

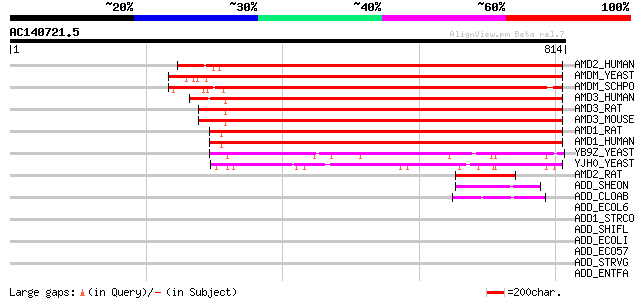
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.5 - phase: 0
(814 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AMD2_HUMAN (Q01433) AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase ... 644 0.0
AMDM_YEAST (P15274) AMP deaminase (EC 3.5.4.6) (Myoadenylate dea... 637 0.0
AMDM_SCHPO (P50998) AMP deaminase (EC 3.5.4.6) (Myoadenylate dea... 623 e-178
AMD3_HUMAN (Q01432) AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase ... 610 e-174
AMD3_RAT (O09178) AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase is... 606 e-173
AMD3_MOUSE (O08739) AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase ... 603 e-172
AMD1_RAT (P10759) AMP deaminase 1 (EC 3.5.4.6) (Myoadenylate dea... 567 e-161
AMD1_HUMAN (P23109) AMP deaminase 1 (EC 3.5.4.6) (Myoadenylate d... 564 e-160
YB9Z_YEAST (P38150) Hypothetical 92.9 kDa protein in SSH1-APE3 i... 217 9e-56
YJH0_YEAST (P40361) Hypothetical 104.3 kDa protein in SMC3-MRPL8... 205 4e-52
AMD2_RAT (Q02356) AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase is... 140 2e-32
ADD_SHEON (Q8E8D4) Adenosine deaminase (EC 3.5.4.4) (Adenosine a... 52 8e-06
ADD_CLOAB (Q97EV1) Adenosine deaminase (EC 3.5.4.4) (Adenosine a... 49 5e-05
ADD_ECOL6 (Q8FH99) Adenosine deaminase (EC 3.5.4.4) (Adenosine a... 44 0.002
ADD1_STRCO (O86737) Probable adenosine deaminase 1 (EC 3.5.4.4) ... 43 0.003
ADD_SHIFL (Q83RC0) Adenosine deaminase (EC 3.5.4.4) (Adenosine a... 42 0.005
ADD_ECOLI (P22333) Adenosine deaminase (EC 3.5.4.4) (Adenosine a... 42 0.005
ADD_ECO57 (Q8X661) Adenosine deaminase (EC 3.5.4.4) (Adenosine a... 42 0.005
ADD_STRVG (P53984) Adenosine deaminase (EC 3.5.4.4) (Adenosine a... 42 0.007
ADD_ENTFA (Q839J4) Adenosine deaminase (EC 3.5.4.4) (Adenosine a... 41 0.015
>AMD2_HUMAN (Q01433) AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase
isoform L)
Length = 879
Score = 644 bits (1662), Expect = 0.0
Identities = 319/587 (54%), Positives = 399/587 (67%), Gaps = 24/587 (4%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTPKPNLEPFFYAPEGKSDHYFE-------- 297
LQ +RY+ + A P E P TP P P H +E
Sbjct: 259 LQSFCPTTRRYLQQLAEKPLETRTYEQGPDTPVSADAPVH--PPALEQHPYEHCEPSTMP 316
Query: 298 --------MQDGVIHVY----PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCH 345
M GV+HVY P+++ +E P D F D++ ++ +I G I++ C+
Sbjct: 317 GDLGLGLRMVRGVVHVYTRREPDEHCSEVELPYPDLQEFVADVNVLMALIINGPIKSFCY 376
Query: 346 HRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKS 405
RL L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK
Sbjct: 377 RRLQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKR 436
