

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146330.6 + phase: 0
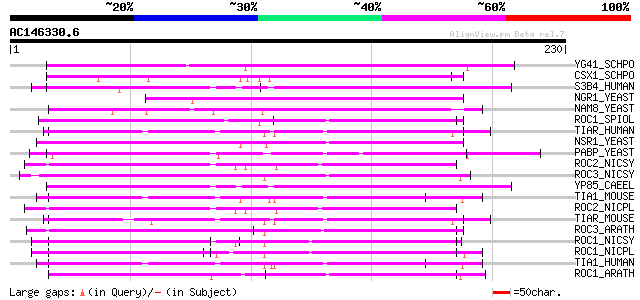
(230 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YG41_SCHPO (O60176) Hypothetical RNA-binding protein C23E6.01c i... 158 9e-39
CSX1_SCHPO (O13759) RNA-binding post-transcriptional regulator csx1 150 3e-36
S3B4_HUMAN (Q15427) Splicing factor 3B subunit 4 (Spliceosome as... 107 2e-23
NGR1_YEAST (P32831) Negative growth regulatory protein NGR1 (RNA... 103 3e-22
NAM8_YEAST (Q00539) NAM8 protein 99 1e-20
ROC1_SPIOL (P28644) 28 kDa ribonucleoprotein, chloroplast (28RNP) 98 1e-20
TIAR_HUMAN (Q01085) Nucleolysin TIAR (TIA-1 related protein) 98 2e-20
NSR1_YEAST (P27476) Nuclear localization sequence binding protei... 96 9e-20
PABP_YEAST (P04147) Polyadenylate-binding protein, cytoplasmic a... 94 2e-19
ROC2_NICSY (Q08937) 29 kDa ribonucleoprotein B, chloroplast prec... 93 5e-19
ROC3_NICSY (P19682) 28 kDa ribonucleoprotein, chloroplast precur... 92 1e-18
YP85_CAEEL (Q09442) Hypothetical RNA-binding protein C08B11.5 in... 92 1e-18
TIA1_MOUSE (P52912) Nucleolysin TIA-1 (RNA-binding protein TIA-1) 92 1e-18
ROC2_NICPL (P49314) 31 kDa ribonucleoprotein, chloroplast precur... 91 3e-18
TIAR_MOUSE (P70318) Nucleolysin TIAR (TIA-1 related protein) 90 4e-18
ROC3_ARATH (Q04836) 31 kDa ribonucleoprotein, chloroplast precur... 90 4e-18
ROC1_NICSY (Q08935) 29 kDa ribonucleoprotein A, chloroplast prec... 90 5e-18
ROC1_NICPL (P49313) 30 kDa ribonucleoprotein, chloroplast precur... 89 7e-18
TIA1_HUMAN (P31483) Nucleolysin TIA-1 (RNA-binding protein TIA-1... 89 9e-18
ROC1_ARATH (Q9ZUU4) Putative ribonucleoprotein At2g37220, chloro... 88 1e-17
>YG41_SCHPO (O60176) Hypothetical RNA-binding protein C23E6.01c in
chromosome II
Length = 473
Score = 158 bits (400), Expect = 9e-39
Identities = 79/204 (38%), Positives = 120/204 (58%), Gaps = 11/204 (5%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
TLW+G+L+ WV E ++ ++ G+ + +K+IRN+ TG GY F+EF S A + +
Sbjct: 94 TLWMGELEPWVTEAFIQQVWNTLGKAVKVKLIRNRYTGMNAGYCFVEFASPHEASSAM-S 152
Query: 76 YNGTQMPGTEQTFRLNWASFG-IGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGS 134
N +PGT F+LNWAS G + E+ ++SIFVGDL+P+V ++ + F + Y S
Sbjct: 153 MNNKPIPGTNHLFKLNWASGGGLREKSISKASEYSIFVGDLSPNVNEFDVYSLFASRYNS 212
Query: 135 VRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPK------ 188
+ AK++TDP T S+GYGFV+F+DE+++ A++EM G C RP+R+ ATPK
Sbjct: 213 CKSAKIMTDPQTNVSRGYGFVRFTDENDQKSALAEMQGQICGDRPIRVGLATPKSKAHVF 272
Query: 189 ---KTTGYQQNPYAAVVAAAPVPK 209
P AA PVP+
Sbjct: 273 SPVNVVPVSMPPVGFYSAAQPVPQ 296
Score = 41.6 bits (96), Expect = 0.002
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 8/77 (10%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G L +V E L + F + GE++ +KI K G GF++FV+ +AE +
Sbjct: 304 TVFVGGLSKFVSEEELKYLFQNFGEIVYVKIPPGK------GCGFVQFVNRQSAEIAINQ 357
Query: 76 YNGTQMPGTEQTFRLNW 92
G P RL+W
Sbjct: 358 LQG--YPLGNSRIRLSW 372
Score = 35.8 bits (81), Expect = 0.087
Identities = 48/211 (22%), Positives = 86/211 (40%), Gaps = 42/211 (19%)
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V+E + F S S KI+ + T GYGF+ F + + L
Sbjct: 187 SIFVGDLSPNVNEFDVYSLFASRYNSCKSAKIMTDPQTNVSRGYGFVRFTDENDQKSALA 246
Query: 75 TYNGTQMPGTEQTFRLNWAS-----------------------FGIGERRPDAGP--DHS 109
G Q+ G ++ R+ A+ + + P + +
Sbjct: 247 EMQG-QICG-DRPIRVGLATPKSKAHVFSPVNVVPVSMPPVGFYSAAQPVPQFADTANST 304
Query: 110 IFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSE 169
+FVG L+ V++ L+ F+ ++G + K+ KG GFV+F + A+++
Sbjct: 305 VFVGGLSKFVSEEELKYLFQ-NFGEIVYVKIPP------GKGCGFVQFVNRQSAEIAINQ 357
Query: 170 MNGVYCSTRPMRISAATPKKTTGYQQNPYAA 200
+ G +R+S G QNP AA
Sbjct: 358 LQGYPLGNSRIRLS-------WGRNQNPIAA 381
>CSX1_SCHPO (O13759) RNA-binding post-transcriptional regulator csx1
Length = 632
Score = 150 bits (378), Expect = 3e-36
Identities = 76/177 (42%), Positives = 107/177 (59%), Gaps = 4/177 (2%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPE--GYGFIEFVSHSAAERVL 73
