

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144617.5 + phase: 0
(428 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
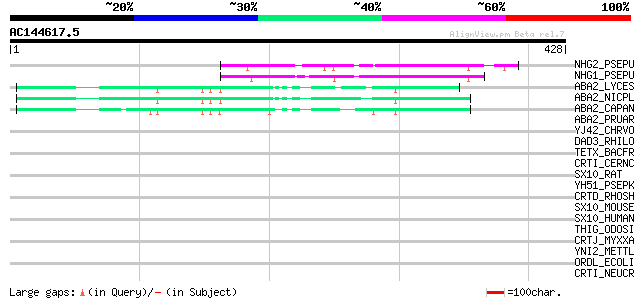
Score E
Sequences producing significant alignments: (bits) Value
NHG2_PSEPU (Q53552) Salicylate hydroxylase (EC 1.14.13.1) (Salic... 51 7e-06
NHG1_PSEPU (P23262) Salicylate hydroxylase (EC 1.14.13.1) (Salic... 50 1e-05
ABA2_LYCES (P93236) Zeaxanthin epoxidase, chloroplast precursor ... 48 4e-05
ABA2_NICPL (Q40412) Zeaxanthin epoxidase, chloroplast precursor ... 48 6e-05
ABA2_CAPAN (Q96375) Zeaxanthin epoxidase, chloroplast precursor ... 47 7e-05
ABA2_PRUAR (O81360) Zeaxanthin epoxidase, chloroplast precursor ... 43 0.001
YJ42_CHRVO (Q7NWN9) Hypothetical UPF0209 protein CV1942 39 0.020
DAD3_RHILO (Q981X2) D-amino acid dehydrogenase 3 small subunit (... 39 0.026
TETX_BACFR (Q01911) Tetracycline resistance protein from transpo... 38 0.044
CRTI_CERNC (P48537) Phytoene dehydrogenase (EC 1.14.99.-) (Phyto... 38 0.044
SX10_RAT (O55170) Transcription factor SOX-10 36 0.22
YH51_PSEPK (Q88M24) Hypothetical UPF0209 protein PP1751 35 0.28
CRTD_RHOSH (Q01671) Methoxyneurosporene dehydrogenase (EC 1.14.9... 35 0.37
SX10_MOUSE (Q04888) Transcription factor SOX-10 (SOX-21) (Transc... 35 0.48
SX10_HUMAN (P56693) Transcription factor SOX-10 35 0.48
THIG_ODOSI (P49570) Thiazole biosynthesis protein thiG 34 0.63
CRTJ_MYXXA (P54979) Phytoene dehydrogenase (EC 1.14.99.-) (Phyto... 34 0.63
YNI2_METTL (P05410) Hypothetical protein in nifH2 3'region (Frag... 33 1.1
ORDL_ECOLI (P37906) Probable oxidoreductase ordL (EC 1.-.-.-) 33 1.1
CRTI_NEUCR (P21334) Phytoene dehydrogenase (EC 1.14.99.-) (Phyto... 33 1.1
>NHG2_PSEPU (Q53552) Salicylate hydroxylase (EC 1.14.13.1)
(Salicylate 1-monooxygenase)
Length = 435
Score = 50.8 bits (120), Expect = 7e-06
Identities = 62/245 (25%), Positives = 103/245 (41%), Gaps = 31/245 (12%)
Query: 163 DLLVAADGCLSSIRRKYLPD-----FKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPD 217
DLL+ DG S++R L + R+SG CA+RG++D ++ + I GI + D
Sbjct: 158 DLLIGRDGIKSALRSYVLEGQGQDHLEPRFSGTCAYRGMVDSLQLREAYRINGIDEHLVD 217
Query: 218 LGKCLYFDLASGTHSVLYELPNKK---LNWIWYV---NQPEPEVKGTSVTTKVTSDMIQK 271
+ + + G + + P +K +N + + +QPEP + + S Q+
Sbjct: 218 VPQ-----MYLGLYGHILTFPVRKGRIVNVVAFTSDRSQPEPTWPADAPWVREAS---QR 269
Query: 272 MHQEAEKIWIPELAKVMKETKEPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRST 331
