

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147472.2 - phase: 0
(363 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
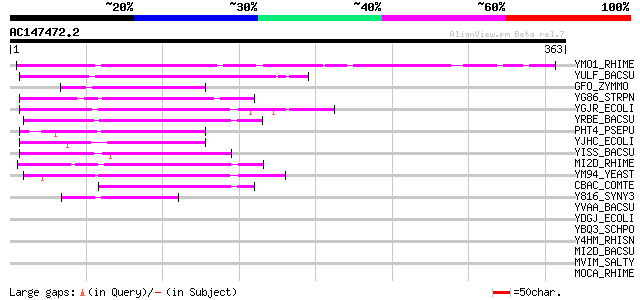
Score E
Sequences producing significant alignments: (bits) Value
YMO1_RHIME (P49305) Hypothetical 36.4 kDa protein in mocC-mocA i... 112 1e-24
YULF_BACSU (O05265) Hypothetical oxidoreductase yulF (EC 1.-.-.-) 70 1e-11
GFO_ZYMMO (Q07982) Glucose--fructose oxidoreductase precursor (E... 65 4e-10
YG86_STRPN (Q54728) Hypothetical oxidoreductase SP1686 (EC 1.-.-.-) 63 1e-09
YGJR_ECOLI (P42599) Hypothetical oxidoreductase ygjR (EC 1.-.-.-) 62 2e-09
YRBE_BACSU (O05389) Hypothetical oxidoreductase yrbE (EC 1.-.-.-) 60 1e-08
PHT4_PSEPU (Q05184) Putative 4,5,-dihydroxyphthalate dehydrogena... 59 3e-08
YJHC_ECOLI (P39353) Hypothetical oxidoreductase yjhC (EC 1.-.-.-) 57 1e-07
YISS_BACSU (P40332) Hypothetical oxidoreductase yisS (EC 1.-.-.-) 54 5e-07
MI2D_RHIME (O68965) Inositol 2-dehydrogenase (EC 1.1.1.18) 50 7e-06
YM94_YEAST (Q04869) Hypothetical 38.2 kDa protein in PRE5-FET4 i... 50 9e-06
CBAC_COMTE (Q44258) 1-carboxy-3-chloro-3,4-dihydroxycyclo hexa-1... 47 6e-05
Y816_SYNY3 (P74041) Hypothetical oxidoreductase sll0816 (EC 1.-.... 44 5e-04
YVAA_BACSU (O32223) Hypothetical oxidoreductase yvaA (EC 1.-.-.-) 43 0.001
YDGJ_ECOLI (P77376) Hypothetical oxidoreductase ydgJ (EC 1.-.-.-) 43 0.001
YBQ3_SCHPO (O42896) Hypothetical oxidoreductase C115.03 (EC 1.-.... 40 0.012
Y4HM_RHISN (P55480) Hypothetical protein y4hM precursor 37 0.060
MI2D_BACSU (P26935) Inositol 2-dehydrogenase (EC 1.1.1.18) 37 0.060
MVIM_SALTY (P37168) Virulence factor mviM 36 0.17
MOCA_RHIME (P49307) Rhizopine catabolism protein mocA (EC 1.-.-.-) 34 0.51
>YMO1_RHIME (P49305) Hypothetical 36.4 kDa protein in mocC-mocA
intergenic region (ORF334)
Length = 334
Score = 112 bits (280), Expect = 1e-24
Identities = 107/355 (30%), Positives = 155/355 (43%), Gaps = 29/355 (8%)
Query: 5 DTVRFGILGCATIA-RKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSY 63
D VRFGI+G A IA K+ ++ + AI SR + +A A + S YGSY
Sbjct: 2 DLVRFGIIGTAAIAVEKVIPSMLSAEGLEVVAIASRDLDRARAAATRFGIGRS---YGSY 58
Query: 64 DEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGV 123
DE++ D ++AVYIPLP LHV WA+ AA KHVL EKP ALDV EL +++ + G
Sbjct: 59 DEILADPEIEAVYIPLPNHLHVHWAIRAAEAGKHVLCEKPLALDVEELSRLIDCRDRTGR 118
Query: 124 QFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELDA 183
+ + M HP+ + D+ S IG V+ I T+ L+ V
Sbjct: 119 RIQEAVMIRAHPQ---WDEIFDIVASGEIGEVRAIQGV----FTEVNLDPKSIVNDASIG 171
Query: 184 LGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVANI 243
GAL DL Y I A+ + P V A+ D+ + TS P A +
Sbjct: 172 GGALYDLGVYPIAAARMVFAAE-PERVFAVSDLDPVFG---IDRLTSAVLLFPGGRQATL 227
