

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148341.11 - phase: 0
(164 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
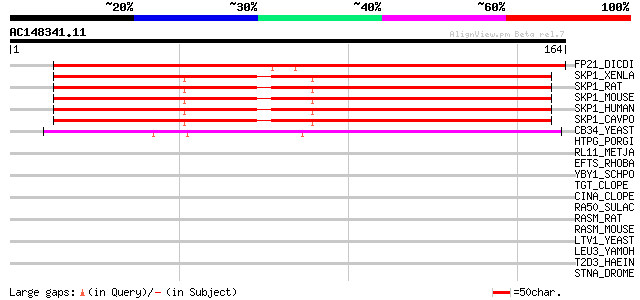
Score E
Sequences producing significant alignments: (bits) Value
FP21_DICDI (P52285) Glycoprotein FP21 precursor 173 1e-43
SKP1_XENLA (Q71U00) S-phase kinase-associated protein 1A (Cyclin... 169 3e-42
SKP1_RAT (Q6PEC4) S-phase kinase-associated protein 1A (Cyclin A... 169 3e-42
SKP1_MOUSE (Q9WTX5) S-phase kinase-associated protein 1A (Cyclin... 169 3e-42
SKP1_HUMAN (P63208) S-phase kinase-associated protein 1A (Cyclin... 169 3e-42
SKP1_CAVPO (P63209) S-phase kinase-associated protein 1A (Cyclin... 169 3e-42
CB34_YEAST (P52286) Centromere DNA-binding protein complex CBF3 ... 140 1e-33
HTPG_PORGI (Q9S3Q2) Chaperone protein htpG (Heat shock protein h... 34 0.13
RL11_METJA (P54030) 50S ribosomal protein L11P 33 0.22
EFTS_RHOBA (Q7UKH3) Elongation factor Ts (EF-Ts) 32 0.64
YBY1_SCHPO (Q10200) Hypothetical protein C11C11.01 in chromosome II 30 1.9
TGT_CLOPE (Q8XJ16) Queuine tRNA-ribosyltransferase (EC 2.4.2.29)... 30 2.4
CINA_CLOPE (Q8XP33) CinA-like protein 30 2.4
RA50_SULAC (O33600) DNA double-strand break repair rad50 ATPase 30 3.2
RASM_RAT (P97538) Ras-related protein M-Ras (Ras-related protein... 29 4.2
RASM_MOUSE (O08989) Ras-related protein M-Ras (Ras-related prote... 29 4.2
LTV1_YEAST (P34078) Low-temperature viability protein LTV1 29 4.2
LEU3_YAMOH (P41926) 3-isopropylmalate dehydrogenase (EC 1.1.1.85... 29 4.2
T2D3_HAEIN (P43870) Type II restriction enzyme HindIII (EC 3.1.2... 29 5.4
STNA_DROME (Q24211) Stoned A protein (StonedA) (Stn-A) 29 5.4
>FP21_DICDI (P52285) Glycoprotein FP21 precursor
Length = 162
Score = 173 bits (439), Expect = 1e-43
Identities = 89/156 (57%), Positives = 115/156 (73%), Gaps = 5/156 (3%)
Query: 14 ITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKHVD 73
+ L+SSD + FEIEK +A S TIK++I+D DS IPLPNVT IL V+++C+ H
Sbjct: 4 VKLESSDEKVFEIEKEIACMSVTIKNMIEDIGESDSPIPLPNVTSTILEKVLDYCRHHHQ 63
Query: 74 ATS--SDEKPSE---DEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
S D+K E D+I +D +F KVDQ TLF+LILAANYL+IK LLD+TCKTVA+MI
Sbjct: 64 HPSPQGDDKKDEKRLDDIPPYDRDFCKVDQPTLFELILAANYLDIKPLLDVTCKTVANMI 123
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFDE 164
+G+TPEEIRK FNI ND+TPEEEE++R E +W D+
Sbjct: 124 RGKTPEEIRKIFNIKNDFTPEEEEQIRKENEWCEDK 159
>SKP1_XENLA (Q71U00) S-phase kinase-associated protein 1A (Cyclin
A/CDK2-associated protein p19) (p19A) (p19skp1)
Length = 162
