

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146559.13 - phase: 0 /pseudo
(661 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
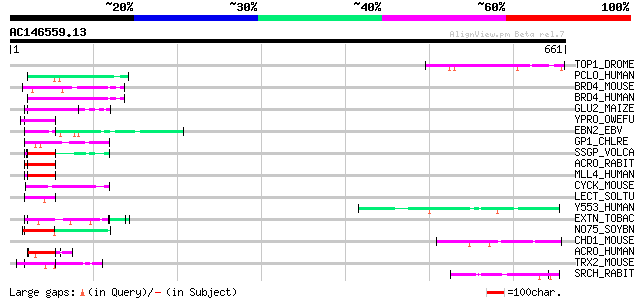
Score E
Sequences producing significant alignments: (bits) Value
TOP1_DROME (P30189) DNA topoisomerase I (EC 5.99.1.2) 57 2e-07
PCLO_HUMAN (Q9Y6V0) Piccolo protein (Aczonin) 56 3e-07
BRD4_MOUSE (Q9ESU6) Bromodomain-containing protein 4 (Mitotic ch... 54 1e-06
BRD4_HUMAN (O60885) Bromodomain-containing protein 4 (HUNK1 prot... 53 2e-06
GLU2_MAIZE (P04706) Glutelin 2 precursor (Zein-gamma) (27 kDa ze... 52 4e-06
YPRO_OWEFU (P21260) Hypothetical proline-rich protein (Fragment) 52 5e-06
EBN2_EBV (P12978) EBNA-2 nuclear protein 52 5e-06
GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor (H... 52 6e-06
SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor ... 51 8e-06
ACRO_RABIT (P48038) Acrosin precursor (EC 3.4.21.10) 51 8e-06
MLL4_HUMAN (Q9UMN6) Myeloid/lymphoid or mixed-lineage leukemia p... 50 1e-05
CYCK_MOUSE (O88874) Cyclin K 50 1e-05
LECT_SOLTU (Q9S8M0) Chitin-binding lectin 1 precursor (PL-I) 50 2e-05
Y553_HUMAN (Q9UKJ3) Hypothetical protein KIAA0553 49 3e-05
EXTN_TOBAC (P13983) Extensin precursor (Cell wall hydroxyproline... 49 3e-05
NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75) (NGM-75) 49 4e-05
CHD1_MOUSE (P40201) Chromodomain-helicase-DNA-binding protein 1 ... 49 4e-05
ACRO_HUMAN (P10323) Acrosin precursor (EC 3.4.21.10) 48 7e-05
TRX2_MOUSE (O08550) Trithorax homolog 2 (WW domain binding prote... 48 9e-05
SRCH_RABIT (P16230) Sarcoplasmic reticulum histidine-rich calciu... 47 1e-04
>TOP1_DROME (P30189) DNA topoisomerase I (EC 5.99.1.2)
Length = 972
Score = 57.0 bits (136), Expect = 2e-07
Identities = 53/198 (26%), Positives = 89/198 (44%), Gaps = 38/198 (19%)
Query: 496 KLLSSHTISTEANNSREVSHDSPAVTI----SNPDHH---EQQSHTNYSDKSKVVHDATS 548
+++ S+ ++T + H S + + S+ D H E++ ++ S S H ++S
Sbjct: 20 EVVQSNGVTTNGHGHHHHHHSSSSSSSKHKSSSKDKHRDREREHKSSNSSSSSKEHKSSS 79
Query: 549 RDYEQRKQGHQGSHHHGGD-ERGTDQENHRSHASTS-PERHRSRSRSHEHRVHHEKQ--- 603
RD ++ K S H D ER +HRS +S+S ++ S S H+ H K+
Sbjct: 80 RDKDRHKSSSSSSKHRDKDKERDGSSNSHRSGSSSSHKDKDGSSSSKHKSSSGHHKRSSK 139
Query: 604 -------------DYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHDI 650
S R K ++SSR K+R + +HK+ +S S S+S S SR H
Sbjct: 140 DKERRDKDKDRGSSSSSRHKSSSSSRDKER-SSSSHKSSSSSSSSKSKHSSSR----HSS 194
Query: 651 SSDDK--------YIKPD 660
SS K ++KP+
Sbjct: 195 SSSSKDHPSYDGVFVKPE 212
Score = 37.7 bits (86), Expect = 0.097
Identities = 24/72 (33%), Positives = 32/72 (44%), Gaps = 2/72 (2%)
Query: 591 SRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEH-D 649
+ H H HH S K+ +SS+ K R + HK+ S S S+ S SR H
Sbjct: 29 TNGHGHH-HHHHSSSSSSSKHKSSSKDKHRDREREHKSSNSSSSSKEHKSSSRDKDRHKS 87
Query: 650 ISSDDKYIKPDK 661
SS K+ DK
Sbjct: 88 SSSSSKHRDKDK 99
>PCLO_HUMAN (Q9Y6V0) Piccolo protein (Aczonin)
Length = 5183
Score = 55.8 bits (133), Expect = 3e-07
Identities = 46/132 (34%), Positives = 52/132 (38%), Gaps = 18/132 (13%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPP---PPLNLN-----TTLSSLTNLLTF--S 71
PPP PPP PPP PPP PPP P P P L TT + L + +T +
Sbjct: 2336 PPPPPPPPPPPPPPPPPPPPPPLPPPTSPKPTILPKKKLTVASPVTTATPLFDAVTTLET 2395
Query: 72 TQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSP--RPLPHIDHLLTSLS 129
T +L + P T P P P L H PS P P P I L
Sbjct: 2396 TAVLRSNGLPVTRICTTAPPPVPPKPSSIPSGLVFTHRPEPSKPPIAPKPVIPQL----- 2450
Query: 130 YPKTLQNPTSIH 141
P T Q PT IH
Sbjct: 2451 -PTTTQKPTDIH 2461
Score = 35.8 bits (81), Expect = 0.37
Identities = 50/210 (23%), Positives = 80/210 (37%), Gaps = 35/210 (16%)
Query: 19 TMNPPPAQLPPPSQLPPPL--SPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILS 76
T PPP P PS +P L + P+ PP+ P P L TT T++ T +
Sbjct: 2411 TTAPPPVP-PKPSSIPSGLVFTHRPEPSKPPIAPKPVIPQLPTTTQKPTDIHPKPTGLSL 2469
Query: 77 TTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPHIDHLLTSLSYPKTLQN 136
T+ + T NL+ + ++ +P P+ L P S + P LT TL +
Sbjct: 2470 TS----SMTLNLVT---SADYKLPSPTSPL----SPHSNKSSPRFSKSLTETYVVITLPS 2518
Query: 137 PTSIHLSSYLDFTNFFYNNCPGVVTFSDANSVAQTATFHLP--DFLSRECSTVCSPTPNH 194
PG T S A+ + P D +S E P
Sbjct: 2519 E-------------------PGTPTDSSASQAITSWPLGSPSKDLVSVEPVFSVVPPVTA 2559
Query: 195 KPSPIVPSEYYYITREIESWNNFPITYSNS 224
PI + +YI+ +++++ P+T +S
Sbjct: 2560 VEIPISSEQTFYISGALQTFSATPVTAPSS 2589
Score = 35.4 bits (80), Expect = 0.48
Identities = 18/35 (51%), Positives = 18/35 (51%), Gaps = 1/35 (2%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPP 52
P PAQ PPSQ P PPPQQ P P P P
Sbjct: 434 PGPTKTPAQTKPPSQQPGSTKPPPQQ-PGPAKPSP 467
Score = 33.1 bits (74), Expect = 2.4
Identities = 19/67 (28%), Positives = 25/67 (36%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
P+ + PP Q P P + P + P P PP P L L+ + Q T
Sbjct: 299 PSPSKPPIQQPTPGKPPAQQPGHEKSQPGPAKPPAQPSGLTKPLAQQPGTVKPPVQPPGT 358
Query: 78 TPQPQTP 84
T P P
Sbjct: 359 TKPPAQP 365
Score = 31.2 bits (69), Expect = 9.1
Identities = 17/38 (44%), Positives = 18/38 (46%), Gaps = 5/38 (13%)
Query: 18 PTMNPPPAQLP----PPSQLPPPLSPPPQQHPPPLLPP 51
P PP+Q P PP Q P P P PQQ P PP
Sbjct: 440 PAQTKPPSQQPGSTKPPPQQPGPAKPSPQQ-PGSTKPP 476
>BRD4_MOUSE (Q9ESU6) Bromodomain-containing protein 4 (Mitotic
chromosome-associated protein) (MCAP)
Length = 1400
Score = 53.9 bits (128), Expect = 1e-06
Identities = 47/132 (35%), Positives = 56/132 (41%), Gaps = 19/132 (14%)
Query: 16 H*PTMNPPPA----QLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTL----SSLTNL 67
H P M P PA Q PPP Q PPP PP QQ P PPPPP T SS
Sbjct: 743 HHPQMQPAPAPVPQQPPPPPQQPPPPPPPQQQQQQPPPPPPPPSMPQQTAPAMKSSPPPF 802
Query: 68 LTFSTQILSTTPQPQTPTTNLIPC--LFNPNHLIPPPSLFLHHLRCPSSPRPLPHID-HL 124
+T +L +PQ P + P P +P P L H + P P PH++ H
Sbjct: 803 ITAQVPVL----EPQLPGSVFDPIGHFTQPILHLPQPELPPHLPQPPEHSTP-PHLNQHA 857
Query: 125 LTSLSYPKTLQN 136
+ S P L N
Sbjct: 858 VVS---PPALHN 866
Score = 39.7 bits (91), Expect = 0.026
Identities = 39/147 (26%), Positives = 53/147 (35%), Gaps = 26/147 (17%)
Query: 23 PPAQLPPP-------SQLPPP--LSPPPQQHPPPL---LPPPPPLNLNTTLSSLTNLLTF 70
PP LPPP QL P PPPQ PPP PPP P++L + + F
Sbjct: 956 PPPPLPPPPHPSVQQQQLQPQPPPPPPPQPQPPPQQQHQPPPRPVHLPS--------MPF 1007
Query: 71 STQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPHIDHLLTSLSY 130
S I P P T+ P P P + H P + P+ S +
Sbjct: 1008 SAHIQQPPPPPGQQPTHPPPGQQPPPPQPAKPQQVIQHHPSPRHHKSDPY------SAGH 1061
Query: 131 PKTLQNPTSIHLSSYLDFTNFFYNNCP 157
+ +P IH F + + + P
Sbjct: 1062 LREAPSPLMIHSPQMPQFQSLTHQSPP 1088
Score = 37.4 bits (85), Expect = 0.13
Identities = 28/92 (30%), Positives = 34/92 (36%), Gaps = 25/92 (27%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTPQP 81
PP + PP L P PPQ PPP P P P+ + I++TTPQP
Sbjct: 237 PPVMTMVPPQPLQTPSPVPPQPPPPP-APVPQPVQSH-------------PPIIATTPQP 282
Query: 82 -----------QTPTTNLIPCLFNPNHLIPPP 102
T T I + P L P P
Sbjct: 283 VKTKKGVKRKADTTTPTTIDPIHEPPSLAPEP 314
Score = 36.6 bits (83), Expect = 0.22
Identities = 22/70 (31%), Positives = 32/70 (45%), Gaps = 8/70 (11%)
Query: 19 TMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFST-----Q 73
T +P P Q PPP P P+ P Q HPP + P P+ + + T +T +
Sbjct: 250 TPSPVPPQPPPP---PAPVPQPVQSHPPIIATTPQPVKTKKGVKRKADTTTPTTIDPIHE 306
Query: 74 ILSTTPQPQT 83
S P+P+T
Sbjct: 307 PPSLAPEPKT 316