TLW+GDL+ W+D ++ ++ E +++K++R+K + Y F++F S +AAER L
Sbjct: 86 TLWMGDLEPWMDATFIQQLWASLNEPVNVKVMRSKASSSETLISYCFVQFSSSAAAERAL 145
Query: 74 QTYNGTQMPGTEQTFRLNWASFGIGERRP--DAGPDHSIFVGDLAPDVTDYLLQETFRTH 131
YN T +PG TF+LNWA+ G + P+ SIFVGDL P D L TFR+
Sbjct: 146 MKYNNTMIPGAHCTFKLNWATGGGIQHNNFVSRDPEFSIFVGDLLPTTEDSDLFMTFRSI 205
Query: 132 YGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPK 188
Y S AK++ DP TG S+ YGFV+FS E E+ A+ M G C RP+RIS A+PK
Sbjct: 206 YPSCTSAKIIVDPVTGLSRKYGFVRFSSEKEQQHALMHMQGYLCQGRPLRISVASPK 262
Score = 47.8 bits (112), Expect = 2e-05
Identities = 49/192 (25%), Positives = 86/192 (44%), Gaps = 33/192 (17%)
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL +++ L F S S KII + +TG YGF+ F S + L
Sbjct: 183 SIFVGDLLPTTEDSDLFMTFRSIYPSCTSAKIIVDPVTGLSRKYGFVRFSSEKEQQHALM 242
Query: 75 TYNGTQMPGTEQTFRLNWAS------------FGI-----GERRPDAGP------DHSIF 111
G G + R++ AS GI R+P+ + ++F
Sbjct: 243 HMQGYLCQG--RPLRISVASPKSRASIAADSALGIVPTSTSNRQPNQDLCSMDPLNTTVF 300
Query: 112 VGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMN 171
VG LA ++++ LQ F+ +G + K+ KG GFV++S++S +A++ M
Sbjct: 301 VGGLASNLSEKDLQVCFQP-FGRILNIKIPF------GKGCGFVQYSEKSAAEKAINTMQ 353
Query: 172 GVYCSTRPMRIS 183
G T +R++
Sbjct: 354 GALVGTSHIRLA 365
>S3B4_HUMAN (Q15427) Splicing factor 3B subunit 4 (Spliceosome
associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA
splicing factor SF3b 49 kDa subunit)
Length = 424
Score = 107 bits (267), Expect = 2e-23
Identities = 63/193 (32%), Positives = 105/193 (53%), Gaps = 6/193 (3%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G L V E L F G V++ + ++++TGQ +GYGF+EF+S A+ ++
Sbjct: 14 TVYVGGLDEKVSEPLLWELFLQAGPVVNTHMPKDRVTGQHQGYGFVEFLSEEDADYAIKI 73
Query: 76 YNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSV 135
N ++ G + R+N AS + D G + IF+G+L P++ + LL +TF +
Sbjct: 74 MNMIKLYG--KPIRVNKAS--AHNKNLDVGAN--IFIGNLDPEIDEKLLYDTFSAFGVIL 127
Query: 136 RGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPKKTTGYQQ 195
+ K++ DP+TG SKGY F+ F+ + A+ MNG Y RP+ +S A K + G +
Sbjct: 128 QTPKIMRDPDTGNSKGYAFINFASFDASDAAIEAMNGQYLCNRPITVSYAFKKDSKGERH 187
Query: 196 NPYAAVVAAAPVP 208
A + AA P
Sbjct: 188 GSAAERLLAAQNP 200
Score = 46.2 bits (108), Expect = 6e-05
Identities = 29/96 (30%), Positives = 46/96 (47%), Gaps = 1/96 (1%)
Query: 10 SLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISI-KIIRNKITGQPEGYGFIEFVSHSA 68
+L+ ++IG+L +DE L FS G ++ KI+R+ TG +GY FI F S A
Sbjct: 95 NLDVGANIFIGNLDPEIDEKLLYDTFSAFGVILQTPKIMRDPDTGNSKGYAFINFASFDA 154
Query: 69 AERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDA 104
++ ++ NG + T + GER A
Sbjct: 155 SDAAIEAMNGQYLCNRPITVSYAFKKDSKGERHGSA 190
>NGR1_YEAST (P32831) Negative growth regulatory protein NGR1
(RNA-binding protein RBP1)
Length = 672
Score = 103 bits (258), Expect = 3e-22
Identities = 57/144 (39%), Positives = 83/144 (57%), Gaps = 12/144 (8%)
Query: 57 GYGFIEFVSHSAAERVLQ------------TYNGTQMPGTEQTFRLNWASFGIGERRPDA 104
GY F+EF + A+ L T N P ++TFRLNWAS + +
Sbjct: 129 GYCFVEFETQKDAKFALSLNATPLPNFYSPTTNSQTNPTFKRTFRLNWASGATLQSSIPS 188
Query: 105 GPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERN 164
P+ S+FVGDL+P T+ L F+T + SV+ +V+TDP TG S+ +GFV+F DE ER
Sbjct: 189 TPEFSLFVGDLSPTATEADLLSLFQTRFKSVKTVRVMTDPLTGSSRCFGFVRFGDEDERR 248
Query: 165 RAMSEMNGVYCSTRPMRISAATPK 188
RA+ EM+G + R +R++ ATP+
Sbjct: 249 RALIEMSGKWFQGRALRVAYATPR 272
Score = 38.1 bits (87), Expect = 0.017
Identities = 22/82 (26%), Positives = 39/82 (46%), Gaps = 7/82 (8%)
Query: 109 SIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMS 168
++FVG L P T++ L+ F+ +G + ++ PN K GFVKF + ++
Sbjct: 361 TVFVGGLVPKTTEFQLRSLFKP-FGPILNVRI---PN---GKNCGFVKFEKRIDAEASIQ 413
Query: 169 EMNGVYCSTRPMRISAATPKKT 190
+ G P+R+S P +
Sbjct: 414 GLQGFIVGGSPIRLSWGRPSSS 435
Score = 32.3 bits (72), Expect = 0.96
Identities = 25/89 (28%), Positives = 34/89 (38%), Gaps = 3/89 (3%)
Query: 10 SLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIR---NKITGQPEGYGFIEFVSH 66