+A W + +++ P L ++D L V L+G+AAH PH
Sbjct: 270 EMLDAFAGW-GDARALLECIPAPTLWALHDLPELPGYVHGRVALIGDAAHAMLPHQGAGA 328
Query: 332 NMTILDAAVLGKCIEKWGTEM--LESALEEYQLTRLPVTSKQVLHARRLGR--IKQGLVL 387
+ DA L + + TE L L Y R P HA R+ R ++ G +
Sbjct: 329 GQGLEDAYFLARLLGDSRTETGNLPELLGAYDDLRRP-------HACRVQRTTVETGELY 381
Query: 388 PDRDP 392
RDP
Sbjct: 382 ELRDP 386
>NHG1_PSEPU (P23262) Salicylate hydroxylase (EC 1.14.13.1)
(Salicylate 1-monooxygenase)
Length = 433
Score = 50.1 bits (118), Expect = 1e-05
Identities = 54/215 (25%), Positives = 92/215 (42%), Gaps = 17/215 (7%)
Query: 163 DLLVAADGCLSSIRRKYLPDFKL-----RYSGYCAWRGVLDFSEIENSETIKGIKKAYPD 217
DLL+ ADG S++R L L R+SG CA+RG++D + + GI + D
Sbjct: 155 DLLIGADGIKSALRSHVLEGQGLAPQVPRFSGTCAYRGMVDSLHLREAYRAHGIDEHLVD 214
Query: 218 LGKCLYFDLASGTHSVLYELPNKKL-NWIWYVN---QPEPEVKGTSVTTKVTSDMIQKMH 273
+ + +Y L H + + + N + N + +++ +P+P + + S Q+
Sbjct: 215 VPQ-MYLGLDG--HILTFPVRNGGIINVVAFISDRSEPKPTWPADAPWVREAS---QREM 268
Query: 274 QEAEKIWIPELAKVMKETKEPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNM 333
+A W +++ P L ++D L VVL+G+AAH PH
Sbjct: 269 LDAFAGWGDAARALLECIPAPTLWALHDLAELPGYVHGRVVLIGDAAHAMLPHQGAGAGQ 328
Query: 334 TILDAAVLGKCIEKWGTEM--LESALEEYQLTRLP 366
+ DA L + + + L LE Y R P
Sbjct: 329 GLEDAYFLARLLGDTQADAGNLAELLEAYDDLRRP 363
>ABA2_LYCES (P93236) Zeaxanthin epoxidase, chloroplast precursor (EC
1.14.-.-)
Length = 669
Score = 48.1 bits (113), Expect = 4e-05
Identities = 89/362 (24%), Positives = 136/362 (36%), Gaps = 58/362 (16%)
Query: 6 RKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW 65
+K K ++ GG IGG+ A A G+DVLV E+ S G G
Sbjct: 84 KKLKVLVAGGGIGGLVFALAAKKRGFDVLVFERDLSAIRGEGQYRG-------------- 129
Query: 66 ISKPPQFLHNTTHPL-TIDQNLVTDSEKKVNRTLTRDESL-NFRAAHWG---DLHGVLYE 120
P Q N L ID ++ D T R L + + +W D E
Sbjct: 130 ---PIQIQSNALAALEAIDLDVAEDIMNAGCITGQRINGLVDGISGNWYCKFDTFTPAVE 186
Query: 121 SLPPQIFLWGHIFLSFHVANEKGPVIV--KAKVVK---TGEVIEIV--------GDLLVA 167
P + + L +A G I+ ++ VV GE + +V GDLLV
Sbjct: 187 RGLPVTRVISRMTLQQILARAVGEEIIMNESNVVDFEDDGEKVTVVLENGQRFTGDLLVG 246
Query: 168 ADGCLSSIRRKYLPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLA 227
ADG S +R + YSGY + G+ DF + +T+ G + LG YF
Sbjct: 247 ADGIRSKVRTNLFGPSEATYSGYTCYTGIADFVPAD-IDTV-GYRVF---LGHKQYF--- 298
Query: 228 SGTHSVLYELPNKKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKV 287
V ++ K+ W + N+P G + + + K+ W + +
Sbjct: 299 -----VSSDVGGGKMQWYAFYNEP----AGGADAPNGKKERLLKIFGG----WCDNVIDL 345
