

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147405.9 + phase: 0
(589 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
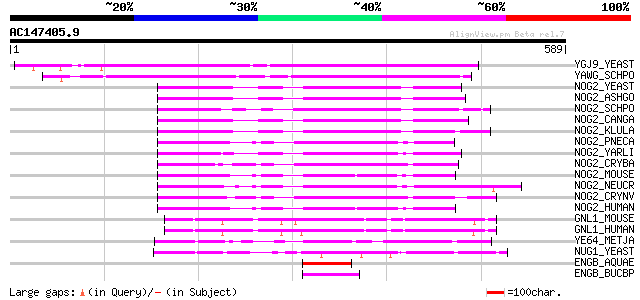
Score E
Sequences producing significant alignments: (bits) Value
YGJ9_YEAST (P53145) Hypothetical GTP-binding protein in SEH1-PRP... 311 4e-84
YAWG_SCHPO (Q10190) Hypothetical GTP-binding protein C3F10.16c i... 304 4e-82
NOG2_YEAST (P53742) Nucleolar GTP-binding protein 2 145 3e-34
NOG2_ASHGO (Q75DA4) Nucleolar GTP-binding protein 2 144 5e-34
NOG2_SCHPO (O14236) Nucleolar GTP-binding protein 2 143 1e-33
NOG2_CANGA (Q6FWS1) Nucleolar GTP-binding protein 2 142 2e-33
NOG2_KLULA (Q6CSP9) Nucleolar GTP-binding protein 2 136 1e-31
NOG2_PNECA (Q9C3Z4) Nucleolar GTP-binding protein 2 (Binding-ind... 134 5e-31
NOG2_YARLI (Q6C036) Nucleolar GTP-binding protein 2 132 2e-30
NOG2_CRYBA (Q6TGJ8) Nucleolar GTP-binding protein 2 131 6e-30
NOG2_MOUSE (Q99LH1) Nucleolar GTP-binding protein 2 130 1e-29
NOG2_NEUCR (Q7SHR8) Nucleolar GTP-binding protein 2 127 8e-29
NOG2_CRYNV (Q8J109) Nucleolar GTP-binding protein 2 127 1e-28
NOG2_HUMAN (Q13823) Nucleolar GTP-binding protein 2 (Autoantigen... 126 2e-28
GNL1_MOUSE (P36916) Guanine nucleotide-binding protein-like 1 (G... 125 3e-28
GNL1_HUMAN (P36915) Guanine nucleotide-binding protein-like 1 (G... 125 3e-28
YE64_METJA (Q58859) Hypothetical GTP-binding protein MJ1464 103 1e-21
NUG1_YEAST (P40010) Nuclear GTP-binding protein NUG1 (Nuclear GT... 97 1e-19
ENGB_AQUAE (O67679) Probable GTP-binding protein engB 52 6e-06
ENGB_BUCBP (Q89AD0) Probable GTP-binding protein engB 51 1e-05
>YGJ9_YEAST (P53145) Hypothetical GTP-binding protein in SEH1-PRP20
intergenic region
Length = 640
Score = 311 bits (796), Expect = 4e-84
Identities = 204/515 (39%), Positives = 289/515 (55%), Gaps = 36/515 (6%)
Query: 6 KTELGRAL--VKQHNQMIQQ--------TKEKGKIYKKKFLESFTEVSDIDAIIEQA--- 52
K ELGRA+ +Q I+ T +K + K L S T+ S +D + A
Sbjct: 26 KLELGRAIKYARQKENAIEYLPDGEMRFTTDKHEANWVK-LRSVTQESALDEFLSTAALA 84
Query: 53 DEDVDEQQLLDIPLPPPTALINLDHASGSDFNGLTVEEMKKEQK----IEEALHASSLRV 108
D+D + ++ + I +D SG+D M EQ+ ++ A L V
Sbjct: 85 DKDFTADRHSNVKI------IRMD--SGNDSATSQGFSMTNEQRGNLNAKQRALAKDLIV 136
Query: 109 PRRPFWSAEMSADELHANETQHFLTWRRSLARLEE-NKKLVLTPFEKNLDIWRQLWRVVE 167
PRRP W+ MS +L E + FL WRR LA L+E N+ L+LTPFE+N+++W+QLWRVVE
Sbjct: 137 PRRPEWNEGMSKFQLDRQEKEAFLEWRRKLAHLQESNEDLLLTPFERNIEVWKQLWRVVE 196
Query: 168 RSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAH 227
RSDL+V +VD+R+PL +R DLE Y KE D K LLLVNKADLL R WA+YF +
Sbjct: 197 RSDLVVQIVDARNPLLFRSVDLERYVKESDDRKANLLLVNKADLLTKKQRIAWAKYFISK 256
Query: 228 DILFIFWSAKAATAVLEGKK-LGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVDM 286
+I F F+SA A +LE +K +G + + D + + DE + ++ +D
Sbjct: 257 NISFTFYSALRANQLLEKQKEMGEDYREQ--DFEEADKEGFDADEKV--MEKVKILSIDQ 312
Query: 287 RRSSGSSKSSDDNA--SVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTK 344
SK+ ++ + + +G VGYPNVGKSSTIN+LVG KK V+STPGKTK
Sbjct: 313 LEELFLSKAPNEPLLPPLPGQPPLINIGLVGYPNVGKSSTINSLVGAKKVSVSSTPGKTK 372
Query: 345 HFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRVPRHV 404
HFQT+ +S+ ++LCDCPGLVFP+F+ ++ E++ GVLPID++ + +VA R+P++
Sbjct: 373 HFQTIKLSDSVMLCDCPGLVFPNFAYNKGELVCNGVLPIDQLRDYIGPAGLVAERIPKYY 432
Query: 405 IEEIYNISL-PKPKSYESQSRPPLASELLRTYCASRGQTTSS-GLPDETRASRQILKDYI 462
IE IY I + K + P A ELL Y +RG T G DE RASR ILKDY+
