

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149206.8 - phase: 0
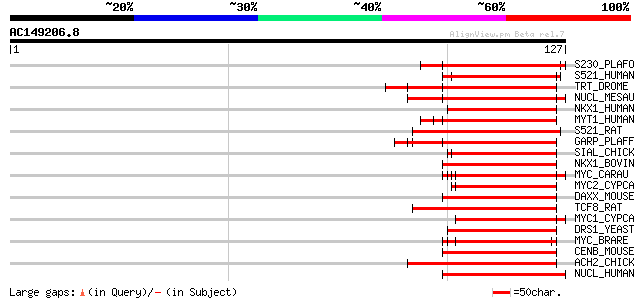
(127 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
S230_PLAFO (Q08372) Transmission-blocking target antigen S230 pr... 45 3e-05
S521_HUMAN (Q96MU7) Putative splicing factor YT521 44 5e-05
TRT_DROME (P19351) Troponin T, skeletal muscle (Upheld protein) ... 44 6e-05
NUCL_MESAU (P08199) Nucleolin (Protein C23) 43 1e-04
NKX1_HUMAN (O60721) Sodium/potassium/calcium exchanger 1 precurs... 43 1e-04
MYT1_HUMAN (Q01538) Myelin transcription factor 1 (MYT1) (MYTI) ... 43 1e-04
S521_RAT (Q9QY02) Putative splicing factor YT521 (RA301-binding ... 42 2e-04
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 42 2e-04
SIAL_CHICK (P79780) Bone sialoprotein II precursor (BSP II) (Cel... 42 2e-04
NKX1_BOVIN (Q28139) Sodium/potassium/calcium exchanger 1 precurs... 42 2e-04
MYC_CARAU (P49709) Myc protein (c-myc) 42 2e-04
MYC2_CYPCA (Q90342) Myc II protein (C-MYC II) 42 2e-04
DAXX_MOUSE (O35613) Death domain-associated protein 6 (Daxx) 42 2e-04
TCF8_RAT (Q62947) Transcription factor 8 (Zinc finger homeodomai... 42 3e-04
MYC1_CYPCA (Q90341) MYC I protein (C-MYC I) 42 3e-04
DRS1_YEAST (P32892) Probable ATP-dependent RNA helicase DRS1 42 3e-04
MYC_BRARE (P52160) Myc protein (c-myc) (zc-Myc) 41 4e-04
CENB_MOUSE (P27790) Major centromere autoantigen B (Centromere p... 41 4e-04
ACH2_CHICK (P09480) Neuronal acetylcholine receptor protein, alp... 41 4e-04
NUCL_HUMAN (P19338) Nucleolin (Protein C23) 41 5e-04
>S230_PLAFO (Q08372) Transmission-blocking target antigen S230
precursor
Length = 3135
Score = 45.1 bits (105), Expect = 3e-05
Identities = 19/32 (59%), Positives = 26/32 (80%)
Query: 95 NHIHYDDDSDSDMEEEEEEEEEEEEEEEQQEY 126
N + D++ + + EEEEEEEEEEEEEEE++EY
Sbjct: 274 NFVIDDEEEEEEEEEEEEEEEEEEEEEEEEEY 305
Score = 41.2 bits (95), Expect = 4e-04
Identities = 17/28 (60%), Positives = 23/28 (81%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEYV 127
+++ + + EEEEEEEEEEEEEEE +YV
Sbjct: 282 EEEEEEEEEEEEEEEEEEEEEEEYDDYV 309
Score = 39.7 bits (91), Expect = 0.001
Identities = 20/37 (54%), Positives = 26/37 (70%)
Query: 89 NKILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
N+ L+ + D+ D EEEEEEEEEEEEEEE++E
Sbjct: 261 NRNLDEEDMSPRDNFVIDDEEEEEEEEEEEEEEEEEE 297
Score = 37.7 bits (86), Expect = 0.005
Identities = 15/26 (57%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE+ +
Sbjct: 281 EEEEEEEEEEEEEEEEEEEEEEEEYD 306
Score = 34.3 bits (77), Expect = 0.050
Identities = 14/27 (51%), Positives = 20/27 (73%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEY 126
+++ + + EEEEEEEEEEEEE + Y
Sbjct: 284 EEEEEEEEEEEEEEEEEEEEEYDDYVY 310
Score = 33.1 bits (74), Expect = 0.11
Identities = 13/34 (38%), Positives = 23/34 (67%)
Query: 89 NKILETNHIHYDDDSDSDMEEEEEEEEEEEEEEE 122
N +++ +++ + + EEEEEEEEEEEE ++
Sbjct: 274 NFVIDDEEEEEEEEEEEEEEEEEEEEEEEEEYDD 307
Score = 32.3 bits (72), Expect = 0.19
Identities = 18/57 (31%), Positives = 31/57 (53%), Gaps = 5/57 (8%)
Query: 74 FFVTAILSWRWRRV-----LNKILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
FF+ + L + + + N + ++ +D S D ++EEEEEEEEEE++E
Sbjct: 236 FFINSSLKYIYMYLTPSDSFNLVRRNRNLDEEDMSPRDNFVIDDEEEEEEEEEEEEE 292
Score = 28.5 bits (62), Expect = 2.7
Identities = 11/26 (42%), Positives = 19/26 (72%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEE ++ +E
Sbjct: 287 EEEEEEEEEEEEEEEEEEYDDYVYEE 312
>S521_HUMAN (Q96MU7) Putative splicing factor YT521
Length = 727
Score = 44.3 bits (103), Expect = 5e-05
Identities = 19/25 (76%), Positives = 22/25 (88%)
Query: 102 DSDSDMEEEEEEEEEEEEEEEQQEY 126
D D + EEEEEEEEEEEEEEE++EY
Sbjct: 226 DEDGEEEEEEEEEEEEEEEEEEEEY 250
Score = 43.5 bits (101), Expect = 8e-05
Identities = 19/26 (73%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ D D EEEEEEEEEEEEEEE++E
Sbjct: 222 DEEVDEDGEEEEEEEEEEEEEEEEEE 247
Score = 42.0 bits (97), Expect = 2e-04
Identities = 18/26 (69%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D+D + + EEEEEEEEEEEEEEE+ E
Sbjct: 226 DEDGEEEEEEEEEEEEEEEEEEEEYE 251
Score = 39.7 bits (91), Expect = 0.001
Identities = 16/33 (48%), Positives = 24/33 (72%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
E + D + D +++E+ EEEEEEEEEEE++E
Sbjct: 211 EDEEVEEDAEEDEEVDEDGEEEEEEEEEEEEEE 243
Score = 39.3 bits (90), Expect = 0.002
Identities = 17/33 (51%), Positives = 23/33 (69%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
E + D + + + EEEEEEEEEEEEEE +Q+
Sbjct: 221 EDEEVDEDGEEEEEEEEEEEEEEEEEEEEYEQD 253
Score = 38.5 bits (88), Expect = 0.003
Identities = 16/26 (61%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
++D + D + EEEEEEEEEEEEE++E
Sbjct: 220 EEDEEVDEDGEEEEEEEEEEEEEEEE 245
Score = 37.0 bits (84), Expect = 0.008
Identities = 16/25 (64%), Positives = 20/25 (80%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
++ + + EEEEEEEEEEEEE EQ E
Sbjct: 230 EEEEEEEEEEEEEEEEEEEEYEQDE 254
Score = 34.3 bits (77), Expect = 0.050
Identities = 13/29 (44%), Positives = 22/29 (75%)
Query: 97 IHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
+ D++ + D EE+EE +E+ EEEEE++E
Sbjct: 209 VEEDEEVEEDAEEDEEVDEDGEEEEEEEE 237
Score = 33.5 bits (75), Expect = 0.085
Identities = 12/26 (46%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEE E+++++
Sbjct: 231 EEEEEEEEEEEEEEEEEEEYEQDERD 256
Score = 33.5 bits (75), Expect = 0.085
Identities = 13/25 (52%), Positives = 20/25 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQ 124
+++ + + EEEEEEEEEEEE E+ +
Sbjct: 230 EEEEEEEEEEEEEEEEEEEEYEQDE 254
Score = 32.7 bits (73), Expect = 0.15
Identities = 16/38 (42%), Positives = 24/38 (63%), Gaps = 5/38 (13%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEE-----EEEEEEQQE 125
E + D + D ++EE+ EE+EE EEEEEE++E
Sbjct: 201 EEEGVEEDVEEDEEVEEDAEEDEEVDEDGEEEEEEEEE 238
Score = 32.7 bits (73), Expect = 0.15
Identities = 12/26 (46%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEE E++E+ +
Sbjct: 232 EEEEEEEEEEEEEEEEEEYEQDERDQ 257
Score = 32.7 bits (73), Expect = 0.15
Identities = 12/26 (46%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEE E++E +Q+E
Sbjct: 234 EEEEEEEEEEEEEEEEYEQDERDQKE 259
Score = 32.0 bits (71), Expect = 0.25
Identities = 12/26 (46%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEE E++E Q+
Sbjct: 233 EEEEEEEEEEEEEEEEEYEQDERDQK 258
Score = 30.4 bits (67), Expect = 0.72
Identities = 10/26 (38%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEE E++E ++++E
Sbjct: 235 EEEEEEEEEEEEEEEYEQDERDQKEE 260
Score = 28.5 bits (62), Expect = 2.7
Identities = 9/27 (33%), Positives = 20/27 (73%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEY 126
+++ + + EEEE E++E +++EE +Y
Sbjct: 238 EEEEEEEEEEEEYEQDERDQKEEGNDY 264
Score = 27.7 bits (60), Expect = 4.7
