sbjct: 194 GEEGHMSRECPQGGGGGRGSGCFKCGEEGHMSRECPQGGGGGRGSGCFKCGEEGHMSREC 253
Query: 119 DRSDRNDRNDRS 130
R+ + ++S
Sbjct: 254 PRNTSGEGGEKS 265
>UniRef100_Q86EQ4 Clone ZZD1536 mRNA sequence [Schistosoma japonicum]
Length = 192
Score = 138 bits (347), Expect = 2e-31
Identities = 73/197 (37%), Positives = 92/197 (46%), Gaps = 53/197 (26%)
Query: 103 GGCYTCGDTGHIARDCDRSDRNDRNDRS-----GGGGGGDRDR------ACYTCGSFEHF 151
G C+ CG GH ARDC R R GGGGG DRD C+ CG +H+
Sbjct: 3 GECFKCGREGHFARDCQAQSRGGRGGGGGYRGRGGGGGRDRDNNDGRRDGCFNCGGLDHY 62
Query: 152 ARDCMRGGGN-NNNGGGGYGGGGT--SCYRCGGVGHIARDCATP----------SSGGGG 198
ARDC G+ GGGGYGG G+ C+ CGGVGH AR+C GGGG
Sbjct: 63 ARDCPNDRGHYGGGGGGGYGGYGSRDKCFNCGGVGHFARECTNDGQRGDSGYNNGGGGGG 122
Query: 199 GGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCF 258
GG CY CG+ GH+ R+C + N R +D + C+
Sbjct: 123 GGRCYNCGQSGHVVRNCPS--------NNR---------------------NDMSEILCY 153
Query: 259 NCGKAGHFARDCVEASG 275
C K GH+A++C E+ G
Sbjct: 154 RCNKYGHYAKECTESGG 170
Score = 127 bits (318), Expect = 4e-28
Identities = 68/151 (45%), Positives = 82/151 (54%), Gaps = 22/151 (14%)
Query: 85 GGGGGGRGFRGG---ERRNGGG---GCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGG-- 136
GGGGG RG GG +R N G GC+ CG H ARDC NDR GGGGGG
Sbjct: 27 GGGGGYRGRGGGGGRDRDNNDGRRDGCFNCGGLDHYARDCP----NDRGHYGGGGGGGYG 82
Query: 137 ---DRDRACYTCGSFEHFARDC----MRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDC 189
RD+ C+ CG HFAR+C RG NNGGG GGGG CY CG GH+ R+C
Sbjct: 83 GYGSRDK-CFNCGGVGHFARECTNDGQRGDSGYNNGGG--GGGGGRCYNCGQSGHVVRNC 139
Query: 190 ATPSSGGGGGGACYKCGEVGHIARDCSNEGG 220
+ + CY+C + GH A++C+ GG
Sbjct: 140 PSNNRNDMSEILCYRCNKYGHYAKECTESGG 170
Score = 99.0 bits (245), Expect = 1e-19
Identities = 56/136 (41%), Positives = 67/136 (49%), Gaps = 17/136 (12%)
Query: 84 HGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDC-DRSDRNDRNDRSGGGGGGDRDRAC 142
H GGGGG G+ G R+ C+ CG GH AR+C + R D +GGGGGG C
Sbjct: 72 HYGGGGGGGYGGYGSRDK---CFNCGGVGHFARECTNDGQRGDSGYNNGGGGGG--GGRC 126
Query: 143 YTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGAC 202
Y CG H R+C NN N CYRC GH A++C + GG G C
Sbjct: 127 YNCGQSGHVVRNC---PSNNRN-----DMSEILCYRCNKYGHYAKEC---TESGGSGPQC 175
Query: 203 YKCGEVGHIARDCSNE 218
YKC GHIA C+ E
Sbjct: 176 YKCRGYGHIASRCNVE 191
Score = 80.9 bits (198), Expect = 3e-14
Identities = 37/110 (33%), Positives = 53/110 (47%), Gaps = 22/110 (20%)
Query: 80 RQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRD 139
R+ + G G G+ G GGG CY CG +GH+ R+C ++RND + +
Sbjct: 101 RECTNDGQRGDSGYNNGGGGGGGGRCYNCGQSGHVVRNCPSNNRNDMS-----------E 149
Query: 140 RACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDC 189
CY C + H+A++C GG+ G CY+C G GHIA C