Query: 244 HCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRPEEVH 303
S ++ I G+ S+ LK+ P + G+K E
Sbjct: 228 VVSTQLALRHNVEIFGTRKSISLKNPFNPTPDDHCRIVLDNGSKLA-------AAAAETR 280
Query: 304 VVNKLPQEALMVQEFAMLVASIRDSGSQP-SIKWPEISRKTQLVVDAVKKSLELG 357
V Q L + F+ A+IR P ++W S T V++A+++S E G
Sbjct: 281 RVAPADQYRLQAERFS---AAIRSGSPLPIELEW---SLGTMKVLNAIQRSAERG 329
>YULF_BACSU (O05265) Hypothetical oxidoreductase yulF (EC 1.-.-.-)
Length = 328
Score = 69.7 bits (169), Expect = 1e-11
Identities = 53/189 (28%), Positives = 85/189 (44%), Gaps = 5/189 (2%)
Query: 7 VRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEV 66
+RF I+G I + + + ++A+ SRS +A +FA +++ + + E+
Sbjct: 2 IRFAIIGTNWITDRFLESAADIEDFQLTAVYSRSAERAGEFAAKHN---AAHAFSDLQEM 58
Query: 67 VEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFM 126
DAVYI P +LH AV N KHVL EKP A + E + ++ A +ANGV M
Sbjct: 59 AASDCFDAVYIASPNALHKEQAVLFMNHGKHVLCEKPFASNTKETEEMISAAKANGVVLM 118
Query: 127 DGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELDALGA 186
+ P +K L + + +S + N +PEL + G+
Sbjct: 119 EAMKTTFLPNFKELKKHLHKIGTVRRFTASYCQYSSRYDAFRSGTVLN-AFQPEL-SNGS 176
Query: 187 LGDLAWYCI 195
L D+ YCI
Sbjct: 177 LMDIGVYCI 185
>GFO_ZYMMO (Q07982) Glucose--fructose oxidoreductase precursor (EC
1.1.99.28) (GFOR)
Length = 439
Score = 64.7 bits (156), Expect = 4e-10
Identities = 33/95 (34%), Positives = 56/95 (58%), Gaps = 3/95 (3%)
Query: 34 SAIGSRSISKAAKFAVENSLPASVRIYGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAAN 93
SA+ ++ AA++ V+ P + Y ++D++ +D +DAVYI LP SLH +A+ +
Sbjct: 115 SAVTEKAKIVAAEYGVD---PRKIYDYSNFDKIAKDPKIDAVYIILPNSLHAEFAIRSFK 171
Query: 94 KKKHVLVEKPAALDVAELDLILEACEANGVQFMDG 128
KHV+ EKP A VA+ +++A +A + M G
Sbjct: 172 AGKHVMCEKPMATSVADCQRMIDAAKAANKKLMIG 206
>YG86_STRPN (Q54728) Hypothetical oxidoreductase SP1686 (EC 1.-.-.-)
Length = 367
Score = 63.2 bits (152), Expect = 1e-09
Identities = 45/154 (29%), Positives = 72/154 (46%), Gaps = 9/154 (5%)
Query: 7 VRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEV 66
V++G++G +LAR + A I+ + ++A + L A V S DE+
Sbjct: 2 VKYGVVGTGYFGAELARYMQKNDGAEITLLYDPDNAEA----IAEELGAKVA--SSLDEL 55
Query: 67 VEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFM 126
V VD V + P +LH + AA K+V EKP AL + +++AC+ N V FM
Sbjct: 56 VSSDEVDCVIVATPNNLHKEPVIKAAQHGKNVFCEKPIALSYQDCREMVDACKENNVTFM 115
Query: 127 DGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHS 160
G + + + H +L N IG V + H+
Sbjct: 116 AGHIMNFF---NGVHHAKELINQGVIGDVLYCHT 146
>YGJR_ECOLI (P42599) Hypothetical oxidoreductase ygjR (EC 1.-.-.-)
Length = 328
Score = 62.0 bits (149), Expect = 2e-09
Identities = 52/210 (24%), Positives = 93/210 (43%), Gaps = 13/210 (6%)
Query: 7 VRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEV 66