S E RTLW+GDL DE + +S + + +K+IR N + S
Sbjct: 29 STEPPRTLWMGDLDPSFDEATIEEIWSKLDKKVIVKLIRAKKNLLIPCSSTSSSNNNTSE 88
Query: 67 SAAERVLQTYNGTQMPGTEQTFRLNWASF 95
AE N T Q +N SF
Sbjct: 89 ENAENQQSASNSTDQLDNSQMININGISF 117
>NAM8_YEAST (Q00539) NAM8 protein
Length = 523
Score = 98.6 bits (244), Expect = 1e-20
Identities = 68/197 (34%), Positives = 103/197 (51%), Gaps = 23/197 (11%)
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEV-ISIKIIRNKITGQP----------EGYGFIEFVS 65
L++GDL D+N + ++ GE I+++++ N +GY F++F S
Sbjct: 56 LYMGDLDPTWDKNTVRQIWASLGEANINVRMMWNNTLNNGSRSSMGPKNNQGYCFVDFPS 115
Query: 66 HSAAERVLQTYNGTQMPG-TEQTFRLNWASFGIGERRPD-----AGPDHSIFVGDLAPDV 119
+ A L NG +P + +LNWA+ +G + SIFVGDLAP+V
Sbjct: 116 STHAANALLK-NGMLIPNFPNKKLKLNWATSSYSNSNNSLNNVKSGNNCSIFVGDLAPNV 174
Query: 120 TDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRP 179
T+ L E F Y S AK+V D TG SKGYGFVKF++ E+ A+SEM GV+ + R
Sbjct: 175 TESQLFELFINRYASTSHAKIVHDQVTGMSKGYGFVKFTNSDEQQLALSEMQGVFLNGRA 234
Query: 180 MRISAATPKKTTGYQQN 196
+++ T+G QQ+
Sbjct: 235 IKVG-----PTSGQQQH 246
Score = 31.2 bits (69), Expect = 2.1
Identities = 22/77 (28%), Positives = 34/77 (43%), Gaps = 8/77 (10%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T++IG L V E+ L F G ++ +KI K GF+++V +AE +
Sbjct: 314 TVFIGGLSSLVTEDELRAYFQPFGTIVYVKIPVGKCC------GFVQYVDRLSAEAAIAG 367
Query: 76 YNGTQMPGTEQTFRLNW 92
G P RL+W
Sbjct: 368 MQG--FPIANSRVRLSW 382
>ROC1_SPIOL (P28644) 28 kDa ribonucleoprotein, chloroplast (28RNP)
Length = 233
Score = 98.2 bits (243), Expect = 1e-20
Identities = 63/175 (36%), Positives = 93/175 (53%), Gaps = 4/175 (2%)
Query: 13 EVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERV 72
E L++G+L Y VD L F G V ++I N+ T + G+GF+ + AE+
Sbjct: 53 EEAKLFVGNLPYDVDSEKLAGIFDAAGVVEIAEVIYNRETDRSRGFGFVTMSTVEEAEKA 112
Query: 73 LQTYNGTQMPGTEQTFRLNWASFGIGERRP--DAGPDHSIFVGDLAPDVTDYLLQETFRT 130
++ NG M G + T A G ER P D P ++VG+L DV L++ F
Sbjct: 113 VELLNGYDMDGRQLTVN-KAAPRGSPERAPRGDFEPSCRVYVGNLPWDVDTSRLEQLFSE 171
Query: 131 HYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAA 185
H G V A+VV+D TGRS+G+GFV S ESE N A++ ++G R +R++ A
Sbjct: 172 H-GKVVSARVVSDRETGRSRGFGFVTMSSESEVNDAIAALDGQTLDGRAVRVNVA 225
Score = 44.3 bits (103), Expect = 2e-04
Identities = 27/79 (34%), Positives = 41/79 (51%), Gaps = 1/79 (1%)
Query: 110 IFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSE 169
+FVG+L DV L F G V A+V+ + T RS+G+GFV S E +A+
Sbjct: 57 LFVGNLPYDVDSEKLAGIFDAA-GVVEIAEVIYNRETDRSRGFGFVTMSTVEEAEKAVEL 115
Query: 170 MNGVYCSTRPMRISAATPK 188
+NG R + ++ A P+
Sbjct: 116 LNGYDMDGRQLTVNKAAPR 134
>TIAR_HUMAN (Q01085) Nucleolysin TIAR (TIA-1 related protein)
Length = 375
Score = 97.8 bits (242), Expect = 2e-20
Identities = 60/174 (34%), Positives = 93/174 (52%), Gaps = 6/174 (3%)
Query: 15 RTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
RTL++G+L V E + FS G S K+I + P Y F+EF H A L
Sbjct: 9 RTLYVGNLSRDVTEVLILQLFSQIGPCKSCKMITEHTSNDP--YCFVEFYEHRDAAAALA 66
Query: 75 TYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGS 134
NG ++ G E ++NWA+ +++ + H +FVGDL+P++T ++ F +G
Sbjct: 67 AMNGRKILGKE--VKVNWATTPSSQKKDTSNHFH-VFVGDLSPEITTEDIKSAFAP-FGK 122
Query: 135 VRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPK 188
+ A+VV D TG+SKGYGFV F ++ + A+ M G + R +R + AT K
Sbjct: 123 ISDARVVKDMATGKSKGYGFVSFYNKLDAENAIVHMGGQWLGGRQIRTNWATRK 176
Score = 64.3 bits (155), Expect = 2e-10
Identities = 48/207 (23%), Positives = 92/207 (44%), Gaps = 38/207 (18%)
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 76
+++GDL + + F+ G++ +++++ TG+ +GYGF+ F + AE +
Sbjct: 99 VFVGDLSPEITTEDIKSAFAPFGKISDARVVKDMATGKSKGYGFVSFYNKLDAENAIVHM 158
Query: 77 NGTQMPGTEQTFRLNWASFGIGERRPDA---------------------GPDH-SIFVGD 114
G + G + R NWA+ R+P A P + +++ G
Sbjct: 159 GGQWLGGRQ--IRTNWAT-----RKPPAPKSTQENNTKQLRFEDVVNQSSPKNCTVYCGG 211
Query: 115 LAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVY 174
+A +TD L+++TF + +G + +V + KGY FV+FS A+ +NG
Sbjct: 212 IASGLTDQLMRQTF-SPFGQIMEIRVFPE------KGYSFVRFSTHESAAHAIVSVNGTT 264