Identities = 11/27 (40%), Positives = 19/27 (69%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQEYV 127
++ + +EE+ EE+EE EE+ E+ E V
Sbjct: 199 ENEEEGVEEDVEEDEEVEEDAEEDEEV 225
Score = 27.3 bits (59), Expect = 6.1
Identities = 13/33 (39%), Positives = 19/33 (57%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
ET D+ ++ E EEE EE+ EE+E+ E
Sbjct: 184 ETGSSGSSDEQGNNTENEEEGVEEDVEEDEEVE 216
>TRT_DROME (P19351) Troponin T, skeletal muscle (Upheld protein)
(Intended thorax protein)
Length = 396
Score = 43.9 bits (102), Expect = 6e-05
Identities = 19/34 (55%), Positives = 26/34 (75%)
Query: 92 LETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
+E + +D+ D + EEEEEEEEEEEEEEE++E
Sbjct: 351 VEEEVVEEEDEEDEEDEEEEEEEEEEEEEEEEEE 384
Score = 43.1 bits (100), Expect = 1e-04
Identities = 18/26 (69%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ + D EEEEEEEEEEEEEEE++E
Sbjct: 360 DEEDEEDEEEEEEEEEEEEEEEEEEE 385
Score = 43.1 bits (100), Expect = 1e-04
Identities = 18/26 (69%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ D + EEEEEEEEEEEEEEE++E
Sbjct: 362 EDEEDEEEEEEEEEEEEEEEEEEEEE 387
Score = 42.7 bits (99), Expect = 1e-04
Identities = 19/39 (48%), Positives = 28/39 (71%)
Query: 87 VLNKILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
V +++E ++D + + EEEEEEEEEEEEEEE++E
Sbjct: 351 VEEEVVEEEDEEDEEDEEEEEEEEEEEEEEEEEEEEEEE 389
Score = 41.6 bits (96), Expect = 3e-04
Identities = 17/26 (65%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ + + EEEEEEEEEEEEEEE++E
Sbjct: 366 DEEEEEEEEEEEEEEEEEEEEEEEEE 391
Score = 41.6 bits (96), Expect = 3e-04
Identities = 17/26 (65%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ + + EEEEEEEEEEEEEEE++E
Sbjct: 365 EDEEEEEEEEEEEEEEEEEEEEEEEE 390
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 370 EEEEEEEEEEEEEEEEEEEEEEEEEE 395
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 371 EEEEEEEEEEEEEEEEEEEEEEEEEE 396
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 368 EEEEEEEEEEEEEEEEEEEEEEEEEE 393
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 369 EEEEEEEEEEEEEEEEEEEEEEEEEE 394
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 367 EEEEEEEEEEEEEEEEEEEEEEEEEE 392
Score = 35.8 bits (81), Expect = 0.017
Identities = 13/26 (50%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ + ++ EEE+EE+EE+EEEE++E
Sbjct: 348 DEEVEEEVVEEEDEEDEEDEEEEEEE 373
Score = 35.4 bits (80), Expect = 0.022
Identities = 16/35 (45%), Positives = 26/35 (73%)
Query: 91 ILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
I+E + ++ + + EE+EE+EEEEEEEEE++E
Sbjct: 344 IVEDDEEVEEEVVEEEDEEDEEDEEEEEEEEEEEE 378
Score = 30.4 bits (67), Expect = 0.72
Identities = 15/33 (45%), Positives = 20/33 (60%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
ET D +D D+ E++EE EEE EEE +E
Sbjct: 330 ETPEGEEDAKADEDIVEDDEEVEEEVVEEEDEE 362
Score = 30.0 bits (66), Expect = 0.94
Identities = 13/26 (50%), Positives = 19/26 (73%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D+D D EE EEE EEE+EE++++
Sbjct: 341 DEDIVEDDEEVEEEVVEEEDEEDEED 366
Score = 29.3 bits (64), Expect = 1.6
Identities = 13/26 (50%), Positives = 18/26 (69%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D + D E EEE EEE+EE+E+ E
Sbjct: 342 EDIVEDDEEVEEEVVEEEDEEDEEDE 367
>NUCL_MESAU (P08199) Nucleolin (Protein C23)
Length = 713
Score = 42.7 bits (99), Expect = 1e-04
Identities = 17/26 (65%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DDD + D +EEE+EEEEE+EEEE++E
Sbjct: 242 DDDEEEDEDEEEDEEEEEDEEEEEEE 267
Score = 42.4 bits (98), Expect = 2e-04
Identities = 17/26 (65%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ D D EE+EEEEE+EEEEEE++E
Sbjct: 244 DEEEDEDEEEDEEEEEDEEEEEEEEE 269
Score = 41.2 bits (95), Expect = 4e-04
Identities = 17/28 (60%), Positives = 24/28 (85%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEYV 127
++D D + +EEEEE+EEEEEEEE++E V
Sbjct: 246 EEDEDEEEDEEEEEDEEEEEEEEEEEPV 273
Score = 40.8 bits (94), Expect = 5e-04
Identities = 15/34 (44%), Positives = 27/34 (79%)
Query: 92 LETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
++ ++ +DD D + +E+EEE+EEEEE+EE++E
Sbjct: 232 VKAKNVAEEDDDDEEEDEDEEEDEEEEEDEEEEE 265
Score = 40.8 bits (94), Expect = 5e-04
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DDD + + E+EEE+EEEEE+EEE++E
Sbjct: 241 DDDDEEEDEDEEEDEEEEEDEEEEEE 266
Score = 40.4 bits (93), Expect = 7e-04
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DD+ + + EEE+EEEEE+EEEEE++E
Sbjct: 243 DDEEEDEDEEEDEEEEEDEEEEEEEE 268
Score = 38.1 bits (87), Expect = 0.003
Identities = 15/36 (41%), Positives = 26/36 (71%)
Query: 90 KILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
K++ + ++ D D EE+E+EEE+EEEEE+++E
Sbjct: 228 KVVPVKAKNVAEEDDDDEEEDEDEEEDEEEEEDEEE 263
Score = 37.4 bits (85), Expect = 0.006
Identities = 14/25 (56%), Positives = 21/25 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQ 124
D+D + D EEEEE+EEEE++ EE++
Sbjct: 189 DEDEEEDEEEEEEDEEEEDDSEEEE 213
Score = 35.8 bits (81), Expect = 0.017
Identities = 14/26 (53%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ D + EEE+EEEE++ EEEE E
Sbjct: 191 DEEEDEEEEEEDEEEEDDSEEEEAME 216
Score = 35.8 bits (81), Expect = 0.017
Identities = 13/26 (50%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ + + EEEEEE+EEEE++ E++E
Sbjct: 188 EDEDEEEDEEEEEEDEEEEDDSEEEE 213
Score = 34.3 bits (77), Expect = 0.050
Identities = 11/27 (40%), Positives = 21/27 (77%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEY 126
D D D D +++E++ +E+EE+EE+ E+
Sbjct: 142 DSDEDEDDDDDEDDSDEDEEDEEEDEF 168
Score = 33.9 bits (76), Expect = 0.065
Identities = 12/26 (46%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D+D D D +E++ +E+EE+EEE++ E
Sbjct: 144 DEDEDDDDDEDDSDEDEEDEEEDEFE 169
Score = 33.5 bits (75), Expect = 0.085
Identities = 13/23 (56%), Positives = 19/23 (82%)
Query: 103 SDSDMEEEEEEEEEEEEEEEQQE 125
+ D +EEE+EEEEEE+EEE+ +
Sbjct: 186 ASEDEDEEEDEEEEEEDEEEEDD 208
Score = 32.0 bits (71), Expect = 0.25
Identities = 9/25 (36%), Positives = 22/25 (88%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
+DSD D +++++E++ +E+EE+++E
Sbjct: 141 EDSDEDEDDDDDEDDSDEDEEDEEE 165
>NKX1_HUMAN (O60721) Sodium/potassium/calcium exchanger 1 precursor
(Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod
Na-Ca+K exchanger)
Length = 1099
Score = 42.7 bits (99), Expect = 1e-04
Identities = 19/25 (76%), Positives = 22/25 (88%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
D DS+ EEEEEEE+EEEEEEE+QE
Sbjct: 855 DGGDSEEEEEEEEEQEEEEEEEEQE 879
Score = 39.7 bits (91), Expect = 0.001
Identities = 15/26 (57%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEE+EEEEEE++E
Sbjct: 862 EEEEEEEQEEEEEEEEQEEEEEEEEE 887
Score = 39.3 bits (90), Expect = 0.002
Identities = 16/24 (66%), Positives = 22/24 (91%)
Query: 102 DSDSDMEEEEEEEEEEEEEEEQQE 125
DS+ + EEEEE+EEEEEEEE+++E
Sbjct: 858 DSEEEEEEEEEQEEEEEEEEQEEE 881