Score = 36.6 bits (83), Expect = 0.22
Identities = 32/104 (30%), Positives = 40/104 (37%), Gaps = 11/104 (10%)
Query: 25 AQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLN-----LNTTLSSLTNLLTFSTQILSTTP 79
A LPP PP +SP Q PPLLP PP + L L + Q+
Sbjct: 880 AALPPKPTRPPAVSPALAQ--PPLLPQPPMVQPPQVLLEDEEPPAPPLTSMQMQLYLQQL 937
Query: 80 QPQTPTTNLIPCL----FNPNHLIPPPSLFLHHLRCPSSPRPLP 119
Q P T L+P + P L PPP + + P P P
Sbjct: 938 QKVQPPTPLLPSVKVQSQPPPPLPPPPHPSVQQQQLQPQPPPPP 981
>BRD4_HUMAN (O60885) Bromodomain-containing protein 4 (HUNK1
protein)
Length = 1362
Score = 53.1 bits (126), Expect = 2e-06
Identities = 40/117 (34%), Positives = 52/117 (44%), Gaps = 6/117 (5%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTF-STQILSTTPQ 80
P P Q PPP Q PPP PP QQ PP PPPP + + ++ F +TQ+ PQ
Sbjct: 752 PVPQQPPPPPQQPPPPPPPQQQQQPPPPPPPPSMPQQAAPAMKSSPPPFIATQVPVLEPQ 811
Query: 81 PQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPHID-HLLTSLSYPKTLQN 136
+ I P +P P L H + P P PH++ H + S P L N
Sbjct: 812 LPGSVFDPIGHFTQPILHLPQPELPPHLPQPPEHSTP-PHLNQHAVVS---PPALHN 864
Score = 41.6 bits (96), Expect = 0.007
Identities = 43/152 (28%), Positives = 54/152 (35%), Gaps = 34/152 (22%)
Query: 18 PTMNPPP--------AQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLT 69
P + PPP Q PPP P P PP QQH PPP P++L +
Sbjct: 956 PPLPPPPHPSVQQQLQQQPPPPPPPQPQPPPQQQHQ----PPPRPVHLQP--------MQ 1003
Query: 70 FSTQILSTTP----QPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPHIDHLL 125
FST I P QP P P P P + HH SPR H
Sbjct: 1004 FSTHIQQPPPPQGQQPPHPPPGQQP---PPPQPAKPQQVIQHH----HSPR---HHKSDP 1053
Query: 126 TSLSYPKTLQNPTSIHLSSYLDFTNFFYNNCP 157
S + + +P IH F + + + P
Sbjct: 1054 YSTGHLREAPSPLMIHSPQMSQFQSLTHQSPP 1085
Score = 37.0 bits (84), Expect = 0.17
Identities = 28/85 (32%), Positives = 37/85 (42%), Gaps = 8/85 (9%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
P M P P P Q PPP+ PPQ PPP P P P+ + + + T + + +
Sbjct: 237 PVMTVVP---PQPLQTPPPV--PPQPQPPP-APAPQPVQSHPPIIAATPQPVKTKKGVKR 290
Query: 78 TPQPQTPTTNLIPCLFNPNHLIPPP 102
TPTT I + P L P P
Sbjct: 291 KADTTTPTT--IDPIHEPPSLPPEP 313
Score = 36.2 bits (82), Expect = 0.28
Identities = 23/72 (31%), Positives = 33/72 (44%), Gaps = 8/72 (11%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPP-QQHPPPLLPPPPPLNLNTTLSSLTNLLTFST---- 72
P PPP +PP Q PP +P P Q HPP + P P+ + + T +T
Sbjct: 246 PLQTPPP--VPPQPQPPPAPAPQPVQSHPPIIAATPQPVKTKKGVKRKADTTTPTTIDPI 303
Query: 73 -QILSTTPQPQT 83
+ S P+P+T
Sbjct: 304 HEPPSLPPEPKT 315
Score = 35.8 bits (81), Expect = 0.37
Identities = 29/101 (28%), Positives = 39/101 (37%), Gaps = 16/101 (15%)
Query: 18 PTMNPPPAQLPP---PSQLPPPL---------SPPPQQHPPPL--LPPPPPLNLNTTLSS 63
P NPPP Q P P+ P + P P Q PPP+ P PPP + S
Sbjct: 211 PQPNPPPVQATPHPFPAVTPDLIVQTPVMTVVPPQPLQTPPPVPPQPQPPPAPAPQPVQS 270
Query: 64 LTNLLTFSTQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSL 104
++ + Q + T + P +P H PPSL
Sbjct: 271 HPPIIAATPQPVKTKKGVKRKADTTTPTTIDPIH--EPPSL 309
Score = 34.3 bits (77), Expect = 1.1
Identities = 35/122 (28%), Positives = 44/122 (35%), Gaps = 32/122 (26%)
Query: 25 AQLPPPSQLPPPLSP----------PPQQHPPPLL-----PPPPPL---NLNTTLSSLTN 66
A LPP PP +SP PP PP +L PP PPL + L L
Sbjct: 878 AALPPKPARPPAVSPALTQTPLLPQPPMAQPPQVLLEDEEPPAPPLTSMQMQLYLQQLQK 937
Query: 67 L-----LTFSTQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPHI 121
+ L S ++ S P P P + P + PPP P P+P P
Sbjct: 938 VQPPTPLLPSVKVQSQPPPPLPPPPH--PSVQQQLQQQPPPP-------PPPQPQPPPQQ 988
Query: 122 DH 123
H
Sbjct: 989 QH 990
Score = 33.5 bits (75), Expect = 1.8
Identities = 21/64 (32%), Positives = 26/64 (39%), Gaps = 1/64 (1%)
Query: 21 NPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTPQ 80
N A PP +Q P P PP Q P P P L + T + ++ T PQ
Sbjct: 198 NTTQASTPPQTQTPQPNPPPVQATPHPFPAVTPDLIVQTPVMTVVPPQPLQTP-PPVPPQ 256
Query: 81 PQTP 84
PQ P
Sbjct: 257 PQPP 260
>GLU2_MAIZE (P04706) Glutelin 2 precursor (Zein-gamma) (27 kDa zein)
(Alcohol-soluble reduced glutelin) (ASG) (Zein ZC2)
Length = 223
Score = 52.4 bits (124), Expect = 4e-06
Identities = 35/99 (35%), Positives = 42/99 (42%), Gaps = 34/99 (34%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTPQP 81
PPP LPPP LPPP+ PP H PP + PPP++L
Sbjct: 31 PPPVHLPPPVHLPPPVHLPPPVHLPPPVHLPPPVHL------------------------ 66
Query: 82 QTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPH 120
P ++ P P HL PPP H+ P PRP PH
Sbjct: 67 -PPPVHVPP----PVHLPPPP---CHYPTQP--PRPQPH 95
Score = 50.4 bits (119), Expect = 1e-05
Identities = 25/65 (38%), Positives = 32/65 (48%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
P PPP LPPP LPPP+ PP H PP + PPP+++ + + TQ
Sbjct: 33 PVHLPPPVHLPPPVHLPPPVHLPPPVHLPPPVHLPPPVHVPPPVHLPPPPCHYPTQPPRP 92
Query: 78 TPQPQ 82
P PQ
Sbjct: 93 QPHPQ 97
Score = 39.7 bits (91), Expect = 0.026
Identities = 20/42 (47%), Positives = 22/42 (51%), Gaps = 5/42 (11%)
Query: 18 PTMNPPPAQLPPPSQLPPPLS-PPPQQH----PPPLLPPPPP 54
P PPP LPPP +PPP+ PPP H PP P P P
Sbjct: 57 PVHLPPPVHLPPPVHVPPPVHLPPPPCHYPTQPPRPQPHPQP 98
Score = 38.9 bits (89), Expect = 0.044
Identities = 22/54 (40%), Positives = 23/54 (41%), Gaps = 11/54 (20%)
Query: 18 PTMNPPPAQLPPPSQLPPP------LSPPPQQHPPP-----LLPPPPPLNLNTT 60
P PPP +PPP LPPP P PQ HP P P P P L T
Sbjct: 63 PVHLPPPVHVPPPVHLPPPPCHYPTQPPRPQPHPQPHPCPCQQPHPSPCQLQGT 116
>YPRO_OWEFU (P21260) Hypothetical proline-rich protein (Fragment)
Length = 141
Score = 52.0 bits (123), Expect = 5e-06
Identities = 23/42 (54%), Positives = 24/42 (56%)
Query: 13 KALH*PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
+ LH T PPP PPP PPP PPP PPP PPPPP
Sbjct: 2 RLLHSLTPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 43
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 9 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 45
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 18 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 54
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 13 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 49
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 11 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 47
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 14 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 50
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 10 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 46
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 12 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 48
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 20 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 56
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 15 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 51
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 16 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 52
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 17 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 53
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 19 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 55
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 21 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 57
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 22 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 58