Query: 175 CSTRPMRI--SAATPKKTTGYQQNPYA 199
++ +P T +QQ Y+
Sbjct: 265 IEGHVVKCYWGKESPDMTKNFQQVDYS 291
Score = 40.8 bits (94), Expect = 0.003
Identities = 23/84 (27%), Positives = 48/84 (56%), Gaps = 3/84 (3%)
Query: 103 DAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESE 162
D G +++VG+L+ DVT+ L+ + F + G + K++T+ + + Y FV+F + +
Sbjct: 4 DDGQPRTLYVGNLSRDVTEVLILQLF-SQIGPCKSCKMITEHTS--NDPYCFVEFYEHRD 60
Query: 163 RNRAMSEMNGVYCSTRPMRISAAT 186
A++ MNG + ++++ AT
Sbjct: 61 AAAALAAMNGRKILGKEVKVNWAT 84
Score = 37.0 bits (84), Expect = 0.039
Identities = 17/68 (25%), Positives = 34/68 (50%), Gaps = 6/68 (8%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T++ G + + + + FS G+++ I++ K GY F+ F +H +A + +
Sbjct: 206 TVYCGGIASGLTDQLMRQTFSPFGQIMEIRVFPEK------GYSFVRFSTHESAAHAIVS 259
Query: 76 YNGTQMPG 83
NGT + G
Sbjct: 260 VNGTTIEG 267
>NSR1_YEAST (P27476) Nuclear localization sequence binding protein
(P67)
Length = 414
Score = 95.5 bits (236), Expect = 9e-20
Identities = 52/183 (28%), Positives = 98/183 (53%), Gaps = 7/183 (3%)
Query: 12 EEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAER 71
EE T+++G L + +D+ +L F H G VI ++I + T + GYG+++F + S AE+
Sbjct: 165 EEPATIFVGRLSWSIDDEWLKKEFEHIGGVIGARVIYERGTDRSRGYGYVDFENKSYAEK 224
Query: 72 VLQTYNGTQMPGTEQTFRLNWAS-FGIGERRPDAG-----PDHSIFVGDLAPDVTDYLLQ 125
+Q G ++ G ++ + G +R G P ++F+G+L+ + +
Sbjct: 225 AIQEMQGKEIDGRPINCDMSTSKPAGNNDRAKKFGDTPSEPSDTLFLGNLSFNADRDAIF 284
Query: 126 ETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAA 185
E F H G V ++ T P T + KG+G+V+FS+ + +A+ + G Y RP+R+ +
Sbjct: 285 ELFAKH-GEVVSVRIPTHPETEQPKGFGYVQFSNMEDAKKALDALQGEYIDNRPVRLDFS 343
Query: 186 TPK 188
+P+
Sbjct: 344 SPR 346
>PABP_YEAST (P04147) Polyadenylate-binding protein, cytoplasmic and
nuclear (Poly(A)-binding protein) (PABP) (ARS consensus
binding protein ACBP-67) (Polyadenylate tail-binding
protein)
Length = 576
Score = 94.4 bits (233), Expect = 2e-19
Identities = 54/174 (31%), Positives = 95/174 (54%), Gaps = 7/174 (4%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
+L++GDL+ V E +L FS G V SI++ R+ IT GY ++ F H A + ++
Sbjct: 38 SLYVGDLEPSVSEAHLYDIFSPIGSVSSIRVCRDAITKTSLGYAYVNFNDHEAGRKAIEQ 97
Query: 76 YNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSV 135
N T + G + R+ W+ R+ +G +IF+ +L PD+ + L +TF +G +
Sbjct: 98 LNYTPIKG--RLCRIMWSQRDPSLRKKGSG---NIFIKNLHPDIDNKALYDTFSV-FGDI 151
Query: 136 RGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPKK 189
+K+ TD N G+SKG+GFV F +E A+ +NG+ + + + ++ +K
Sbjct: 152 LSSKIATDEN-GKSKGFGFVHFEEEGAAKEAIDALNGMLLNGQEIYVAPHLSRK 204
Score = 67.4 bits (163), Expect = 3e-11
Identities = 56/243 (23%), Positives = 108/243 (44%), Gaps = 36/243 (14%)
Query: 9 ASLEEVRT----LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFV 64
+ LEE + L++ ++ + F+ G ++S + ++ G+ +G+GF+ +
Sbjct: 208 SQLEETKAHYTNLYVKNINSETTDEQFQELFAKFGPIVSASLEKDA-DGKLKGFGFVNYE 266
Query: 65 SHSAAERVLQTYNGTQMPGTE-------------QTFRLNWASFGIGERRPDAGPDHSIF 111
H A + ++ N +++ G + + + ++ + + G + +F
Sbjct: 267 KHEDAVKAVEALNDSELNGEKLYVGRAQKKNERMHVLKKQYEAYRLEKMAKYQGVN--LF 324
Query: 112 VGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMN 171
V +L V D L+E F YG++ AKV+ N G+SKG+GFV FS E +A++E N
Sbjct: 325 VKNLDDSVDDEKLEEEFAP-YGTITSAKVMRTEN-GKSKGFGFVCFSTPEEATKAITEKN 382
Query: 172 GVYCSTRPMRISAATPK--------------KTTGYQQNPYAAVVAAAPVPKGITKPYTF 217
+ +P+ ++ A K YQQ AA AAA +P P +
Sbjct: 383 QQIVAGKPLYVAIAQRKDVRRSQLAQQIQARNQMRYQQATAAAAAAAAGMPGQFMPPMFY 442
Query: 218 FII 220
++
Sbjct: 443 GVM 445
>ROC2_NICSY (Q08937) 29 kDa ribonucleoprotein B, chloroplast
precursor (CP29B)
Length = 291
Score = 93.2 bits (230), Expect = 5e-19
Identities = 67/208 (32%), Positives = 105/208 (50%), Gaps = 33/208 (15%)
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
QP E+++ L++G+L + VD L F G V +++I +K+TG+ G+GF+ +
Sbjct: 80 QPRFSEDLK-LFVGNLPFSVDSAALAGLFERAGNVEMVEVIYDKLTGRSRGFGFVTMSTK 138
Query: 67 SAAERVLQTYNGTQMPGTEQTFRLNW---------ASFG------------------IGE 99