Sbjct: 150 ILCYRCNKYGHYAKECTESGGS-----------GPQCYKCRGYGHIASRC 188
>UniRef100_Q7S753 Hypothetical protein [Neurospora crassa]
Length = 183
Score = 137 bits (344), Expect = 4e-31
Identities = 74/182 (40%), Positives = 91/182 (49%), Gaps = 36/182 (19%)
Query: 101 GGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGG 160
G C+TCG T H ARDC CY CG+ H +RDC G
Sbjct: 9 GTRACFTCGQTTHQARDCPNKGA----------------AKCYNCGNEGHMSRDCPEGPK 52
Query: 161 NNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGG 220
+N +CYRCG GHI+RDC+ G G CYKCGEVGHIAR+CS G
Sbjct: 53 DN----------ARTCYRCGQTGHISRDCSQSGGGQSSGAECYKCGEVGHIARNCSKGGA 102
Query: 221 RFDGGNGRYDDGNGRFGGGNRR-FGSGGGGH------DGGKGTCFNCGKAGHFARDCVEA 273
+ GG G G FGG + + GG GH +G K C+NCG++GHF+RDC +
Sbjct: 103 SYGGGYQNSGYGGG-FGGPQKTCYSCGGIGHMSRDCVNGSK--CYNCGESGHFSRDCPKD 159
Query: 274 SG 275
SG
Sbjct: 160 SG 161
Score = 96.3 bits (238), Expect = 8e-19
Identities = 50/146 (34%), Positives = 69/146 (47%), Gaps = 31/146 (21%)
Query: 74 GEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRN----DRNDR 129
G+ + +D GGG ++ G CY CG+ GHIAR+C + + +N
Sbjct: 62 GQTGHISRDCSQSGGG---------QSSGAECYKCGEVGHIARNCSKGGASYGGGYQNSG 112
Query: 130 SGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDC 189
GGG GG + + CY+CG H +RDC+ G+ CY CG GH +RDC
Sbjct: 113 YGGGFGGPQ-KTCYSCGGIGHMSRDCV---------------NGSKCYNCGESGHFSRDC 156
Query: 190 ATPSSGGGGGGACYKCGEVGHIARDC 215
P G G CYKC + GH+ C
Sbjct: 157 --PKDSGSGEKICYKCQQPGHVQSQC 180
>UniRef100_Q9GV13 Vasa-related protein CnVAS1 [Hydra magnipapillata]
Length = 797
Score = 136 bits (342), Expect = 7e-31
Identities = 88/241 (36%), Positives = 113/241 (46%), Gaps = 65/241 (26%)
Query: 82 DNHGGGGGGRGFRGGERRNGGGG--CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRD 139
D GG G G GG NGGGG C+ CG GH++R+C GGGGGG R
Sbjct: 46 DTSVGGFGSSGGFGGSSFNGGGGRACHKCGKEGHMSRECP----------DGGGGGGGR- 94
Query: 140 RACYTCGSFEHFARDCMRGGGNNNN-----GGGGY--------GGGGTSCYRCGGVGHIA 186
AC+ C H +RDC +GG G G+ GGGG +C++C GH++
Sbjct: 95 -ACFKCKQEGHMSRDCPQGGSGGGRACHKCGKEGHMSRECPDGGGGGRACFKCKQEGHMS 153
Query: 187 RDCATPSSGGGGGGACYKCGEVGHIARDC---SNEGGRF---DGGNGRYDDGNGRFG-GG 239
+DC SGGGG C+KCG+ GH++R+C S GG F GG G + G FG G
Sbjct: 154 KDCPQ-GSGGGGSRTCHKCGKEGHMSRECPDGSGGGGGFGEKSGGGGFGEKSGGGFGASG 212
Query: 240 NRRFGSGGG-------------GHDGGKGT-----------------CFNCGKAGHFARD 269
FG+GGG G+ GG G C C ++GHFA+D
Sbjct: 213 GGGFGAGGGGFGTISTGSNSFEGNGGGFGDDAAGGGGFGASEKRDDGCRICKQSGHFAKD 272
Query: 270 C 270
C
Sbjct: 273 C 273
Score = 80.9 bits (198), Expect = 3e-14
Identities = 54/179 (30%), Positives = 70/179 (38%), Gaps = 69/179 (38%)
Query: 160 GNNNNGGGGYG-----------------------------------------GGGTSCYR 178