Sbjct: 433 IEAIYGIHIQTKSRDEGGNGDIPTAQELLVAYARARGYMTQGYGSADEPRASRYILKDYV 492
Query: 463 DGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSD 497
+GKL + PP L + + E + +V D
Sbjct: 493 NGKLLYVNPPPHLEDDTPYTREECEEFNKDLYVFD 527
>YAWG_SCHPO (Q10190) Hypothetical GTP-binding protein C3F10.16c in
chromosome I
Length = 616
Score = 304 bits (779), Expect = 4e-82
Identities = 177/460 (38%), Positives = 265/460 (57%), Gaps = 28/460 (6%)
Query: 36 LESFTEVSDIDAIIEQAD----EDVDEQQLLDIPLPPPTALINLDHASGSDFNGLTVEEM 91
L S T +D+D + A+ E + E+Q + + + + + F E
Sbjct: 46 LRSVTHETDLDEFLNTAELGEVEFIAEKQNVTV----------IQNPEQNPFLLSKEEAA 95
Query: 92 KKEQKIEEALHASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLTP 151
+ +QK E+ + L +PRRP W +A EL E + FL WRR+LA+L++ + ++TP
Sbjct: 96 RSKQKQEK--NKDRLTIPRRPHWDQTTTAVELDRMERESFLNWRRNLAQLQDVEGFIVTP 153
Query: 152 FEKNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADL 211
FE+NL+IWRQLWRV+ERSD++V +VD+R+PLF+R LE Y KEV K LLVNKAD+
Sbjct: 154 FERNLEIWRQLWRVIERSDVVVQIVDARNPLFFRSAHLEQYVKEVGPSKKNFLLVNKADM 213
Query: 212 LPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDE 271
L R W+ YF ++I F+F+SA+ A E + G +++ N + DE
Sbjct: 214 LTEEQRNYWSSYFNENNIPFLFFSARMAA---EANERGEDLETYESTSSNEIPESLQADE 270
Query: 272 LLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQ 331
+ I ++ G + +++ + + G VGYPNVGKSSTINALVG
Sbjct: 271 ----NDVHSSRIATLKVLEGIFEKF--ASTLPDGKTKMTFGLVGYPNVGKSSTINALVGS 324
Query: 332 KKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRE 391
KK V+STPGKTKHFQT+ +SEK+ L DCPGLVFPSF++++ +++ GVLPID++ ++
Sbjct: 325 KKVSVSSTPGKTKHFQTINLSEKVSLLDCPGLVFPSFATTQADLVLDGVLPIDQLREYTG 384
Query: 392 CVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRG-QTTSSGLPDE 450
++A R+P+ V+E +Y I + E + P A E+L + SRG G PD+
Sbjct: 385 PSALMAERIPKEVLETLYTIRIRIKPIEEGGTGVPSAQEVLFPFARSRGFMRAHHGTPDD 444
Query: 451 TRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQ 490
+RA+R +LKDY++GKL + PP SE + EH Q
Sbjct: 445 SRAARILLKDYVNGKLLYVHPPPNYPNS--GSEFNKEHHQ 482
>NOG2_YEAST (P53742) Nucleolar GTP-binding protein 2
Length = 486
Score = 145 bits (366), Expect = 3e-34
Identities = 91/322 (28%), Positives = 151/322 (46%), Gaps = 57/322 (17%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++V++ SD+++ V+D+RDPL RC +E Y K+ HK+ + ++NK DL+P V
Sbjct: 211 IWNELYKVIDSSDVVIHVLDARDPLGTRCKSVEEYMKKETPHKHLIYVLNKCDLVPTWVA 270
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
W ++ F ++ T +G+ L+ L+
Sbjct: 271 AAWVKHLSKERPTLAFHASI--------------------------TNSFGKGSLIQLLR 304
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
++ D ++ S VGF+GYPN GKSS IN L +K V
Sbjct: 305 QFSQLHTDRKQIS--------------------VGFIGYPNTGKSSIINTLRKKKVCQVA 344
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
PG+TK +Q + + +++ L DCPG+V PS S +++ GV+ ++ +T + + V
Sbjct: 345 PIPGETKVWQYITLMKRIFLIDCPGIVPPSSKDSEEDILFRGVVRVEHVTHPEQYIPGVL 404
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
R +E Y IS K A+E + +G+ G PDE+ S+QI
Sbjct: 405 KRCQVKHLERTYEISGWKD-----------ATEFIEILARKQGRLLKGGEPDESGVSKQI 453
Query: 458 LKDYIDGKLPHYEMPPGLSTQE 479
L D+ GK+P + +PP +E
Sbjct: 454 LNDFNRGKIPWFVLPPEKEGEE 475
>NOG2_ASHGO (Q75DA4) Nucleolar GTP-binding protein 2
Length = 502
Score = 144 bits (364), Expect = 5e-34
Identities = 89/326 (27%), Positives = 152/326 (46%), Gaps = 57/326 (17%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++V++ SD+++ V+D+RDPL RC +E Y K+ HK+ + ++NK DL+P +