Score = 169 bits (427), Expect = 3e-42
Identities = 91/160 (56%), Positives = 110/160 (67%), Gaps = 17/160 (10%)
Query: 14 ITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSG----IPLPNVTGKILAMVIEHCK 69
I L+SSD E FE++ +A +S TIK +++D DD G +PLPNV IL VI+ C
Sbjct: 3 IKLQSSDGEIFEVDVEIAKQSVTIKTMLEDLGMDDEGDDDPVPLPNVNAAILKKVIQWCT 62
Query: 70 KHVDATSSDEKPSEDEINK---------WDTEFVKVDQDTLFDLILAANYLNIKSLLDLT 120
H D D P ED+ NK WD EF+KVDQ TLF+LILAANYL+IK LLD+T
Sbjct: 63 HHKD----DPPPPEDDENKEKRTDDIPVWDQEFLKVDQGTLFELILAANYLDIKGLLDVT 118
Query: 121 CKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQW 160
CKTVA+MIKG+TPEEIRKTFNI ND+T EEE +VR E QW
Sbjct: 119 CKTVANMIKGKTPEEIRKTFNIKNDFTEEEEAQVRKENQW 158
>SKP1_RAT (Q6PEC4) S-phase kinase-associated protein 1A (Cyclin
A/CDK2-associated protein p19) (p19A) (p19skp1)
Length = 162
Score = 169 bits (427), Expect = 3e-42
Identities = 91/160 (56%), Positives = 110/160 (67%), Gaps = 17/160 (10%)
Query: 14 ITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSG----IPLPNVTGKILAMVIEHCK 69
I L+SSD E FE++ +A +S TIK +++D DD G +PLPNV IL VI+ C
Sbjct: 3 IKLQSSDGEIFEVDVEIAKQSVTIKTMLEDLGMDDEGDDDPVPLPNVNAAILKKVIQWCT 62
Query: 70 KHVDATSSDEKPSEDEINK---------WDTEFVKVDQDTLFDLILAANYLNIKSLLDLT 120
H D D P ED+ NK WD EF+KVDQ TLF+LILAANYL+IK LLD+T
Sbjct: 63 HHKD----DPPPPEDDENKEKRTDDIPVWDQEFLKVDQGTLFELILAANYLDIKGLLDVT 118
Query: 121 CKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQW 160
CKTVA+MIKG+TPEEIRKTFNI ND+T EEE +VR E QW
Sbjct: 119 CKTVANMIKGKTPEEIRKTFNIKNDFTEEEEAQVRKENQW 158
>SKP1_MOUSE (Q9WTX5) S-phase kinase-associated protein 1A (Cyclin
A/CDK2-associated protein p19) (p19A) (p19skp1)
Length = 162
Score = 169 bits (427), Expect = 3e-42
Identities = 91/160 (56%), Positives = 110/160 (67%), Gaps = 17/160 (10%)
Query: 14 ITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSG----IPLPNVTGKILAMVIEHCK 69
I L+SSD E FE++ +A +S TIK +++D DD G +PLPNV IL VI+ C
Sbjct: 3 IKLQSSDGEIFEVDVEIAKQSVTIKTMLEDLGMDDEGDDDPVPLPNVNAAILKKVIQWCT 62
Query: 70 KHVDATSSDEKPSEDEINK---------WDTEFVKVDQDTLFDLILAANYLNIKSLLDLT 120
H D D P ED+ NK WD EF+KVDQ TLF+LILAANYL+IK LLD+T
Sbjct: 63 HHKD----DPPPPEDDENKEKRTDDIPVWDQEFLKVDQGTLFELILAANYLDIKGLLDVT 118
Query: 121 CKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQW 160
CKTVA+MIKG+TPEEIRKTFNI ND+T EEE +VR E QW
Sbjct: 119 CKTVANMIKGKTPEEIRKTFNIKNDFTEEEEAQVRKENQW 158
>SKP1_HUMAN (P63208) S-phase kinase-associated protein 1A (Cyclin
A/CDK2-associated protein p19) (p19A) (p19skp1) (RNA
polymerase II elongation factor-like protein) (Organ of