+RF ++G I R+ A + ++A+ SRS+ +A FA + S+ ++ S + +
Sbjct: 2 IRFAVIGTNWITRQFVEAAHESGKYKLTAVYSRSLEQAQHFANDFSVE---HLFTSLEAM 58
Query: 67 VEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFM 126
E +DAVYI P SLH + K +V+ EKP A ++AE+D + N V
Sbjct: 59 AESDAIDAVYIASPNSLHFSQTQLFLSHKINVICEKPLASNLAEVDAAIACARENQVVLF 118
Query: 127 DGCMWLHHPRTSNMKHLLDLSNSDGIGPVQ--FIHSTSTMPTTQEFL--ENNIRVKPELD 182
+ P ++ L +G ++ F + Q +L EN P
Sbjct: 119 EAFKTACLPNFHLLRQAL-----PKVGKLRKVFFNYCQYSSRYQRYLDGENPNTFNPAF- 172
Query: 183 ALGALGDLAWYCIGASLWAKGYKLPTTVTA 212
+ G++ D+ +YC+ +++ G TA
Sbjct: 173 SNGSIMDIGFYCLASAVALFGEPKSVQATA 202
>YRBE_BACSU (O05389) Hypothetical oxidoreductase yrbE (EC 1.-.-.-)
Length = 341
Score = 59.7 bits (143), Expect = 1e-08
Identities = 41/156 (26%), Positives = 69/156 (43%), Gaps = 6/156 (3%)
Query: 10 GILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEVVED 69
GI+G I + + I+ P+ I AI S+ +A + + I Y +++ D
Sbjct: 8 GIIGAGRIGKLHVQNISRIPHMKIKAISDIQASRIKSWADSHQIEY---ITSDYRDLLHD 64
Query: 70 SGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFMDGC 129
+DA++I PT++H + AA KKH+ EKP + + E L A +GV G
Sbjct: 65 PDIDAIFICSPTAVHAQMIKEAAEAKKHIFCEKPVSFSLDETSEALAAVRKHGVTLQVGF 124
Query: 130 MWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMP 165
P +K +++ + IG + TS P
Sbjct: 125 NRRFDPHFKKIKTIVE---NGEIGTPHLLKITSRDP 157
>PHT4_PSEPU (Q05184) Putative 4,5,-dihydroxyphthalate dehydrogenase
(EC 1.-.-.-) (DHP dehydrogenase)
Length = 410
Score = 58.5 bits (140), Expect = 3e-08
Identities = 40/127 (31%), Positives = 60/127 (46%), Gaps = 14/127 (11%)
Query: 7 VRFGILGCATIARKLARAITLT-----PNATISAIGSRSISKAAKFAVENSLPASVRIYG 61
+R G++G L RA TL + + +G+ + A+ E A Y
Sbjct: 12 LRLGVVG-------LGRAFTLMLPTFLADRRVQLVGACDPREQARRQFERDFDAPA--YE 62
Query: 62 SYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEAN 121
+ +++ DS VDA+YI P H AA +KHVLVEKP AL + E D ++ C
Sbjct: 63 TIEDLAADSNVDALYIASPHQFHAEHTRIAAANRKHVLVEKPMALSLDECDRMIADCAEA 122
Query: 122 GVQFMDG 128
GV+ + G
Sbjct: 123 GVKLIVG 129
>YJHC_ECOLI (P39353) Hypothetical oxidoreductase yjhC (EC 1.-.-.-)
Length = 372
Score = 56.6 bits (135), Expect = 1e-07
Identities = 37/126 (29%), Positives = 59/126 (46%), Gaps = 14/126 (11%)
Query: 7 VRFGILGCATIARKLARAITLTPNATISAI----GSRSISKAAKFAVENSLPASVRIYGS 62
+ +G++G +LAR + + NA I+ + +I++ + +SL
Sbjct: 2 INYGVVGVGYFGAELARFMNMHDNAKITCVYDPENGENIARELQCINMSSL--------- 52
Query: 63 YDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANG 122
D +V VD V + P LH + AA KKHV EKP AL + +++AC+ G
Sbjct: 53 -DALVSSKLVDCVIVATPNYLHKEPVIKAAKNKKHVFCEKPIALSYEDCVDMVKACKEAG 111
Query: 123 VQFMDG 128
V FM G
Sbjct: 112 VTFMAG 117
>YISS_BACSU (P40332) Hypothetical oxidoreductase yisS (EC 1.-.-.-)