Sbjct: 210 IWNELYKVIDSSDVVIHVLDARDPLGTRCKSVEEYMKKETPHKHLIYVLNKCDLVPTWLA 269
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
W ++ F ++ T +G+ L+ L+
Sbjct: 270 AAWVKHLSKDRPTLAFHASI--------------------------TNSFGKGSLIQLLR 303
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
++ D ++ S VGF+GYPN GKSS IN L +K V
Sbjct: 304 QFSQLHKDRQQIS--------------------VGFIGYPNTGKSSIINTLRKKKVCQVA 343
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
PG+TK +Q + + +++ L DCPG+V PS + +++ GV+ ++ ++ + + V
Sbjct: 344 PIPGETKVWQYITLMKRIFLIDCPGIVPPSAKDTEEDILFRGVVRVEHVSHPEQYIPAVL 403
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
R RH +E Y IS A+E + +G+ G PDET ++Q+
Sbjct: 404 RRCKRHHLERTYEISGWAD-----------ATEFIEMLARKQGRLLKGGEPDETGVAKQV 452
Query: 458 LKDYIDGKLPHYEMPPGLSTQELASE 483
L D+ GK+P + PP Q S+
Sbjct: 453 LNDFNRGKIPWFVSPPDRDPQPETSK 478
>NOG2_SCHPO (O14236) Nucleolar GTP-binding protein 2
Length = 537
Score = 143 bits (361), Expect = 1e-33
Identities = 96/353 (27%), Positives = 164/353 (46%), Gaps = 61/353 (17%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++V++ SD+L+ V+D+RDP+ RC +E Y + HK+ +L++NK DL+P SV
Sbjct: 206 IWNELYKVIDSSDVLIQVLDARDPVGTRCGTVERYLRNEASHKHMILVLNKVDLVPTSVA 265
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
W + AK + + +S +G+ L+ L+
Sbjct: 266 AAWVKIL-----------AKEYPTIAFHASINNS---------------FGKGSLIQILR 299
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
A S+ S + VG +G+PN GKSS IN L +K V
Sbjct: 300 QFA--------------------SLHSDKKQISVGLIGFPNAGKSSIINTLRKKKVCNVA 339
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
PG+TK +Q + + +++ L DCPG+V PS + S E++ GV+ ++ ++ + V
Sbjct: 340 PIPGETKVWQYVALMKRIFLIDCPGIVPPSSNDSDAELLLKGVVRVENVSNPEAYIPTVL 399
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
+R +E Y IS ++E L G+ G PDE ++ +
Sbjct: 400 SRCKVKHLERTYEISGWND-----------STEFLAKLAKKGGRLLKGGEPDEASVAKMV 448
Query: 458 LKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASDIEDSSVVETE 510
L D++ GK+P + P GLS+ ++++ N +V ++ SD +D E E
Sbjct: 449 LNDFMRGKIPWFIGPKGLSS---SNDEINSSQKVATQQTEGSD-QDGEEAEEE 497
>NOG2_CANGA (Q6FWS1) Nucleolar GTP-binding protein 2
Length = 494
Score = 142 bits (359), Expect = 2e-33
Identities = 90/330 (27%), Positives = 152/330 (45%), Gaps = 57/330 (17%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++V++ SD+++ V+D+RDPL RC +E Y K+ HK+ + ++NK DL+P V
Sbjct: 211 IWNELYKVIDSSDVVIHVLDARDPLGTRCKSVEEYMKKETPHKHLIYVLNKCDLVPTWVA 270
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
W ++ F ++ T +G+ L+ L+
Sbjct: 271 AAWVKHLSKDRPTLAFHASI--------------------------TNSFGKGSLIQLLR 304
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
++ D ++ S VGF+GYPN GKSS IN L +K V
Sbjct: 305 QFSQLHTDRKQIS--------------------VGFIGYPNTGKSSIINTLRKKKVCQVA 344
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
PG+TK +Q + + +++ L DCPG+V PS S +++ GV+ ++ ++ + + V
Sbjct: 345 PIPGETKVWQYITLMKRIFLIDCPGIVPPSTKDSEEDILFRGVVRVEHVSHPEQYIPGVL 404
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
R +E Y IS K A++ + +G+ G PDE+ S+QI
Sbjct: 405 KRCQTKHLERTYEISGWKD-----------ATDFIEMLARKQGRLLKGGEPDESGVSKQI 453
Query: 458 LKDYIDGKLPHYEMPPGLSTQELASEDSNE 487
L D+ GK+P + +PP + D E
Sbjct: 454 LNDFNRGKIPWFVIPPEKEEDKKRKLDEEE 483
>NOG2_KLULA (Q6CSP9) Nucleolar GTP-binding protein 2
Length = 513