Score = 38.9 bits (89), Expect = 0.002
Identities = 15/26 (57%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEE+EEEEEEEE++E
Sbjct: 864 EEEEEQEEEEEEEEQEEEEEEEEEEE 889
Score = 38.5 bits (88), Expect = 0.003
Identities = 17/26 (65%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D + EEEEEE+EEEEEEEEQ+E
Sbjct: 855 DGGDSEEEEEEEEEQEEEEEEEEQEE 880
Score = 38.5 bits (88), Expect = 0.003
Identities = 15/26 (57%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D + + + EEE+EEEEEEEE+EE++E
Sbjct: 858 DSEEEEEEEEEQEEEEEEEEQEEEEE 883
Score = 38.1 bits (87), Expect = 0.003
Identities = 15/26 (57%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + EEEEEEE+EEEEEEE++E
Sbjct: 863 EEEEEEQEEEEEEEEQEEEEEEEEEE 888
Score = 38.1 bits (87), Expect = 0.003
Identities = 15/26 (57%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + EEEEE+EEEEEEEEE++E
Sbjct: 865 EEEEQEEEEEEEEQEEEEEEEEEEEE 890
Score = 37.7 bits (86), Expect = 0.005
Identities = 14/26 (53%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + +EEEEEEEE+EEEEE++E
Sbjct: 861 EEEEEEEEQEEEEEEEEQEEEEEEEE 886
Score = 37.7 bits (86), Expect = 0.005
Identities = 14/26 (53%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + E+EEEEEEEE+EEEE++E
Sbjct: 860 EEEEEEEEEQEEEEEEEEQEEEEEEE 885
Score = 37.4 bits (85), Expect = 0.006
Identities = 14/26 (53%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEE+EEEEEEEEEE+++
Sbjct: 866 EEEQEEEEEEEEQEEEEEEEEEEEEK 891
Score = 36.2 bits (82), Expect = 0.013
Identities = 16/23 (69%), Positives = 19/23 (82%)
Query: 103 SDSDMEEEEEEEEEEEEEEEQQE 125
SD EEEEEEEEE+EEEE++E
Sbjct: 854 SDGGDSEEEEEEEEEQEEEEEEE 876
Score = 35.0 bits (79), Expect = 0.029
Identities = 15/26 (57%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D SD + EEEEEEEEE+EE++E
Sbjct: 849 EDGGGSDGGDSEEEEEEEEEQEEEEE 874
Score = 34.3 bits (77), Expect = 0.050
Identities = 14/25 (56%), Positives = 20/25 (80%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
++ + + E+EEEEEEEEEEEE+ E
Sbjct: 870 EEEEEEEEQEEEEEEEEEEEEKGNE 894
Score = 33.9 bits (76), Expect = 0.065
Identities = 13/26 (50%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + +EEEEEEEEEEEE+ +E
Sbjct: 870 EEEEEEEEQEEEEEEEEEEEEKGNEE 895
>MYT1_HUMAN (Q01538) Myelin transcription factor 1 (MYT1) (MYTI)
(Proteolipid protein binding protein) (PLPB1)
Length = 1121
Score = 42.7 bits (99), Expect = 1e-04
Identities = 17/28 (60%), Positives = 24/28 (85%)
Query: 98 HYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
H ++D + + EEEEEEE+EEEEEEE++E
Sbjct: 257 HEEEDEEEEEEEEEEEEDEEEEEEEEEE 284
Score = 42.0 bits (97), Expect = 2e-04
Identities = 17/31 (54%), Positives = 25/31 (79%)
Query: 95 NHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
+H D++ + + EEEEE+EEEEEEEEE++E
Sbjct: 256 SHEEEDEEEEEEEEEEEEDEEEEEEEEEEEE 286
Score = 41.6 bits (96), Expect = 3e-04
Identities = 17/26 (65%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ D + EEEEEEEEEEEEEEE++E
Sbjct: 270 EEEEDEEEEEEEEEEEEEEEEEEEEE 295
Score = 41.6 bits (96), Expect = 3e-04
Identities = 17/26 (65%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ + + EEEEEEEEEEEEEEE++E
Sbjct: 274 DEEEEEEEEEEEEEEEEEEEEEEEEE 299
Score = 41.6 bits (96), Expect = 3e-04
Identities = 17/26 (65%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
++D + + EEEEEEEEEEEEEEE++E
Sbjct: 272 EEDEEEEEEEEEEEEEEEEEEEEEEE 297
Score = 41.6 bits (96), Expect = 3e-04
Identities = 17/26 (65%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + D EEEEEEEEEEEEEEE++E
Sbjct: 268 EEEEEEDEEEEEEEEEEEEEEEEEEE 293
Score = 41.6 bits (96), Expect = 3e-04
Identities = 17/26 (65%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ + + EEEEEEEEEEEEEEE++E
Sbjct: 273 EDEEEEEEEEEEEEEEEEEEEEEEEE 298
Score = 40.0 bits (92), Expect = 0.001
Identities = 19/45 (42%), Positives = 30/45 (66%)
Query: 81 SWRWRRVLNKILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
S + + +L+ E +++ + + EEEEEEEEEEEEEEE++E
Sbjct: 248 SSKQKGILSHEEEDEEEEEEEEEEEEDEEEEEEEEEEEEEEEEEE 292
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 276 EEEEEEEEEEEEEEEEEEEEEEEEEE 301
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 280 EEEEEEEEEEEEEEEEEEEEEEEEEE 305
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 269 EEEEEDEEEEEEEEEEEEEEEEEEEE 294
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 271 EEEDEEEEEEEEEEEEEEEEEEEEEE 296
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 278 EEEEEEEEEEEEEEEEEEEEEEEEEE 303
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 279 EEEEEEEEEEEEEEEEEEEEEEEEEE 304
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 277 EEEEEEEEEEEEEEEEEEEEEEEEEE 302
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 281 EEEEEEEEEEEEEEEEEEEEEEEEEE 306
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE++E
Sbjct: 275 EEEEEEEEEEEEEEEEEEEEEEEEEE 300
Score = 35.0 bits (79), Expect = 0.029
Identities = 14/28 (50%), Positives = 21/28 (75%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEYV 127
+++ + + EEEEEEEEEEEEEE + +
Sbjct: 285 EEEEEEEEEEEEEEEEEEEEEEAAPDVI 312
>S521_RAT (Q9QY02) Putative splicing factor YT521 (RA301-binding
protein)
Length = 738
Score = 42.4 bits (98), Expect = 2e-04
Identities = 17/34 (50%), Positives = 25/34 (73%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQEY 126
+ + D + + D EE+EEEE+EEEEEEE++EY
Sbjct: 220 DDEEVDEDAEEEEDEEEDEEEEDEEEEEEEEEEY 253
Score = 39.3 bits (90), Expect = 0.002
Identities = 16/26 (61%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
++D + D EEE+EEEEEEEEEE +Q+
Sbjct: 231 EEDEEEDEEEEDEEEEEEEEEEYEQD 256
Score = 38.9 bits (89), Expect = 0.002
Identities = 16/26 (61%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DDD + D + EEEE+EEE+EEEE +E
Sbjct: 219 DDDEEVDEDAEEEEDEEEDEEEEDEE 244
Score = 38.1 bits (87), Expect = 0.003
Identities = 16/26 (61%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ + + EE+EEEEEEEEEE EQ E
Sbjct: 232 EDEEEDEEEEDEEEEEEEEEEYEQDE 257
Score = 38.1 bits (87), Expect = 0.003
Identities = 15/26 (57%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ D + +EEEE+EEEEEEEEE+ E
Sbjct: 229 EEEEDEEEDEEEEDEEEEEEEEEEYE 254
Score = 38.1 bits (87), Expect = 0.003
Identities = 14/33 (42%), Positives = 24/33 (72%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
E + D D D +++E+ EEEE+EEE+EE+++
Sbjct: 210 EDEEVDEDGDDDEEVDEDAEEEEDEEEDEEEED 242
Score = 36.2 bits (82), Expect = 0.013
Identities = 14/26 (53%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEE+EEEEEEEEEE ++
Sbjct: 230 EEEDEEEDEEEEDEEEEEEEEEEYEQ 255
Score = 35.0 bits (79), Expect = 0.029
Identities = 14/26 (53%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ + D EEEEEEEEE E++E Q+
Sbjct: 236 EDEEEEDEEEEEEEEEEYEQDERDQK 261
Score = 34.3 bits (77), Expect = 0.050
Identities = 13/26 (50%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ + + EEEEEEEE E++E +Q+E