Query: 406 KLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNP 465
+++ +E+V G TLREVFES++LT YDL+VD LDVHAD++TFHRFDKFN KYNP
Sbjct: 437 AMKRHLEEIVHVEQGREQTLREVFESMNLTAYDLSVDTLDVHADRNTFHRFDKFNAKYNP 496
Query: 466 CGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLAS 525
G+S LREIF+K DN + G++ + K+V SDLE SKYQ AE R+SIYGR + EWD+LA
Sbjct: 497 IGESVLREIFIKTDNRVSGKYFAHIIKEVMSDLEESKYQNAELRLSIYGRSRDEWDKLAR 556
Query: 526 WIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHV 585
W V + ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+
Sbjct: 557 WAVMHRVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHL 616
Query: 586 FLKQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRES 644
FL+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR
Sbjct: 617 FLEHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQ 676
Query: 645 KGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLS 704
+G T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLS
Sbjct: 677 RGFHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLS 736
Query: 705 NNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCE 764
NNSLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE
Sbjct: 737 NNSLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCE 796
Query: 765 IARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
+ARNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 797 LARNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 843
>AMDM_YEAST (P15274) AMP deaminase (EC 3.5.4.6) (Myoadenylate
deaminase)
Length = 810
Score = 637 bits (1644), Expect = 0.0
Identities = 321/605 (53%), Positives = 413/605 (68%), Gaps = 26/605 (4%)
Query: 233 MEAPSPDEIESYVILQECLEMRKRY----IFKEAVAPWEK------------EVISD--- 273
++ PS + I+ Y + EC +R +Y + + P K SD
Sbjct: 182 LDDPSSELIDLYSKVAECRNLRAKYQTISVQNDDQNPKNKPGWVVYPPPPKPSYNSDTKT 241
Query: 274 --PSTPKPNLEPFFYA----PEGKSDHYFEMQDGVIHVYPNKNSNEELFP-VADATTFFT 326
P T KP+ E F + P D F + D +V +EL + ++
Sbjct: 242 VVPVTNKPDAEVFDFTKCEIPGEDPDWEFTLNDDDSYVVHRSGKTDELIAQIPTLRDYYL 301
Query: 327 DLHQILRVIAAGNIRTLCHHRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDT 386
DL +++ + + G ++ + RL LE ++NL+ +LN +E K PHRDFYNVRKVDT
Sbjct: 302 DLEKMISISSDGPAKSFAYRRLQYLEARWNLYYLLNEYQETSVSKRNPHRDFYNVRKVDT 361
Query: 387 HVHHSACMNQKHLLRFIKSKLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDV 446
HVHHSACMNQKHLLRFIK KLR DE VIFRDG LTL EVF SL LTGYDL++D LD+
Sbjct: 362 HVHHSACMNQKHLLRFIKHKLRHSKDEKVIFRDGKLLTLDEVFRSLHLTGYDLSIDTLDM 421
Query: 447 HADKSTFHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMA 506
HA K TFHRFDKFNLKYNP G+SRLREIFLK +N I+G +L ++TKQV DLE SKYQ