Query: 288 MKETKEPFL--NFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCI 345
+ T E + IYD P V L+G++ H P+ + M I D+ L +
Sbjct: 346 LVATDEDAILRRDIYDRPPTFSWGRGRVTLLGDSVHAMQPNLGQGGCMAIEDSYQLALEL 405
Query: 346 EK 347
EK
Sbjct: 406 EK 407
>ABA2_NICPL (Q40412) Zeaxanthin epoxidase, chloroplast precursor (EC
1.14.-.-)
Length = 663
Score = 47.8 bits (112), Expect = 6e-05
Identities = 89/370 (24%), Positives = 137/370 (36%), Gaps = 58/370 (15%)
Query: 6 RKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW 65
+K K ++ GG IGG+ A A G+DVLV E+ S G G
Sbjct: 78 KKLKVLVAGGGIGGLVFALAAKKRGFDVLVFERDLSAIRGEGQYRG-------------- 123
Query: 66 ISKPPQFLHNTTHPL-TIDQNLVTDSEKKVNRTLTRDESL-NFRAAHWG---DLHGVLYE 120
P Q N L ID ++ D T R L + + +W D E
Sbjct: 124 ---PIQIQSNALAALEAIDMDVAEDIMNAGCITGQRINGLVDGVSGNWYCKFDTFTPAVE 180
Query: 121 SLPPQIFLWGHIFLSFHVANEKGPVIV--KAKVVK---TGEVIEIV--------GDLLVA 167
P + + L ++A G I+ ++ VV GE + + GDLLV
Sbjct: 181 RGLPVTRVISRMTLQQNLARAVGEDIIMNESNVVNFEDDGEKVTVTLEDGQQYTGDLLVG 240
Query: 168 ADGCLSSIRRKYLPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLA 227
ADG S +R + YSGY + G+ DF + ET+ G + LG YF
Sbjct: 241 ADGIRSKVRTNLFGPSDVTYSGYTCYTGIADFVPAD-IETV-GYRVF---LGHKQYF--- 292
Query: 228 SGTHSVLYELPNKKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKV 287
V ++ K+ W + N+P V + + + W + +
Sbjct: 293 -----VSSDVGGGKMQWYAFHNEPAGGVDDPNGKKARLLKIFEG--------WCDNVIDL 339
Query: 288 MKETKEPFL--NFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCI 345
+ T E + IYD P V L+G++ H P+ + M I D+ L +
Sbjct: 340 LVATDEDAILRRDIYDRPPTFSWGKGRVTLLGDSVHAMQPNLGQGGCMAIEDSYQLALEL 399
Query: 346 EKWGTEMLES 355
+K + ES
Sbjct: 400 DKALSRSAES 409
>ABA2_CAPAN (Q96375) Zeaxanthin epoxidase, chloroplast precursor
(Xanthophyll epoxidase) (Beta-cyclohexenyl epoxidase)
Length = 660
Score = 47.4 bits (111), Expect = 7e-05
Identities = 91/378 (24%), Positives = 142/378 (37%), Gaps = 74/378 (19%)
Query: 6 RKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW 65
+K K ++ GG IGG+ A A G+DVLV E+ S G G
Sbjct: 76 KKLKVLVAGGGIGGLVFALAGKKRGFDVLVFERDISAIRGEGQYRG-------------- 121
Query: 66 ISKPPQFLHNTTHPL-TIDQNLVTDSEKKVNRTLTRDESLNFR----AAHWG---DLHGV 117
P Q N L ID ++ +E+ +N + +N + +W D
Sbjct: 122 ---PIQIQSNALAALEAIDMDV---AEEIMNAGCITGQRINGLVDGISGNWYCKFDTFTP 175
Query: 118 LYESLPPQIFLWGHIFLSFHVANEKGPVIV--KAKVVK---TGEVIEI--------VGDL 164
E P + + L +A +G ++ ++ VV GE + + GDL