Sbjct: 237 DEEEEDEEEEEEEEEEYEQDERDQKE 262
Score = 33.9 bits (76), Expect = 0.065
Identities = 13/26 (50%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
++D + + EEEEEEEEEE E++E+ +
Sbjct: 235 EEDEEEEDEEEEEEEEEEYEQDERDQ 260
Score = 32.3 bits (72), Expect = 0.19
Identities = 11/26 (42%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + +EEEEEEEEEE E+++++
Sbjct: 234 EEEDEEEEDEEEEEEEEEEYEQDERD 259
Score = 32.0 bits (71), Expect = 0.25
Identities = 11/26 (42%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ D + EEEEEEE E++E ++++E
Sbjct: 238 EEEEDEEEEEEEEEEYEQDERDQKEE 263
Score = 32.0 bits (71), Expect = 0.25
Identities = 11/29 (37%), Positives = 22/29 (74%)
Query: 97 IHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
+ D++ D D +++EE +E+ EEEE+++E
Sbjct: 208 VEEDEEVDEDGDDDEEVDEDAEEEEDEEE 236
Score = 30.0 bits (66), Expect = 0.94
Identities = 10/27 (37%), Positives = 20/27 (74%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEY 126
+D+ + + EEEE E++E +++EE +Y
Sbjct: 241 EDEEEEEEEEEEYEQDERDQKEEGNDY 267
Score = 28.1 bits (61), Expect = 3.6
Identities = 14/41 (34%), Positives = 23/41 (55%), Gaps = 15/41 (36%)
Query: 100 DDDSDSDMEEEEE---------------EEEEEEEEEEQQE 125
++ + D+EE+EE EEEE+EEE+E++E
Sbjct: 201 EEGGEEDVEEDEEVDEDGDDDEEVDEDAEEEEDEEEDEEEE 241
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 42.4 bits (98), Expect = 2e-04
Identities = 18/37 (48%), Positives = 28/37 (75%)
Query: 89 NKILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
+K ++ + ++D + + EEEEEEEEEEEEEEE++E
Sbjct: 562 SKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEE 598
Score = 42.4 bits (98), Expect = 2e-04
Identities = 17/33 (51%), Positives = 25/33 (75%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
+ + D++ + + EEEEEEEEEEEEEEE++E
Sbjct: 568 DEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEE 600
Score = 42.0 bits (97), Expect = 2e-04
Identities = 18/33 (54%), Positives = 25/33 (75%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
E+ + D++ + EEEEEEEEEEEEEEE++E
Sbjct: 561 ESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEE 593
Score = 41.6 bits (96), Expect = 3e-04
Identities = 17/26 (65%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DDD D D +EE+EEEEEEEEEE +++
Sbjct: 642 DDDEDEDEDEEDEEEEEEEEEESEKK 667
Score = 41.6 bits (96), Expect = 3e-04
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DD+ D D +E+E+EE+EEEEEEE++E
Sbjct: 638 DDEEDDDEDEDEDEEDEEEEEEEEEE 663
Score = 41.2 bits (95), Expect = 4e-04
Identities = 16/26 (61%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DDD + D E+E+E+EE+EEEEEE++E
Sbjct: 637 DDDEEDDDEDEDEDEEDEEEEEEEEE 662
Score = 40.4 bits (93), Expect = 7e-04
Identities = 17/34 (50%), Positives = 26/34 (76%)
Query: 92 LETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
+E + +++ + + EEEEEEEEEEEEEEE++E
Sbjct: 572 VEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 605
Score = 40.4 bits (93), Expect = 7e-04
Identities = 16/26 (61%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
++D D D +E+EE+EEEEEEEEE+ E
Sbjct: 640 EEDDDEDEDEDEEDEEEEEEEEEESE 665
Score = 39.3 bits (90), Expect = 0.002
Identities = 16/26 (61%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE+ E
Sbjct: 582 EEEEEEEEEEEEEEEEEEEEEEEEDE 607
Score = 38.9 bits (89), Expect = 0.002
Identities = 15/26 (57%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE+++
Sbjct: 581 EEEEEEEEEEEEEEEEEEEEEEEEED 606
Score = 38.5 bits (88), Expect = 0.003
Identities = 14/26 (53%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+DD + D +E+E+E+EE+EEEEE++E
Sbjct: 636 EDDDEEDDDEDEDEDEEDEEEEEEEE 661
Score = 38.1 bits (87), Expect = 0.003
Identities = 15/26 (57%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEE++ E
Sbjct: 584 EEEEEEEEEEEEEEEEEEEEEEDEDE 609
Score = 38.1 bits (87), Expect = 0.003
Identities = 15/26 (57%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEEEE ++
Sbjct: 583 EEEEEEEEEEEEEEEEEEEEEEEDED 608
Score = 38.1 bits (87), Expect = 0.003
Identities = 13/26 (50%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DD+ D D E+++E+E+E+EE+EE++E
Sbjct: 633 DDEEDDDEEDDDEDEDEDEEDEEEEE 658
Score = 38.1 bits (87), Expect = 0.003
Identities = 15/26 (57%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEEE+E +E
Sbjct: 585 EEEEEEEEEEEEEEEEEEEEEDEDEE 610
Score = 37.7 bits (86), Expect = 0.005
Identities = 13/26 (50%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DDD + D EE+++E+E+E+EE+E++E
Sbjct: 632 DDDEEDDDEEDDDEDEDEDEEDEEEE 657
Score = 37.0 bits (84), Expect = 0.008
Identities = 14/26 (53%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEE+E+E+ E
Sbjct: 587 EEEEEEEEEEEEEEEEEEEDEDEEDE 612
Score = 36.6 bits (83), Expect = 0.010
Identities = 13/26 (50%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEEE+E++++
Sbjct: 586 EEEEEEEEEEEEEEEEEEEEDEDEED 611
Score = 35.8 bits (81), Expect = 0.017
Identities = 13/26 (50%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEE+E+EE ++
Sbjct: 588 EEEEEEEEEEEEEEEEEEDEDEEDED 613
Score = 35.8 bits (81), Expect = 0.017
Identities = 12/26 (46%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
++D D + ++E+E+E+EE+EEEE++E
Sbjct: 635 EEDDDEEDDDEDEDEDEEDEEEEEEE 660
Score = 35.0 bits (79), Expect = 0.029
Identities = 11/26 (42%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+DD + D +EE+++E+E+E+EE+++E
Sbjct: 631 EDDDEEDDDEEDDDEDEDEDEEDEEE 656
Score = 35.0 bits (79), Expect = 0.029
Identities = 11/26 (42%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ D + +++E+E+E+EE+EEE++E
Sbjct: 634 DEEDDDEEDDDEDEDEDEEDEEEEEE 659
Score = 34.7 bits (78), Expect = 0.038
Identities = 12/26 (46%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEE+E+EE++ +
Sbjct: 589 EEEEEEEEEEEEEEEEEDEDEEDEDD 614
Score = 34.7 bits (78), Expect = 0.038
Identities = 15/33 (45%), Positives = 22/33 (66%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
E+ + + + EEE EE+EEEEEEEE++E
Sbjct: 554 ESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEE 586
Score = 33.5 bits (75), Expect = 0.085
Identities = 12/26 (46%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEE+E+EE+E+ E
Sbjct: 591 EEEEEEEEEEEEEEEDEDEEDEDDAE 616
Score = 33.5 bits (75), Expect = 0.085
Identities = 10/26 (38%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+DD++ D +EE+++EE+++E+E++ E
Sbjct: 626 EDDAEEDDDEEDDDEEDDDEDEDEDE 651
Score = 33.1 bits (74), Expect = 0.11
Identities = 11/26 (42%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEE+E+EE+E++ +E
Sbjct: 592 EEEEEEEEEEEEEEDEDEEDEDDAEE 617
Score = 32.7 bits (73), Expect = 0.15
Identities = 11/26 (42%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DD + D EE+++EE+++E+E+E +E
Sbjct: 627 DDAEEDDDEEDDDEEDDDEDEDEDEE 652
Score = 32.7 bits (73), Expect = 0.15