Score = 136 bits (343), Expect = 1e-31
Identities = 91/353 (25%), Positives = 157/353 (43%), Gaps = 61/353 (17%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++V++ SD+++ V+D+RDPL RC + Y HK+ + ++NK DL+P V
Sbjct: 210 IWNELYKVIDSSDVVIHVLDARDPLGTRCKSVTDYMTNETPHKHLIYVLNKCDLVPTWVA 269
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
W ++ F ++ T +G+ L+ L+
Sbjct: 270 AAWVKHLSKERPTLAFHASI--------------------------TNSFGKGSLIQLLR 303
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
++ D + S VGF+GYPN GKSS IN L +K V
Sbjct: 304 QFSQLHKDRHQIS--------------------VGFIGYPNTGKSSIINTLRKKKVCQVA 343
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
PG+TK +Q + + +++ L DCPG+V PS S +++ GV+ ++ ++ + + +
Sbjct: 344 PIPGETKVWQYITLMKRIFLIDCPGIVPPSSKDSEEDILFRGVVRVEHVSHPEQYIPGIL 403
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
R R +E Y IS K + + + +G+ G PDE+ S+QI
Sbjct: 404 KRCKRQHLERTYEISGWKD-----------SVDFIEMIARKQGRLLKGGEPDESGVSKQI 452
Query: 458 LKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASDIEDSSVVETE 510
L D+ GK+P + PP ++ E+ D+ P + D E+ + E
Sbjct: 453 LNDFNRGKIPWFVPPP----EKDKIEEKEPGDKKRPAEENQEDQEEEEKEQEE 501
>NOG2_PNECA (Q9C3Z4) Nucleolar GTP-binding protein 2
(Binding-inducible GTPase)
Length = 483
Score = 134 bits (338), Expect = 5e-31
Identities = 83/315 (26%), Positives = 148/315 (46%), Gaps = 56/315 (17%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++ ++ SD+++ ++D+R+PL RC +E Y K+ HK+ +LL+NK DL+P
Sbjct: 188 IWNELYKXIDSSDVIIQLLDARNPLGTRCKHVEEYLKKEKPHKHMILLLNKCDLIPTWCT 247
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
+W + F AS +NP +G+ L+ L+
Sbjct: 248 REWIKQLSKEYPTLAFH----------------------ASINNP----FGKGSLIQLLR 281
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
++ + S+ + VGF+GYPN GKSS IN L +K
Sbjct: 282 QFSK--------------------LHSNRRQISVGFIGYPNTGKSSVINTLRSKKVCNTA 321
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
PG+TK +Q + ++ K+ + DCPG+V P+ + S E+I G L I++++ + + +
Sbjct: 322 PIPGETKVWQYVRMTSKIFMIDCPGIVPPNSNDSETEIIIKGALRIEKVSNPEQYIHAIL 381
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
N +E Y IS +E+ S ++ + G+ G DE+ ++ +
Sbjct: 382 NLCETKHLERTYQIS-----GWENDS-----TKFIELLARKTGKLLKGGEVDESSIAKMV 431
Query: 458 LKDYIDGKLPHYEMP 472
+ D+I GK+P + P
Sbjct: 432 INDFIRGKIPWFIAP 446
>NOG2_YARLI (Q6C036) Nucleolar GTP-binding protein 2
Length = 509
Score = 132 bits (333), Expect = 2e-30
Identities = 86/322 (26%), Positives = 148/322 (45%), Gaps = 57/322 (17%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++V++ SD+++ V+D+RDPL RC +E Y K+ HK+ + ++NK DL+P V
Sbjct: 203 IWNELYKVIDSSDVVIHVLDARDPLGTRCTSVEQYIKKEAPHKHLIFVLNKCDLVPTWVA 262
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
W ++ + D + + A T +G+ L+ L+
Sbjct: 263 AAWVKHL-SQDYPTLAFHASI-------------------------TNSFGKGSLIQLLR 296
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
+ D ++ S VGF+GYPN GKSS IN L +K
Sbjct: 297 QYSALHPDRQQIS--------------------VGFIGYPNTGKSSIINTLRKKKVCKTA 336
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
PG+TK +Q + + +++ L DCPG+V PS S +++ GV+ ++ ++ + + +
Sbjct: 337 PIPGETKVWQYITLMKRIFLIDCPGIVPPSQKDSETDILFRGVVRVEHVSYPEQYIPALL 396
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
R +E Y +S + + A+E L G+ G PDE+ ++ I
Sbjct: 397 ERCETKHLERTYEVS-----GWSN------ATEFLEKIARKHGRLLKGGEPDESGIAKLI 445
Query: 458 LKDYIDGKLPHYEMPPGLSTQE 479
L D+ GK+P + PP E