G +G GG GGG +C++
Sbjct: 14 GGFGSGAGGSSSFGKSFNSGGGFGSSSAGGFGDTSVGGFGSSGGFGGSSFNGGGGRACHK 73
Query: 179 CGGVGHIARDCATPSSGGGGGG-ACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFG 237
CG GH++R+C P GGGGGG AC+KC + GH++RDC G G GR G+ G
Sbjct: 74 CGKEGHMSREC--PDGGGGGGGRACFKCKQEGHMSRDCPQGG----SGGGRACHKCGKEG 127
Query: 238 GGNRRFGSGGGG---------------------HDGGKGTCFNCGKAGHFARDCVEASG 275
+R GGGG GG TC CGK GH +R+C + SG
Sbjct: 128 HMSRECPDGGGGGRACFKCKQEGHMSKDCPQGSGGGGSRTCHKCGKEGHMSRECPDGSG 186
Score = 58.2 bits (139), Expect = 2e-07
Identities = 54/212 (25%), Positives = 67/212 (31%), Gaps = 104/212 (49%)
Query: 73 KGEPLQVRQDNHGGGGGGR--------GFRGGERRNGGGG---CYTCGDTGHIARDCDRS 121
K E R GG GGGR G E +GGGG C+ C GH+++DC +
Sbjct: 100 KQEGHMSRDCPQGGSGGGRACHKCGKEGHMSRECPDGGGGGRACFKCKQEGHMSKDCPQ- 158
Query: 122 DRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDC---------------------MRGGG 160
G GGGG R C+ CG H +R+C GGG
Sbjct: 159 ---------GSGGGGSR--TCHKCGKEGHMSRECPDGSGGGGGFGEKSGGGGFGEKSGGG 207
Query: 161 NNNNGGGGY-----------------------------GGGG------------------ 173
+GGGG+ GGGG
Sbjct: 208 FGASGGGGFGAGGGGFGTISTGSNSFEGNGGGFGDDAAGGGGFGASEKRDDGCRICKQSG 267
Query: 174 -------------TSCYRCGGVGHIARDCATP 192
+C RCG GH A+DC P
Sbjct: 268 HFAKDCPDKKPRDDTCRRCGESGHFAKDCEAP 299
>UniRef100_Q9C1E7 Putative DNA binding protein [Schizophyllum commune]
Length = 146
Score = 122 bits (307), Expect = 8e-27
Identities = 69/175 (39%), Positives = 79/175 (44%), Gaps = 50/175 (28%)
Query: 105 CYTCGDTGHIARDCDRSDRNDRNDRSGGG-GGGDRDRACYTCGSFEHFARDCMR--GGGN 161
CY CG GHI+RDC +D SGGG GGG R CY CG H AR C GGN
Sbjct: 6 CYKCGGEGHISRDCSSADAGGAGGYSGGGFGGGARGGECYRCGKAGHMARACPEPAPGGN 65
Query: 162 NNNGGG---GYGGG---GTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDC 215
+ GGG GYGGG SCY CGGVGH+++DC G CY C E GHI+RDC
Sbjct: 66 ASYGGGGSYGYGGGFQSQKSCYTCGGVGHLSKDCVQ-------GQRCYNCSETGHISRDC 118
Query: 216 SNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
N K C++CG H +RDC
Sbjct: 119 PNP----------------------------------QKKACYSCGSESHISRDC 139
Score = 102 bits (254), Expect = 1e-20
Identities = 55/138 (39%), Positives = 64/138 (45%), Gaps = 30/138 (21%)
Query: 85 GGGGG--GRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGG-----GGGGD 137
GG GG G GF GG R GG CY CG GH+AR C GG GGG
Sbjct: 25 GGAGGYSGGGFGGGAR---GGECYRCGKAGHMARACPEPAPGGNASYGGGGSYGYGGGFQ 81
Query: 138 RDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGG 197
++CYTCG H ++DC++ G CY C GHI+RDC P
Sbjct: 82 SQKSCYTCGGVGHLSKDCVQ---------------GQRCYNCSETGHISRDCPNPQK--- 123
Query: 198 GGGACYKCGEVGHIARDC 215
ACY CG HI+RDC
Sbjct: 124 --KACYSCGSESHISRDC 139
Score = 89.0 bits (219), Expect = 1e-16