Identities = 10/26 (38%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
++D D + ++EE+++E+E+E+EE +E
Sbjct: 630 EEDDDEEDDDEEDDDEDEDEDEEDEE 655
Score = 31.6 bits (70), Expect = 0.32
Identities = 9/26 (34%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D + D D E+++EE+++E+E+E++++
Sbjct: 628 DAEEDDDEEDDDEEDDDEDEDEDEED 653
Score = 31.2 bits (69), Expect = 0.42
Identities = 10/26 (38%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEE+E+EE+E++ E+ E
Sbjct: 594 EEEEEEEEEEEEDEDEEDEDDAEEDE 619
Score = 31.2 bits (69), Expect = 0.42
Identities = 8/26 (30%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ D++ +++EE+++EE+++E++ E
Sbjct: 624 EDEDDAEEDDDEEDDDEEDDDEDEDE 649
Score = 30.8 bits (68), Expect = 0.55
Identities = 9/26 (34%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+DD++ D ++ EE+++EE+++EE +
Sbjct: 619 EDDAEEDEDDAEEDDDEEDDDEEDDD 644
Score = 30.8 bits (68), Expect = 0.55
Identities = 9/26 (34%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEE+E+EE+E++ +++
Sbjct: 593 EEEEEEEEEEEEEDEDEEDEDDAEED 618
Score = 30.0 bits (66), Expect = 0.94
Identities = 9/26 (34%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ + D EE+E++ EE+E++ E+ +
Sbjct: 608 DEEDEDDAEEDEDDAEEDEDDAEEDD 633
Score = 30.0 bits (66), Expect = 0.94
Identities = 9/26 (34%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEE+E+EE+E++ EE ++
Sbjct: 595 EEEEEEEEEEEDEDEEDEDDAEEDED 620
Score = 30.0 bits (66), Expect = 0.94
Identities = 8/26 (30%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+DD++ D ++ EE+E++ EE++++++
Sbjct: 612 EDDAEEDEDDAEEDEDDAEEDDDEED 637
Score = 30.0 bits (66), Expect = 0.94
Identities = 8/25 (32%), Positives = 21/25 (84%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
++ + D EE+++EE+++EE++++ E
Sbjct: 623 EEDEDDAEEDDDEEDDDEEDDDEDE 647
Score = 29.6 bits (65), Expect = 1.2
Identities = 9/26 (34%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ D D E+E++ EE+E++ EE ++
Sbjct: 602 EEEEDEDEEDEDDAEEDEDDAEEDED 627
Score = 29.6 bits (65), Expect = 1.2
Identities = 7/26 (26%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ D++ +E++ EE+++EE++++++
Sbjct: 617 EDEDDAEEDEDDAEEDDDEEDDDEED 642
Score = 29.3 bits (64), Expect = 1.6
Identities = 9/26 (34%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D + D D EE+++EE+++EE++ ++
Sbjct: 621 DAEEDEDDAEEDDDEEDDDEEDDDED 646
Score = 29.3 bits (64), Expect = 1.6
Identities = 9/26 (34%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
++D D E+E++ EE+++EE++ +E
Sbjct: 616 EEDEDDAEEDEDDAEEDDDEEDDDEE 641
Score = 29.3 bits (64), Expect = 1.6
Identities = 8/26 (30%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
++D D E+++EE+++EE+++E ++
Sbjct: 623 EEDEDDAEEDDDEEDDDEEDDDEDED 648
Score = 29.3 bits (64), Expect = 1.6
Identities = 9/26 (34%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ D++ +E++ EE+E++ EE+ E
Sbjct: 610 EDEDDAEEDEDDAEEDEDDAEEDDDE 635
Score = 29.3 bits (64), Expect = 1.6
Identities = 9/26 (34%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ D D EE+E++ EE+E++ +++
Sbjct: 607 EDEEDEDDAEEDEDDAEEDEDDAEED 632
Score = 28.9 bits (63), Expect = 2.1
Identities = 9/26 (34%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D+D + + + EE+E++ EE+E++ +E
Sbjct: 606 DEDEEDEDDAEEDEDDAEEDEDDAEE 631
Score = 28.9 bits (63), Expect = 2.1
Identities = 8/26 (30%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EE+E+EE+E++ EE++ +
Sbjct: 596 EEEEEEEEEEDEDEEDEDDAEEDEDD 621
Score = 28.9 bits (63), Expect = 2.1
Identities = 10/36 (27%), Positives = 23/36 (63%)
Query: 90 KILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
++ + H+ ++D + +E +EE +E +E+EE+ E
Sbjct: 538 ELQKQKHVDKEEDKKEESKEVQEESKEVQEDEEEVE 573
Score = 28.5 bits (62), Expect = 2.7
Identities = 10/26 (38%), Positives = 18/26 (68%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D + D D EE+E++ EE+++EE +
Sbjct: 614 DAEEDEDDAEEDEDDAEEDDDEEDDD 639
Score = 28.5 bits (62), Expect = 2.7
Identities = 9/26 (34%), Positives = 19/26 (72%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ + D ++ EE+E++ EE+E+ E
Sbjct: 605 EDEDEEDEDDAEEDEDDAEEDEDDAE 630
Score = 28.1 bits (61), Expect = 3.6
Identities = 8/26 (30%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + E+EE+E++ EE+E++ +E
Sbjct: 599 EEEEEEEDEDEEDEDDAEEDEDDAEE 624
Score = 27.7 bits (60), Expect = 4.7
Identities = 7/26 (26%), Positives = 21/26 (79%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + D +EE+E++ EE+E++ +++
Sbjct: 600 EEEEEEDEDEEDEDDAEEDEDDAEED 625
Score = 27.7 bits (60), Expect = 4.7
Identities = 8/26 (30%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + +E+EE+E++ EE+E+ E
Sbjct: 598 EEEEEEEEDEDEEDEDDAEEDEDDAE 623
Score = 27.3 bits (59), Expect = 6.1
Identities = 8/26 (30%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EE+E++ EE+E++ E+ E
Sbjct: 601 EEEEEDEDEEDEDDAEEDEDDAEEDE 626
>SIAL_CHICK (P79780) Bone sialoprotein II precursor (BSP II)
(Cell-binding sialoprotein) (Integrin-binding
sialoprotein)
Length = 276
Score = 42.0 bits (97), Expect = 2e-04
Identities = 19/24 (79%), Positives = 21/24 (87%)
Query: 102 DSDSDMEEEEEEEEEEEEEEEQQE 125
D DSD EEEEEEEEEEEEE E+Q+
Sbjct: 121 DEDSDEEEEEEEEEEEEEEVEEQD 144
Score = 40.8 bits (94), Expect = 5e-04
Identities = 18/25 (72%), Positives = 20/25 (80%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
D D D +EEEEEEEEEEEEEE +E
Sbjct: 118 DSGDEDSDEEEEEEEEEEEEEEVEE 142
Score = 38.9 bits (89), Expect = 0.002
Identities = 17/23 (73%), Positives = 20/23 (86%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEE 122
D+DSD + EEEEEEEEEEE EE+
Sbjct: 121 DEDSDEEEEEEEEEEEEEEVEEQ 143
Score = 34.7 bits (78), Expect = 0.038
Identities = 16/36 (44%), Positives = 24/36 (66%)
Query: 89 NKILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQ 124
N ET H + ++ + +++EEEEEEEEEEEE +
Sbjct: 153 NTTAETPHGNNTVAAEEEEDDDEEEEEEEEEEEEAE 188
Score = 33.5 bits (75), Expect = 0.085
Identities = 13/18 (72%), Positives = 18/18 (99%)
Query: 108 EEEEEEEEEEEEEEEQQE 125
EEEE+++EEEEEEEE++E
Sbjct: 168 EEEEDDDEEEEEEEEEEE 185
>NKX1_BOVIN (Q28139) Sodium/potassium/calcium exchanger 1 precursor
(Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod
Na-Ca+K exchanger)
Length = 1216
Score = 42.0 bits (97), Expect = 2e-04
Identities = 17/26 (65%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ + D EEE+EEEEEEEEEEE++E
Sbjct: 984 EDEEEEDEEEEDEEEEEEEEEEEEEE 1009
Score = 40.0 bits (92), Expect = 0.001
Identities = 17/25 (68%), Positives = 22/25 (88%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
D DS+ EEEE+EEEE+EEEEE++E
Sbjct: 979 DGGDSEDEEEEDEEEEDEEEEEEEE 1003
Score = 39.7 bits (91), Expect = 0.001
Identities = 17/24 (70%), Positives = 20/24 (82%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQ 123
+D+ + D EEEEEEEEEEEEE EQ
Sbjct: 989 EDEEEEDEEEEEEEEEEEEEENEQ 1012
Score = 39.7 bits (91), Expect = 0.001