Sbjct: 422 HAHKDTFHRFDKFNLKYNPIGESRLREIFLKTNNYIKGTYLADITKQVIFDLENSKYQNC 481
Query: 507 EYRISIYGRKQSEWDQLASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNI 566
EYRIS+YGR EWD+LASW+++N + S NV WL+Q+PRLY+IYK GIV SFQ++ N+
Sbjct: 482 EYRISVYGRSLDEWDKLASWVIDNKVISHNVRWLVQIPRLYDIYKKTGIVQSFQDICKNL 541
Query: 567 FIPLFEVTVDPDSHPQLHVFLKQVVGLDLVDDESKPERRPTKHMPTPAQWTNVFNPAFSY 626
F PLFEVT +P SHP+LHVFL++V+G D VDDESK +RR + P P+ W NP +SY
Sbjct: 542 FQPLFEVTKNPQSHPKLHVFLQRVIGFDSVDDESKVDRRFHRKYPKPSLWEAPQNPPYSY 601
Query: 627 YVYYCYANLYTLNKLRESKGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPV 686
Y+YY Y+N+ +LN+ R +G T+ RPH GEAGD +HL + +L AH I+HGI LRK P
Sbjct: 602 YLYYLYSNVASLNQWRAKRGFNTLVLRPHCGEAGDPEHLVSAYLLAHGISHGILLRKVPF 661
Query: 687 LQYLYYLAQIGLAMSPLSNNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVE 746
+QYLYYL Q+G+AMSPLSNN+LFL Y +NP P +F RGLNVSLSTDDPLQ T+EPL+E
Sbjct: 662 VQYLYYLDQVGIAMSPLSNNALFLTYDKNPFPRYFKRGLNVSLSTDDPLQFSYTREPLIE 721
Query: 747 EYSIAASVWKLSSCDLCEIARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHI 806
EYS+AA ++KLS+ D+CE+ARNSV QSG+ +K HWIGK++ K G GND+ RTNVP I
Sbjct: 722 EYSVAAQIYKLSNVDMCELARNSVLQSGWEAQIKKHWIGKDFDKSGVEGNDVVRTNVPDI 781
Query: 807 RLEFR 811
R+ +R
Sbjct: 782 RINYR 786
>AMDM_SCHPO (P50998) AMP deaminase (EC 3.5.4.6) (Myoadenylate
deaminase)
Length = 846
Score = 623 bits (1607), Expect = e-178
Identities = 317/599 (52%), Positives = 412/599 (67%), Gaps = 28/599 (4%)
Query: 233 MEAPSP---DEI-ESYVILQECLEMRKRYIFKEAVAPWEKEVISDPSTPKPNLEP----- 283
ME P DE+ E Y+ + +C++MR +YI + + D P+ +
Sbjct: 161 METTDPLFSDELAEIYLSIHKCMDMRHKYIRVSLQGELDNPIDDDSWIIYPDCKEGEDDT 220
Query: 284 --FFYAP------EGKSDHYFEMQDGVIHVYPNKNS---NEELFPVADATTFFTDLHQIL 332
F +A E + +++ + Q G+ VY N ++ F + ++ DL +L
Sbjct: 221 GLFNFADCKIPGIENEMEYHMDHQ-GIFQVYENDSAYIAGTPSFHIPTIRDYYIDLEFLL 279
Query: 333 RVIAAGNIRTLCHHRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSA 392
+ G ++ RL LE ++N++++LN +E K PHRDFYNVRKVDTHVHHSA
Sbjct: 280 SASSDGPSKSFSFRRLQYLEGRWNMYMLLNEYQELADTKKVPHRDFYNVRKVDTHVHHSA 339
Query: 393 CMNQKHLLRFIKSKLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKST 452
NQKHLLRFIK+KLRK P+E VI+RDG +LTL+EVF+SL LT YDL++D LD+HA T
Sbjct: 340 LANQKHLLRFIKAKLRKCPNEKVIWRDGKFLTLQEVFDSLKLTSYDLSIDTLDMHAHTDT 399
Query: 453 FHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISI 512
FHRFDKFNLKYNP G+SRLR IFLK DN I GR+L ELTK+VF+DL KYQMAEYRISI
Sbjct: 400 FHRFDKFNLKYNPIGESRLRTIFLKTDNDINGRYLAELTKEVFTDLRTQKYQMAEYRISI 459
Query: 513 YGRKQSEWDQLASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFE 572
YGR + EWD+LA+WI++N+L+S NV WLIQ+PRLY++YK GIV +F+ ++ N+F PLFE