E Q +NG ++ G + R+N +SFG G
Sbjct: 139 EEVEAAEQQFNGYEIDG--RAIRVNAGPAPAKRENSSFGGGRGGNSSYGGGRDGNSSFGG 196
Query: 100 RRPDAGPDHS--IFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKF 157
R D S ++VG+L+ V D L+E F + G+V AKVV D ++GRS+G+GFV +
Sbjct: 197 ARGGRSVDSSNRVYVGNLSWGVDDLALKELF-SEQGNVVDAKVVYDRDSGRSRGFGFVTY 255
Query: 158 SDESERNRAMSEMNGVYCSTRPMRISAA 185
S E N A+ +NGV R +R+SAA
Sbjct: 256 SSSKEVNDAIDSLNGVDLDGRSIRVSAA 283
>ROC3_NICSY (P19682) 28 kDa ribonucleoprotein, chloroplast precursor
(28RNP)
Length = 276
Score = 92.0 bits (227), Expect = 1e-18
Identities = 58/190 (30%), Positives = 99/190 (51%), Gaps = 7/190 (3%)
Query: 5 YHQPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFV 64
Y +P+ E L++G+L Y +D L F G V ++I N+ T + G+GF+
Sbjct: 90 YQEPS---EDAKLFVGNLPYDIDSEGLAQLFQQAGVVEIAEVIYNRETDRSRGFGFVTMS 146
Query: 65 SHSAAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDA-GPDHSIFVGDLAPDVTDYL 123
+ A++ ++ Y+ + G T ER P P + I+VG++ D+ D
Sbjct: 147 TVEEADKAVELYSQYDLNGRLLTVNKAAPRGSRPERAPRTFQPTYRIYVGNIPWDIDDAR 206
Query: 124 LQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRIS 183
L++ F H G V A+VV D +GRS+G+GFV S E+E + A++ ++G R +R++
Sbjct: 207 LEQVFSEH-GKVVSARVVFDRESGRSRGFGFVTMSSEAEMSEAIANLDGQTLDGRTIRVN 265
Query: 184 AA--TPKKTT 191
AA P++ T
Sbjct: 266 AAEERPRRNT 275
>YP85_CAEEL (Q09442) Hypothetical RNA-binding protein C08B11.5 in
chromosome II
Length = 388
Score = 91.7 bits (226), Expect = 1e-18
Identities = 55/193 (28%), Positives = 102/193 (52%), Gaps = 6/193 (3%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G L V E+ L G V+S+ + ++++T +G+GF+EF+ A+ ++
Sbjct: 14 TIYVGGLDEKVSESILWELMVQAGPVVSVNMPKDRVTANHQGFGFVEFMGEEDADYAIKI 73
Query: 76 YNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSV 135
N ++ G + ++N AS E+ D G + IFVG+L P+V + LL +TF +
Sbjct: 74 LNMIKLYG--KPIKVNKAS--AHEKNMDVGAN--IFVGNLDPEVDEKLLYDTFSAFGVIL 127
Query: 136 RGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPKKTTGYQQ 195
+ K++ D ++G SKG+ F+ F+ + A+ MNG + R + +S A + + G +
Sbjct: 128 QVPKIMRDVDSGTSKGFAFINFASFEASDTALEAMNGQFLCNRAITVSYAFKRDSKGERH 187
Query: 196 NPYAAVVAAAPVP 208
A + AA P
Sbjct: 188 GTAAERMLAAQNP 200
>TIA1_MOUSE (P52912) Nucleolysin TIA-1 (RNA-binding protein TIA-1)
Length = 386
Score = 91.7 bits (226), Expect = 1e-18
Identities = 62/198 (31%), Positives = 100/198 (50%), Gaps = 18/198 (9%)
Query: 12 EEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAER 71
E +TL++G+L V E + FS G + K+I + P Y F+EF H A
Sbjct: 4 EMPKTLYVGNLSRDVTEALILQLFSQIGPCKNCKMIMDTAGNDP--YCFVEFHEHRHAAA 61
Query: 72 VLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGP---------DH-SIFVGDLAPDVTD 121
L NG ++ G E ++NWA+ +++ + DH +FVGDL+P++T
Sbjct: 62 ALAAMNGRKIMGKE--VKVNWATTPSSQKKDTSSSTVVSTQRSQDHFHVFVGDLSPEITT 119
Query: 122 YLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMR 181
++ F +G + A+VV D TG+SKGYGFV F ++ + A+ +M G + R +R
Sbjct: 120 EDIKAAFAP-FGRISDARVVKDMATGKSKGYGFVSFFNKWDAENAIQQMGGQWLGGRQIR 178
Query: 182 ISAAT---PKKTTGYQQN 196
+ AT P + Y+ N
Sbjct: 179 TNWATRKPPAPKSTYESN 196
Score = 63.2 bits (152), Expect = 5e-10
Identities = 41/173 (23%), Positives = 80/173 (45%), Gaps = 26/173 (15%)
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 76
+++GDL + + F+ G + +++++ TG+ +GYGF+ F + AE +Q
Sbjct: 108 VFVGDLSPEITTEDIKAAFAPFGRISDARVVKDMATGKSKGYGFVSFFNKWDAENAIQQM 167
Query: 77 NGTQMPGTEQTFRLNWAS----------------FGIGERRPDAGPDH-SIFVGDLAPDV 119
G + G + R NWA+ E + P++ +++ G + +
Sbjct: 168 GGQWLGGRQ--IRTNWATRKPPAPKSTYESNTKQLSYDEVVSQSSPNNCTVYCGGVTSGL 225
Query: 120 TDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNG 172
T+ L+++TF + +G + +V D KGY FV+FS A+ +NG
Sbjct: 226 TEQLMRQTF-SPFGQIMEIRVFPD------KGYSFVRFSSHESAAHAIVSVNG 271
Score = 39.3 bits (90), Expect = 0.008
Identities = 19/68 (27%), Positives = 35/68 (50%), Gaps = 6/68 (8%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T++ G + + E + FS G+++ I++ +K GY F+ F SH +A + +
Sbjct: 215 TVYCGGVTSGLTEQLMRQTFSPFGQIMEIRVFPDK------GYSFVRFSSHESAAHAIVS 268