Corti protein 2) (OCP-II protein) (OCP-2) (Transcription
elongation factor B) (S
Length = 162
Score = 169 bits (427), Expect = 3e-42
Identities = 91/160 (56%), Positives = 110/160 (67%), Gaps = 17/160 (10%)
Query: 14 ITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSG----IPLPNVTGKILAMVIEHCK 69
I L+SSD E FE++ +A +S TIK +++D DD G +PLPNV IL VI+ C
Sbjct: 3 IKLQSSDGEIFEVDVEIAKQSVTIKTMLEDLGMDDEGDDDPVPLPNVNAAILKKVIQWCT 62
Query: 70 KHVDATSSDEKPSEDEINK---------WDTEFVKVDQDTLFDLILAANYLNIKSLLDLT 120
H D D P ED+ NK WD EF+KVDQ TLF+LILAANYL+IK LLD+T
Sbjct: 63 HHKD----DPPPPEDDENKEKRTDDIPVWDQEFLKVDQGTLFELILAANYLDIKGLLDVT 118
Query: 121 CKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQW 160
CKTVA+MIKG+TPEEIRKTFNI ND+T EEE +VR E QW
Sbjct: 119 CKTVANMIKGKTPEEIRKTFNIKNDFTEEEEAQVRKENQW 158
>SKP1_CAVPO (P63209) S-phase kinase-associated protein 1A (Cyclin
A/CDK2-associated protein p19) (p19A) (p19skp1) (Organ
of Corti protein 2) (OCP-II protein) (OCP-2)
Length = 162
Score = 169 bits (427), Expect = 3e-42
Identities = 91/160 (56%), Positives = 110/160 (67%), Gaps = 17/160 (10%)
Query: 14 ITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSG----IPLPNVTGKILAMVIEHCK 69
I L+SSD E FE++ +A +S TIK +++D DD G +PLPNV IL VI+ C
Sbjct: 3 IKLQSSDGEIFEVDVEIAKQSVTIKTMLEDLGMDDEGDDDPVPLPNVNAAILKKVIQWCT 62
Query: 70 KHVDATSSDEKPSEDEINK---------WDTEFVKVDQDTLFDLILAANYLNIKSLLDLT 120
H D D P ED+ NK WD EF+KVDQ TLF+LILAANYL+IK LLD+T
Sbjct: 63 HHKD----DPPPPEDDENKEKRTDDIPVWDQEFLKVDQGTLFELILAANYLDIKGLLDVT 118
Query: 121 CKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQW 160
CKTVA+MIKG+TPEEIRKTFNI ND+T EEE +VR E QW
Sbjct: 119 CKTVANMIKGKTPEEIRKTFNIKNDFTEEEEAQVRKENQW 158
>CB34_YEAST (P52286) Centromere DNA-binding protein complex CBF3
subunit D (Suppressor of kinetochore protein 1)
Length = 193
Score = 140 bits (353), Expect = 1e-33
Identities = 77/191 (40%), Positives = 106/191 (55%), Gaps = 38/191 (19%)
Query: 11 TKKITLKSSDNETFEIEKAVALESQTIKHLI----------------------------- 41
T + L S + E F ++K +A S +K+ +
Sbjct: 2 TSNVVLVSGEGERFTVDKKIAERSLLLKNYLNDMHDSNLQNNSDSESDSDSETNHKSKDN 61
Query: 42 ---DDNCADDSGI--PLPNVTGKILAMVIEHCKKHVDATSSDEKPSEDE----INKWDTE 92
DD+ DD I P+PNV +L VIE + H D+ DE + ++ WD E
Sbjct: 62 NNGDDDDEDDDEIVMPVPNVRSSVLQKVIEWAEHHRDSNFPDEDDDDSRKSAPVDSWDRE 121
Query: 93 FVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEE 152
F+KVDQ+ L+++ILAANYLNIK LLD CK VA+MI+GR+PEEIR+TFNI+ND+TPEEE
Sbjct: 122 FLKVDQEMLYEIILAANYLNIKPLLDAGCKVVAEMIRGRSPEEIRRTFNIVNDFTPEEEA 181
Query: 153 EVRSETQWAFD 163
+R E +WA D
