

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
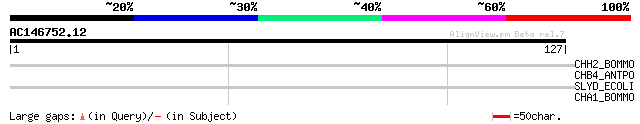
Query= AC146752.12 + phase: 2 /pseudo
(127 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CHH2_BOMMO (P05687) Chorion class high-cysteine HCA protein 12 p... 29 2.1
CHB4_ANTPO (P02847) Chorion class B protein PC401 precursor (Fra... 28 4.7
SLYD_ECOLI (P30856) FKBP-type peptidyl-prolyl cis-trans isomeras... 27 6.1
CHA1_BOMMO (P08826) Chorion class A protein L11 precursor 27 6.1
>CHH2_BOMMO (P05687) Chorion class high-cysteine HCA protein 12
precursor (HC-A.12)
Length = 124
Score = 28.9 bits (63), Expect = 2.1
Identities = 14/24 (58%), Positives = 14/24 (58%), Gaps = 1/24 (4%)
Query: 1 CSFLVGGKGVVVGNGVGGCGCGRR 24
C GG G G G GGCGCGRR
Sbjct: 98 CGCGCGGCGCGCG-GCGGCGCGRR 120
>CHB4_ANTPO (P02847) Chorion class B protein PC401 precursor
(Fragment)
Length = 171
Score = 27.7 bits (60), Expect = 4.7
Identities = 12/19 (63%), Positives = 14/19 (73%), Gaps = 1/19 (5%)
Query: 5 VGGKGVVVGN-GVGGCGCG 22
VGG G+ G G+GGCGCG
Sbjct: 148 VGGYGLGYGGYGLGGCGCG 166
>SLYD_ECOLI (P30856) FKBP-type peptidyl-prolyl cis-trans isomerase
slyD (EC 5.2.1.8) (PPIase) (Rotamase) (Histidine-rich
protein) (WHP)
Length = 196
Score = 27.3 bits (59), Expect = 6.1
Identities = 10/16 (62%), Positives = 11/16 (68%)
Query: 6 GGKGVVVGNGVGGCGC 21
GG+G G G GGCGC
Sbjct: 180 GGEGCCGGKGNGGCGC 195
>CHA1_BOMMO (P08826) Chorion class A protein L11 precursor
Length = 129
Score = 27.3 bits (59), Expect = 6.1
Identities = 9/14 (64%), Positives = 11/14 (78%)
Query: 9 GVVVGNGVGGCGCG 22
G +G G+GGCGCG
Sbjct: 21 GQCLGRGLGGCGCG 34
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.358 0.167 0.583
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,244,593
Number of Sequences: 164201
Number of extensions: 463083
Number of successful extensions: 2442
Number of sequences better than 10.0: 4
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 2434
Number of HSP's gapped (non-prelim): 8
length of query: 127
length of database: 59,974,054
effective HSP length: 103
effective length of query: 24
effective length of database: 43,061,351
effective search space: 1033472424
effective search space used: 1033472424
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146752.12