Length = 342
Score = 54.3 bits (129), Expect = 5e-07
Identities = 44/142 (30%), Positives = 62/142 (42%), Gaps = 8/142 (5%)
Query: 7 VRFGILGCATIARKLAR-AITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYD- 64
VR +LG + A+ +T P A + +G +A + A E + S D
Sbjct: 5 VRCAVLGLGRLGYYHAKNLVTSVPGAKLVCVGDPLKGRAEQVARELGIEK-----WSEDP 59
Query: 65 -EVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGV 123
EV+ED G+DAV I PTS H + AA K + VEKP L + E E + GV
Sbjct: 60 YEVLEDPGIDAVIIVTPTSTHGDMIIKAAENGKQIFVEKPLTLSLEESKAASEKVKETGV 119
Query: 124 QFMDGCMWLHHPRTSNMKHLLD 145
G M P ++ K +D
Sbjct: 120 ICQVGFMRRFDPAYADAKRRID 141
>MI2D_RHIME (O68965) Inositol 2-dehydrogenase (EC 1.1.1.18)
Length = 330
Score = 50.4 bits (119), Expect = 7e-06
Identities = 45/162 (27%), Positives = 69/162 (41%), Gaps = 10/162 (6%)
Query: 6 TVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDE 65
TVRFG+LG I + A+A++ +A + A+ AA A+ + VR + D
Sbjct: 2 TVRFGLLGAGRIGKVHAKAVSGNADARLVAVADAF--PAAAEAIAGAYGCEVR---TIDA 56
Query: 66 VVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQF 125
+ + +DAV I PT H A K + EKP LD + L+ +
Sbjct: 57 IEAAADIDAVVICTPTDTHADLIERFARAGKAIFCEKPIDLDAERVRACLKVVSDTKAKL 116
Query: 126 MDGCMWLHHPRTSNMKHLLDLSNSDG-IGPVQFIHSTSTMPT 166
M G P ++ +D DG IG V+ + TS P+
Sbjct: 117 MVGFNRRFDPHFMAVRKAID----DGRIGEVEMVTITSRDPS 154
>YM94_YEAST (Q04869) Hypothetical 38.2 kDa protein in PRE5-FET4
intergenic region
Length = 349
Score = 50.1 bits (118), Expect = 9e-06
Identities = 48/175 (27%), Positives = 75/175 (42%), Gaps = 10/175 (5%)
Query: 10 GILGCATIARK--LARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEVV 67
GI+G AR L + A +R +KA FA +P + ++Y + DE++
Sbjct: 7 GIVGTGIFARDRHLPSYQEFPDKFKVIAAFNRHKAKALDFAKVADIPEN-KVYDNLDEIL 65
Query: 68 EDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFMD 127
D VD + LP + A K V++EKP A ++ + I++ E+ +
Sbjct: 66 NDPHVDYIDALLPAQFNADIVEKAVKAGKPVILEKPIAANLDQAKEIVKIAESTPLPVGV 125
Query: 128 GCMWLHHPRTSNMKHLLDLSNSDGIGP-VQFIH-STSTMPTTQEFLENNIRVKPE 180
WL+ P K + + IGP V F H ST T ++L R KPE
Sbjct: 126 AENWLYLPCIKIAKEQI-----EKIGPVVAFTHNSTGPFVTQNKYLTTTWRQKPE 175
>CBAC_COMTE (Q44258) 1-carboxy-3-chloro-3,4-dihydroxycyclo
hexa-1,5-diene dehydrogenase (EC 1.-.-.-)
Length = 397
Score = 47.4 bits (111), Expect = 6e-05
Identities = 32/102 (31%), Positives = 47/102 (45%), Gaps = 3/102 (2%)
Query: 59 IYGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEAC 118
++G + + ++ V+ VYI P H A A N KHVLVEKP A+ + + +
Sbjct: 50 VHGDIESLCKNPDVEVVYIGTPHQFHAVHAEIALNAGKHVLVEKPMAVTLEDCCRMNACA 109
Query: 119 EANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHS 160
+ G + G S K L+D S G V+FIHS
Sbjct: 110 QRAGKYLIVGHSHSFDHPISRAKELID---SGRYGRVRFIHS 148
>Y816_SYNY3 (P74041) Hypothetical oxidoreductase sll0816 (EC