Sbjct: 182 AIRRENEWAED 192
>HTPG_PORGI (Q9S3Q2) Chaperone protein htpG (Heat shock protein
htpG) (High temperature protein G)
Length = 684
Score = 34.3 bits (77), Expect = 0.13
Identities = 20/64 (31%), Positives = 32/64 (49%), Gaps = 1/64 (1%)
Query: 91 TEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEE 150
T FV+VD DT+ +LI +K L D T+ + + R P + +K FN+ + E
Sbjct: 470 THFVRVDSDTINNLIRKEERAEVK-LSDTERATLVKLFEARLPRDEKKHFNVAFESLGAE 528
Query: 151 EEEV 154
E +
Sbjct: 529 GEAI 532
>RL11_METJA (P54030) 50S ribosomal protein L11P
Length = 161
Score = 33.5 bits (75), Expect = 0.22
Identities = 33/150 (22%), Positives = 66/150 (44%), Gaps = 20/150 (13%)
Query: 4 GVESSTATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAM 63
GV K+I K+ D E ++ V ++++T K I+ + +P T I
Sbjct: 29 GVNVMQVVKEINEKTKDYEGMQVPVKVIVDTETRKFEIE--------VGIPPTTALI--- 77
Query: 64 VIEHCKKHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLN-IKSLLDLTCK 122
KK + ++ +P + + E V D +L+ N +K +L TC
Sbjct: 78 -----KKELGIETAAHEPRHEVVGNLTLEQVIKIAKMKKDAMLSYTLKNAVKEVLG-TCG 131
Query: 123 TVADMIKGRTPEEIRKTFN--IINDYTPEE 150
++ ++G+ P+E++K + + ++Y EE
Sbjct: 132 SMGVTVEGKDPKEVQKEIDAGVYDEYFKEE 161
>EFTS_RHOBA (Q7UKH3) Elongation factor Ts (EF-Ts)
Length = 326
Score = 32.0 bits (71), Expect = 0.64
Identities = 28/83 (33%), Positives = 37/83 (43%), Gaps = 5/83 (6%)
Query: 80 KPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKGR--TPEEIR 137
KPS N++D EFV+ + + L I A N LN K L K V R TPE +
Sbjct: 188 KPSILHPNEFDEEFVQKETEALQAQINAENELNEKENLGKPMKNVPQFASRRQLTPEVLA 247
Query: 138 KTFNIINDYTPEEEEEVRSETQW 160
T I + E + E + E W
Sbjct: 248 ATEEAIKE---ELKAEGKPEKIW 267
>YBY1_SCHPO (Q10200) Hypothetical protein C11C11.01 in chromosome II
Length = 493
Score = 30.4 bits (67), Expect = 1.9
Identities = 27/115 (23%), Positives = 51/115 (43%), Gaps = 23/115 (20%)
Query: 2 SDGVESSTATKKITLKSSDNETFEIEKAVALESQTIKHLIDD-NCADDSGIPLPNVTGKI 60
++ +ESST TK +E+ + +K L+ ++ LI NC S G +
Sbjct: 217 AEHLESSTKTK--------DESQDKDKLTKLDRAKLEWLIQGLNCEKGS-------IGNL 261
Query: 61 LAMVIEHCKKHVDATSS------DEKPSEDEINKWDTEFVKVDQDTLFDLILAAN 109
L +EH HV+ T + +++P+ D+ +D +++ + LI N
Sbjct: 262 LCFAVEHVNNHVEITDAFLKEFFNDQPT-DDTKVYDDDYINERGEKKLSLIYLMN 315
>TGT_CLOPE (Q8XJ16) Queuine tRNA-ribosyltransferase (EC 2.4.2.29)
(tRNA-guanine transglycosylase) (Guanine insertion
enzyme)
Length = 380
Score = 30.0 bits (66), Expect = 2.4
Identities = 21/87 (24%), Positives = 43/87 (49%), Gaps = 6/87 (6%)
Query: 65 IEHCKKHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTV 124