Score = 33.1 bits (74), Expect = 2.4
Identities = 13/26 (50%), Positives = 15/26 (57%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQ 43
P PPP PPP PPP PPP++
Sbjct: 35 PPPPPPPPPPPPPPPPPPPPPPPPRR 60
>EBN2_EBV (P12978) EBNA-2 nuclear protein
Length = 487
Score = 52.0 bits (123), Expect = 5e-06
Identities = 56/234 (23%), Positives = 77/234 (31%), Gaps = 56/234 (23%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNT------------------ 59
P PPP PPP PPP PPP PPP PPPPP +
Sbjct: 64 PPPPPPPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPQRRDAWTQEPSPLDRDPLGYDVG 123
Query: 60 --TLSSLTNLLTFSTQIL---------------STTPQ---------PQTPTTNLIPCLF 93
L+S +L + I+ T PQ P PT I
Sbjct: 124 HGPLASAMRMLWMANYIVRQSRGDRGLILPQGPQTAPQARLVQPHVPPLRPTAPTILSPL 183
Query: 94 NPNHLIPPPSLFLHHLRCPSSPRPLPHIDHLLTSLSYPKTLQNPTSIHLSSYLDFTNFFY 153
+ L PP L + P+ P PLP +L+ P PT++ + L
Sbjct: 184 SQPRLTPPQPLMMPPR--PTPPTPLPP-----ATLTVPPRPTRPTTLPPTPLLTVL---- 232
Query: 154 NNCPGVVTFSDANSVAQTATFHLPDFLSRECSTVCSPTPNHKPSPIVPSEYYYI 207
P + + + H+PD + +P P P P+ +Y I
Sbjct: 233 -QRPTELQPTPSPPRMHLPVLHVPDQSMHPLTHQSTPNDPDSPEPRSPTVFYNI 285
Score = 50.1 bits (118), Expect = 2e-05
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 59 PPPLPPPPPPPPPPPPPPPPPPPPPPPPPPSPPPPPP 95
Score = 48.5 bits (114), Expect = 5e-05
Identities = 20/37 (54%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P + PPP PPP PPP PPP PP PPPPP
Sbjct: 60 PPLPPPPPPPPPPPPPPPPPPPPPPPPPPSPPPPPPP 96
Score = 47.8 bits (112), Expect = 9e-05
Identities = 21/37 (56%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP PPP PPP PPP PPPPP
Sbjct: 63 PPPPPPPPPPPPPPPPPPPPPPPPPPSPPP--PPPPP 97
Score = 43.1 bits (100), Expect = 0.002
Identities = 17/28 (60%), Positives = 18/28 (63%)
Query: 27 LPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
+PPP PPP PPP PPP PPPPP
Sbjct: 58 VPPPLPPPPPPPPPPPPPPPPPPPPPPP 85
Score = 35.8 bits (81), Expect = 0.37
Identities = 40/131 (30%), Positives = 51/131 (38%), Gaps = 32/131 (24%)
Query: 16 H*PTMNPP-PAQLPPPSQLPPPLSPP-PQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQ 73
H P + P P L P SQ P L+PP P PP PP P
Sbjct: 168 HVPPLRPTAPTILSPLSQ--PRLTPPQPLMMPPRPTPPTP----------------LPPA 209
Query: 74 ILSTTPQPQTPT----TNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPHI-DHLLTSL 128
L+ P+P PT T L+ L P L P PS HL P+ H+ D + L
Sbjct: 210 TLTVPPRPTRPTTLPPTPLLTVLQRPTELQPTPSPPRMHL-------PVLHVPDQSMHPL 262
Query: 129 SYPKTLQNPTS 139
++ T +P S
Sbjct: 263 THQSTPNDPDS 273
Score = 32.3 bits (72), Expect = 4.1
Identities = 17/58 (29%), Positives = 24/58 (41%), Gaps = 7/58 (12%)
Query: 4 HFKILELKNKALH*PTMNPPPAQLPPPSQLPP-------PLSPPPQQHPPPLLPPPPP 54
H +L + ++++H T P P P P+ PP Q PPP P PP
Sbjct: 248 HLPVLHVPDQSMHPLTHQSTPNDPDSPEPRSPTVFYNIPPMPLPPSQLPPPAAPAQPP 305
>GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor
(Hydroxyproline-rich glycoprotein 1)
Length = 555
Score = 51.6 bits (122), Expect = 6e-06
Identities = 37/109 (33%), Positives = 46/109 (41%), Gaps = 17/109 (15%)
Query: 18 PTMNPPPAQLPP----PSQLPP---PLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTF 70
P+ PP+ +PP PS PP P +PPP PPP PP PP NT +
Sbjct: 236 PSPPAPPSPVPPSPAPPSPAPPSPKPPAPPPPPSPPPPPPPRPPFPANTPMPP------- 288
Query: 71 STQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
S +P P TP T P +P +PP + P SP P P
Sbjct: 289 SPPSPPPSPAPPTPPTPPSP---SPPSPVPPSPAPVPPSPAPPSPAPSP 334
Score = 39.7 bits (91), Expect = 0.026
Identities = 31/98 (31%), Positives = 37/98 (37%), Gaps = 17/98 (17%)
Query: 22 PPPAQLPPPSQLPPPLSP--PPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTP 79
P PA PPS PP SP PP PP PP PP + S P
Sbjct: 114 PSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSPAPPSPS---------------PP 158
Query: 80 QPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRP 117
P +P+ + P P+ P PS + P SP P
Sbjct: 159 VPPSPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAP 196
Score = 37.7 bits (86), Expect = 0.097
Identities = 25/74 (33%), Positives = 33/74 (43%), Gaps = 2/74 (2%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSP-PPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILS 76
P P P PPS +PP +P PP PP P PPP T S + + S
Sbjct: 299 PPTPPTPPSPSPPSPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSPSPSPSPSPS 358
Query: 77 TTPQPQTPTTNLIP 90
+P P +P+ + IP
Sbjct: 359 PSPSP-SPSPSPIP 371
Score = 37.4 bits (85), Expect = 0.13
Identities = 31/105 (29%), Positives = 37/105 (34%), Gaps = 35/105 (33%)
Query: 18 PTMNPP-PAQLPPPSQLPPPLSP--PPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQI 74
P+ +PP P PPS PP SP PP PP PP PP
Sbjct: 161 PSPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPP-------------------- 200
Query: 75 LSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
+P P +P + P P+ P P PS P P P
Sbjct: 201 ---SPAPPSPAPPVPPSPAPPSPPSPAP---------PSPPSPAP 233
Score = 35.0 bits (79), Expect = 0.63
Identities = 31/104 (29%), Positives = 36/104 (33%), Gaps = 27/104 (25%)
Query: 18 PTMNPP-PAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILS 76
P+ PP PA PPS PP +PP P P P P P
Sbjct: 66 PSPGPPSPAPPSPPSPAPPSPAPPSPAPPSPAPPSPAP-----------------PSPAP 108
Query: 77 TTPQPQTPTTNLIPCLFNPNHLIP-PPSLFLHHLRCPSSPRPLP 119
+P P +P P P+ P PPS PS P P P
Sbjct: 109 PSPAPPSPAPPSPPSPAPPSPSPPAPPS--------PSPPSPAP 144
Score = 34.7 bits (78), Expect = 0.82
Identities = 26/102 (25%), Positives = 36/102 (34%), Gaps = 23/102 (22%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
P PP+ PP P P SP P PP+ P P P + +
Sbjct: 168 PPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAPPS--------------PAPPVPP 213
Query: 78 TPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
+P P +P + P +P P P P+ P P+P
Sbjct: 214 SPAPPSPPSPAPPSPPSPAPPSPSP---------PAPPSPVP 246
Score = 33.9 bits (76), Expect = 1.4
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPP 53
PT +P P+ P PS P P SP P P P+ P P
Sbjct: 341 PTPSPSPSPSPSPSPSPSP-SPSPSPSPSPIPSPSP 375
Score = 33.5 bits (75), Expect = 1.8
Identities = 31/104 (29%), Positives = 35/104 (32%), Gaps = 6/104 (5%)
Query: 18 PTMNPPPAQLPPPSQLPP----PLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQ 73
P + P PA PPS PP P P P PP PP P + S
Sbjct: 209 PPVPPSPAPPSPPSPAPPSPPSPAPPSPSPPAPPSPVPPSPAPPSPAPPSPKPPAPPPPP 268
Query: 74 ILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRP 117
P P+ P P P+ PPPS P SP P
Sbjct: 269 SPPPPPPPRPPFPANTP--MPPSPPSPPPSPAPPTPPTPPSPSP 310
Score = 33.5 bits (75), Expect = 1.8
Identities = 16/38 (42%), Positives = 18/38 (47%), Gaps = 4/38 (10%)
Query: 21 NPPPAQLPPPSQLPPPLSPP----PQQHPPPLLPPPPP 54
+P P PPS PP +PP P PP PP PP
Sbjct: 42 SPAPPSPAPPSPAPPSPAPPSPAPPSPGPPSPAPPSPP 79
Score = 31.6 bits (70), Expect = 6.9
Identities = 14/36 (38%), Positives = 16/36 (43%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPP 53
P+ PP P P+ PP PP PP PP P
Sbjct: 61 PSPAPPSPGPPSPAPPSPPSPAPPSPAPPSPAPPSP 96
Score = 31.6 bits (70), Expect = 6.9
Identities = 26/99 (26%), Positives = 34/99 (34%), Gaps = 23/99 (23%)
Query: 21 NPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTPQ 80
N PP+ PP S PP +PP P P P P P +P
Sbjct: 38 NCPPSPAPP-SPAPPSPAPPSPAPPSPAPPSPGP----------------------PSPA 74
Query: 81 PQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
P +P + P P+ P P+ P+ P P P