Sbjct: 176 AVERGLPVTRVISRMTLQQILARLQGEDVIMNESHVVNFADDGETVTVNPELCQQYTGDL 235
Query: 165 LVAADGCLSSIRRKYLPDFKLRYSGYCAWRGVLDF--SEIENSETIKGIKKAYPDLGKCL 222
LV ADG S +R +L YSGY + G+ DF ++I+ + G + LG
Sbjct: 236 LVGADGIRSKVRTNLFGPSELTYSGYTCYTGIADFVPADIDTA----GYRVF---LGHKQ 288
Query: 223 YFDLASGTHSVLYELPNKKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKI--- 279
YF V ++ K+ W + N+P V D + KI
Sbjct: 289 YF--------VSSDVGGGKMQWYAFHNEPAGGV-----------DAPNGKKERLLKIFGG 329
Query: 280 WIPELAKVMKETKEPFL--NFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILD 337
W + + T E + IYD P V L+G++ H P+ + M I D
Sbjct: 330 WCDNVIDLSVATDEDAILRRDIYDRPPTFSWGKGRVTLLGDSVHAMQPNLGQGGCMAIED 389
Query: 338 AAVLGKCIEKWGTEMLES 355
+ L +EK + ES
Sbjct: 390 SYQLALELEKAWSRSAES 407
>ABA2_PRUAR (O81360) Zeaxanthin epoxidase, chloroplast precursor (EC
1.14.-.-) (PA-ZE)
Length = 661
Score = 43.1 bits (100), Expect = 0.001
Identities = 83/389 (21%), Positives = 153/389 (38%), Gaps = 70/389 (17%)
Query: 6 RKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGS---------PTGAGLGLNS 56
+K + ++ GG IGG+ A A G+DV+V EK S G + A L +
Sbjct: 79 KKLRILVAGGGIGGLVFALAAKKKGFDVVVFEKDLSAVRGEGQYRGPIQIQSNALAALEA 138
Query: 57 LSQQIIQSWI-------SKPPQFLHNTTHPLTIDQNLVTDSEKK---VNRTLTRDESLNF 106
+ + + + + + + + + T + ++ V R ++R
Sbjct: 139 IDMDVAEEVMRVGCVTGDRINGLVDGVSGTWYVKFDTFTPAVERGLPVTRVISRIA---- 194
Query: 107 RAAHWGDLHGVLYESLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLV 166
L +L ++ +I + ++F +K VI++ G+ E GD+LV
Sbjct: 195 -------LQQILARAVGEEIIINDSNVVNFEDLGDKVNVILE-----NGQRYE--GDMLV 240
Query: 167 AADGCLSSIRRKYLPDFKLRYSGYCAWRGVLDFSEIE-NSETIKGIKKAYPDLGKCLYFD 225
ADG S +R+ + YSGY + G+ DF + NS + LG YF
Sbjct: 241 GADGIWSKVRKNLFGLNEAVYSGYTCYTGIADFVPADINSVGYRVF------LGHKQYF- 293
Query: 226 LASGTHSVLYELPNKKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELA 285
V ++ K+ W + E G + + + K+ + W +
Sbjct: 294 -------VSSDVGGGKMQWYAF----HKESPGGVDSPNGKKERLLKIFEG----WCDNVI 338
Query: 286 KVMKETKEPFL--NFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGK 343
++ T+E + IYD P+ +V L+G++ H P+ + M I D L
Sbjct: 339 DLLLATEEDAILRRDIYDRTPILTWGKGHVTLLGDSVHAMQPNMGQGGCMAIEDGYQLAL 398
Query: 344 CIEK-W------GTEM-LESALEEYQLTR 364
++K W GT + + S+L Y+ +R
Sbjct: 399 ELDKAWKKSSETGTPVDVASSLRSYENSR 427
>YJ42_CHRVO (Q7NWN9) Hypothetical UPF0209 protein CV1942
Length = 660
Score = 39.3 bits (90), Expect = 0.020
Identities = 18/47 (38%), Positives = 29/47 (61%), Gaps = 3/47 (6%)