1.-.-.-)
Length = 371
Score = 44.3 bits (103), Expect = 5e-04
Identities = 30/76 (39%), Positives = 39/76 (50%), Gaps = 3/76 (3%)
Query: 35 AIGSRSISKAAKFAVENSLPASVRIYGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANK 94
AI R ++KA + A N L S Y + +E++ + V AV I P LH A A
Sbjct: 35 AIYHRDLAKAQEVAKSNDLAYS---YNNLEELLANPEVQAVTIASPPFLHYEMAKQAILA 91
Query: 95 KKHVLVEKPAALDVAE 110
KHVL+EKP L V E
Sbjct: 92 GKHVLLEKPMTLRVEE 107
>YVAA_BACSU (O32223) Hypothetical oxidoreductase yvaA (EC 1.-.-.-)
Length = 358
Score = 42.7 bits (99), Expect = 0.001
Identities = 51/215 (23%), Positives = 84/215 (38%), Gaps = 27/215 (12%)
Query: 5 DTVRFGILGCATIARKLARAITLTPNATISAIGSRSISK---AAKFAVENSLPASVRIYG 61
DT++ GILG L+ ++ P + + ISK + V+ P +
Sbjct: 12 DTIKVGILGYG-----LSGSVFHGP--LLDVLDEYQISKIMTSRTEEVKRDFP-DAEVVH 63
Query: 62 SYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEAN 121
+E+ D ++ V + P+ LH +A KHV++EKP E + + A +
Sbjct: 64 ELEEITNDPAIELVIVTTPSGLHYEHTMACIQAGKHVVMEKPMTATAEEGETLKRAADEK 123
Query: 122 GVQFMDGCMWLHHPRTSN----MKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRV 177
GV H+ R N +K L+ + + I Q ++ + E
Sbjct: 124 GVLL----SVYHNRRWDNDFLTIKKLISEGSLEDINTYQVSYNRYRPEVQARWREK---- 175
Query: 178 KPELDALGALGDLAWYCIGASLWAKGYKLPTTVTA 212
E A G L DL + I +L G +P VTA
Sbjct: 176 --EGTATGTLYDLGSHIIDQTLHLFG--MPKAVTA 206
>YDGJ_ECOLI (P77376) Hypothetical oxidoreductase ydgJ (EC 1.-.-.-)
Length = 346
Score = 42.7 bits (99), Expect = 0.001
Identities = 32/117 (27%), Positives = 54/117 (45%), Gaps = 10/117 (8%)
Query: 5 DTVRFGILGCATIARKL-ARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSY 63
D +R G++G ++ A I TP ++ I S +K V+ P +V +
Sbjct: 3 DNIRVGLIGYGYASKTFHAPLIAGTPGQELAVISSSDETK-----VKADWP-TVTVVSEP 56
Query: 64 DEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVA---ELDLILEA 117
+ D +D + IP P H A AA KHV+V+KP + ++ ELD + ++
Sbjct: 57 KHLFNDPNIDLIVIPTPNDTHFPLAKAALEAGKHVVVDKPFTVTLSQARELDALAKS 113
>YBQ3_SCHPO (O42896) Hypothetical oxidoreductase C115.03 (EC
1.-.-.-)
Length = 368
Score = 39.7 bits (91), Expect = 0.012
Identities = 19/68 (27%), Positives = 34/68 (49%)
Query: 56 SVRIYGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLIL 115
++ +Y + DE++ D ++ V + LP ++H A N KHV+ EKP E +
Sbjct: 53 NILVYTNLDELLADVNIELVVVSLPPNVHSEIVKKALNAGKHVVCEKPFTPTYEEAKELY 112
Query: 116 EACEANGV 123
E E+ +
Sbjct: 113 ELAESKSL 120
>Y4HM_RHISN (P55480) Hypothetical protein y4hM precursor
Length = 403
Score = 37.4 bits (85), Expect = 0.060
Identities = 32/140 (22%), Positives = 57/140 (39%), Gaps = 12/140 (8%)
Query: 65 EVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQ 124
E D G++AV I P H R A A V+++KP V++ ++E A