Sbjct: 446 LNDFNRGKIPWFVPPPQAEDME 467
>NOG2_CRYBA (Q6TGJ8) Nucleolar GTP-binding protein 2
Length = 731
Score = 131 bits (329), Expect = 6e-30
Identities = 88/319 (27%), Positives = 149/319 (46%), Gaps = 38/319 (11%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++V++ SD+++ V+D+RDPL RC + Y ++ HK+ + ++NK DL+P V
Sbjct: 222 IWGELYKVLDSSDVVIHVLDARDPLGTRCKPVVEYLRKEKAHKHLVYVLNKVDLVPTWVT 281
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
Y A+ A ++ L + AS +N +G+ L+ L+
Sbjct: 282 S--GPYAYAY--------ANGPARWVKHLSLSAPTIAFHASINNS----FGKGSLIQLLR 327
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
+ + S + VGF+GYPN GKSS IN L +K V
Sbjct: 328 QFSV--------------------LHSDKKQISVGFIGYPNTGKSSIINTLKKKKVCTVA 367
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
PG+TK +Q + + ++ L DCPG+V S S + + GV+ ++ + E + +
Sbjct: 368 PIPGETKVWQYITLMRRIYLIDCPGIVPVSAKDSDTDTVLKGVVRVENLATPAEHIPALL 427
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
RV +E YN+ + + Q A+ +L G+ G PD+ A++ +
Sbjct: 428 ERVRPEYLERTYNLEHVEGGWHGEQG----ATVILTAIAKKSGKLLKGGEPDQEAAAKMV 483
Query: 458 LKDYIDGKLPHYEMPPGLS 476
L D+I GK+P + PP S
Sbjct: 484 LNDWIRGKIPFFVAPPAKS 502
>NOG2_MOUSE (Q99LH1) Nucleolar GTP-binding protein 2
Length = 728
Score = 130 bits (327), Expect = 1e-29
Identities = 86/316 (27%), Positives = 145/316 (45%), Gaps = 58/316 (18%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++V++ SD++V V+D+RDP+ R P +EAY K+ K+ + ++NK DL+P
Sbjct: 206 IWGELYKVIDSSDVVVQVLDARDPMGTRSPHIEAYLKKEKPWKHLIFVLNKCDLVPTWAT 265
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
++W F AS NP +G+ + L+
Sbjct: 266 KRWVAVLSQDYPTLAFH----------------------ASLTNP----FGKGAFIQLLR 299
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
+ D ++ S VGF+GYPNVGKSS IN L +K V
Sbjct: 300 QFGKLHTDKKQIS--------------------VGFIGYPNVGKSSVINTLRSKKVCNVA 339
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
G+TK +Q + + ++ L DCPG+V+PS S +++ GV+ ++++ ++ + V
Sbjct: 340 PIAGETKVWQYITLMRRIFLIDCPGVVYPS-EDSETDIVLKGVVQVEKIKAPQDHIGAVL 398
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
R I + Y I +S+E+ A + L G+ G PD S+ +
Sbjct: 399 ERAKPEYISKTYKI-----ESWEN------AEDFLEKLALRTGKLLKGGEPDMLTVSKMV 447
Query: 458 LKDYIDGKLPHYEMPP 473
L D+ G++P + PP
Sbjct: 448 LNDWQRGRIPFFVKPP 463
>NOG2_NEUCR (Q7SHR8) Nucleolar GTP-binding protein 2
Length = 619
Score = 127 bits (319), Expect = 8e-29
Identities = 96/389 (24%), Positives = 167/389 (42%), Gaps = 60/389 (15%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++V++ SD+++ V+D+RDPL RC +E Y HK+ + ++NK DL+P+
Sbjct: 221 IWNELYKVIDSSDVILHVIDARDPLGTRCRHVEKYLATEAPHKHLIFVLNKIDLVPSKTA 280
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
W + T + +S NP +GR L+ L+
Sbjct: 281 AAWIRVLQKDH----------PTCAMR------------SSIKNP----FGRGSLIDLLR 314
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
+ D ++ S VG VGYPNVGKSS INAL G+ V
Sbjct: 315 QFSILHKDRKQIS--------------------VGLVGYPNVGKSSIINALRGKPVAKVA 354
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
PG+TK +Q + + ++ L DCPG+V P+ + + +++ GV+ ++ + + + V
Sbjct: 355 PIPGETKVWQYVTLMRRIYLIDCPGIVPPNQNDTPQDLLLRGVVRVENVDNPEQYIPAVL 414
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
N+V H +E Y + K ++ L G+ G PD ++ +
Sbjct: 415 NKVKPHHMERTYEL-----KGWKDH------IHFLEMLARKGGRLLKGGEPDVDGVAKMV 463
Query: 458 LKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASDIEDSSVVETELA---PK 514