Query: 76 YNGTQMPG 83
NGT + G
Sbjct: 269 VNGTTIEG 276
>ROC2_NICPL (P49314) 31 kDa ribonucleoprotein, chloroplast precursor
(CP-RBP31)
Length = 292
Score = 90.5 bits (223), Expect = 3e-18
Identities = 65/208 (31%), Positives = 105/208 (50%), Gaps = 33/208 (15%)
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
QP E+++ L++G+L + VD L F G V +++I +K++G+ G+GF+ +
Sbjct: 81 QPRFSEDLK-LFVGNLPFSVDSAALAGLFERAGNVEIVEVIYDKLSGRSRGFGFVTMSTK 139
Query: 67 SAAERVLQTYNGTQMPGTEQTFRLNW---------ASFG------------------IGE 99
E Q +NG ++ G + R+N +SFG G
Sbjct: 140 EEVEAAEQQFNGYEIDG--RAIRVNAGPAPAKRENSSFGGGRGGNSSYGGGRDGNSSFGG 197
Query: 100 RRPDAGPDHS--IFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKF 157
R D S ++VG+L+ V D L+E F + G+V AKVV D ++GRS+G+GFV +
Sbjct: 198 ARGGRSVDSSNRVYVGNLSWGVDDLALKELF-SEQGNVVDAKVVYDRDSGRSRGFGFVTY 256
Query: 158 SDESERNRAMSEMNGVYCSTRPMRISAA 185
S E N A+ +NG+ R +R+SAA
Sbjct: 257 SSAKEVNDAIDSLNGIDLDGRSIRVSAA 284
>TIAR_MOUSE (P70318) Nucleolysin TIAR (TIA-1 related protein)
Length = 392
Score = 90.1 bits (222), Expect = 4e-18
Identities = 62/193 (32%), Positives = 95/193 (49%), Gaps = 27/193 (13%)
Query: 15 RTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEG----------------- 57
RTL++G+L V E + FS G S K+I T QP+
Sbjct: 9 RTLYVGNLSRDVTEVLILQLFSQIGPCKSCKMI----TEQPDSRRVNSSVGFSVLQHTSN 64
Query: 58 --YGFIEFVSHSAAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDL 115
Y F+EF H A L NG ++ G E ++NWA+ +++ + H +FVGDL
Sbjct: 65 DPYCFVEFYEHRDAAAALAAMNGRKILGKE--VKVNWATTPSSQKKDTSNHFH-VFVGDL 121
Query: 116 APDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYC 175
+P++T ++ F +G + A+VV D TG+SKGYGFV F ++ + A+ M G +
Sbjct: 122 SPEITTEDIKSAFAP-FGKISDARVVKDMATGKSKGYGFVSFYNKLDAENAIVHMGGQWL 180
Query: 176 STRPMRISAATPK 188
R +R + AT K
Sbjct: 181 GGRQIRTNWATRK 193
Score = 64.3 bits (155), Expect = 2e-10
Identities = 48/207 (23%), Positives = 92/207 (44%), Gaps = 38/207 (18%)
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 76
+++GDL + + F+ G++ +++++ TG+ +GYGF+ F + AE +
Sbjct: 116 VFVGDLSPEITTEDIKSAFAPFGKISDARVVKDMATGKSKGYGFVSFYNKLDAENAIVHM 175
Query: 77 NGTQMPGTEQTFRLNWASFGIGERRPDA---------------------GPDH-SIFVGD 114
G + G + R NWA+ R+P A P + +++ G
Sbjct: 176 GGQWLGGRQ--IRTNWAT-----RKPPAPKSTQETNTKQLRFEDVVNQSSPKNCTVYCGG 228
Query: 115 LAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVY 174
+A +TD L+++TF + +G + +V + KGY FV+FS A+ +NG
Sbjct: 229 IASGLTDQLMRQTF-SPFGQIMEIRVFPE------KGYSFVRFSTHESAAHAIVSVNGTT 281
Query: 175 CSTRPMRI--SAATPKKTTGYQQNPYA 199
++ +P T +QQ Y+
Sbjct: 282 IEGHVVKCYWGKESPDMTKNFQQVDYS 308
Score = 37.0 bits (84), Expect = 0.039
Identities = 17/68 (25%), Positives = 34/68 (50%), Gaps = 6/68 (8%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T++ G + + + + FS G+++ I++ K GY F+ F +H +A + +
Sbjct: 223 TVYCGGIASGLTDQLMRQTFSPFGQIMEIRVFPEK------GYSFVRFSTHESAAHAIVS 276
Query: 76 YNGTQMPG 83
NGT + G
Sbjct: 277 VNGTTIEG 284
Score = 36.6 bits (83), Expect = 0.051
Identities = 24/99 (24%), Positives = 48/99 (48%), Gaps = 16/99 (16%)
Query: 103 DAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGR-------------- 148
D G +++VG+L+ DVT+ L+ + F + G + K++T+ R
Sbjct: 4 DDGQPRTLYVGNLSRDVTEVLILQLF-SQIGPCKSCKMITEQPDSRRVNSSVGFSVLQHT 62
Query: 149 -SKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAAT 186
+ Y FV+F + + A++ MNG + ++++ AT
Sbjct: 63 SNDPYCFVEFYEHRDAAAALAAMNGRKILGKEVKVNWAT 101
>ROC3_ARATH (Q04836) 31 kDa ribonucleoprotein, chloroplast precursor
(RNA-binding protein RNP-T) (RNA-binding protein 1/2/3)
(AtRBP33) (RNA-binding protein cp31)
Length = 329
Score = 90.1 bits (222), Expect = 4e-18
Identities = 61/179 (34%), Positives = 90/179 (50%), Gaps = 3/179 (1%)
Query: 8 PASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHS 67
P EE + L++G+L Y V+ L F G V ++I N+ T Q G+GF+ S
Sbjct: 144 PEPSEEAK-LFVGNLAYDVNSQALAMLFEQAGTVEIAEVIYNRETDQSRGFGFVTMSSVD 202
Query: 68 AAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDA-GPDHSIFVGDLAPDVTDYLLQE 126