Sbjct: 75 PPSPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAP 113
>SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor
(SSG 185)
Length = 485
Score = 51.2 bits (121), Expect = 8e-06
Identities = 21/37 (56%), Positives = 22/37 (58%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P+ PPP PPP PPP PPP PPP PPPPP
Sbjct: 258 PSPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPP 294
Score = 51.2 bits (121), Expect = 8e-06
Identities = 21/33 (63%), Positives = 21/33 (63%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
PPP PPP PPP SPPP PPP PPPPP
Sbjct: 261 PPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPP 293
Score = 49.7 bits (117), Expect = 2e-05
Identities = 34/102 (33%), Positives = 41/102 (39%), Gaps = 36/102 (35%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
P+ +PPP PPP PPP PPP PPP PPPPP
Sbjct: 256 PSPSPPPPPPPPP---PPPPPPPPSPPPPPPPPPPPP----------------------P 290
Query: 78 TPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
P P +P+ P +P+ +PPP PS P LP
Sbjct: 291 PPPPPSPSPPRKPP--SPSPPVPPP---------PSPPSVLP 321
Score = 48.5 bits (114), Expect = 5e-05
Identities = 20/37 (54%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP+ PPP PPP PPP PP PPPPP
Sbjct: 250 PPSPPPPSPSPPPPPPPPPPPPPPPPPSPPPPPPPPP 286
Score = 48.5 bits (114), Expect = 5e-05
Identities = 20/37 (54%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P+ PP PPP PPP PPP PPP PPPPP
Sbjct: 251 PSPPPPSPSPPPPPPPPPPPPPPPPPSPPPPPPPPPP 287
Score = 47.8 bits (112), Expect = 9e-05
Identities = 20/37 (54%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P+ PP P PS PPP PPP PPP PPPPP
Sbjct: 246 PSPRPPSPPPPSPSPPPPPPPPPPPPPPPPPSPPPPP 282
Score = 47.4 bits (111), Expect = 1e-04
Identities = 20/37 (54%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P +P P PPPS PPP PPP PPP P PPP
Sbjct: 244 PPPSPRPPSPPPPSPSPPPPPPPPPPPPPPPPPSPPP 280
Score = 45.1 bits (105), Expect = 6e-04
Identities = 18/34 (52%), Positives = 20/34 (57%)
Query: 21 NPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
+PPP+ PP P P PPP PPP PPPPP
Sbjct: 243 SPPPSPRPPSPPPPSPSPPPPPPPPPPPPPPPPP 276
Score = 42.7 bits (99), Expect = 0.003
Identities = 19/44 (43%), Positives = 24/44 (54%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLT 65
PPP PPP PPP PP++ P P P PPP + + L + T
Sbjct: 281 PPPPPPPPPPPPPPPSPSPPRKPPSPSPPVPPPPSPPSVLPAAT 324
Score = 39.3 bits (90), Expect = 0.033
Identities = 20/37 (54%), Positives = 21/37 (56%), Gaps = 3/37 (8%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
PT + P PPPS PP SPPP PP PPPPP
Sbjct: 235 PTASSRPPS-PPPSPRPP--SPPPPSPSPPPPPPPPP 268
Score = 38.5 bits (88), Expect = 0.057
Identities = 20/40 (50%), Positives = 20/40 (50%), Gaps = 5/40 (12%)
Query: 18 PTMNPPPAQLPPPSQLP---PPLSPPPQQHPPPLLPPPPP 54
P P A PPS P PP PPP PPP PPPPP
Sbjct: 230 PPSPQPTASSRPPSPPPSPRPPSPPPPSPSPPP--PPPPP 267
Score = 35.0 bits (79), Expect = 0.63
Identities = 20/44 (45%), Positives = 22/44 (49%), Gaps = 9/44 (20%)
Query: 18 PTMNP-PPAQLPPPSQLPPPLSPPPQQHPP------PLLPPPPP 54
P +P PP+ P S PP SPPP PP P PPPPP
Sbjct: 224 PNNSPLPPSPQPTASSRPP--SPPPSPRPPSPPPPSPSPPPPPP 265
>ACRO_RABIT (P48038) Acrosin precursor (EC 3.4.21.10)
Length = 431
Score = 51.2 bits (121), Expect = 8e-06
Identities = 21/33 (63%), Positives = 21/33 (63%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
PP AQ PPP PPP PPP PPP PPPPP
Sbjct: 346 PPAAQAPPPPPPPPPPPPPPPPPPPPPPPPPPP 378
Score = 47.0 bits (110), Expect = 2e-04
Identities = 21/37 (56%), Positives = 23/37 (61%), Gaps = 1/37 (2%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P +P P Q PP +Q PPP PPP PPP PPPPP
Sbjct: 337 PHPHPRPPQ-PPAAQAPPPPPPPPPPPPPPPPPPPPP 372
Score = 43.5 bits (101), Expect = 0.002
Identities = 20/47 (42%), Positives = 22/47 (46%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLL 68
PPP PPP PPP PPP PP PP L+ L L +L
Sbjct: 354 PPPPPPPPPPPPPPPPPPPPPPPPPASTKPPQALSFAKRLQQLVEVL 400
Score = 40.8 bits (94), Expect = 0.011
Identities = 18/37 (48%), Positives = 20/37 (53%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P +P P P P Q P +PPP PPP PPPPP
Sbjct: 331 PHPHPHPHPHPRPPQPPAAQAPPPPPPPPPPPPPPPP 367
Score = 38.1 bits (87), Expect = 0.074
Identities = 17/33 (51%), Positives = 17/33 (51%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P P P P P P PP Q PPP PPPPP
Sbjct: 329 PHPHPHPHPHPHPRPPQPPAAQAPPPPPPPPPP 361
>MLL4_HUMAN (Q9UMN6) Myeloid/lymphoid or mixed-lineage leukemia
protein 4 (Trithorax homolog 2)
Length = 2715
Score = 50.4 bits (119), Expect = 1e-05
Identities = 20/33 (60%), Positives = 20/33 (60%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
PPP PP PPPL PP PPPL PPPPP
Sbjct: 402 PPPPLTPPAPSPPPPLPPPSTSPPPPLCPPPPP 434
Score = 47.0 bits (110), Expect = 2e-04
Identities = 21/39 (53%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQ--HPPPLLPPPPP 54
P +PPP PP + PPPL PPP PPPL PPPP
Sbjct: 409 PAPSPPPPLPPPSTSPPPPLCPPPPPPVSPPPLPSPPPP 447
Score = 41.6 bits (96), Expect = 0.007
Identities = 23/46 (50%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query: 11 KNKALH*PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLP-PPPPL 55
K +A P PPA PPP PP SPPP PPP P PPPL
Sbjct: 396 KEEAKLPPPPLTPPAPSPPPPLPPPSTSPPPPLCPPPPPPVSPPPL 441
Score = 41.2 bits (95), Expect = 0.009
Identities = 36/127 (28%), Positives = 47/127 (36%), Gaps = 31/127 (24%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQIL-- 75
P + PP PP Q P P+ PP+ PP P P P + L T T T+ L
Sbjct: 561 PVLRPPITTSPPVPQEPAPVPSPPRAPTPPSTPVPLPEKRRSILREPTFRWTSLTRELPP 620
Query: 76 ---------STTPQPQTPTTNLIPCL-----FNPNH---------LIPPPSLFLHHLRCP 112
+ +P P T++ P L F P+ L PPP L P
Sbjct: 621 PPPAPPPPPAPSPPPAPATSSRRPLLLRAPQFTPSEAHLKIYESVLTPPP------LGAP 674
Query: 113 SSPRPLP 119
+P P P
Sbjct: 675 EAPEPEP 681
Score = 36.2 bits (82), Expect = 0.28
Identities = 19/43 (44%), Positives = 25/43 (57%), Gaps = 10/43 (23%)
Query: 18 PTMNPPPAQLPPPSQ--LPPPL-SPPP-------QQHPPPLLP 50
P+ +PPP PPP PPPL SPPP ++ PPP++P
Sbjct: 420 PSTSPPPPLCPPPPPPVSPPPLPSPPPPPAQEEQEESPPPVVP 462
Score = 32.3 bits (72), Expect = 4.1
Identities = 23/58 (39%), Positives = 25/58 (42%), Gaps = 19/58 (32%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQH----PPPLL------------PPPPPLNLNTTLSS 63
PPP + PP LPP +SP P PLL PPPPP L LSS
Sbjct: 2212 PPPVKQPP---LPPTISPTAPTSWTLPPGPLLGVLPVVGVVRPAPPPPPPPLTLVLSS 2266
>CYCK_MOUSE (O88874) Cyclin K
Length = 554
Score = 50.4 bits (119), Expect = 1e-05
Identities = 32/102 (31%), Positives = 48/102 (46%), Gaps = 12/102 (11%)
Query: 19 TMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLT-FSTQILST 77
T + P Q+PPP+ P PPP H P PPPPP + T +S+ ++ ++ Q L +
Sbjct: 395 TSDLPKVQIPPPAHPAPVHQPPPLPHRP---PPPPPSSYMTGMSTTSSYMSGEGYQSLQS 451
Query: 78 TPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
+ + P+ +P P IPPP+ P P P P
Sbjct: 452 MMKTEGPSYGALPPASFPPPTIPPPT--------PGYPPPPP 485
Score = 38.9 bits (89), Expect = 0.044
Identities = 19/38 (50%), Positives = 23/38 (60%), Gaps = 3/38 (7%)
Query: 21 NPPPAQLPPPSQLPPPLSPPPQQHP-PPLLPPP--PPL 55
+PPP PP+ PPP PP Q P PP +PPP PP+
Sbjct: 507 HPPPGLGLPPASYPPPAVPPGGQPPVPPPIPPPGMPPV 544
Score = 38.5 bits (88), Expect = 0.057
Identities = 18/33 (54%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Query: 23 PPAQLPPPSQLPP-PLSPPPQQHPPPLLPPPPP 54
PPA PPP+ PP P PPP P PPPPP