Query: 6 RKPKAVIVGGSIGGISSAHALILAGWDVLVLEK---TTSPPTGSPTG 49
R+ A++VGG + G +SA +L L GW V ++E+ S +G+P G
Sbjct: 253 RERSAIVVGGGLAGAASARSLALRGWQVTLIERMPQLASAASGNPQG 299
>DAD3_RHILO (Q981X2) D-amino acid dehydrogenase 3 small subunit
(EC 1.4.99.1)
Length = 412
Score = 38.9 bits (89), Expect = 0.026
Identities = 18/58 (31%), Positives = 33/58 (56%)
Query: 8 PKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW 65
PK V++G I G+S+A+AL+ G+DV V+E+ + G L++ + ++ W
Sbjct: 2 PKIVVIGAGIAGVSTAYALLEQGYDVTVVERRRYAAMETSFANGGQLSASNAEVWNHW 59
>TETX_BACFR (Q01911) Tetracycline resistance protein from transposon
Tn4351/Tn4400
Length = 388
Score = 38.1 bits (87), Expect = 0.044
Identities = 78/368 (21%), Positives = 140/368 (37%), Gaps = 52/368 (14%)
Query: 12 IVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWISKPPQ 71
I+GG G++ A L G DV V E+ G L L+ S Q
Sbjct: 21 IIGGGPVGLTMAKLLQQNGIDVSVYERDNDREARI-FGGTLDLHKGSGQEAMKKAG---- 75
Query: 72 FLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFR----AAHWGDLHGVLYESLPPQIF 127
L T + L + V ++KK N T++ R + DL +L SL
Sbjct: 76 -LLQTYYDLALPMG-VNIADKKGNILSTKNVKPENRFDNPEINRNDLRAILLNSLENDTV 133
Query: 128 LWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRY 187
+W + +K + + K +T DL++ A+G +S +R K++ D
Sbjct: 134 IWDRKLVMLEPGKKKWTLTFENKPSETA-------DLVILANGGMSKVR-KFVTD----- 180
Query: 188 SGYCAWRGVLDFSEIENSETIKGIKKAY-PDLGKCLYFDLASGTH-------SVLYELPN 239
+E+E + T + P++ +F L +G ++L+ PN
Sbjct: 181 ------------TEVEETGTFNIQADIHQPEINCPGFFQLCNGNRLMASHQGNLLFANPN 228
Query: 240 KK--LNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLN 297
L++ P+ T V + + ++ + +E W E K + T F+
Sbjct: 229 NNGALHFGISFKTPDEWKNQTQVDFQNRNSVVDFLLKEFSD-W-DERYKELIHTTLSFVG 286
Query: 298 FIYDSDPLEKIFWDN----VVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEML 353
PLEK + + ++G+AAH P + N ++DA +L + +
Sbjct: 287 LATRIFPLEKPWKSKRPLPITMIGDAAHLMPPFAGQGVNSGLVDALILSDNLADGKFNSI 346
Query: 354 ESALEEYQ 361
E A++ Y+
Sbjct: 347 EEAVKNYE 354
>CRTI_CERNC (P48537) Phytoene dehydrogenase (EC 1.14.99.-)
(Phytoene desaturase)
Length = 621
Score = 38.1 bits (87), Expect = 0.044
Identities = 16/38 (42%), Positives = 25/38 (65%)
Query: 1 MVGEGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEK 38
M ++P A+++G +GG+S+A L AG+ V VLEK
Sbjct: 1 MPSTSKRPTAIVIGSGVGGVSTAARLARAGFHVTVLEK 38
>SX10_RAT (O55170) Transcription factor SOX-10
Length = 466
Score = 35.8 bits (81), Expect = 0.22