AE ++ +N + G T ER P P ++VG+L DV + L++
Sbjct: 203 EAETAVEKFNRYDLNGRLLTVNKAAPRGSRPERAPRVYEPAFRVYVGNLPWDVDNGRLEQ 262
Query: 127 TFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAA 185
F H G V A+VV D TGRS+G+GFV SD E N A+S ++G R +R++ A
Sbjct: 263 LFSEH-GKVVEARVVYDRETGRSRGFGFVTMSDVDELNEAISALDGQNLEGRAIRVNVA 320
Score = 47.0 bits (110), Expect = 4e-05
Identities = 27/87 (31%), Positives = 45/87 (51%), Gaps = 1/87 (1%)
Query: 102 PDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDES 161
P+ + +FVG+LA DV L F G+V A+V+ + T +S+G+GFV S
Sbjct: 144 PEPSEEAKLFVGNLAYDVNSQALAMLFE-QAGTVEIAEVIYNRETDQSRGFGFVTMSSVD 202
Query: 162 ERNRAMSEMNGVYCSTRPMRISAATPK 188
E A+ + N + R + ++ A P+
Sbjct: 203 EAETAVEKFNRYDLNGRLLTVNKAAPR 229
>ROC1_NICSY (Q08935) 29 kDa ribonucleoprotein A, chloroplast
precursor (CP29A)
Length = 273
Score = 89.7 bits (221), Expect = 5e-18
Identities = 58/179 (32%), Positives = 94/179 (52%), Gaps = 13/179 (7%)
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 76
+++G+L + D L F G V +++I +K+TG+ G+GF+ S E Q +
Sbjct: 89 IFVGNLPFSADSAALAELFERAGNVEMVEVIYDKLTGRSRGFGFVTMSSKEEVEAACQQF 148
Query: 77 NGTQMPGTEQTFRLNW---------ASFGIGERRPDA-GPDHSIFVGDLAPDVTDYLLQE 126
NG ++ G + R+N +SF G R + + ++VG+LA V L E
Sbjct: 149 NGYELDG--RALRVNSGPPPEKRENSSFRGGSRGGGSFDSSNRVYVGNLAWGVDQDAL-E 205
Query: 127 TFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAA 185
T + G V AKVV D ++GRS+G+GFV +S E N A+ ++GV + R +R+S A
Sbjct: 206 TLFSEQGKVVDAKVVYDRDSGRSRGFGFVTYSSAEEVNNAIESLDGVDLNGRAIRVSPA 264
Score = 54.7 bits (130), Expect = 2e-07
Identities = 33/92 (35%), Positives = 47/92 (50%), Gaps = 2/92 (2%)
Query: 96 GIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFV 155
G+ E R + PD IFVG+L L E F G+V +V+ D TGRS+G+GFV
Sbjct: 76 GVEEER-NFSPDLKIFVGNLPFSADSAALAELFE-RAGNVEMVEVIYDKLTGRSRGFGFV 133
Query: 156 KFSDESERNRAMSEMNGVYCSTRPMRISAATP 187
S + E A + NG R +R+++ P
Sbjct: 134 TMSSKEEVEAACQQFNGYELDGRALRVNSGPP 165
Score = 47.0 bits (110), Expect = 4e-05
Identities = 18/74 (24%), Positives = 42/74 (56%)
Query: 10 SLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAA 69
S + +++G+L + VD++ L FS G+V+ K++ ++ +G+ G+GF+ + S
Sbjct: 183 SFDSSNRVYVGNLAWGVDQDALETLFSEQGKVVDAKVVYDRDSGRSRGFGFVTYSSAEEV 242
Query: 70 ERVLQTYNGTQMPG 83
+++ +G + G
Sbjct: 243 NNAIESLDGVDLNG 256
>ROC1_NICPL (P49313) 30 kDa ribonucleoprotein, chloroplast precursor
(CP-RBP30)
Length = 279
Score = 89.4 bits (220), Expect = 7e-18
Identities = 59/184 (32%), Positives = 94/184 (51%), Gaps = 17/184 (9%)
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 76
+++G+L + D L F G V +++I +K+TG+ G+GF+ S E Q +
Sbjct: 89 IFVGNLLFSADSAALAELFERAGNVEMVEVIYDKLTGRSRGFGFVTMSSKEEVEAACQQF 148
Query: 77 NGTQMPGT--------------EQTFRLNWASFGIGERRPDA-GPDHSIFVGDLAPDVTD 121
NG ++ G +FR N +SF G R + + ++VG+LA V
Sbjct: 149 NGYELDGRALRVNSGPPPEKRENSSFREN-SSFRGGSRGGGSFDSSNRVYVGNLAWGVDQ 207
Query: 122 YLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMR 181
L ET + G V AKVV D ++GRS+G+GFV +S E N A+ ++GV + R +R
Sbjct: 208 DAL-ETLFSEQGKVVDAKVVYDRDSGRSRGFGFVTYSSAEEVNNAIESLDGVDLNGRAIR 266
Query: 182 ISAA 185
+S A
Sbjct: 267 VSPA 270
Score = 55.8 bits (133), Expect = 8e-08
Identities = 36/119 (30%), Positives = 59/119 (49%), Gaps = 5/119 (4%)
Query: 81 MPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKV 140
+P +Q + G+ E R + PD IFVG+L L E F G+V +V
Sbjct: 61 LPDFDQIEDVEDGDEGVEEER-NFSPDLKIFVGNLLFSADSAALAELFE-RAGNVEMVEV 118
Query: 141 VTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATP---KKTTGYQQN 196
+ D TGRS+G+GFV S + E A + NG R +R+++ P ++ + +++N
Sbjct: 119 IYDKLTGRSRGFGFVTMSSKEEVEAACQQFNGYELDGRALRVNSGPPPEKRENSSFREN 177
Score = 47.0 bits (110), Expect = 4e-05
Identities = 18/74 (24%), Positives = 42/74 (56%)
Query: 10 SLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAA 69
S + +++G+L + VD++ L FS G+V+ K++ ++ +G+ G+GF+ + S
Sbjct: 189 SFDSSNRVYVGNLAWGVDQDALETLFSEQGKVVDAKVVYDRDSGRSRGFGFVTYSSAEEV 248
Query: 70 ERVLQTYNGTQMPG 83