Sbjct: 464 PPASFPPPTIPPPTPGYPPPPPTYNPNFPPPPP 496
Score = 37.7 bits (86), Expect = 0.097
Identities = 21/52 (40%), Positives = 25/52 (47%), Gaps = 14/52 (26%)
Query: 18 PTMNP---PPAQLPPPSQLPPPLSP-----PPQQHPPPLLPP------PPPL 55
PT NP PP PP+ PP P PP +PPP +PP PPP+
Sbjct: 485 PTYNPNFPPPPPRLPPTHAVPPHPPPGLGLPPASYPPPAVPPGGQPPVPPPI 536
Score = 36.2 bits (82), Expect = 0.28
Identities = 19/42 (45%), Positives = 22/42 (52%), Gaps = 2/42 (4%)
Query: 18 PTMNPP-PAQLPPPSQLPPPLSPPPQQHPPP-LLPPPPPLNL 57
PT+ PP P PPP P PPP + PP +PP PP L
Sbjct: 471 PTIPPPTPGYPPPPPTYNPNFPPPPPRLPPTHAVPPHPPPGL 512
Score = 34.7 bits (78), Expect = 0.82
Identities = 15/34 (44%), Positives = 18/34 (52%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPP 51
P + PPA PPP+ P P P PPP +PP
Sbjct: 510 PGLGLPPASYPPPAVPPGGQPPVPPPIPPPGMPP 543
Score = 34.7 bits (78), Expect = 0.82
Identities = 25/77 (32%), Positives = 31/77 (39%), Gaps = 17/77 (22%)
Query: 28 PPPSQLPPPLSPPPQQHPP-PLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTPQPQTPTT 86
P LPP PPP PP P PPPPP T++ P P+ P T
Sbjct: 458 PSYGALPPASFPPPTIPPPTPGYPPPPP--------------TYNPNF--PPPPPRLPPT 501
Query: 87 NLIPCLFNPNHLIPPPS 103
+ +P P +PP S
Sbjct: 502 HAVPPHPPPGLGLPPAS 518
>LECT_SOLTU (Q9S8M0) Chitin-binding lectin 1 precursor (PL-I)
Length = 323
Score = 49.7 bits (117), Expect = 2e-05
Identities = 22/40 (55%), Positives = 23/40 (57%), Gaps = 3/40 (7%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPP---PQQHPPPLLPPPPP 54
P PPP+ PPP PPP SPP P PPP PPPPP
Sbjct: 166 PPSPPPPSPPPPPPPSPPPPSPPPPSPSPPPPPASPPPPP 205
Score = 44.3 bits (103), Expect = 0.001
Identities = 22/46 (47%), Positives = 22/46 (47%), Gaps = 9/46 (19%)
Query: 18 PTMNPPPAQLPPPS---------QLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPPS PPP SPPP PPP PPPP
Sbjct: 152 PPPPPPPPSPPPPSPPSPPPPSPPPPPPPSPPPPSPPPPSPSPPPP 197
Score = 43.9 bits (102), Expect = 0.001
Identities = 22/38 (57%), Positives = 23/38 (59%), Gaps = 4/38 (10%)
Query: 18 PTMNPPPAQLPPPSQLPP-PLSPPPQQHPPPLLPPPPP 54
P+ PPP PPPS PP P SPPP PPP P PPP
Sbjct: 150 PSPPPPP---PPPSPPPPSPPSPPPPSPPPPPPPSPPP 184
Score = 42.0 bits (97), Expect = 0.005
Identities = 21/37 (56%), Positives = 22/37 (58%), Gaps = 5/37 (13%)
Query: 18 PTMNPPPAQLPPPSQL--PPPLSPPPQQHPPPLLPPP 52
P +PPP PPPS PPP SPPP PPP LP P
Sbjct: 178 PPPSPPPPSPPPPSPSPPPPPASPPP---PPPALPYP 211
Score = 40.4 bits (93), Expect = 0.015
Identities = 20/35 (57%), Positives = 20/35 (57%), Gaps = 4/35 (11%)
Query: 22 PPPAQLPPPSQLPP-PLSPPPQQHPPPLLPPPPPL 55
PPP PPPS PP P PPP PP PPPP L
Sbjct: 177 PPPPSPPPPSPPPPSPSPPPPPASPP---PPPPAL 208
Score = 40.0 bits (92), Expect = 0.020
Identities = 18/31 (58%), Positives = 19/31 (61%), Gaps = 2/31 (6%)
Query: 26 QLPPPSQLPPPLSPPPQQ--HPPPLLPPPPP 54
+LP P PPP SPPP PPP PPPPP
Sbjct: 148 KLPSPPPPPPPPSPPPPSPPSPPPPSPPPPP 178
>Y553_HUMAN (Q9UKJ3) Hypothetical protein KIAA0553
Length = 1089
Score = 49.3 bits (116), Expect = 3e-05
Identities = 61/258 (23%), Positives = 90/258 (34%), Gaps = 42/258 (16%)
Query: 416 HQQHSHHQETQKEKTREELLAEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIKLA 475
H++ S H+ K T E+ E K ++ R +K+N+S+ A
Sbjct: 268 HKKSSKHKRKPKADTEEKSSKAESGEKSKKRKKRKRKKNKSSAP---------------A 312
Query: 476 AGVKSPVNVSEDSGMPPPPPKLL----SSHTISTEANNSREVSHDSPAVTISNPDHHEQQ 531
+ P SG P PP + S S A D S+ DH ++
Sbjct: 313 DSERGPKPEPPGSGSPAPPRRRRRAQDDSQRRSLPAEEGSSGKKDEGGGGSSSQDHGGRK 372
Query: 532 SHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQENHRSH------------ 579
S T R + H+ S GDE D +HR H
Sbjct: 373 HKGELPPSSCQRRAGTKR---SSRSSHR-SQPSSGDEDSDDASSHRLHQKSPSQYSEEEE 428
Query: 580 -ASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSS-RTKDRWQNDTHKNHTSDSFS-R 636
+ E RSRSRS H S R+ Y++SS + D+ ++++ DS+S
Sbjct: 429 EEDSGSEHSRSRSRSGRRHSSHR----SSRRSYSSSSDASSDQSCYSRQRSYSDDSYSDY 484
Query: 637 SDPSESRGVGEHDISSDD 654
SD S HD D
Sbjct: 485 SDRSRRHSKRSHDSDDSD 502
Score = 40.4 bits (93), Expect = 0.015
Identities = 47/179 (26%), Positives = 72/179 (39%), Gaps = 26/179 (14%)
Query: 479 KSPVNVSE-----DSGMPPPPPKLLSSHTISTEANNSREVSHDSPAVT----ISNPDHHE 529
KSP SE DSG + S S+ ++ R S S A + S +
Sbjct: 418 KSPSQYSEEEEEEDSGSEHSRSRSRSGRRHSSHRSSRRSYSSSSDASSDQSCYSRQRSYS 477
Query: 530 QQSHTNYSDKS----KVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQENHRSHASTSPE 585
S+++YSD+S K HD+ DY K H+ H + + +++ S
Sbjct: 478 DDSYSDYSDRSRRHSKRSHDSDDSDYASSK--HRSKRH----KYSSSDDDYSLSCS---- 527
Query: 586 RHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDT-HKNHTSDSFSRSDPSESR 643
+SRSRS H + R + SR+K R ++ T H S S+SR +R
Sbjct: 528 --QSRSRSRSHTRERSRSRGRSRSSSCSRSRSKRRSRSTTAHSWQRSRSYSRDRSRSTR 584
>EXTN_TOBAC (P13983) Extensin precursor (Cell wall
hydroxyproline-rich glycoprotein)
Length = 620
Score = 49.3 bits (116), Expect = 3e-05
Identities = 40/127 (31%), Positives = 50/127 (38%), Gaps = 23/127 (18%)
Query: 18 PTMNPPPAQL--PPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQIL 75
PT +PPP PPP+ PP PP PPP PPPP + + + Q
Sbjct: 407 PTYSPPPPTYSPPPPTYAQPPPLPPTYSPPPPAYSPPPPPTYSPPPPTYSPPPPAYAQ-- 464
Query: 76 STTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPHIDHLLTSLSYPKTLQ 135
P P PT + P ++P PPPS P P P + L + S P
Sbjct: 465 ---PPPPPPTYSPPPPAYSP----PPPS--------PIYSPPPPQVQPLPPTFSPPP--- 506
Query: 136 NPTSIHL 142
P IHL
Sbjct: 507 -PRRIHL 512
Score = 48.1 bits (113), Expect = 7e-05
Identities = 38/100 (38%), Positives = 42/100 (42%), Gaps = 20/100 (20%)
Query: 22 PPPAQLPPPSQLPPPL-SPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTPQ 80
PPP PP P P+ SP PQ PPP PPPP ++ T S S Q
Sbjct: 179 PPPTYAQPP---PTPIYSPSPQVQPPPTYSPPPPTHVQPTPSP-----------PSRGHQ 224
Query: 81 PQTPTTNLIPCLFNPNHLIPPPSLFLHHLR-CPSSPRPLP 119
PQ PT P P H PP+ LR P SPR P
Sbjct: 225 PQPPTHRHAP----PTHRHAPPTHQPSPLRHLPPSPRRQP 260
Score = 47.4 bits (111), Expect = 1e-04
Identities = 42/134 (31%), Positives = 52/134 (38%), Gaps = 23/134 (17%)
Query: 18 PTMNPPPAQL--PPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLL--TFS-- 71
PT +PPP PPP+ PP PP PPP PPPP + + L TFS
Sbjct: 446 PTYSPPPPTYSPPPPAYAQPPPPPPTYSPPPPAYSPPPPSPIYSPPPPQVQPLPPTFSPP 505
Query: 72 -------TQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPHIDHL 124
P+P TPT P +P PPP +H SP P PH
Sbjct: 506 PPRRIHLPPPPHRQPRPPTPTYGQPP---SPPTFSPPPPRQIH------SPPP-PHWQPR 555
Query: 125 LTSLSYPKTLQNPT 138
+ +Y + PT
Sbjct: 556 TPTPTYGQPPSPPT 569
Score = 46.2 bits (108), Expect = 3e-04
Identities = 36/109 (33%), Positives = 48/109 (44%), Gaps = 18/109 (16%)
Query: 18 PTMNPPPAQL--PPPSQL----PPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFS 71
PT +PPP PPPS + PP SP P P P PPPP + + T+S
Sbjct: 280 PTYSPPPPTYSPPPPSPIYSPPPPAYSPSPPPTPTPTFSPPPP--------AYSPPPTYS 331
Query: 72 TQILSTTPQPQTPTTNLIPCLFN---PNHLIPPPSLFLHHLRCPSSPRP 117
+ P P +P + P +++ P PPP +L PSSP P
Sbjct: 332 PPPPTYLPLPSSPIYSPPPPVYSPPPPPSYSPPPPTYLPP-PPPSSPPP 379
Score = 43.5 bits (101), Expect = 0.002
Identities = 39/116 (33%), Positives = 46/116 (39%), Gaps = 14/116 (12%)
Query: 18 PTMNPPP-AQLPPPSQLPPPL------SPPPQQHPPPLL-PPPPPLNLNTTLSSLTNLLT 69
PT +PPP A PPP+ PPP S P PPP+ PPPPP + L
Sbjct: 315 PTFSPPPPAYSPPPTYSPPPPTYLPLPSSPIYSPPPPVYSPPPPPSYSPPPPTYLPPPPP 374
Query: 70 FSTQILSTTPQPQT------PTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
S S +P P T P P L P PPP + + P PLP