+E CKK +D +S +D +NK F ++Q +++ I + I+ + DL +
Sbjct: 170 LERCKKEMDRLNS----LDDTVNKEQMLF-GINQGGVYEDIRIEHAKTIREM-DLDGYAI 223
Query: 125 ADMIKGRTPEEIRKTFNIINDYTPEEE 151
+ G T EE+ + + + + PE++
Sbjct: 224 GGLAVGETHEEMYRVIDAVVPHLPEDK 250
>CINA_CLOPE (Q8XP33) CinA-like protein
Length = 412
Score = 30.0 bits (66), Expect = 2.4
Identities = 17/49 (34%), Positives = 25/49 (50%), Gaps = 2/49 (4%)
Query: 13 KITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKIL 61
+IT K D E E EK +A + I+ + DN + + L V GK+L
Sbjct: 219 RITAKGKDKE--EAEKLIAPMEKEIRKRLGDNIYGEGEVTLEEVVGKLL 265
>RA50_SULAC (O33600) DNA double-strand break repair rad50 ATPase
Length = 886
Score = 29.6 bits (65), Expect = 3.2
Identities = 21/89 (23%), Positives = 42/89 (46%), Gaps = 8/89 (8%)
Query: 65 IEHCKKHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTV 124
IE K ++ + K EDE+N+++TEF ++ + + Y + L + K +
Sbjct: 205 IEEIKVKLENIEREAKEKEDELNQYNTEFNRIKE-------IKVQYDILSGELSVVNKKI 257
Query: 125 ADM-IKGRTPEEIRKTFNIINDYTPEEEE 152
++ ++ + EE K +N I E +E
Sbjct: 258 EEIALRLKDFEEKEKRYNKIETEVKELDE 286
>RASM_RAT (P97538) Ras-related protein M-Ras (Ras-related protein
R-Ras3)
Length = 208
Score = 29.3 bits (64), Expect = 4.2
Identities = 22/103 (21%), Positives = 45/103 (43%), Gaps = 15/103 (14%)
Query: 76 SSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKGRT--- 132
S +K S + ++++ ++V F +IL AN +++ L +T +M
Sbjct: 93 SVTDKASFEHVDRFHQLILRVKDRESFPMILVANKVDLMHLRKVTRDQGKEMATKYNIPY 152
Query: 133 --------PEEIRKTFN----IINDYTPEEEEEVRSETQWAFD 163
P + KTF+ +I PE+ ++ + +T+W D
Sbjct: 153 IETSAKDPPLNVDKTFHDLVRVIRQQVPEKNQKKKKKTKWRGD 195
>RASM_MOUSE (O08989) Ras-related protein M-Ras (Ras-related protein
R-Ras3) (Muscle and microspikes Ras) (X-Ras)
Length = 208
Score = 29.3 bits (64), Expect = 4.2
Identities = 22/103 (21%), Positives = 45/103 (43%), Gaps = 15/103 (14%)
Query: 76 SSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKGRT--- 132
S +K S + ++++ ++V F +IL AN +++ L +T +M
Sbjct: 93 SVTDKASFEHVDRFHQLILRVKDRESFPMILVANKVDLMHLRKVTRDQGKEMATKYNIPY 152
Query: 133 --------PEEIRKTFN----IINDYTPEEEEEVRSETQWAFD 163
P + KTF+ +I PE+ ++ + +T+W D
Sbjct: 153 IETSAKDPPLNVDKTFHDLVRVIRQQVPEKNQKKKKKTKWRGD 195
>LTV1_YEAST (P34078) Low-temperature viability protein LTV1
Length = 463
Score = 29.3 bits (64), Expect = 4.2
Identities = 25/91 (27%), Positives = 45/91 (48%), Gaps = 10/91 (10%)
Query: 76 SSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDL-TCKTVADMIKGRTPE 134
+ DE EDE ++WD + V+ +D + + A + NI++L DL AD+ + +
Sbjct: 222 AKDEDEFEDEFDEWDIDNVENFEDENY-VKEMAQFDNIENLEDLENIDYQADVRRFQKDN 280