Sbjct: 460 YGRNREEWDKLAAWIIDNELFSPNVRWLIQVPRLYDVYKKSGIVETFEEVVRNVFEPLFE 519
Query: 573 VTVDPDSHPQLHVFLKQVVGLDLVDDESKPERRPTKHMPTPAQWTNVFNPAFSYYVYYCY 632
VT DP +HP+LHVFL++V+G D VDDESKPERR + P P W NP +SY++YY Y
Sbjct: 520 VTKDPRTHPKLHVFLQRVIGFDSVDDESKPERRTFRKFPYPKHWDINLNPPYSYWLYYMY 579
Query: 633 ANLYTLNKLRESKGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYY 692
AN+ +LN R+ +G T RPH GEAGD DHLA+ FL +H I HGI LRK P LQYL+Y
Sbjct: 580 ANMTSLNSWRKIRGFNTFVLRPHCGEAGDTDHLASAFLLSHGINHGILLRKVPFLQYLWY 639
Query: 693 LAQIGLAMSPLSNNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAA 752
L QI +AMSPLSNN+LFL Y +NP +F RGLNVSLSTDDPLQ T+EPL+EEY++AA
Sbjct: 640 LDQIPIAMSPLSNNALFLAYDKNPFLTYFKRGLNVSLSTDDPLQFAFTREPLIEEYAVAA 699
Query: 753 SVWKLSSCDLCEIARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
++KLS+ D+CE+ARNSV QSGF LK W+G ++ DI RTNVP IRL +R
Sbjct: 700 QIYKLSAVDMCELARNSVLQSGFERQLKERWLGVDF-------QDIDRTNVPIIRLAYR 751
>AMD3_HUMAN (Q01432) AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase
isoform E) (Erythrocyte AMP deaminase)
Length = 767
Score = 610 bits (1572), Expect = e-174
Identities = 302/555 (54%), Positives = 381/555 (68%), Gaps = 8/555 (1%)
Query: 264 APWEKEVISDPSTPKPNLEPFFY--APEGKSDHYFEMQDGVIHVYPNKNSNEEL----FP 317
AP E+ + P P +P+ AP D+ MQ G++ VY NK E P
Sbjct: 189 APPEEGLPDFHPPPLPQEDPYCLDDAPPNL-DYLVHMQGGILFVYDNKKMLEHQEPHSLP 247
Query: 318 VADATTFFTDLHQILRVIAAGNIRTLCHHRLNLLEQKFNLHLMLNADREFLAQKSAPHRD 377
D T+ D+ IL +I G +T CH RLN LE KF+LH MLN EF KS PHRD
Sbjct: 248 YPDLETYTVDMSHILALITDGPTKTYCHRRLNFLESKFSLHEMLNEMSEFKELKSNPHRD 307
Query: 378 FYNVRKVDTHVHHSACMNQKHLLRFIKSKLRKEPDEVVIFRDGTYLTLREVFESLDLTGY 437
FYNVRKVDTH+H +ACMNQKHLLRFIK + EPD V + G +TLR+VF+ L + Y
Sbjct: 308 FYNVRKVDTHIHAAACMNQKHLLRFIKHTYQTEPDRTVAEKRGRKITLRQVFDGLHMDPY 367
Query: 438 DLNVDLLDVHADKSTFHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGRFLGELTKQVFSD 497
DL VD LDVHA + TFHRFDKFN KYNP G S LR+++LK +N + G + + K+V +
Sbjct: 368 DLTVDSLDVHAGRQTFHRFDKFNSKYNPVGASELRDLYLKTENYLGGEYFARMVKEVARE 427
Query: 498 LEASKYQMAEYRISIYGRKQSEWDQLASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVT 557
LE SKYQ +E R+SIYGR EW LA W + + +YS N+ W+IQ+PR+Y+I++ ++
Sbjct: 428 LEESKYQYSEPRLSIYGRSPEEWPNLAYWFIQHKVYSPNMRWIIQVPRIYDIFRSKKLLP 487
Query: 558 SFQNMLDNIFIPLFEVTVDPDSHPQLHVFLKQVVGLDLVDDESK-PERRPTKHMPTPAQW 616
+F ML+NIF+PLF+ T++P H +LH+FLK V G D VDDESK + + P P W
Sbjct: 488 NFGKMLENIFLPLFKATINPQDHRELHLFLKYVTGFDSVDDESKHSDHMFSDKSPNPDVW 547
Query: 617 TNVFNPAFSYYVYYCYANLYTLNKLRESKGMTTIKFRPHAGEAGDIDHLAATFLTAHNIA 676
T+ NP +SYY+YY YAN+ LN LR +G++T FRPH GEAG I HL + FLTA NI+
Sbjct: 548 TSEQNPPYSYYLYYMYANIMVLNNLRRERGLSTFLFRPHCGEAGSITHLVSAFLTADNIS 607