Sbjct: 375 SSPPPPSFSPPPPTYEQSPPPPPAYSPPLPAPPTYSPPPPTYSPPPPTYAQPPPLP 430
Score = 43.1 bits (100), Expect = 0.002
Identities = 30/92 (32%), Positives = 39/92 (41%), Gaps = 10/92 (10%)
Query: 18 PTMNPPP---AQLPPPSQLPPPLSPPPQQH----PPPLLPPPPPLNLNTTLSSLTNLLTF 70
PT +PPP AQ P PS P SPPP + P P+ PPPP + + T +
Sbjct: 263 PTYSPPPPAYAQSPQPS---PTYSPPPPTYSPPPPSPIYSPPPPAYSPSPPPTPTPTFSP 319
Query: 71 STQILSTTPQPQTPTTNLIPCLFNPNHLIPPP 102
S P P +P +P + PPP
Sbjct: 320 PPPAYSPPPTYSPPPPTYLPLPSSPIYSPPPP 351
Score = 41.6 bits (96), Expect = 0.007
Identities = 40/129 (31%), Positives = 47/129 (36%), Gaps = 27/129 (20%)
Query: 18 PTMNPPPAQL----PPPSQLPPPLSPPPQQHP-PPLLPPPPPLNLNTTLSSLTNLLTFST 72
PT +PPP + PPP + P P +P Q P PP PPPP
Sbjct: 500 PTFSPPPPRRIHLPPPPHRQPRPPTPTYGQPPSPPTFSPPPP-----------------R 542
Query: 73 QILSTTP---QPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPHIDHLLTSLS 129
QI S P QP+TPT P PPP H P +P P S
Sbjct: 543 QIHSPPPPHWQPRTPTPTYGQPPSPPTFSAPPPRQI--HSPPPPHRQPRPPTPTYGQPPS 600
Query: 130 YPKTLQNPT 138
P T P+
Sbjct: 601 PPTTYSPPS 609
Score = 40.4 bits (93), Expect = 0.015
Identities = 37/112 (33%), Positives = 48/112 (42%), Gaps = 23/112 (20%)
Query: 18 PTMNPPPAQLPPPSQL----PPPLSPPPQQH-----PPPLLPPPPPLNLNTTLSSLTNLL 68
PT P P + PPS PP SPPP + P P PPPP T S
Sbjct: 242 PTHQPSPLRHLPPSPRRQPQPPTYSPPPPAYAQSPQPSPTYSPPPP-----TYSPPPPSP 296
Query: 69 TFSTQILSTTPQP---QTPTTNLIPCLFN--PNHLIPPPSLFLHHLRCPSSP 115
+S + +P P TPT + P ++ P + PPP+ +L PSSP
Sbjct: 297 IYSPPPPAYSPSPPPTPTPTFSPPPPAYSPPPTYSPPPPT----YLPLPSSP 344
Score = 37.0 bits (84), Expect = 0.17
Identities = 35/121 (28%), Positives = 47/121 (37%), Gaps = 24/121 (19%)
Query: 18 PTMNPPPAQLPPP--------------SQLPPPL---SPPPQQHPPPLLPPPPPLNLNTT 60
P + PPP PPP Q PP +PP +H PP P P +L +
Sbjct: 196 PQVQPPPTYSPPPPTHVQPTPSPPSRGHQPQPPTHRHAPPTHRHAPPTHQPSPLRHLPPS 255
Query: 61 LSSLTNLLTFS--TQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPL 118
T+S + +PQP +PT + P ++P PPPS SP P
Sbjct: 256 PRRQPQPPTYSPPPPAYAQSPQP-SPTYSPPPPTYSP----PPPSPIYSPPPPAYSPSPP 310
Query: 119 P 119
P
Sbjct: 311 P 311
Score = 36.6 bits (83), Expect = 0.22
Identities = 35/125 (28%), Positives = 49/125 (39%), Gaps = 22/125 (17%)
Query: 21 NPPPAQLPPPSQLPPPLSPPPQQHPPPLLP-----PPPPLNLNTTLSSLTNLLTFSTQIL 75
NPPP+ + PS PP PP H P LP PP P + + S
Sbjct: 106 NPPPSPVISPSHPPPSYGAPPPSHGPGHLPSHGQRPPSPSHGHAPPSG-----------G 154
Query: 76 STTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSP--RPLPHIDHLLTSLSYPKT 133
T P+ Q P ++ P + + PPP+ + + P +P P P + T P T
Sbjct: 155 HTPPRGQHPPSHRRPSPPSRHGHPPPPT----YAQPPPTPIYSPSPQVQPPPTYSPPPPT 210
Query: 134 LQNPT 138
PT
Sbjct: 211 HVQPT 215
Score = 36.2 bits (82), Expect = 0.28
Identities = 32/123 (26%), Positives = 44/123 (35%), Gaps = 37/123 (30%)
Query: 18 PTMNPPPAQLPPPSQLPPPLS----------------PPPQQHPPPLLPPPPPLNLNTTL 61
P +P P PPP+ PPP + PP +H PP PP + + L
Sbjct: 190 PIYSPSPQVQPPPTYSPPPPTHVQPTPSPPSRGHQPQPPTHRHAPPTHRHAPPTHQPSPL 249
Query: 62 SSLTNLLTFSTQILSTTPQPQTPTTNLIPCLF------NPNHLIPPPSLFLHHLRCPSSP 115
L S QPQ PT + P + +P + PPP+ P P
Sbjct: 250 RHLPP---------SPRRQPQPPTYSPPPPAYAQSPQPSPTYSPPPPTY------SPPPP 294
Query: 116 RPL 118
P+
Sbjct: 295 SPI 297
Score = 35.4 bits (80), Expect = 0.48
Identities = 35/107 (32%), Positives = 40/107 (36%), Gaps = 25/107 (23%)
Query: 18 PTMNPPPAQLPPPS----------QLPPPLS-----------PPPQQHPPPLLPPPPPLN 56
P+ PP PPPS Q PP S PP QHPP P PP +
Sbjct: 115 PSHPPPSYGAPPPSHGPGHLPSHGQRPPSPSHGHAPPSGGHTPPRGQHPPSHRRPSPP-S 173
Query: 57 LNTTLSSLTNLLTFSTQILSTTPQPQTPTTNLIPCLFNPNHLIPPPS 103
+ T T I S +PQ Q P T P P H+ P PS
Sbjct: 174 RHGHPPPPTYAQPPPTPIYSPSPQVQPPPTYSPP---PPTHVQPTPS 217
Score = 32.0 bits (71), Expect = 5.3
Identities = 20/48 (41%), Positives = 24/48 (49%), Gaps = 8/48 (16%)
Query: 18 PTMN-PPPAQL---PPPSQLPPPLSPPPQQHPPPLL----PPPPPLNL 57
PT + PPP Q+ PPP + P P +P Q P P P PPP L
Sbjct: 568 PTFSAPPPRQIHSPPPPHRQPRPPTPTYGQPPSPPTTYSPPSPPPYGL 615
>NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75) (NGM-75)
Length = 309
Score = 48.9 bits (115), Expect = 4e-05
Identities = 34/106 (32%), Positives = 39/106 (36%), Gaps = 20/106 (18%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPP---PPPLNLNTTLSSLTNLLTFSTQI 74
P PPP + PPP LPP PPP+ PP PP PPP Q
Sbjct: 75 PEYLPPPHEKPPPEYLPPHEKPPPEYQPPHEKPPHENPPP----------------EHQP 118
Query: 75 LSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPH 120
P P P + P H PPP H + P +P PH
Sbjct: 119 PHEKPPEHQPPHEKPPPEYEPPHEKPPPEYQPPHEKPPPEYQP-PH 163
Score = 47.0 bits (110), Expect = 2e-04
Identities = 18/38 (47%), Positives = 23/38 (60%)
Query: 16 H*PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPP 53
H P PP PPP + PPP+ PP + PPP++ PPP
Sbjct: 240 HQPPHEKPPPVYPPPYEKPPPVYEPPYEKPPPVVYPPP 277
Score = 45.1 bits (105), Expect = 6e-04
Identities = 34/107 (31%), Positives = 41/107 (37%), Gaps = 5/107 (4%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPP---PPLNLNTTLSSLTNLLTFSTQI 74
P PP LPPP + PPP PP + PPP PP PP + Q
Sbjct: 69 PHEKTPPEYLPPPHEKPPPEYLPPHEKPPPEYQPPHEKPPHENPPPEHQPPHEKPPEHQP 128
Query: 75 LSTTPQPQ-TPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPH 120
P P+ P P + P H PPP H + P +P PH
Sbjct: 129 PHEKPPPEYEPPHEKPPPEYQPPHEKPPPEYQPPHEKPPPEYQP-PH 174
Score = 43.9 bits (102), Expect = 0.001
Identities = 16/35 (45%), Positives = 21/35 (59%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPP 52
P PP + PP + PPP+ PPP + PPP+ PP
Sbjct: 231 PPHEKPPPEHQPPHEKPPPVYPPPYEKPPPVYEPP 265
Score = 43.9 bits (102), Expect = 0.001
Identities = 16/35 (45%), Positives = 20/35 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPP 52
P PP + PP + PPP PP + PPP+ PPP
Sbjct: 220 PPHEKPPPEYQPPHEKPPPEHQPPHEKPPPVYPPP 254
Score = 43.5 bits (101), Expect = 0.002
Identities = 20/45 (44%), Positives = 22/45 (48%), Gaps = 4/45 (8%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPP----PPLNLN 58
P PPP PPP + PP PPP + PP PPP PP N
Sbjct: 265 PYEKPPPVVYPPPHEKPPIYEPPPLEKPPVYNPPPYGRYPPSKKN 309
Score = 42.4 bits (98), Expect = 0.004
Identities = 30/106 (28%), Positives = 37/106 (34%), Gaps = 20/106 (18%)
Query: 16 H*PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPP----PPPLNLNTTLSSLTNLLTFS 71
H P PP + PP + PPP PP + PPP P PPP
Sbjct: 126 HQPPHEKPPPEYEPPHEKPPPEYQPPHEKPPPEYQPPHEKPPP----------------E 169
Query: 72 TQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRP 117
Q P P+ + P P H PPP H + P +P
Sbjct: 170 YQPPHEKPPPEHQPPHEKPPEHQPPHEKPPPEYQPPHEKPPPEYQP 215
Score = 42.4 bits (98), Expect = 0.004
Identities = 35/112 (31%), Positives = 42/112 (37%), Gaps = 10/112 (8%)
Query: 18 PTMNPPP----AQLPPPSQLPPP-LSPPPQQHPPPLLPP----PPPLNLNTTLSSLTNLL 68
PT PPP PPP PPP PP ++ PP LPP PPP L
Sbjct: 42 PTYEPPPFYKPPYYPPPVHHPPPEYQPPHEKTPPEYLPPPHEKPPPEYLPPHEKPPPEYQ 101
Query: 69 TFSTQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPH 120
+ P P+ + P P H PPP H + P +P PH
Sbjct: 102 PPHEKPPHENPPPEHQPPHEKPPEHQPPHEKPPPEYEPPHEKPPPEYQP-PH 152
Score = 42.4 bits (98), Expect = 0.004
Identities = 30/103 (29%), Positives = 35/103 (33%), Gaps = 25/103 (24%)
Query: 22 PPPAQLPPPSQLPP----PLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
PPP + PP + PP P PPP HPPP PP T
Sbjct: 35 PPPIEKPPTYEPPPFYKPPYYPPPVHHPPPEYQPPHE---------------------KT 73
Query: 78 TPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPH 120