Query: 135 EIRKTFNIINDYT--------PEEEEEVRSE 157
I + N ++++ P EEE+V E
Sbjct: 281 SILEKHNSDDEFSNAGLDSVNPSEEEDVLGE 311
>LEU3_YAMOH (P41926) 3-isopropylmalate dehydrogenase (EC 1.1.1.85)
(Beta-IPM dehydrogenase) (IMDH) (3-IPM-DH)
Length = 368
Score = 29.3 bits (64), Expect = 4.2
Identities = 42/174 (24%), Positives = 72/174 (41%), Gaps = 36/174 (20%)
Query: 10 ATKKITLKSSDNETFEI-----------EKAVALESQTIKHLIDDNCADDSGIPLPNVTG 58
+TK IT+ D+ EI EK ++ + HLI D +G+PLP+ T
Sbjct: 2 STKTITVVPGDHVGTEICDEAIKVLKAIEKVSPVKFEFKHHLIGGAAIDATGVPLPDET- 60
Query: 59 KILAMVIEHCKKHVDATSSDEKPSEDEIN--KWDTEFVKVDQDTLFDLILAANYLNIK-- 114
++A S + + KW T V+ +Q L Y N++
Sbjct: 61 -------------LEAAKSSDAVLLGAVGGPKWGTGAVRPEQGLLKIRKELNLYANLRPC 107
Query: 115 -----SLLDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
SLL+L+ ++++KG +R+ I + +E+E S+ Q A+D
Sbjct: 108 NFVSDSLLELS-PLKSEIVKGTNFTVVRELVGGIY-FGERQEQEESSDGQSAWD 159
>T2D3_HAEIN (P43870) Type II restriction enzyme HindIII (EC
3.1.21.4) (Endonuclease HindIII) (R.HindIII)
Length = 300
Score = 28.9 bits (63), Expect = 5.4
Identities = 20/70 (28%), Positives = 34/70 (48%), Gaps = 15/70 (21%)
Query: 85 EINKWDTEFVKVDQDTLFDLI-LAANYLNIKSLLDLTCKTVADMIKGRTPEEIRKTFNII 143
+ NK+ + K+D+DTL L+ N++ +SL++ E +K NII
Sbjct: 215 DFNKYFMDLFKIDKDTLNQLLQKEINFIEERSLIE--------------KEYWKKQINII 260
Query: 144 NDYTPEEEEE 153
++T EE E
Sbjct: 261 KNFTREEAIE 270
>STNA_DROME (Q24211) Stoned A protein (StonedA) (Stn-A)
Length = 850
Score = 28.9 bits (63), Expect = 5.4
Identities = 29/104 (27%), Positives = 42/104 (39%), Gaps = 10/104 (9%)
Query: 17 KSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKHVDATS 76
K SD E IEK + ++H + D D P P V ++L V E+ V +
Sbjct: 677 KLSDIELKHIEKDLISVPANLRHSLSDPDFDPRAPPTP-VPAEVLLAVEENINIKVLTPA 735
Query: 77 SDEK---------PSEDEINKWDTEFVKVDQDTLFDLILAANYL 111
D K SE++I+ +DT Q +L L N L
Sbjct: 736 QDRKKLTNSGGSGKSEEDIDPFDTSIAANLQPGQTELKLLENEL 779
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.310 0.129 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,898,922
Number of Sequences: 164201
Number of extensions: 738667
Number of successful extensions: 1898
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 1871
Number of HSP's gapped (non-prelim): 35
length of query: 164
length of database: 59,974,054
effective HSP length: 102
effective length of query: 62
effective length of database: 43,225,552
effective search space: 2679984224
effective search space used: 2679984224
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148341.11