Identities = 16/26 (61%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ D + E+EEEEEEEEEEEEE+ E
Sbjct: 986 EEEEDEEEEDEEEEEEEEEEEEEENE 1011
Score = 39.3 bits (90), Expect = 0.002
Identities = 16/25 (64%), Positives = 21/25 (84%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
D D + E+EEEE+EEEEEEEE++E
Sbjct: 982 DSEDEEEEDEEEEDEEEEEEEEEEE 1006
Score = 38.5 bits (88), Expect = 0.003
Identities = 16/25 (64%), Positives = 21/25 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQ 124
++D + + EEEEEEEEEEEEEE +Q
Sbjct: 988 EEDEEEEDEEEEEEEEEEEEEENEQ 1012
Score = 38.5 bits (88), Expect = 0.003
Identities = 15/26 (57%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D + + + +EEEE+EEEEEEEEE++E
Sbjct: 982 DSEDEEEEDEEEEDEEEEEEEEEEEE 1007
Score = 37.7 bits (86), Expect = 0.005
Identities = 16/26 (61%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D D EEE+EEEE+EEEEEE++E
Sbjct: 979 DGGDSEDEEEEDEEEEDEEEEEEEEE 1004
Score = 37.0 bits (84), Expect = 0.008
Identities = 14/26 (53%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + +EEEEEEEEEEEEEE ++
Sbjct: 987 EEEDEEEEDEEEEEEEEEEEEEENEQ 1012
Score = 31.2 bits (69), Expect = 0.42
Identities = 12/26 (46%), Positives = 19/26 (72%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D + D + E+EEEE+EEEE+++E
Sbjct: 973 DGEGGGDGGDSEDEEEEDEEEEDEEE 998
Score = 30.0 bits (66), Expect = 0.94
Identities = 12/27 (44%), Positives = 19/27 (69%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEY 126
+++ + + EEEEEEEEEE E+ E+
Sbjct: 992 EEEDEEEEEEEEEEEEEENEQPLSLEW 1018
>MYC_CARAU (P49709) Myc protein (c-myc)
Length = 399
Score = 42.0 bits (97), Expect = 2e-04
Identities = 19/23 (82%), Positives = 22/23 (95%)
Query: 103 SDSDMEEEEEEEEEEEEEEEQQE 125
SDS+ EEEEEEEEEEEEEEE++E
Sbjct: 204 SDSEDEEEEEEEEEEEEEEEEEE 226
Score = 41.6 bits (96), Expect = 3e-04
Identities = 18/25 (72%), Positives = 21/25 (84%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
D D + EEEEEEEEEEEEEEE++E
Sbjct: 205 DSEDEEEEEEEEEEEEEEEEEEEEE 229
Score = 41.6 bits (96), Expect = 3e-04
Identities = 18/24 (75%), Positives = 22/24 (91%)
Query: 102 DSDSDMEEEEEEEEEEEEEEEQQE 125
DS+ + EEEEEEEEEEEEEEE++E
Sbjct: 205 DSEDEEEEEEEEEEEEEEEEEEEE 228
Score = 41.2 bits (95), Expect = 4e-04
Identities = 17/28 (60%), Positives = 23/28 (81%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEYV 127
D + + + EEEEEEEEEEEEEEE++E +
Sbjct: 205 DSEDEEEEEEEEEEEEEEEEEEEEEEEI 232
Score = 40.4 bits (93), Expect = 7e-04
Identities = 18/23 (78%), Positives = 21/23 (91%)
Query: 103 SDSDMEEEEEEEEEEEEEEEQQE 125
S SD E+EEEEEEEEEEEEE++E
Sbjct: 202 SGSDSEDEEEEEEEEEEEEEEEE 224
Score = 39.7 bits (91), Expect = 0.001
Identities = 17/28 (60%), Positives = 23/28 (81%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEYV 127
D++ + + EEEEEEEEEEEEEEE+ + V
Sbjct: 208 DEEEEEEEEEEEEEEEEEEEEEEEIDVV 235
Score = 35.4 bits (80), Expect = 0.022
Identities = 15/26 (57%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+ S S + E+EEEEEEEEEEE++E
Sbjct: 197 NSSSSSGSDSEDEEEEEEEEEEEEEE 222
Score = 26.9 bits (58), Expect = 8.0
Identities = 11/26 (42%), Positives = 16/26 (61%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D +S + E+EEEEEEE++E
Sbjct: 193 DTPPNSSSSSGSDSEDEEEEEEEEEE 218
>MYC2_CYPCA (Q90342) Myc II protein (C-MYC II)
Length = 401
Score = 42.0 bits (97), Expect = 2e-04
Identities = 19/23 (82%), Positives = 22/23 (95%)
Query: 103 SDSDMEEEEEEEEEEEEEEEQQE 125
SDS+ EEEEEEEEEEEEEEE++E
Sbjct: 204 SDSEDEEEEEEEEEEEEEEEEEE 226
Score = 41.6 bits (96), Expect = 3e-04
Identities = 18/24 (75%), Positives = 22/24 (91%)
Query: 102 DSDSDMEEEEEEEEEEEEEEEQQE 125
DS+ + EEEEEEEEEEEEEEE++E
Sbjct: 205 DSEDEEEEEEEEEEEEEEEEEEEE 228
Score = 40.4 bits (93), Expect = 7e-04
Identities = 18/23 (78%), Positives = 21/23 (91%)
Query: 103 SDSDMEEEEEEEEEEEEEEEQQE 125
S SD E+EEEEEEEEEEEEE++E
Sbjct: 202 SGSDSEDEEEEEEEEEEEEEEEE 224
Score = 39.7 bits (91), Expect = 0.001
Identities = 17/24 (70%), Positives = 20/24 (82%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQ 124
D D + EEEEEEEEEEEEEEE++
Sbjct: 205 DSEDEEEEEEEEEEEEEEEEEEEE 228
Score = 38.9 bits (89), Expect = 0.002
Identities = 17/28 (60%), Positives = 22/28 (77%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEYV 127
D + + + EEEEEEEEEEEEEEE+ + V
Sbjct: 205 DSEDEEEEEEEEEEEEEEEEEEEEIDVV 232
Score = 35.4 bits (80), Expect = 0.022
Identities = 15/26 (57%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+ S S + E+EEEEEEEEEEE++E
Sbjct: 197 NSSSSSGSDSEDEEEEEEEEEEEEEE 222
Score = 26.9 bits (58), Expect = 8.0
Identities = 11/26 (42%), Positives = 16/26 (61%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D +S + E+EEEEEEE++E
Sbjct: 193 DTPPNSSSSSGSDSEDEEEEEEEEEE 218
>DAXX_MOUSE (O35613) Death domain-associated protein 6 (Daxx)
Length = 739
Score = 42.0 bits (97), Expect = 2e-04
Identities = 19/26 (73%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DDD D D EE EEEEEEEEEE+E E
Sbjct: 448 DDDDDEDNEESEEEEEEEEEEKEATE 473
Score = 42.0 bits (97), Expect = 2e-04
Identities = 18/26 (69%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DDD D D E+ EE EEEEEEEEE++E
Sbjct: 445 DDDDDDDDEDNEESEEEEEEEEEEKE 470
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/26 (61%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DDD D D ++E+ EE EEEEEEE++E
Sbjct: 443 DDDDDDDDDDEDNEESEEEEEEEEEE 468
Score = 39.7 bits (91), Expect = 0.001
Identities = 16/26 (61%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DDD D D +E+ EE EEEEEEEE+++
Sbjct: 444 DDDDDDDDDEDNEESEEEEEEEEEEK 469
Score = 39.7 bits (91), Expect = 0.001
Identities = 16/32 (50%), Positives = 23/32 (71%)
Query: 94 TNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
T+ DDD D D +++E+ EE EEEEEE++E
Sbjct: 436 TSKAETDDDDDDDDDDDEDNEESEEEEEEEEE 467
Score = 38.9 bits (89), Expect = 0.002
Identities = 18/31 (58%), Positives = 22/31 (70%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQ 123
ET+ DDD D + EE EEEEEEEEEE++
Sbjct: 440 ETDDDDDDDDDDDEDNEESEEEEEEEEEEKE 470
Score = 37.7 bits (86), Expect = 0.005
Identities = 16/26 (61%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DDD D++ EEEEEEEEEE+E + E
Sbjct: 450 DDDEDNEESEEEEEEEEEEKEATEDE 475
Score = 35.0 bits (79), Expect = 0.029
Identities = 14/26 (53%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
DD+ + + EEEEEEEEEE+E E ++
Sbjct: 451 DDEDNEESEEEEEEEEEEKEATEDED 476
Score = 34.7 bits (78), Expect = 0.038
Identities = 14/26 (53%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D+D++ EEEEEEEEE+E E++ E
Sbjct: 452 DEDNEESEEEEEEEEEEKEATEDEDE 477
Score = 33.5 bits (75), Expect = 0.085
Identities = 13/26 (50%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ +S+ EEEEEEEE+E E+E ++
Sbjct: 453 EDNEESEEEEEEEEEEKEATEDEDED 478
Score = 30.4 bits (67), Expect = 0.72