L D++ GK+P + P E + + E D +DS+ T +
Sbjct: 464 LNDFMRGKIPWFTPAPEKEEGETDTMEGREGRYGEMSKKRKRDEDDSAPATTPASAGEDA 523
Query: 515 LEHVLDDLSSFDMANGLASNKVAPKKTKE 543
E ++ + FD + + A +K +E
Sbjct: 524 KEEDPENFAGFDSDSDSEVEEAAEEKGEE 552
>NOG2_CRYNV (Q8J109) Nucleolar GTP-binding protein 2
Length = 708
Score = 127 bits (318), Expect = 1e-28
Identities = 93/359 (25%), Positives = 161/359 (43%), Gaps = 46/359 (12%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++V++ SD+++ V+D+RDPL RC + Y ++ HK+ + ++NK DL+P V
Sbjct: 197 IWGELYKVLDSSDVVIHVLDARDPLGTRCKPVVEYLRKEKAHKHLVYVLNKVDLVPTWVT 256
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
Y A+ F ++ L + AS +N +G+ L+ L+
Sbjct: 257 SGPYAYPYANGPHF-------GARWVKHLSLSAPTIAFHASINNS----FGKGSLIQLLR 305
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
+ + S + VGF+GYPN GKSS IN L +K V
Sbjct: 306 QFSV--------------------LHSDKKQISVGFIGYPNTGKSSIINTLKKKKVCTVA 345
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
PG+TK +Q + + ++ L DCPG+V S S + + GV+ ++ + E + +
Sbjct: 346 PIPGETKVWQYITLMRRIYLIDCPGIVPVSAKDSDTDTVLKGVVRVENLATPAEHIPALL 405
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
RV +E Y + + + + A+ +L G+ G PD+ A++ +
Sbjct: 406 ERVRPEYLERTYGLEHVEGGWHGEEG----ATFVLTAIAKKSGKLLKGGEPDQEAAAKMV 461
Query: 458 LKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASDIEDSSVVETELAPKLE 516
L D+I GK+P + PP + + HVS A+ + + ELA + E
Sbjct: 462 LNDWIRGKVPFFVAPP-----------TKPESGADAHVSSATTEKVQEQEQKELAEEKE 509
>NOG2_HUMAN (Q13823) Nucleolar GTP-binding protein 2 (Autoantigen
NGP-1)
Length = 731
Score = 126 bits (316), Expect = 2e-28
Identities = 84/316 (26%), Positives = 141/316 (44%), Gaps = 58/316 (18%)
Query: 158 IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVR 217
IW +L++V++ SD++V V+D+RDP+ R P +E Y K+ K+ + ++NK DL+P
Sbjct: 206 IWGELYKVIDSSDVVVQVLDARDPMGTRSPHIETYLKKEKPWKHLIFVLNKCDLVPTWAT 265
Query: 218 EKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQ 277
++W F AS NP +G+ + L+
Sbjct: 266 KRWVAVLSQDYPTLAFH----------------------ASLTNP----FGKGAFIQLLR 299
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
+ D ++ S VGF+GYPNVGKSS IN L +K V
Sbjct: 300 QFGKLHTDKKQIS--------------------VGFIGYPNVGKSSVINTLRSKKVCNVA 339
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
G+TK +Q + + ++ L DCPG+V+PS S +++ GV+ ++++ + + V
Sbjct: 340 PIAGETKVWQYITLMRRIFLIDCPGVVYPS-EDSETDIVLKGVVQVEKIKSPEDHIGAVL 398
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQI 457
R I + Y I S+E+ A + L G+ G PD + +
Sbjct: 399 ERAKPEYISKTYKID-----SWEN------AEDFLEKLAFRTGKLLKGGEPDLQTVGKMV 447
Query: 458 LKDYIDGKLPHYEMPP 473
L D+ G++P + PP
Sbjct: 448 LNDWQRGRIPFFVKPP 463
>GNL1_MOUSE (P36916) Guanine nucleotide-binding protein-like 1
(GTP-binding protein MMR1)
Length = 430
Score = 125 bits (314), Expect = 3e-28
Identities = 105/395 (26%), Positives = 176/395 (43%), Gaps = 56/395 (14%)
Query: 165 VVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYF 224
V+E SD+++++ D R P+ P L Y ++ +L++NK DL P ++ W YF
Sbjct: 7 VLEMSDIVLLITDIRHPVVNFPPALYEYVTG-ELGLALVLVLNKVDLAPPALVVAWKHYF 65
Query: 225 -----RAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSE 279
+ H +LF + T G L ++ T+ G ++LL ++
Sbjct: 66 HQRYPQLHIVLFTSFPRDTRTPQEPGGVLKKNRRRGKGW-----TRALGPEQLLRACEAI 120
Query: 280 AEAIVDMR----------------RSSGSSKSSDDNASV------------------SSS 305
VD+ SG + +D +V
Sbjct: 121 TVGKVDLSSWREQIARDVAGASWGNVSGEEEEEEDGPAVLVEQLTDSAMEPTGPSRERYK 180