Identities = 43/110 (39%), Positives = 59/110 (53%), Gaps = 25/110 (22%)
Query: 175 SCYRCGGVGHIARDCATPSSGGGGG------------GACYKCGEVGHIARDCSNEGGRF 222
+CY+CGG GHI+RDC++ +GG GG G CY+CG+ GH+AR C
Sbjct: 5 TCYKCGGEGHISRDCSSADAGGAGGYSGGGFGGGARGGECYRCGKAGHMARACPEPA--- 61
Query: 223 DGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDCVE 272
GGN Y GG +G GGG + +C+ CG GH ++DCV+
Sbjct: 62 PGGNASY--------GGGGSYGYGGGFQS--QKSCYTCGGVGHLSKDCVQ 101
>UniRef100_UPI00004301EE UPI00004301EE UniRef100 entry
Length = 204
Score = 122 bits (306), Expect = 1e-26
Identities = 70/191 (36%), Positives = 91/191 (46%), Gaps = 62/191 (32%)
Query: 105 CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNN 164
CY CG +GH++R+C + +++ACYTCG H + C +G +
Sbjct: 30 CYNCGLSGHLSRECPQP----------------KNKACYTCGQEGHLSSACPQG-----S 68
Query: 165 GGGGYGG--GGTSCYRCGGVGHIARDCATPSS------------GGGGGGA------CYK 204
G GG+GG GG CYRCG GHIAR C GG GGGA CY
Sbjct: 69 GAGGFGGASGGGECYRCGKPGHIARMCPESGDAAAGGFGGAGGYGGFGGGAGFGNKSCYT 128
Query: 205 CGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAG 264
CG VGHI+R+C + G +R FG GGGG GG C+NCG+ G
Sbjct: 129 CGGVGHISRECPS--------------------GASRGFGGGGGGF-GGPRKCYNCGQDG 167
Query: 265 HFARDCVEASG 275
H +R+C + G
Sbjct: 168 HISRECPQEQG 178
Score = 114 bits (286), Expect = 2e-24
Identities = 63/164 (38%), Positives = 73/164 (44%), Gaps = 19/164 (11%)
Query: 63 KTKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSD 122
K KA G +G G GG G +GGG CY CG GHIAR C S
Sbjct: 47 KNKACYTCGQEGHLSSACPQGSGAGGFGGA-------SGGGECYRCGKPGHIARMCPESG 99
Query: 123 RNDRNDRSG-------GGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTS 175
G GGG G +++CYTCG H +R+C G GGGG GG
Sbjct: 100 DAAAGGFGGAGGYGGFGGGAGFGNKSCYTCGGVGHISRECPSGASRGFGGGGGGFGGPRK 159
Query: 176 CYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEG 219
CY CG GHI+R+C G CY CG+ GHIA C G
Sbjct: 160 CYNCGQDGHISRECPQEQ-----GKTCYSCGQPGHIASACPGAG 198
Score = 50.1 bits (118), Expect = 6e-05
Identities = 33/96 (34%), Positives = 39/96 (40%), Gaps = 17/96 (17%)
Query: 195 GGGGGGACYKCGEVGHIARDCSNEGGR-----FDGGNGR----------YDDGNGRFGGG 239
G G +C+KCG+ GH+A C E G R Y G
Sbjct: 3 GAPRGSSCFKCGQQGHVAAACPAEAPTCYNCGLSGHLSRECPQPKNKACYTCGQEGHLSS 62
Query: 240 NRRFGSGGGGHDG--GKGTCFNCGKAGHFARDCVEA 273
GSG GG G G G C+ CGK GH AR C E+
Sbjct: 63 ACPQGSGAGGFGGASGGGECYRCGKPGHIARMCPES 98
>UniRef100_P90606 Nucleic acid binding protein [Trypanosoma equiperdum]
Length = 270
Score = 121 bits (303), Expect = 2e-26
Identities = 71/186 (38%), Positives = 89/186 (47%), Gaps = 27/186 (14%)
Query: 98 RRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMR 157
R GG C+ CG GH AR+C N G G DRACYTCG +H +RDC