Identities = 31/98 (31%), Positives = 41/98 (41%), Gaps = 9/98 (9%)
Query: 2 VGEGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSP------TGAGLGLN 55
+GEG KP IG IS + +DV L++ PP G P + AG GL+
Sbjct: 263 LGEGGKPHIDFGNVDIGEISHEVMSNMETFDVTELDQYL-PPNGHPGHVGSYSAAGYGLS 321
Query: 56 SLSQQII--QSWISKPPQFLHNTTHPLTIDQNLVTDSE 91
S +WISKPP T P +D +E
Sbjct: 322 SALAVASGHSAWISKPPGVALPTVSPPAVDAKAQVKTE 359
>YH51_PSEPK (Q88M24) Hypothetical UPF0209 protein PP1751
Length = 654
Score = 35.4 bits (80), Expect = 0.28
Identities = 17/44 (38%), Positives = 27/44 (60%), Gaps = 3/44 (6%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSP---PTGSPTG 49
+A+++G + G SSA +L GW V VLE+ +P +G+P G
Sbjct: 258 EALVIGAGLAGSSSAASLARRGWQVTVLERHEAPAQEASGNPQG 301
>CRTD_RHOSH (Q01671) Methoxyneurosporene dehydrogenase (EC
1.14.99.-)
Length = 495
Score = 35.0 bits (79), Expect = 0.37
Identities = 42/138 (30%), Positives = 54/138 (38%), Gaps = 28/138 (20%)
Query: 8 PKAVIVGGSIGGISSAHALILAGWDVLVLE--------KTTSPPTGSPTGAG---LGLNS 56
PK V+VG +GG++SA L AG +V +LE T P P AG L L
Sbjct: 6 PKVVVVGAGMGGLASAIRLARAGCEVTLLEAREAPGGRMRTLPSVAGPVDAGPTVLTLRE 65
Query: 57 LSQQIIQSWISKPPQFLHNTTHPL----------TIDQNLVTDSEKKVNRTLT---RDES 103
+ I + K L PL T+D L TD E V E+
Sbjct: 66 VFDDIFEVCGQKLDHHLTLLPQPLLARHWWLDGSTLD--LTTDLEANVEAVAAFAGAREA 123
Query: 104 LNFRAAHWGDLHGVLYES 121
FR H DL LY++
Sbjct: 124 PAFRRFH--DLSARLYDA 139
>SX10_MOUSE (Q04888) Transcription factor SOX-10 (SOX-21)
(Transcription factor SOX-M)
Length = 466
Score = 34.7 bits (78), Expect = 0.48
Identities = 31/98 (31%), Positives = 40/98 (40%), Gaps = 9/98 (9%)
Query: 2 VGEGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSP------TGAGLGLN 55
+GEG KP IG IS + +DV L++ PP G P + AG GL
Sbjct: 263 LGEGGKPHIDFGNVDIGEISHEVMSNMETFDVTELDQYL-PPNGHPGHVGSYSAAGYGLG 321
Query: 56 SLSQQII--QSWISKPPQFLHNTTHPLTIDQNLVTDSE 91
S +WISKPP T P +D +E
Sbjct: 322 SALAVASGHSAWISKPPGVALPTVSPPGVDAKAQVKTE 359
>SX10_HUMAN (P56693) Transcription factor SOX-10
Length = 466
Score = 34.7 bits (78), Expect = 0.48
Identities = 31/98 (31%), Positives = 40/98 (40%), Gaps = 9/98 (9%)
Query: 2 VGEGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSP------TGAGLGLN 55
+GEG KP IG IS + +DV L++ PP G P + AG GL
Sbjct: 263 MGEGGKPHIDFGNVDIGEISHEVMSNMETFDVAELDQYL-PPNGHPGHVSSYSAAGYGLG 321
Query: 56 SLSQQII--QSWISKPPQFLHNTTHPLTIDQNLVTDSE 91
S +WISKPP T P +D +E
Sbjct: 322 SALAVASGHSAWISKPPGVALPTVSPPGVDAKAQVKTE 359