Query: 306 SSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVF 365
V +G +G+PNVGKSS IN LVG+K V+ TPG T++FQT ++ + LCDCPGL+F
Sbjct: 181 DGVVTIGCIGFPNVGKSSLINGLVGRKVVSVSRTPGHTRYFQTYFLTPSVKLCDCPGLIF 240
Query: 366 PSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRP 425
PS + +++ G+ PI ++ + V +A+R+P ++ + ++ P+ + S P
Sbjct: 241 PSLLPRQLQVL-AGIYPIAQIQEPYTSVGYLASRIP---VQALLHLRHPEAED-PSAEHP 295
Query: 426 PLASELLRTYCASRG-QTTSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASED 484
A ++ + RG +T + D RA+ +L+ +DG+L PPG S Q E
Sbjct: 296 WCAWDICEAWAEKRGYKTRKAARNDVYRAANSLLRLAVDGRLSLCFYPPGYSEQRGTWES 355
Query: 485 SNEHDQV---NPHVSDASDIEDSSVVETELAPKLE 516
E ++ V A D E+ E EL+ E
Sbjct: 356 HPETAELVLSQGRVGPAGDEEEEE--EEELSSSCE 388
>GNL1_HUMAN (P36915) Guanine nucleotide-binding protein-like 1
(GTP-binding protein HSR1)
Length = 430
Score = 125 bits (314), Expect = 3e-28
Identities = 105/395 (26%), Positives = 175/395 (43%), Gaps = 56/395 (14%)
Query: 165 VVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYF 224
V+E SD+++++ D R P+ P L Y ++ +L++NK DL P + W YF
Sbjct: 7 VLEMSDIVLLITDIRHPVVNFPPALYEYVTG-ELGLALVLVLNKVDLAPRRLVVAWKHYF 65
Query: 225 -----RAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSE 279
+ H +LF + T L S+ T+ G ++LL ++
Sbjct: 66 HQHYPQLHVVLFTSFPRDPRTPQDPSSVLKKSRRRGRGW-----TRALGPEQLLRACEAI 120
Query: 280 AEAIVDMR---------------RSSGSSKSSDDNASVSSSSSH---------------- 308
VD+ SG + +D +V
Sbjct: 121 TVGKVDLSSWREKIARDAGATWGNGSGEEEEEEDGPAVLVEQQQTDSAMEPTGPTQERYK 180
Query: 309 ---VVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVF 365
V +G VG+PNVGKSS IN LVG+K V+ TPG T++FQT ++ + LCDCPGL+F
Sbjct: 181 DGVVTIGCVGFPNVGKSSLINGLVGRKVVSVSRTPGHTRYFQTYFLTPSVKLCDCPGLIF 240
Query: 366 PSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRP 425
PS + +++ G+ PI ++ + V +A+R+P ++ + ++ P+ + S P
Sbjct: 241 PSLLPRQLQVL-AGIYPIAQIQEPYTAVGYLASRIP---VQALLHLRHPEAED-PSAEHP 295
Query: 426 PLASELLRTYCASRG-QTTSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASED 484
A ++ + RG +T + D RA+ +L+ +DG++ PPG S Q+ E
Sbjct: 296 WCAWDICEAWAEKRGYKTAKAARNDVYRAANSLLRLAVDGRVSLCFHPPGYSEQKGTWES 355
Query: 485 SNEHDQ---VNPHVSDASDIEDSSVVETELAPKLE 516
E + + V A D E+ E EL+ E
Sbjct: 356 HPETTELVVLQGRVGPAGDEEEEE--EEELSSSCE 388
>YE64_METJA (Q58859) Hypothetical GTP-binding protein MJ1464
Length = 373
Score = 103 bits (258), Expect = 1e-21
Identities = 90/360 (25%), Positives = 157/360 (43%), Gaps = 72/360 (20%)
Query: 154 KNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLP 213
K + + + + ++++ D++++V+D+RDP R +LE K K + ++NKADL+P
Sbjct: 9 KKVPVKKIVNKIIDECDVILLVLDARDPEMTRNRELEKKIKAKG--KKLIYVLNKADLVP 66
Query: 214 ASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELL 273
+ EKW E F + + F SAK ++LG+ M
Sbjct: 67 KDILEKWKEVFGENTV---FVSAK--------RRLGTKILREMIK--------------- 100
Query: 274 ARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKK 333
+++ +M + G VG VGYPNVGKSS INAL G++K
Sbjct: 101 -------QSLKEMGKKEGK------------------VGIVGYPNVGKSSIINALTGKRK 135
Query: 334 TGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECV 393
S G TK Q + +++ + L D PG++ +++ G L +++
Sbjct: 136 ALTGSVAGLTKGEQWVRLTKNIKLMDTPGVL---EMRDEDDLVISGALRLEK-------- 184
Query: 394 QVVANRVP--RHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDET 451
V N +P ++ I N K Y + ELL+ R T G D