Query: 677 HGINLRKSPVLQYLYYLAQIGLAMSPLSNNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQ 736
HG+ L+KSPVLQYLYYLAQI +AMSPLSNNSLFL+Y +NPL F +GL+VSLSTDDP+Q
Sbjct: 608 HGLLLKKSPVLQYLYYLAQIPIAMSPLSNNSLFLEYSKNPLREFLHKGLHVSLSTDDPMQ 667
Query: 737 IHLTKEPLVEEYSIAASVWKLSSCDLCEIARNSVYQSGFSHALKSHWIGKEYYKRGPNGN 796
H TKE L+EEY+IAA VWKLS+CDLCEIARNSV QSG SH K ++G+ YYK GP GN
Sbjct: 668 FHYTKEALMEEYAIAAQVWKLSTCDLCEIARNSVLQSGLSHQEKQKFLGQNYYKEGPEGN 727
Query: 797 DIHRTNVPHIRLEFR 811
DI +TNV IR+ FR
Sbjct: 728 DIRKTNVAQIRMAFR 742
>AMD3_RAT (O09178) AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase
isoform E)
Length = 765
Score = 606 bits (1563), Expect = e-173
Identities = 298/541 (55%), Positives = 375/541 (69%), Gaps = 6/541 (1%)
Query: 277 PKPNLEPFFYAPEGKSDHYF-EMQDGVIHVYPNKNSNEEL----FPVADATTFFTDLHQI 331
P P +P+ + Y MQ GV+ VY N+ E P D T+ D+ I
Sbjct: 200 PLPQEDPYCLDDAPPNLGYLVRMQGGVLFVYDNQTMLERQEPHSLPYPDLETYIVDMSHI 259
Query: 332 LRVIAAGNIRTLCHHRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHS 391
L +I G +T CH RLN LE KF+LH MLN EF KS PHRDFYNVRKVDTH+H +
Sbjct: 260 LALITDGPTKTYCHRRLNFLESKFSLHEMLNEMSEFKELKSNPHRDFYNVRKVDTHIHAA 319
Query: 392 ACMNQKHLLRFIKSKLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKS 451
ACMNQKHLLRFIK + EPD V + G +TLR+VF+SL + YDL VD LDVHA +
Sbjct: 320 ACMNQKHLLRFIKYTYQTEPDRTVAEKLGRKITLRQVFDSLHMDPYDLTVDSLDVHAGRQ 379
Query: 452 TFHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRIS 511
TFH FDKFN KYNP G S LR+++LK +N + G + + K+V +LE SKYQ +E R+S
Sbjct: 380 TFHGFDKFNSKYNPVGASELRDLYLKTENYLGGEYFARMVKEVARELEDSKYQYSEPRLS 439
Query: 512 IYGRKQSEWDQLASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLF 571
IYGR EW LA W + + +YS N+ W+IQ+PR+Y+I++ ++ SF ML+NIF+PLF
Sbjct: 440 IYGRSPKEWSSLARWFIQHKVYSPNMRWIIQVPRIYDIFRSKKLLPSFGKMLENIFLPLF 499
Query: 572 EVTVDPDSHPQLHVFLKQVVGLDLVDDESK-PERRPTKHMPTPAQWTNVFNPAFSYYVYY 630
+ T++P H +LH+FLK V G D VDDESK + + P+P WT+ NP +SYY+YY
Sbjct: 500 QATINPQDHRELHLFLKYVTGFDSVDDESKHSDHMFSDKSPSPDLWTSEQNPPYSYYLYY 559
Query: 631 CYANLYTLNKLRESKGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYL 690
YAN+ LN LR +G++T FRPH GEAG I HL + FLTA NI+HG+ L+KSPVLQYL
Sbjct: 560 MYANIMVLNNLRRERGLSTFLFRPHCGEAGSITHLVSAFLTADNISHGLLLKKSPVLQYL 619
Query: 691 YYLAQIGLAMSPLSNNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSI 750
YYLAQI +AMSPLSNNSLFL+Y +NPL F +GL+VSLSTDDP+Q H TKE L+EEY+I
Sbjct: 620 YYLAQIPIAMSPLSNNSLFLEYSKNPLREFLHKGLHVSLSTDDPMQFHYTKEALMEEYAI 679
Query: 751 AASVWKLSSCDLCEIARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEF 810
AA VWKLS+CDLCEIARNSV QSG SH K ++G+ YYK GP GNDI +TNV IR+ F