Identities = 12/26 (46%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++S+ + EEEEEE+E E+E+E E
Sbjct: 455 NEESEEEEEEEEEEKEATEDEDEDLE 480
>TCF8_RAT (Q62947) Transcription factor 8 (Zinc finger homeodomain
enhancer-binding protein) (Zfhep)
Length = 1109
Score = 41.6 bits (96), Expect = 3e-04
Identities = 18/33 (54%), Positives = 24/33 (72%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
E + D + ++ EEEEEEEEEEEEEEE++E
Sbjct: 1027 EMEELQEDKECENPQEEEEEEEEEEEEEEEEEE 1059
Score = 37.7 bits (86), Expect = 0.005
Identities = 16/24 (66%), Positives = 21/24 (86%)
Query: 102 DSDSDMEEEEEEEEEEEEEEEQQE 125
++ + EEEEEEEEEEEEEEE++E
Sbjct: 1038 ENPQEEEEEEEEEEEEEEEEEEEE 1061
Score = 37.4 bits (85), Expect = 0.006
Identities = 16/26 (61%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
++ + + EEEEEEEEEEEEEEE+ E
Sbjct: 1038 ENPQEEEEEEEEEEEEEEEEEEEEAE 1063
Score = 36.6 bits (83), Expect = 0.010
Identities = 15/26 (57%), Positives = 21/26 (80%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQEY 126
++ + + EEEEEEEEEEEEE E+ E+
Sbjct: 1042 EEEEEEEEEEEEEEEEEEEEAEEAEH 1067
Score = 35.4 bits (80), Expect = 0.022
Identities = 15/37 (40%), Positives = 23/37 (61%)
Query: 88 LNKILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQ 124
+ ++ E + + + EEEEEEEEEEEEEE ++
Sbjct: 1028 MEELQEDKECENPQEEEEEEEEEEEEEEEEEEEEAEE 1064
Score = 34.7 bits (78), Expect = 0.038
Identities = 14/26 (53%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEEEE EE + E
Sbjct: 1043 EEEEEEEEEEEEEEEEEEEAEEAEHE 1068
Score = 34.7 bits (78), Expect = 0.038
Identities = 14/25 (56%), Positives = 20/25 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQ 124
+++ + + EEEEEEEEEEEE EE +
Sbjct: 1042 EEEEEEEEEEEEEEEEEEEEAEEAE 1066
Score = 34.7 bits (78), Expect = 0.038
Identities = 20/50 (40%), Positives = 26/50 (52%), Gaps = 17/50 (34%)
Query: 93 ETNHIHYDDDSDSDMEEEEE-----------------EEEEEEEEEEQQE 125
E + ++D DS+ EEEEE EEEEEEEEEE++E
Sbjct: 1005 ERESLTREEDEDSEKEEEEEDKEMEELQEDKECENPQEEEEEEEEEEEEE 1054
Score = 30.4 bits (67), Expect = 0.72
Identities = 12/26 (46%), Positives = 17/26 (65%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D D + EE+E+ E+EEEEE +E
Sbjct: 1002 DSDERESLTREEDEDSEKEEEEEDKE 1027
Score = 28.5 bits (62), Expect = 2.7
Identities = 11/26 (42%), Positives = 18/26 (68%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D+ EE+E+ E+EEEEE+++ E
Sbjct: 1004 DERESLTREEDEDSEKEEEEEDKEME 1029
>MYC1_CYPCA (Q90341) MYC I protein (C-MYC I)
Length = 394
Score = 41.6 bits (96), Expect = 3e-04
Identities = 19/23 (82%), Positives = 21/23 (90%)
Query: 103 SDSDMEEEEEEEEEEEEEEEQQE 125
S SD EEEEEEEEEEEEEEE++E
Sbjct: 202 SGSDSEEEEEEEEEEEEEEEEEE 224
Score = 40.4 bits (93), Expect = 7e-04
Identities = 19/25 (76%), Positives = 22/25 (88%)
Query: 103 SDSDMEEEEEEEEEEEEEEEQQEYV 127
SDS+ EEEEEEEEEEEEEEE+ + V
Sbjct: 204 SDSEEEEEEEEEEEEEEEEEEIDVV 228
Score = 36.6 bits (83), Expect = 0.010
Identities = 16/26 (61%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+ S S + EEEEEEEEEEEEE++E
Sbjct: 197 NSSSSSGSDSEEEEEEEEEEEEEEEE 222
Score = 36.2 bits (82), Expect = 0.013
Identities = 16/20 (80%), Positives = 18/20 (90%)
Query: 102 DSDSDMEEEEEEEEEEEEEE 121
DS+ + EEEEEEEEEEEEEE
Sbjct: 205 DSEEEEEEEEEEEEEEEEEE 224
Score = 32.7 bits (73), Expect = 0.15
Identities = 14/20 (70%), Positives = 16/20 (80%)
Query: 101 DDSDSDMEEEEEEEEEEEEE 120
D + + EEEEEEEEEEEEE
Sbjct: 205 DSEEEEEEEEEEEEEEEEEE 224
Score = 31.6 bits (70), Expect = 0.32
Identities = 13/20 (65%), Positives = 16/20 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEE 119
D + + + EEEEEEEEEEEE
Sbjct: 205 DSEEEEEEEEEEEEEEEEEE 224
Score = 28.1 bits (61), Expect = 3.6
Identities = 12/26 (46%), Positives = 16/26 (61%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D +S + EEEEEEEEE++E
Sbjct: 193 DTPPNSSSSSGSDSEEEEEEEEEEEE 218
>DRS1_YEAST (P32892) Probable ATP-dependent RNA helicase DRS1
Length = 752
Score = 41.6 bits (96), Expect = 3e-04
Identities = 18/25 (72%), Positives = 23/25 (92%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ S+ EEEEEE+EEEEEEEE+QE
Sbjct: 165 DENQSEEEEEEEEKEEEEEEEEEQE 189
Score = 40.0 bits (92), Expect = 0.001
Identities = 17/26 (65%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ + EEEEE+EEEEEEEEEQ+E
Sbjct: 165 DENQSEEEEEEEEKEEEEEEEEEQEE 190
Score = 37.0 bits (84), Expect = 0.008
Identities = 15/25 (60%), Positives = 21/25 (84%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
++ D + EEEEEEEE+EEEEE++E
Sbjct: 162 NNGDENQSEEEEEEEEKEEEEEEEE 186
Score = 36.6 bits (83), Expect = 0.010
Identities = 15/26 (57%), Positives = 22/26 (83%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
++ ++ EEEEEEEE+EEEEEE++E
Sbjct: 162 NNGDENQSEEEEEEEEKEEEEEEEEE 187
Score = 27.7 bits (60), Expect = 4.7
Identities = 11/26 (42%), Positives = 17/26 (65%)
Query: 93 ETNHIHYDDDSDSDMEEEEEEEEEEE 118
+ N +++ + EEEEEEEE+EE
Sbjct: 165 DENQSEEEEEEEEKEEEEEEEEEQEE 190
>MYC_BRARE (P52160) Myc protein (c-myc) (zc-Myc)
Length = 408
Score = 41.2 bits (95), Expect = 4e-04
Identities = 17/25 (68%), Positives = 22/25 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQ 124
+D+ + D EEEEEEEEEEEEEEE++
Sbjct: 208 EDEEEEDEEEEEEEEEEEEEEEEEE 232
Score = 40.8 bits (94), Expect = 5e-04
Identities = 18/23 (78%), Positives = 22/23 (95%)
Query: 103 SDSDMEEEEEEEEEEEEEEEQQE 125
SDS+ EEEE+EEEEEEEEEE++E
Sbjct: 205 SDSEDEEEEDEEEEEEEEEEEEE 227
Score = 40.4 bits (93), Expect = 7e-04
Identities = 17/25 (68%), Positives = 22/25 (88%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
+D + + EEEEEEEEEEEEEEE++E
Sbjct: 208 EDEEEEDEEEEEEEEEEEEEEEEEE 232
Score = 40.4 bits (93), Expect = 7e-04
Identities = 17/25 (68%), Positives = 21/25 (84%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
D D + E+EEEEEEEEEEEEE++E
Sbjct: 206 DSEDEEEEDEEEEEEEEEEEEEEEE 230
Score = 40.0 bits (92), Expect = 0.001
Identities = 16/28 (57%), Positives = 23/28 (82%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEYV 127
D + + + +EEEEEEEEEEEEEE++E +
Sbjct: 206 DSEDEEEEDEEEEEEEEEEEEEEEEEEI 233
Score = 39.7 bits (91), Expect = 0.001
Identities = 17/28 (60%), Positives = 23/28 (81%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEYV 127
D++ + + EEEEEEEEEEEEEEE+ + V
Sbjct: 209 DEEEEDEEEEEEEEEEEEEEEEEEIDVV 236
Score = 39.3 bits (90), Expect = 0.002
Identities = 17/23 (73%), Positives = 21/23 (90%)
Query: 103 SDSDMEEEEEEEEEEEEEEEQQE 125
S SD E+EEEE+EEEEEEEE++E
Sbjct: 203 SGSDSEDEEEEDEEEEEEEEEEE 225
Score = 34.3 bits (77), Expect = 0.050
Identities = 14/26 (53%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+ S S + E+EEEE+EEEEEE++E
Sbjct: 198 NSSSSSGSDSEDEEEEDEEEEEEEEE 223
>CENB_MOUSE (P27790) Major centromere autoantigen B (Centromere
protein B) (CENP-B)
Length = 599
Score = 41.2 bits (95), Expect = 4e-04
Identities = 17/26 (65%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D DSDSD EE++EEE+EE+E+EE E