Sbjct: 12 RAEGGNNCHRCGQPGHFARECP-------NVPPGAMG----DRACYTCGQPDHLSRDC-- 58
Query: 158 GGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGG------GGGGACYKCGEVGHI 211
+N G GGG +CY CG GH +R+C G GGG ACY C + GH
Sbjct: 59 ----PSNRGTAPMGGGRACYNCGQPGHFSRECPNMRGGPMGGAPMGGGRACYNCVQPGHF 114
Query: 212 ARDCSN-EGGRFDG---GNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFA 267
+R+C N GG G G GR G+ G +R + G + GG C+ C + GH A
Sbjct: 115 SRECPNMRGGPMGGAPMGGGRACYHCGQPGHFSRECPNMRGANMGGGRECYQCRQEGHIA 174
Query: 268 RDCVEA 273
+C A
Sbjct: 175 SECPNA 180
Score = 120 bits (300), Expect = 5e-26
Identities = 71/183 (38%), Positives = 89/183 (47%), Gaps = 26/183 (14%)
Query: 101 GGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDC--MRG 158
G CYTCG H++RDC ++R GGG RACY CG HF+R+C MRG
Sbjct: 41 GDRACYTCGQPDHLSRDCP----SNRGTAPMGGG-----RACYNCGQPGHFSRECPNMRG 91
Query: 159 GGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGG------GGGGACYKCGEVGHIA 212
G GG GGG +CY C GH +R+C G GGG ACY CG+ GH +
Sbjct: 92 GPM----GGAPMGGGRACYNCVQPGHFSRECPNMRGGPMGGAPMGGGRACYHCGQPGHFS 147
Query: 213 RDCSN-EGGRFDGGNGRY---DDGNGRFGGGNRRFGSGGGGHDGGKG-TCFNCGKAGHFA 267
R+C N G GG Y +G+ N + GG G G C+ CG+ GH +
Sbjct: 148 RECPNMRGANMGGGRECYQCRQEGHIASECPNAPDDAAAGGTAAGGGRACYKCGQPGHLS 207
Query: 268 RDC 270
R C
Sbjct: 208 RAC 210
Score = 99.8 bits (247), Expect = 7e-20
Identities = 56/133 (42%), Positives = 66/133 (49%), Gaps = 23/133 (17%)
Query: 92 GFRGGERRNGGGGCYTCGDTGHIARDCDRSDRNDRNDRSGGG--GGGDRDRACYTCGSFE 149
G GG GG CY C GH +R+C N R GG GGG RACY CG
Sbjct: 92 GPMGGAPMGGGRACYNCVQPGHFSRECP----NMRGGPMGGAPMGGG---RACYHCGQPG 144
Query: 150 HFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDC-------ATPSSGGGGGGAC 202
HF+R+C G N GGG CY+C GHIA +C A + GGG AC
Sbjct: 145 HFSRECPNMRGANM-------GGGRECYQCRQEGHIASECPNAPDDAAAGGTAAGGGRAC 197
Query: 203 YKCGEVGHIARDC 215
YKCG+ GH++R C
Sbjct: 198 YKCGQPGHLSRAC 210
Score = 89.7 bits (221), Expect = 7e-17
Identities = 46/109 (42%), Positives = 58/109 (53%), Gaps = 5/109 (4%)
Query: 172 GGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDCSNEGGRFDGGNGR--Y 229
GG +C+RCG GH AR+C G G ACY CG+ H++RDC + G G GR Y
Sbjct: 15 GGNNCHRCGQPGHFARECPNVPPGAMGDRACYTCGQPDHLSRDCPSNRGTAPMGGGRACY 74
Query: 230 DDGN-GRFGG--GNRRFGSGGGGHDGGKGTCFNCGKAGHFARDCVEASG 275
+ G G F N R G GG GG C+NC + GHF+R+C G
Sbjct: 75 NCGQPGHFSRECPNMRGGPMGGAPMGGGRACYNCVQPGHFSRECPNMRG 123
>UniRef100_Q966I7 Hypothetical protein K08D12.3 [Caenorhabditis elegans]
Length = 151
Score = 120 bits (301), Expect = 4e-26
Identities = 63/149 (42%), Positives = 78/149 (52%), Gaps = 19/149 (12%)
Query: 80 RQDNHGGGGGGRGFRGGERRNGGGG--CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGD 137