Sbjct: 680 AAQVWKLSTCDLCEIARNSVLQSGLSHQEKQKFLGQNYYKEGPEGNDIRKTNVAQIRMAF 739
Query: 811 R 811
R
Sbjct: 740 R 740
>AMD3_MOUSE (O08739) AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase
isoform E) (AMP deaminase H-type) (Heart-type AMPD)
Length = 766
Score = 603 bits (1556), Expect = e-172
Identities = 296/541 (54%), Positives = 374/541 (68%), Gaps = 6/541 (1%)
Query: 277 PKPNLEPFFYAPEGKSDHYF-EMQDGVIHVYPNKNSNEEL----FPVADATTFFTDLHQI 331
P P +P+ + Y M GV+ VY N+ E P D T+ D+ I
Sbjct: 201 PLPQEDPYCLDDAPPNLGYLVRMHGGVLFVYDNQTMLERQEPHSLPYPDLKTYIVDMSHI 260
Query: 332 LRVIAAGNIRTLCHHRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHS 391
L +I G +T CH RLN LE KF+LH MLN EF KS PHRDFYNVRKVDTH+H +
Sbjct: 261 LALITDGPTKTYCHRRLNFLESKFSLHEMLNEMSEFKELKSNPHRDFYNVRKVDTHIHAA 320
Query: 392 ACMNQKHLLRFIKSKLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKS 451
ACMNQKHLLRFIK + EPD V + G +TLR+VF+SL + YDL VD LDVHA +
Sbjct: 321 ACMNQKHLLRFIKHTYQTEPDRTVAEKLGRKITLRQVFDSLHMDPYDLTVDSLDVHAGRQ 380
Query: 452 TFHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRIS 511
TFHRFDKFN KYNP G S LR+++LK +N + G + + K+V +LE SKYQ +E R+S
Sbjct: 381 TFHRFDKFNSKYNPVGASELRDLYLKTENYLGGEYFARMVKEVARELEDSKYQYSEPRLS 440
Query: 512 IYGRKQSEWDQLASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLF 571
IYGR EW LA W + + +YS N+ W+IQ+PR+Y+I++ ++ +F ML+NIF+PLF
Sbjct: 441 IYGRSPKEWSSLARWFIQHKVYSPNMRWIIQVPRIYDIFRSKKLLPNFGKMLENIFLPLF 500
Query: 572 EVTVDPDSHPQLHVFLKQVVGLDLVDDESK-PERRPTKHMPTPAQWTNVFNPAFSYYVYY 630
+ T++P H +LH+FLK V G D VDDESK + + P+P WT+ NP +SYY+YY
Sbjct: 501 KATINPQDHRELHLFLKYVTGFDSVDDESKHSDHMFSDKSPSPDLWTSEQNPPYSYYLYY 560
Query: 631 CYANLYTLNKLRESKGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYL 690
YAN+ LN LR +G++T FRPH GEAG I HL + FLTA NI+HG+ L+KSPVLQYL
Sbjct: 561 MYANIMVLNNLRRERGLSTFLFRPHCGEAGSITHLVSAFLTADNISHGLLLKKSPVLQYL 620
Query: 691 YYLAQIGLAMSPLSNNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSI 750
YYLAQI +AMSPLSNNSLFL+Y + PL F +GL+VSLSTDDP+Q H TKE L+EEY+I
Sbjct: 621 YYLAQIPIAMSPLSNNSLFLEYSKKPLREFLHKGLHVSLSTDDPMQFHYTKEALMEEYAI 680
Query: 751 AASVWKLSSCDLCEIARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEF 810
AA VWKLS+CDLCEIARNSV QSG SH K ++G+ YYK GP GNDI +TNV IR+ F
Sbjct: 681 AAQVWKLSTCDLCEIARNSVLQSGLSHQEKQKFLGQNYYKEGPEGNDIRKTNVAQIRMAF 740
Query: 811 R 811
R
Sbjct: 741 R 741
>AMD1_RAT (P10759) AMP deaminase 1 (EC 3.5.4.6) (Myoadenylate
deaminase) (AMP deaminase isoform M)
Length = 747
Score = 567 bits (1461), Expect = e-161
Identities = 271/523 (51%), Positives = 359/523 (67%), Gaps = 5/523 (0%)