Sbjct: 185 --VENPIPPALKILSRINNFDNSIIKEYFGVDYEEVDEELLKKIGNKRSYLTKGGEVDLV 242
Query: 452 RASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASDIEDSSVVETEL 511
R ++ I+K+Y DGKL +Y+ + ++ + + + ++ D IED+ ++ T L
Sbjct: 243 RTAKTIIKEYQDGKLNYYK----VDLKKYGQDRERDISFITKYLKDFPFIEDAKMIVTHL 298
>NUG1_YEAST (P40010) Nuclear GTP-binding protein NUG1 (Nuclear
GTPase 1)
Length = 520
Score = 97.1 bits (240), Expect = 1e-19
Identities = 100/395 (25%), Positives = 172/395 (43%), Gaps = 64/395 (16%)
Query: 153 EKNLDIWRQLWR-VVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADL 211
EK+ + ++++ V++ SD+++ V+D+RDP R +E + K +L++NK DL
Sbjct: 159 EKSRKAYDKIFKSVIDASDVILYVLDARDPESTRSRKVEEAVLQSQ-GKRLILILNKVDL 217
Query: 212 LPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDE 271
+P V E+W Y ++ F +A++ + G +
Sbjct: 218 IPPHVLEQWLNYLKSS---FPTIPLRASSGAVNGTSFNRKLSQT---------------- 258
Query: 272 LLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALV-- 329
+ A A+++ S K+ +N+++ S +VVG +GYPNVGKSS INAL+
Sbjct: 259 ------TTASALLE------SLKTYSNNSNLKRS---IVVGVIGYPNVGKSSVINALLAR 303
Query: 330 --GQKKT-GVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSR------YEMITCGV 380
GQ K V + G T + + I KL + D PG+ FPS + R E+
Sbjct: 304 RGGQSKACPVGNEAGVTTSLREIKIDNKLKILDSPGICFPSENKKRSKVEHEAELALLNA 363
Query: 381 LPIDRMTQHRECVQVVANRVPR-----HVIEEIYNISLPKPKSYESQSRPPLASELLRTY 435
LP + V ++ R+ + +++Y I P P + L
Sbjct: 364 LPAKHIVDPYPAVLMLVKRLAKSDEMTESFKKLYEIP-PIP----ANDADTFTKHFLIHV 418
Query: 436 CASRGQTTSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELAS-EDSNEHDQVNPH 494
RG+ G+P+ A +L D+ DGK+ + +P +T AS +D +N
Sbjct: 419 ARKRGRLGKGGIPNLASAGLSVLNDWRDGKILGWVLP---NTSAAASQQDKQNLSTINTG 475
Query: 495 VSDASDIEDSSVVETELAPKLEHVLDDL-SSFDMA 528
A + S + +E + + + LD L SS D A
Sbjct: 476 TKQAPIAANESTIVSEWSKEFD--LDGLFSSLDKA 508
>ENGB_AQUAE (O67679) Probable GTP-binding protein engB
Length = 183
Score = 51.6 bits (122), Expect = 6e-06
Identities = 25/52 (48%), Positives = 33/52 (63%)
Query: 311 VGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPG 362
V FVG NVGKSS +N +VG K V+ TPG+T+ ++ +KL L D PG
Sbjct: 21 VVFVGRSNVGKSSLLNMVVGSKVAKVSKTPGRTRAVNYFLLDKKLYLVDVPG 72
>ENGB_BUCBP (Q89AD0) Probable GTP-binding protein engB
Length = 228
Score = 50.8 bits (120), Expect = 1e-05
Identities = 27/62 (43%), Positives = 36/62 (57%), Gaps = 1/62 (1%)
Query: 311 VGFVGYPNVGKSSTINALVGQKKTG-VTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFS 369
V FVGY N GKSS INAL QKK ++ TPG+T+ ++ ++ L D PG + S
Sbjct: 52 VAFVGYSNSGKSSIINALTNQKKLAKISRTPGRTRLINIFSVTSEIRLVDFPGYGYAQVS 111
Query: 370 SS 371
S
Sbjct: 112 RS 113
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.130 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 69,019,292
Number of Sequences: 164201
Number of extensions: 2897355
Number of successful extensions: 8572
Number of sequences better than 10.0: 405
Number of HSP's better than 10.0 without gapping: 296
Number of HSP's successfully gapped in prelim test: 109
Number of HSP's that attempted gapping in prelim test: 7974
Number of HSP's gapped (non-prelim): 634
length of query: 589
length of database: 59,974,054
effective HSP length: 116
effective length of query: 473
effective length of database: 40,926,738
effective search space: 19358347074
effective search space used: 19358347074
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC147405.9