+Q H G G RR GGGG CY C +TGH +RDC + + R GGGGGG
Sbjct: 10 QQPGHISRNCPNGESDGGRRGGGGGSTCYNCQETGHFSRDCPKGG-SGGGQRGGGGGGG- 67
Query: 138 RDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGG--------GGTSCYRCGGVGHIARDC 189
+CY CG H++RDC + G GGYGG GG CY CG GHI+R+C
Sbjct: 68 ---SCYNCGGRGHYSRDC--PSARSEGGSGGYGGRGGEGRSFGGQKCYNCGRSGHISREC 122
Query: 190 ATPSSGGGGGGACYKCGEVGHIARDCSNE 218
SG CY+C E GHI+RDC ++
Sbjct: 123 T--ESGSAEEKRCYQCQETGHISRDCPSQ 149
Score = 118 bits (296), Expect = 1e-25
Identities = 63/180 (35%), Positives = 78/180 (43%), Gaps = 53/180 (29%)
Query: 105 CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNN 164
CY C GHI+R+C + + R GGGGG CY C HF+RDC +GG
Sbjct: 6 CYKCQQPGHISRNCPNGESD--GGRRGGGGGS----TCYNCQETGHFSRDCPKGGSGGGQ 59
Query: 165 GGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGG--------------ACYKCGEVGH 210
GGG GGG SCY CGG GH +RDC + S GG GG CY CG GH
Sbjct: 60 RGGG--GGGGSCYNCGGRGHYSRDCPSARSEGGSGGYGGRGGEGRSFGGQKCYNCGRSGH 117
Query: 211 IARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDC 270
I+R+C+ G + C+ C + GH +RDC
Sbjct: 118 ISRECTESG-------------------------------SAEEKRCYQCQETGHISRDC 146
Score = 116 bits (291), Expect = 6e-25
Identities = 58/141 (41%), Positives = 76/141 (53%), Gaps = 24/141 (17%)
Query: 139 DRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGG- 197
DR CY C H +R+C G ++GG GGGG++CY C GH +RDC SGGG
Sbjct: 3 DRNCYKCQQPGHISRNCPNG---ESDGGRRGGGGGSTCYNCQETGHFSRDCPKGGSGGGQ 59
Query: 198 -----GGGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGHDG 252
GGG+CY CG GH +RDC + R +GG+G Y G GG G
Sbjct: 60 RGGGGGGGSCYNCGGRGHYSRDCPS--ARSEGGSGGYG-------------GRGGEGRSF 104
Query: 253 GKGTCFNCGKAGHFARDCVEA 273
G C+NCG++GH +R+C E+
Sbjct: 105 GGQKCYNCGRSGHISRECTES 125
>UniRef100_UPI00003C130E UPI00003C130E UniRef100 entry
Length = 189
Score = 119 bits (298), Expect = 9e-26
Identities = 51/111 (45%), Positives = 66/111 (58%), Gaps = 9/111 (8%)
Query: 105 CYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNN 164
CY C +TGHI+R+C + + GG GG+ CY CG H AR C GG++
Sbjct: 62 CYKCSETGHISRECPTNPAP-----AAGGPGGE----CYKCGQHGHIARACPTAGGSSRG 112
Query: 165 GGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDC 215
G GG GG SCY CGGVGH++R+C +P+ GG CY C E GHI+R+C
Sbjct: 113 GFGGARSGGRSCYNCGGVGHLSRECTSPAGAAAGGQRCYNCNESGHISREC 163
Score = 95.1 bits (235), Expect = 2e-18
Identities = 51/143 (35%), Positives = 65/143 (44%), Gaps = 31/143 (21%)
Query: 141 ACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGHIARDCAT--PSSGGGG 198
+CY CG H + C G +CY+C GHI+R+C T + GG
Sbjct: 39 SCYNCGQQGHISSQC------------GMEAQPKTCYKCSETGHISRECPTNPAPAAGGP 86
Query: 199 GGACYKCGEVGHIARDCSNEGGRFDGGNGRYDDGNGRFGGGNRRFGSGGGGH-------- 250