+++ +G + G
Sbjct: 249 NNAIESLDGVDLNG 262
>TIA1_HUMAN (P31483) Nucleolysin TIA-1 (RNA-binding protein TIA-1)
(p40-TIA-1) [Contains: Nucleolysin TIA-1 isoform p15
(p15-TIA-1)]
Length = 386
Score = 89.0 bits (219), Expect = 9e-18
Identities = 60/198 (30%), Positives = 97/198 (48%), Gaps = 18/198 (9%)
Query: 12 EEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAER 71
E +TL++G+L V E + FS G + K+I + P Y F+EF H A
Sbjct: 4 EMPKTLYVGNLSRDVTEALILQLFSQIGPCKNCKMIMDTAGNDP--YCFVEFHEHRHAAA 61
Query: 72 VLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPD----------HSIFVGDLAPDVTD 121
L NG ++ G E ++NWA+ +++ + +FVGDL+P +T
Sbjct: 62 ALAAMNGRKIMGKE--VKVNWATTPSSQKKDTSSSTVVSTQRSQNHFHVFVGDLSPQITT 119
Query: 122 YLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMR 181
++ F +G + A+VV D TG+SKGYGFV F ++ + A+ +M G + R +R
Sbjct: 120 EDIKAAFAP-FGRISDARVVKDMATGKSKGYGFVSFFNKWDAENAIQQMGGQWLGGRQIR 178
Query: 182 ISAAT---PKKTTGYQQN 196
+ AT P + Y+ N
Sbjct: 179 TNWATRKPPAPKSTYESN 196
Score = 61.6 bits (148), Expect = 1e-09
Identities = 42/178 (23%), Positives = 81/178 (44%), Gaps = 36/178 (20%)
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 76
+++GDL + + F+ G + +++++ TG+ +GYGF+ F + AE +Q
Sbjct: 108 VFVGDLSPQITTEDIKAAFAPFGRISDARVVKDMATGKSKGYGFVSFFNKWDAENAIQQM 167
Query: 77 NGTQMPGTEQTFRLNWASFGIGERRPDA---------------------GPDH-SIFVGD 114
G + G + R NWA+ R+P A P + +++ G
Sbjct: 168 GGQWLGGRQ--IRTNWAT-----RKPPAPKSTYESNTKQLSYDEVVNQSSPSNCTVYCGG 220
Query: 115 LAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNG 172
+ +T+ L+++TF + +G + +V D KGY FV+F+ A+ +NG
Sbjct: 221 VTSGLTEQLMRQTF-SPFGQIMEIRVFPD------KGYSFVRFNSHESAAHAIVSVNG 271
Score = 38.9 bits (89), Expect = 0.010
Identities = 19/68 (27%), Positives = 35/68 (50%), Gaps = 6/68 (8%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T++ G + + E + FS G+++ I++ +K GY F+ F SH +A + +
Sbjct: 215 TVYCGGVTSGLTEQLMRQTFSPFGQIMEIRVFPDK------GYSFVRFNSHESAAHAIVS 268
Query: 76 YNGTQMPG 83
NGT + G
Sbjct: 269 VNGTTIEG 276
>ROC1_ARATH (Q9ZUU4) Putative ribonucleoprotein At2g37220,
chloroplast precursor
Length = 289
Score = 88.2 bits (217), Expect = 1e-17
Identities = 56/190 (29%), Positives = 92/190 (47%), Gaps = 23/190 (12%)
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 76
L++G+L + VD L F G V +++I +KITG+ G+GF+ S S E Q +
Sbjct: 93 LFVGNLPFNVDSAQLAQLFESAGNVEMVEVIYDKITGRSRGFGFVTMSSVSEVEAAAQQF 152
Query: 77 NGTQMP---------------------GTEQTFRLNWASFGIGERRPDAGPDHSIFVGDL 115
NG ++ G +F + + +G G AG + ++VG+L
Sbjct: 153 NGYELDGRPLRVNAGPPPPKREDGFSRGPRSSFGSSGSGYG-GGGGSGAGSGNRVYVGNL 211
Query: 116 APDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYC 175
+ V D L+ F + G V A+V+ D ++GRSKG+GFV + E A+ ++G
Sbjct: 212 SWGVDDMALESLF-SEQGKVVEARVIYDRDSGRSKGFGFVTYDSSQEVQNAIKSLDGADL 270
Query: 176 STRPMRISAA 185
R +R+S A
Sbjct: 271 DGRQIRVSEA 280
Score = 58.5 bits (140), Expect = 1e-08
Identities = 34/93 (36%), Positives = 51/93 (54%), Gaps = 3/93 (3%)
Query: 107 DHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRA 166
D +FVG+L +V L + F + G+V +V+ D TGRS+G+GFV S SE A
Sbjct: 90 DLKLFVGNLPFNVDSAQLAQLFESA-GNVEMVEVIYDKITGRSRGFGFVTMSSVSEVEAA 148
Query: 167 MSEMNGVYCSTRPMRISAA--TPKKTTGYQQNP 197
+ NG RP+R++A PK+ G+ + P
Sbjct: 149 AQQFNGYELDGRPLRVNAGPPPPKREDGFSRGP 181
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,010,818
Number of Sequences: 164201
Number of extensions: 1199622
Number of successful extensions: 3586
Number of sequences better than 10.0: 312
Number of HSP's better than 10.0 without gapping: 238
Number of HSP's successfully gapped in prelim test: 74
Number of HSP's that attempted gapping in prelim test: 2577
Number of HSP's gapped (non-prelim): 695
length of query: 230
length of database: 59,974,054
effective HSP length: 107
effective length of query: 123
effective length of database: 42,404,547
effective search space: 5215759281
effective search space used: 5215759281
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146330.6