P+ P P + P H PPP H + P P H
Sbjct: 74 PPEYLPPPHEKPPPEYLPPHEKPPPEYQPPHEKPPHENPPPEH 116
Score = 42.0 bits (97), Expect = 0.005
Identities = 30/108 (27%), Positives = 38/108 (34%), Gaps = 23/108 (21%)
Query: 16 H*PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPP---PPLNLNTTLSSLTNLLTFST 72
H P PP + PP + PPP PPQ+ PP PPP PP
Sbjct: 191 HQPPHEKPPPEYQPPHEKPPPEYQPPQEKPPHEKPPPEYQPP------------------ 232
Query: 73 QILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPH 120
P P P ++ P + PPP + + P P PH
Sbjct: 233 --HEKPPPEHQPPHEKPPPVYPPPYEKPPPVYEPPYEKPPPVVYPPPH 278
Score = 42.0 bits (97), Expect = 0.005
Identities = 20/42 (47%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPP----PPPL 55
P + PPP + PPP PP PPP +PPP P PPPL
Sbjct: 248 PPVYPPPYEKPPPVYEPPYEKPPPVVYPPPHEKPPIYEPPPL 289
Score = 40.8 bits (94), Expect = 0.011
Identities = 17/35 (48%), Positives = 19/35 (53%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPP 52
P + PP + PPP PPP PP PPPL PP
Sbjct: 259 PPVYEPPYEKPPPVVYPPPHEKPPIYEPPPLEKPP 293
>CHD1_MOUSE (P40201) Chromodomain-helicase-DNA-binding protein 1
(CHD-1)
Length = 1711
Score = 48.9 bits (115), Expect = 4e-05
Identities = 43/157 (27%), Positives = 69/157 (43%), Gaps = 12/157 (7%)
Query: 509 NSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDA---TSRDYEQRKQGHQGSHHHG 565
NS + S+ + I NPD + +TN+ D S+ + + S+ ++ K HQG +
Sbjct: 1510 NSDQNSNVATTHVIRNPDMERLKENTNHDDSSRDSYSSDRHLSQYHDHHKDRHQGDSYKK 1569
Query: 566 GDERG---TDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNS--SRTKDR 620
D R + N + H + +R SR + R H K D +++ S KDR
Sbjct: 1570 SDSRKRPYSSFSNGKDHREW--DHYRQDSRYYSDREKHRKLDDHRSREHRPSLEGGLKDR 1627
Query: 621 WQNDTHKNHTSDSFSRSDPSESRGVGEHDISSDDKYI 657
+D H++H SD SD S H S D +Y+
Sbjct: 1628 CHSD-HRSH-SDHRMHSDHRSSSEHTHHKSSRDYRYL 1662
Score = 38.1 bits (87), Expect = 0.074
Identities = 38/138 (27%), Positives = 48/138 (34%), Gaps = 18/138 (13%)
Query: 514 SHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQ 573
S P + SN H + H Y S+ D R+ ++ H+ H E G
Sbjct: 1572 SRKRPYSSFSNGKDHREWDH--YRQDSRYYSD---REKHRKLDDHRSREHRPSLEGGLKD 1626
Query: 574 ENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDS 633
H H S S R S RS HH K + R WQ D H+ +S
Sbjct: 1627 RCHSDHRSHSDHRMHSDHRSSSEHTHH---------KSSRDYRYLSDWQLD-HRAASSGP 1676
Query: 634 FSRSD---PSESRGVGEH 648
S D P SR EH
Sbjct: 1677 RSPLDQRSPYGSRSPFEH 1694
>ACRO_HUMAN (P10323) Acrosin precursor (EC 3.4.21.10)
Length = 421
Score = 48.1 bits (113), Expect = 7e-05
Identities = 21/38 (55%), Positives = 22/38 (57%)
Query: 23 PPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTT 60
PPA PPP PPP PPP PP PPPPP +TT
Sbjct: 339 PPAAQPPPPPSPPPPPPPPASPLPPPPPPPPPTPSSTT 376
Score = 46.2 bits (108), Expect = 3e-04
Identities = 24/53 (45%), Positives = 31/53 (58%), Gaps = 3/53 (5%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQI 74
PP AQ PPP PPP PP PPP PPPPP ++T + L L+F+ ++
Sbjct: 339 PPAAQPPPPPSPPPPPPPPASPLPPP--PPPPPPTPSST-TKLPQGLSFAKRL 388
Score = 46.2 bits (108), Expect = 3e-04
Identities = 23/38 (60%), Positives = 24/38 (62%), Gaps = 5/38 (13%)
Query: 22 PPPAQLPP---PSQLPPPLSPPPQQHPP--PLLPPPPP 54
PPP LPP +Q PPP SPPP PP PL PPPPP
Sbjct: 330 PPPRPLPPRPPAAQPPPPPSPPPPPPPPASPLPPPPPP 367
Score = 38.5 bits (88), Expect = 0.057
Identities = 20/51 (39%), Positives = 22/51 (42%), Gaps = 14/51 (27%)
Query: 18 PTMNPPPAQLP--------------PPSQLPPPLSPPPQQHPPPLLPPPPP 54
PT PPP + P PP + PP P Q PPP PPPPP
Sbjct: 305 PTTRPPPIRPPFSHPISAHLPWYFQPPPRPLPPRPPAAQPPPPPSPPPPPP 355
Score = 37.4 bits (85), Expect = 0.13
Identities = 25/77 (32%), Positives = 32/77 (41%), Gaps = 8/77 (10%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQI--- 74
P +PPP PP S LPPP PPP P P L+ L L +L T
Sbjct: 346 PPPSPPPPPPPPASPLPPP-PPPPPPTPSSTTKLPQGLSFAKRLQQLIEVLKGKTYSDGK 404
Query: 75 ----LSTTPQPQTPTTN 87
+ TT P+ +T+
Sbjct: 405 NHYDMETTELPELTSTS 421
Score = 34.7 bits (78), Expect = 0.82
Identities = 20/54 (37%), Positives = 21/54 (38%), Gaps = 23/54 (42%)
Query: 22 PPPAQLPPPSQLPPPLS---------------------PPPQQHPPPLLPPPPP 54
PPP PPP + PP S PP Q PPP PPPPP
Sbjct: 303 PPPTTRPPP--IRPPFSHPISAHLPWYFQPPPRPLPPRPPAAQPPPPPSPPPPP 354
Score = 31.2 bits (69), Expect = 9.1
Identities = 34/127 (26%), Positives = 41/127 (31%), Gaps = 59/127 (46%)
Query: 28 PPPSQLPPPLSPPPQQHP-------------------PPLLPPPPPLNLNTTLSSLTNLL 68
PPP+ PPP+ P P HP PP PPPP
Sbjct: 303 PPPTTRPPPIRP-PFSHPISAHLPWYFQPPPRPLPPRPPAAQPPPP-------------- 347
Query: 69 TFSTQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPHIDHLLTSL 128
+ P P P + +P P PPP+ PSS LP L
Sbjct: 348 -------PSPPPPPPPPASPLP----PPPPPPPPT--------PSSTTKLPQ------GL 382
Query: 129 SYPKTLQ 135
S+ K LQ
Sbjct: 383 SFAKRLQ 389
>TRX2_MOUSE (O08550) Trithorax homolog 2 (WW domain binding protein
7) (Fragment)
Length = 290
Score = 47.8 bits (112), Expect = 9e-05
Identities = 37/108 (34%), Positives = 52/108 (47%), Gaps = 19/108 (17%)
Query: 18 PTMNPPP--AQLPPPSQLPPPLSPPP------QQHPPPLLPPP-------PPLNLNTTLS 62
P+ +PPP + LPPP PPPLSPPP Q+ PPL+P PPL + +
Sbjct: 57 PSTSPPPPASPLPPPVSPPPPLSPPPYPAPEKQEESPPLVPATCSRKRGRPPLT-PSQRA 115
Query: 63 SLTNLLTFSTQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLR 110
+ LS TP P T T + P +P ++P + FL ++R
Sbjct: 116 EREAARSGPEGTLSPTPNPSTTTGS--PLEDSPT-VVPKSTTFLKNIR 160
Score = 46.6 bits (109), Expect = 2e-04
Identities = 23/48 (47%), Positives = 27/48 (55%), Gaps = 2/48 (4%)
Query: 9 ELKNKALH*PTMNPPPAQLPPPSQLPPPLS--PPPQQHPPPLLPPPPP 54
+L + L P +PPP PP + PPP S PPP PPPL PPP P
Sbjct: 37 KLPSPPLTPPVPSPPPPLPPPSTSPPPPASPLPPPVSPPPPLSPPPYP 84
Score = 42.7 bits (99), Expect = 0.003
Identities = 19/35 (54%), Positives = 21/35 (59%), Gaps = 1/35 (2%)
Query: 23 PPAQLPPPSQLPPPLSPPPQQHP-PPLLPPPPPLN 56
PP PPP PP SPPP P PP + PPPPL+
Sbjct: 45 PPVPSPPPPLPPPSTSPPPPASPLPPPVSPPPPLS 79
Score = 42.0 bits (97), Expect = 0.005
Identities = 19/41 (46%), Positives = 21/41 (50%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLS 62
P P PP PPPL PP PPP P PPP++ LS
Sbjct: 39 PSPPLTPPVPSPPPPLPPPSTSPPPPASPLPPPVSPPPPLS 79
Score = 40.4 bits (93), Expect = 0.015
Identities = 25/77 (32%), Positives = 32/77 (41%)
Query: 20 MNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTP 79
+ PP A PP Q P P+S PP+ PP P P P + L T T T+ L P
Sbjct: 197 VRPPVATSPPAPQEPVPVSSPPRVPTPPSTPVPLPEKRRSILREPTFRWTSLTRELPPPP 256
Query: 80 QPQTPTTNLIPCLFNPN 96
P + P P+
Sbjct: 257 PAPPPAPSPPPAPATPS 273
Score = 37.4 bits (85), Expect = 0.13
Identities = 16/29 (55%), Positives = 17/29 (58%)
Query: 25 AQLPPPSQLPPPLSPPPQQHPPPLLPPPP 53
A+LP P PP SPPP PP PPPP
Sbjct: 36 AKLPSPPLTPPVPSPPPPLPPPSTSPPPP 64
Score = 32.0 bits (71), Expect = 5.3
Identities = 13/30 (43%), Positives = 15/30 (49%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPP 51
PPP PPP+ PPP P + P L P
Sbjct: 253 PPPPPAPPPAPSPPPAPATPSRRPLLLRAP 282
>SRCH_RABIT (P16230) Sarcoplasmic reticulum histidine-rich
calcium-binding protein precursor
Length = 852
Score = 47.4 bits (111), Expect = 1e-04
Identities = 37/141 (26%), Positives = 58/141 (40%), Gaps = 19/141 (13%)
Query: 526 DHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQENHRSHASTSPE 585
DHH+ H + ++ D D + + HQ +H H G D+++ ST +
Sbjct: 249 DHHQAHRHRGHEEEEDEEDDDDEGDSTESDR-HQ-AHRHRGHREEEDEDDDDEGDSTESD 306