>THIG_ODOSI (P49570) Thiazole biosynthesis protein thiG
Length = 261
Score = 34.3 bits (77), Expect = 0.63
Identities = 31/100 (31%), Positives = 44/100 (44%), Gaps = 25/100 (25%)
Query: 17 IGGISSAHALILAGWDVLV--------------LEKTTSPPTGSPTGAGLGLNSLSQ-QI 61
IG + +A L+ G+ VL L T P GSP G+G GLN++S QI
Sbjct: 124 IGTLKAAEFLVKKGFTVLPYINADPMLALHLEDLGCATVMPLGSPIGSGQGLNNISNLQI 183
Query: 62 IQSWISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRD 101
I + N++ P+ ID + T SE + L D
Sbjct: 184 I----------IENSSIPVIIDAGIGTPSEAAKSMELGAD 213
>CRTJ_MYXXA (P54979) Phytoene dehydrogenase (EC 1.14.99.-)
(Phytoene desaturase)
Length = 517
Score = 34.3 bits (77), Expect = 0.63
Identities = 15/34 (44%), Positives = 22/34 (64%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSP 42
+ V+VG +GG+++A L G+DV V EKT P
Sbjct: 9 RIVVVGAGVGGLAAAARLAHQGFDVQVFEKTQGP 42
>YNI2_METTL (P05410) Hypothetical protein in nifH2 3'region
(Fragment)
Length = 102
Score = 33.5 bits (75), Expect = 1.1
Identities = 18/30 (60%), Positives = 20/30 (66%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEK 38
K V+VGG G+ SA AL G DVLVLEK
Sbjct: 2 KIVVVGGGTSGLLSALALEKEGHDVLVLEK 31
>ORDL_ECOLI (P37906) Probable oxidoreductase ordL (EC 1.-.-.-)
Length = 426
Score = 33.5 bits (75), Expect = 1.1
Identities = 21/50 (42%), Positives = 28/50 (56%)
Query: 12 IVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQI 61
+VGG G+SSA L AG+DV+VLE + S G +NS S+ I
Sbjct: 32 VVGGGYTGLSSALHLAEAGFDVVVLEASRIGFGASGRNGGQLVNSYSRDI 81
>CRTI_NEUCR (P21334) Phytoene dehydrogenase (EC 1.14.99.-)
(Phytoene desaturase) (Albino-1 protein)
Length = 595
Score = 33.5 bits (75), Expect = 1.1
Identities = 19/38 (50%), Positives = 25/38 (65%), Gaps = 1/38 (2%)
Query: 2 VGEGRKPK-AVIVGGSIGGISSAHALILAGWDVLVLEK 38
+ E ++P+ A+IVG GGI+ A L AG DV VLEK
Sbjct: 1 MAETQRPRSAIIVGAGAGGIAVAARLAKAGVDVTVLEK 38
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,663,165
Number of Sequences: 164201
Number of extensions: 2288793
Number of successful extensions: 5988
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 5948
Number of HSP's gapped (non-prelim): 52
length of query: 428
length of database: 59,974,054
effective HSP length: 113
effective length of query: 315
effective length of database: 41,419,341
effective search space: 13047092415
effective search space used: 13047092415
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC144617.5