Sbjct: 509 DSDSDSDEEEDDEEEDEEDEDEEDDE 534
Score = 39.3 bits (90), Expect = 0.002
Identities = 14/25 (56%), Positives = 24/25 (96%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
+DSDSD +EEE++EEE+EE+E++++
Sbjct: 508 EDSDSDSDEEEDDEEEDEEDEDEED 532
Score = 36.6 bits (83), Expect = 0.010
Identities = 17/38 (44%), Positives = 25/38 (65%)
Query: 88 LNKILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
LN + T+ ++ + + EEEEEEEEEE E EE++E
Sbjct: 393 LNATITTSFKSEGEEEEEEEEEEEEEEEEEGEGEEEEE 430
Score = 34.7 bits (78), Expect = 0.038
Identities = 14/26 (53%), Positives = 21/26 (79%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEE E EEEEE++E
Sbjct: 408 EEEEEEEEEEEEEEEGEGEEEEEEEE 433
Score = 34.7 bits (78), Expect = 0.038
Identities = 14/26 (53%), Positives = 21/26 (79%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEE E EEE++E
Sbjct: 406 EEEEEEEEEEEEEEEEEGEGEEEEEE 431
Score = 34.3 bits (77), Expect = 0.050
Identities = 13/26 (50%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D D + D EEE+EE+E+EE++E+ E
Sbjct: 513 DSDEEEDDEEEDEEDEDEEDDEDGDE 538
Score = 32.0 bits (71), Expect = 0.25
Identities = 13/26 (50%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EE E EEEEEEEEE ++E
Sbjct: 413 EEEEEEEEEEGEGEEEEEEEEEGEEE 438
Score = 31.2 bits (69), Expect = 0.42
Identities = 14/26 (53%), Positives = 19/26 (72%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + E EEEEEEEEE EEE E
Sbjct: 416 EEEEEEEGEGEEEEEEEEEGEEEGGE 441
Score = 30.0 bits (66), Expect = 0.94
Identities = 13/26 (50%), Positives = 19/26 (73%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEEEE EEE + E
Sbjct: 418 EEEEEGEGEEEEEEEEEGEEEGGEGE 443
Score = 29.3 bits (64), Expect = 1.6
Identities = 12/26 (46%), Positives = 19/26 (72%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+++ + + EEEEEEE EEE E ++E
Sbjct: 420 EEEGEGEEEEEEEEEGEEEGGEGEEE 445
Score = 28.5 bits (62), Expect = 2.7
Identities = 16/32 (50%), Positives = 20/32 (62%), Gaps = 4/32 (12%)
Query: 100 DDDSDSDMEEEEEEEEEEEE----EEEQQEYV 127
+++ EEEEEEEE EEE EEE +E V
Sbjct: 419 EEEEGEGEEEEEEEEEGEEEGGEGEEEGEEEV 450
Score = 26.9 bits (58), Expect = 8.0
Identities = 9/23 (39%), Positives = 18/23 (78%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEE 122
+++ + ++EEE E ++ +EEEEE
Sbjct: 443 EEEGEEEVEEEGEVDDSDEEEEE 465
>ACH2_CHICK (P09480) Neuronal acetylcholine receptor protein,
alpha-2 chain precursor
Length = 528
Score = 41.2 bits (95), Expect = 4e-04
Identities = 19/34 (55%), Positives = 24/34 (69%)
Query: 92 LETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
L T+ + D D EEEEEEEEEEEEEEE+++
Sbjct: 378 LSTSRCWLETDVDDKWEEEEEEEEEEEEEEEEEK 411
Score = 38.1 bits (87), Expect = 0.003
Identities = 18/26 (69%), Positives = 20/26 (76%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQEY 126
D D EEEEEEEEEEEEEEE++ Y
Sbjct: 388 DVDDKWEEEEEEEEEEEEEEEEEKAY 413
>NUCL_HUMAN (P19338) Nucleolin (Protein C23)
Length = 706
Score = 40.8 bits (94), Expect = 5e-04
Identities = 16/28 (57%), Positives = 24/28 (85%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEYV 127
DDD D D E++++E++EEEEEEE++E V
Sbjct: 245 DDDDDEDDEDDDDEDDEEEEEEEEEEPV 272
Score = 37.0 bits (84), Expect = 0.008
Identities = 12/26 (46%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D+D D D ++E++++E++EEEEE++E
Sbjct: 243 DEDDDDDEDDEDDDDEDDEEEEEEEE 268
Score = 36.6 bits (83), Expect = 0.010
Identities = 15/26 (57%), Positives = 21/26 (80%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D+D + D +E++EEEEEEEEEE +E
Sbjct: 249 DEDDEDDDDEDDEEEEEEEEEEPVKE 274
Score = 36.2 bits (82), Expect = 0.013
Identities = 12/26 (46%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+DD D + +E++++E++EEEEEE++E
Sbjct: 244 EDDDDDEDDEDDDDEDDEEEEEEEEE 269
Score = 35.4 bits (80), Expect = 0.022
Identities = 11/26 (42%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D+D D D +E++E++++E++EEE++E
Sbjct: 241 DEDEDDDDDEDDEDDDDEDDEEEEEE 266
Score = 35.0 bits (79), Expect = 0.029
Identities = 11/26 (42%), Positives = 23/26 (88%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+D+ D D E++E++++E++EEEE++E
Sbjct: 242 EDEDDDDDEDDEDDDDEDDEEEEEEE 267
Score = 35.0 bits (79), Expect = 0.029
Identities = 12/26 (46%), Positives = 22/26 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D++ D D EE+EE++E+E+E+E++ E
Sbjct: 145 DEEEDDDSEEDEEDDEDEDEDEDEIE 170
Score = 33.1 bits (74), Expect = 0.11
Identities = 9/26 (34%), Positives = 23/26 (87%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+DD D D +++E++E++++E++E++E
Sbjct: 239 EDDEDEDDDDDEDDEDDDDEDDEEEE 264
Score = 32.7 bits (73), Expect = 0.15
Identities = 11/26 (42%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
D D + D + EE+EE++E+E+E++ E
Sbjct: 143 DSDEEEDDDSEEDEEDDEDEDEDEDE 168
Score = 32.0 bits (71), Expect = 0.25
Identities = 10/25 (40%), Positives = 21/25 (84%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
+DSD + +++ EE+EE++E+E++ E
Sbjct: 142 EDSDEEEDDDSEEDEEDDEDEDEDE 166
Score = 31.2 bits (69), Expect = 0.42
Identities = 10/26 (38%), Positives = 20/26 (76%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQE 125
+DD D + +E+++++EE++ EEE E
Sbjct: 186 EDDEDDEDDEDDDDDEEDDSEEEAME 211
Score = 30.0 bits (66), Expect = 0.94
Identities = 8/25 (32%), Positives = 21/25 (84%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQ 124
+D+ D D E++E+++++EE++ E++
Sbjct: 184 EDEDDEDDEDDEDDDDDEEDDSEEE 208
Score = 28.9 bits (63), Expect = 2.1
Identities = 8/28 (28%), Positives = 21/28 (74%)
Query: 100 DDDSDSDMEEEEEEEEEEEEEEEQQEYV 127
+D + + ++ EE+EE++E+E+E ++ +
Sbjct: 142 EDSDEEEDDDSEEDEEDDEDEDEDEDEI 169
Score = 28.5 bits (62), Expect = 2.7
Identities = 7/25 (28%), Positives = 21/25 (84%)
Query: 101 DDSDSDMEEEEEEEEEEEEEEEQQE 125
+D D + +E++E+++++EE++ ++E
Sbjct: 184 EDEDDEDDEDDEDDDDDEEDDSEEE 208
Score = 27.3 bits (59), Expect = 6.1
Identities = 7/36 (19%), Positives = 24/36 (66%)
Query: 90 KILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQE 125
K++ + +D D + ++E+E+++++E++E+ +
Sbjct: 222 KVVPVKAKNVAEDEDEEEDDEDEDDDDDEDDEDDDD 257
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,842,354
Number of Sequences: 164201
Number of extensions: 730814
Number of successful extensions: 34743
Number of sequences better than 10.0: 1136
Number of HSP's better than 10.0 without gapping: 1081
Number of HSP's successfully gapped in prelim test: 63
Number of HSP's that attempted gapping in prelim test: 13003
Number of HSP's gapped (non-prelim): 9609
length of query: 127
length of database: 59,974,054
effective HSP length: 103
effective length of query: 24
effective length of database: 43,061,351
effective search space: 1033472424
effective search space used: 1033472424
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149206.8