Sbjct: 84 EAARDDGIEAVSIVTPNWTHHRIATAFLKAGIDVILDKPMTTTVSDARELVELQRATDRL 143
Query: 125 FMDGCMWLHHPRTSNMKHLLDLSNSDGIGP-----VQFIHSTSTMPTTQEFLENNIRVKP 179
+ + HH KH++ + +G V+++ +T P + + R P
Sbjct: 144 VIMTYPYAHHAMVRQAKHMI---RNGAVGTVRQAHVEYVQEWATAPFSSDAKGAMWRQDP 200
Query: 180 E----LDALGALGDLAWYCI 195
E A G +G A++ +
Sbjct: 201 EKVGRASATGDIGTHAYHLL 220
>MI2D_BACSU (P26935) Inositol 2-dehydrogenase (EC 1.1.1.18)
Length = 344
Score = 37.4 bits (85), Expect = 0.060
Identities = 29/101 (28%), Positives = 46/101 (44%), Gaps = 3/101 (2%)
Query: 6 TVRFGILGCATIARKLARAIT-LTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYD 64
++R G++G I ++ IT A I A+ + A K + L A+V Y + D
Sbjct: 2 SLRIGVIGTGAIGKEHINRITNKLSGAEIVAVTDVNQEAAQKVVEQYQLNATV--YPNDD 59
Query: 65 EVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAA 105
++ D VDAV + H + A +K+V EKP A
Sbjct: 60 SLLADENVDAVLVTSWGPAHESSVLKAIKAQKYVFCEKPLA 100
>MVIM_SALTY (P37168) Virulence factor mviM
Length = 307
Score = 35.8 bits (81), Expect = 0.17
Identities = 39/160 (24%), Positives = 63/160 (39%), Gaps = 19/160 (11%)
Query: 6 TVRFGILGCATIARKL-ARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYD 64
T+R GI+G IA+K +T T T+ S S KA + +P Y
Sbjct: 3 TLRIGIVGLGGIAQKAWLPVLTNTAGWTLQGAWSPSRDKALRICESWRIP--------YV 54
Query: 65 EVVED--SGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANG 122
+ + + S DAV++ T+ H N HV V+KP A ++ + + ++
Sbjct: 55 DSLANLASSCDAVFVHSSTASHYAVVSELLNAGVHVCVDKPLAENLRDAERLVALAAQKK 114
Query: 123 VQFMDGCMWLHHPRTSNMKHLL--------DLSNSDGIGP 154
+ M G P +K L D +D +GP
Sbjct: 115 LTLMVGFNRRFAPLYRELKTRLGTAASLRMDKHRTDSVGP 154
>MOCA_RHIME (P49307) Rhizopine catabolism protein mocA (EC 1.-.-.-)
Length = 317
Score = 34.3 bits (77), Expect = 0.51
Identities = 26/121 (21%), Positives = 46/121 (37%), Gaps = 5/121 (4%)
Query: 8 RFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEVV 67
R G++G + + RA + I+A+ + A ++ Y + +++
Sbjct: 5 RLGLVGAGRMGQVHVRAAAESSLVEIAAVADPIAASRLNLAGNG-----IKTYETAGDMI 59
Query: 68 EDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFMD 127
E VD V I P++ HV A + +L EKP + E + E V
Sbjct: 60 EAGEVDGVLIATPSNTHVDTVADIAARGLPILCEKPCGVTAEEARKAADVAERYKVHLQI 119
Query: 128 G 128
G
Sbjct: 120 G 120
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,639,009
Number of Sequences: 164201
Number of extensions: 1630831
Number of successful extensions: 3970
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 3938
Number of HSP's gapped (non-prelim): 35
length of query: 363
length of database: 59,974,054
effective HSP length: 111
effective length of query: 252
effective length of database: 41,747,743
effective search space: 10520431236
effective search space used: 10520431236
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC147472.2