Query: 294 HYFEMQDGVIHVYPNK----NSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCHHRLN 349
++ +M+ GVI++YP++ + +P + F D++ +L +IA G ++T H RL
Sbjct: 206 YHLKMKGGVIYIYPDEAAASRDEPKPYPYPNLDDFLDDMNFLLALIAQGPVKTYTHRRLK 265
Query: 350 LLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLRK 409
L KF +H MLN E K+ PHRDFYN RKVDTH+H +ACMNQKHLLRFIK
Sbjct: 266 FLSSKFQVHQMLNEMDELKELKNNPHRDFYNCRKVDTHIHAAACMNQKHLLRFIKKSYHI 325
Query: 410 EPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQS 469
+ D VV LTL+E+F L++ YDL VD LDVHA + TF RFDKFN KYNP G S
Sbjct: 326 DADRVVYSTKEKNLTLKELFAQLNMHPYDLTVDSLDVHAGRQTFQRFDKFNDKYNPVGAS 385
Query: 470 RLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLASWIVN 529
LR+++LK DN I G + + K+V +DL +KYQ AE R+SIYGR EW +L+SW V
Sbjct: 386 ELRDLYLKTDNYINGEYFATIIKEVGADLVDAKYQHAEPRLSIYGRSPDEWSKLSSWFVG 445
Query: 530 NDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHVFLKQ 589
N +Y N+ W+IQ+PR+Y++++ + F ML+NIF+P+FE T++P +HP L VFLK
Sbjct: 446 NRIYCPNMTWMIQVPRIYDVFRSKNFLPHFGKMLENIFLPVFEATINPQTHPDLSVFLKH 505
Query: 590 VVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRESKGMT 648
+ G D VDDESK + P P +WT NP+++YY YY YAN+ LN LR+ +GM
Sbjct: 506 ITGFDSVDDESKHSGHMFSSKSPKPEEWTMENNPSYTYYAYYMYANIMVLNCLRKERGMN 565
Query: 649 TIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNNSL 708
T FRPH GEAG + HL F+ A NI+HG+NL+KSPVLQYL++LAQI +AMSPLSNNSL
Sbjct: 566 TFLFRPHCGEAGALTHLMTAFMIADNISHGLNLKKSPVLQYLFFLAQIPIAMSPLSNNSL 625
Query: 709 FLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCEIARN 768
FL+Y +NP F +GL +SLSTDDP+Q H TKEPL+EEY+IAA V+KLS+CD+CE+ARN
Sbjct: 626 FLEYAKNPFLDFLQKGLMISLSTDDPMQFHFTKEPLMEEYAIAAQVFKLSTCDMCEVARN 685
Query: 769 SVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
SV Q G SH K+ ++G Y + GP GNDI RTNV IR+ +R
Sbjct: 686 SVLQCGISHEEKAKFLGNNYLEEGPVGNDIRRTNVAQIRMAYR 728
>AMD1_HUMAN (P23109) AMP deaminase 1 (EC 3.5.4.6) (Myoadenylate
deaminase) (AMP deaminase isoform M)
Length = 747
Score = 564 bits (1454), Expect = e-160
Identities = 271/523 (51%), Positives = 359/523 (67%), Gaps = 5/523 (0%)
Query: 294 HYFEMQDGVIHVYPNK----NSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCHHRLN 349
++ +M+DGV++VYPN+ + P + TF D++ +L +IA G ++T H RL
Sbjct: 206 YHLKMKDGVVYVYPNEAAVSKDEPKPLPYPNLDTFLDDMNFLLALIAQGPVKTYTHRRLK 265
Query: 350 LLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLRK 409
L KF +H MLN E K+ PHRDFYN RKVDTH+H +ACMNQKHLLRFIK +
Sbjct: 266 FLSSKFQVHQMLNEMDELKELKNNPHRDFYNCRKVDTHIHAAACMNQKHLLRFIKKSYQI 325
Query: 410 EPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQS 469
+ D VV LTL+E+F L + YDL VD LDVHA + TF RFDKFN KYNP G S
Sbjct: 326 DADRVVYSTKEKNLTLKELFAKLKMHPYDLTVDSLDVHAGRQTFQRFDKFNDKYNPVGAS 385