Query: 586 RHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNH----TSDSFSRSDPSE 641
RH ++H HR H E++D +S+ + DR Q H+ H D D +E
Sbjct: 307 RH----QAHRHRGHREEEDDDDDDDEGDSTES-DRHQAHRHRGHREEEDEDDDDEGDSTE 361
Query: 642 S--------RGVGEHDISSDD 654
S RG E + DD
Sbjct: 362 SDRHQAHRHRGHREEEDEDDD 382
Score = 45.8 bits (107), Expect = 4e-04
Identities = 29/117 (24%), Positives = 47/117 (39%), Gaps = 9/117 (7%)
Query: 526 DHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQENHRSHA-STSP 584
D H+ H + ++ D + + + H +H H G E D+E+ ST
Sbjct: 220 DRHQAHRHRGHREEED--EDDDDDEGDSTESDHHQAHRHRGHEEEEDEEDDDDEGDSTES 277
Query: 585 ERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSE 641
+RH ++H HR H E++D + S DR Q H+ H + D E
Sbjct: 278 DRH----QAHRHRGHREEEDEDDDDE--GDSTESDRHQAHRHRGHREEEDDDDDDDE 328
Score = 40.0 bits (92), Expect = 0.020
Identities = 49/255 (19%), Positives = 94/255 (36%), Gaps = 26/255 (10%)
Query: 416 HQQHSH--HQETQKEKTREELLAEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIK 473
HQ H H H+E + E +E + E D R ++R + + D G+ E +
Sbjct: 280 HQAHRHRGHREEEDEDDDDEGDSTESD---RHQAHRHRGHREEEDDDDDDDEGDSTESDR 336
Query: 474 LAA----GVKSPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSHDSPAVTISNPDHHE 529
A G + + +D +H E D T S D H+
Sbjct: 337 HQAHRHRGHREEEDEDDDDEGDSTESDRHQAHRHRGHREEEDEDDDDEGDSTES--DRHQ 394
Query: 530 QQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQENHRSHASTSPERHRS 589
H + ++ D + + H +H H G D+E+ ST +RH++
Sbjct: 395 AHRHRGHREEEDEDDDDEG---DSTESDHHQAHRHRGHREEEDEEDDDEGDSTESDRHQA 451
Query: 590 RSRSHEHRVHHEKQDYSGRKK--------YNNSSRTKDRWQNDTHKNHTSDSFSRSDPSE 641
H HR H E++D + + +D +D + ++++ ++ +
Sbjct: 452 ----HRHRGHGEEEDEDDDDEGEHHHVPHRGHRGHEEDDGGDDDDGDDSTENGHQAHRHQ 507
Query: 642 SRGVGEHDISSDDKY 656
G E +++SD+ +
Sbjct: 508 GHGKEEAEVTSDEHH 522
Score = 40.0 bits (92), Expect = 0.020
Identities = 35/154 (22%), Positives = 56/154 (35%), Gaps = 17/154 (11%)
Query: 514 SHDSPAVT---ISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERG 570
SH SP +S + H SH ++ + D SR+Y + QGH+ GDE
Sbjct: 64 SHRSPGEENEDVSMENGHHFWSHRDHGETD----DEVSREYGHQPQGHRYHSPEAGDESV 119
Query: 571 TDQENHRSHASTSP---------ERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRW 621
+++ HR A +P + SH H E +D ++ + R R
Sbjct: 120 SEEGVHREQARQAPGHGGHGEAGAEDLAEHGSHGHGHEEEDEDVISSERPRHVLRRAPRG 179
Query: 622 QNDTHKNHTSDSFSRSDPS-ESRGVGEHDISSDD 654
+ + P RG G+ D +D
Sbjct: 180 HGGEEEGEEEEEEEEVSPEHRHRGHGKEDEEDED 213
Score = 39.3 bits (90), Expect = 0.033
Identities = 29/129 (22%), Positives = 45/129 (34%), Gaps = 21/129 (16%)
Query: 528 HEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQE---------NHRS 578
H H + + V+ R +R HGG+E G ++E HR
Sbjct: 149 HGSHGHGHEEEDEDVISSERPRHVLRR-----APRGHGGEEEGEEEEEEEEVSPEHRHRG 203
Query: 579 HASTSPE------RHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSD 632
H E R ++H HR H E++D +S+ + D Q H+ H +
Sbjct: 204 HGKEDEEDEDDDSTESDRHQAHRHRGHREEEDEDDDDDEGDSTES-DHHQAHRHRGHEEE 262
Query: 633 SFSRSDPSE 641
D E
Sbjct: 263 EDEEDDDDE 271
Score = 38.5 bits (88), Expect = 0.057
Identities = 29/141 (20%), Positives = 52/141 (36%), Gaps = 16/141 (11%)
Query: 526 DHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQENHRSHASTSPE 585
+ E++ + + D D + + +H H G D+++ ++
Sbjct: 189 EEEEEEVSPEHRHRGHGKEDEEDEDDDSTESDRHQAHRHRGHREEEDEDDDDDEGDSTES 248
Query: 586 RHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNH----TSDSFSRSDPSE 641
H ++H HR H E++D +S+ + DR Q H+ H D D +E
Sbjct: 249 DHH---QAHRHRGHEEEEDEEDDDDEGDSTES-DRHQAHRHRGHREEEDEDDDDEGDSTE 304
Query: 642 S--------RGVGEHDISSDD 654
S RG E + DD
Sbjct: 305 SDRHQAHRHRGHREEEDDDDD 325
Score = 37.0 bits (84), Expect = 0.17
Identities = 51/249 (20%), Positives = 80/249 (31%), Gaps = 17/249 (6%)
Query: 416 HQQHSH--HQETQKEKTREELLAEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIK 473
HQ H H H+E + E +E + E D + + +RG + + D G+ E +
Sbjct: 337 HQAHRHRGHREEEDEDDDDEGDSTESD-RHQAHRHRGHREEEDEDD---DDEGDSTESDR 392
Query: 474 LAAGVKSPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVS--HDSPAVTISNPDHHEQQ 531
A ED S H + RE D + D H+
Sbjct: 393 HQAHRHRGHREEEDEDDDDEGDSTESDHHQAHRHRGHREEEDEEDDDEGDSTESDRHQAH 452
Query: 532 SHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGG--DERGTDQ-----ENHRSHASTSP 584
H + ++ D + +GH+G G D+ G D + HR
Sbjct: 453 RHRGHGEEEDEDDDDEGEHHHVPHRGHRGHEEDDGGDDDDGDDSTENGHQAHRHQGHGKE 512
Query: 585 ERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRW-QNDTHKNHTSDSFSRSDPSESR 643
E + H H H Q + G K+ + D W Q H +H E
Sbjct: 513 EAEVTSDEHHHHVPDHGHQGH-GDKEGEEEGVSTDHWHQVPRHAHHGPGGEEEGGEEELT 571
Query: 644 GVGEHDISS 652
H ++S
Sbjct: 572 VKAGHHVAS 580
Score = 35.4 bits (80), Expect = 0.48
Identities = 38/165 (23%), Positives = 54/165 (32%), Gaps = 40/165 (24%)
Query: 526 DHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQENHRSHASTSPE 585
D H+ H + ++ D E + HQ H G E D ++ ST +
Sbjct: 278 DRHQAHRHRGHREEEDEDDDDEGDSTESDR--HQAHRHRGHREEEDDDDDDDEGDSTESD 335
Query: 586 RH---RSRS---------------------RSHEHRVHHEKQDYSGRKKYNNSSRTKDRW 621
RH R R ++H HR H E++D + S DR
Sbjct: 336 RHQAHRHRGHREEEDEDDDDEGDSTESDRHQAHRHRGHREEEDEDDDDE--GDSTESDRH 393
Query: 622 QNDTHKNH----TSDSFSRSDPSES--------RGVGEHDISSDD 654
Q H+ H D D +ES RG E + DD
Sbjct: 394 QAHRHRGHREEEDEDDDDEGDSTESDHHQAHRHRGHREEEDEEDD 438
Score = 33.5 bits (75), Expect = 1.8
Identities = 38/155 (24%), Positives = 56/155 (35%), Gaps = 17/155 (10%)
Query: 511 REVSHDSPAVTISNPDHHEQ---QSHTNYSDKSKVVHDATSRDYEQ--RKQGH-QGSHHH 564
R H +++ +HH H + DK ++ + Q R H G
Sbjct: 505 RHQGHGKEEAEVTSDEHHHHVPDHGHQGHGDKEGEEEGVSTDHWHQVPRHAHHGPGGEEE 564
Query: 565 GGDERGTDQENHRSHASTSPERHRSR-SRSHEHRV----HHEKQ----DYSGRKKYNNSS 615
GG+E T + H AS P HRSR + EH+ HH+ + D S + + SS
Sbjct: 565 GGEEELTVKAGHHV-ASHPPPGHRSREGHAEEHQTEVPGHHQHRMGDTDTSAERGHPASS 623
Query: 616 -RTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHD 649
R + DT +H P G D
Sbjct: 624 PRQQGHPPEDTVHHHRGSLKEEVGPESPGPAGVKD 658
Score = 31.2 bits (69), Expect = 9.1
Identities = 31/139 (22%), Positives = 54/139 (38%), Gaps = 20/139 (14%)
Query: 528 HEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQENHRSHASTSPER- 586
H SH +S+ H + +++ GH H GD TD R H ++SP +
Sbjct: 576 HHVASHPPPGHRSREGH---AEEHQTEVPGHH--QHRMGD---TDTSAERGHPASSPRQQ 627
Query: 587 -HRSRSRSHEHRVHHEKQ---DYSGRKKYNNSSRTK------DRWQNDTHKNHTSDSFSR 636
H H HR +++ + G + SR K + Q TH +H+ +
Sbjct: 628 GHPPEDTVHHHRGSLKEEVGPESPGPAGVKDGSRVKRGGSEEEEEQKGTH-HHSLEDEED 686
Query: 637 SDPSESRGVGEHDISSDDK 655
+ R + + D +D+
Sbjct: 687 EEEGHGRSLSQEDQEEEDR 705
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.132 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 87,333,716
Number of Sequences: 164201
Number of extensions: 4231568
Number of successful extensions: 46679
Number of sequences better than 10.0: 959
Number of HSP's better than 10.0 without gapping: 370
Number of HSP's successfully gapped in prelim test: 634
Number of HSP's that attempted gapping in prelim test: 27099
Number of HSP's gapped (non-prelim): 7957
length of query: 661
length of database: 59,974,054
effective HSP length: 117
effective length of query: 544
effective length of database: 40,762,537
effective search space: 22174820128
effective search space used: 22174820128
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146559.13