GG CYKCG+ GHIAR C GG GG G G GG + GG GH
Sbjct: 87 GGECYKCGQHGHIARACPTAGGSSRGGFG------GARSGGRSCYNCGGVGHLSRECTSP 140
Query: 251 ---DGGKGTCFNCGKAGHFARDC 270
G C+NC ++GH +R+C
Sbjct: 141 AGAAAGGQRCYNCNESGHISREC 163
Score = 91.3 bits (225), Expect = 2e-17
Identities = 43/114 (37%), Positives = 56/114 (48%), Gaps = 16/114 (14%)
Query: 102 GGGCYTCGDTGHIARDCDRSDRNDRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGN 161
GG CY CG GHIAR C + + R G GG R+CY CG H +R+C
Sbjct: 87 GGECYKCGQHGHIARACPTAGGSSRGGFGGARSGG---RSCYNCGGVGHLSREC------ 137
Query: 162 NNNGGGGYGGGGTSCYRCGGVGHIARDCATPSSGGGGGGACYKCGEVGHIARDC 215
G GG CY C GHI+R+C P + +CY+CG+ GH++ C
Sbjct: 138 --TSPAGAAAGGQRCYNCNESGHISRECPKPQT-----KSCYRCGDEGHLSAAC 184
Score = 69.7 bits (169), Expect = 8e-11
Identities = 42/125 (33%), Positives = 53/125 (41%), Gaps = 31/125 (24%)
Query: 71 GPKGEPLQVRQDNH------GGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDRN 124
GP GE + Q H GG RG GG R +GG CY CG GH++R+C
Sbjct: 85 GPGGECYKCGQHGHIARACPTAGGSSRGGFGGAR-SGGRSCYNCGGVGHLSREC------ 137
Query: 125 DRNDRSGGGGGGDRDRACYTCGSFEHFARDCMRGGGNNNNGGGGYGGGGTSCYRCGGVGH 184
+G GG R CY C H +R+C + SCYRCG GH
Sbjct: 138 --TSPAGAAAGGQR---CYNCNESGHISRECPKPQTK-------------SCYRCGDEGH 179
Query: 185 IARDC 189
++ C
Sbjct: 180 LSAAC 184
Score = 53.5 bits (127), Expect = 6e-06
Identities = 38/124 (30%), Positives = 49/124 (38%), Gaps = 38/124 (30%)
Query: 183 GHIARDCATPSSGGGGGGACYKCG----------------------EVGHIARDCSNEGG 220
GH A C T G +CY CG E GHI+R+C
Sbjct: 26 GHNAAACPT-----AGNPSCYNCGQQGHISSQCGMEAQPKTCYKCSETGHISRECPTNPA 80
Query: 221 RFDGGNGRYDDGNGRFG---------GGNRRFGSGGGGHDGGKGTCFNCGKAGHFARDCV 271
GG G G+ G GG+ R G GG GG+ +C+NCG GH +R+C
Sbjct: 81 PAAGGPGGECYKCGQHGHIARACPTAGGSSR-GGFGGARSGGR-SCYNCGGVGHLSRECT 138
Query: 272 EASG 275
+G
Sbjct: 139 SPAG 142
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.145 0.476
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 639,520,012
Number of Sequences: 2790947
Number of extensions: 39742097
Number of successful extensions: 479349
Number of sequences better than 10.0: 9987
Number of HSP's better than 10.0 without gapping: 3132
Number of HSP's successfully gapped in prelim test: 7095
Number of HSP's that attempted gapping in prelim test: 195726
Number of HSP's gapped (non-prelim): 73206
length of query: 275
length of database: 848,049,833
effective HSP length: 125
effective length of query: 150
effective length of database: 499,181,458
effective search space: 74877218700
effective search space used: 74877218700
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 74 (33.1 bits)
Medicago: description of AC121233.14