Query: 470 RLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLASWIVN 529
LR+++LK DN I G + + K+V +DL +KYQ AE R+SIYGR EW +L+SW V
Sbjct: 386 ELRDLYLKTDNYINGEYFATIIKEVGADLVEAKYQHAEPRLSIYGRSPDEWSKLSSWFVC 445
Query: 530 NDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHVFLKQ 589
N ++ N+ W+IQ+PR+Y++++ + F ML+NIF+P+FE T++P + P+L VFLK
Sbjct: 446 NRIHCPNMTWMIQVPRIYDVFRSKNFLPHFGKMLENIFMPVFEATINPQADPELSVFLKH 505
Query: 590 VVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRESKGMT 648
+ G D VDDESK + P P +WT NP+++YY YY YAN+ LN LR+ +GM
Sbjct: 506 ITGFDSVDDESKHSGHMFSSKSPKPQEWTLEKNPSYTYYAYYMYANIMVLNSLRKERGMN 565
Query: 649 TIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNNSL 708
T FRPH GEAG + HL F+ A +I+HG+NL+KSPVLQYL++LAQI +AMSPLSNNSL
Sbjct: 566 TFLFRPHCGEAGALTHLMTAFMIADDISHGLNLKKSPVLQYLFFLAQIPIAMSPLSNNSL 625
Query: 709 FLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCEIARN 768
FL+Y +NP F +GL +SLSTDDP+Q H TKEPL+EEY+IAA V+KLS+CD+CE+ARN
Sbjct: 626 FLEYAKNPFLDFLQKGLMISLSTDDPMQFHFTKEPLMEEYAIAAQVFKLSTCDMCEVARN 685
Query: 769 SVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
SV Q G SH K ++G Y + GP GNDI RTNV IR+ +R
Sbjct: 686 SVLQCGISHEEKVKFLGDNYLEEGPAGNDIRRTNVAQIRMAYR 728
>YB9Z_YEAST (P38150) Hypothetical 92.9 kDa protein in SSH1-APE3
intergenic region
Length = 797
Score = 217 bits (553), Expect = 9e-56
Identities = 186/613 (30%), Positives = 291/613 (47%), Gaps = 103/613 (16%)
Query: 294 HYFEMQDG-VIHVYPNKNSNEELFP----VADATTFFTDLHQILRVIAAGNIRTLCHHRL 348
H+ ++++ +++ + N ELF + F D L++I ++ RL
Sbjct: 184 HFLDLEESNTVNLLQDGNYMTELFNSQINIPTFKEFREDFEWCLKIIRDRSLSRFSEKRL 243
Query: 349 NLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLR 408
L KF + L++ E K PH+DFYN RK+D ++ S C +Q L FI +KLR
Sbjct: 244 QYLVNKFPVFQHLHSKEEMRQSKKVPHKDFYNCRKIDLNLLLSGCFSQWQLTEFIWTKLR 303
Query: 409 KEPDEVVIFR-DGTYLTLREVFE-SLDLTGYDLN-VDLLD---------VHADKSTFHRF 456
KEPD V+ +G+++TL ++F+ + + TG N + ++D ++ K +H
Sbjct: 304 KEPDRVIHQAFNGSHITLSQLFKVNFEETGQFFNGLKIIDDSFLEWYKVIYLAK--YHLV 361
Query: 457 -DKFNLKYNPCGQSR----LREIFLKQDNLIQGRFLGELTKQ-VFSDLEASKYQMAEYRI 510
D+ + G+ + + FL+ DN I G +L EL K + E SKYQ+ + +
Sbjct: 362 NDEMEIHTGSHGKQLRYYLIAKTFLEFDNYINGEYLAELLKTFLIKPQEESKYQLCQLSV 421
Query: 511 SI-----YGRKQSE--WDQLASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNML 563
Y + W A+W+ + +++S N+ W I++ R+Y G V +FQ L
Sbjct: 422 DFQFYLHYDNSDVDNWWMVFANWLNHYNIFSNNIRWNIRISRIYPELYHTGKVKNFQEYL 481
Query: 564 DNIFIPLF--EVTVDPDSHPQLHVFLKQVVGLDLV--DDESKPERRPTKHMPTPAQWTNV 619
+ IF PLF E + P L FL QV +DL D ++ + T P WT+
Sbjct: 482 NLIFKPLFNAENYLHKSLGPILLKFLSQVSSIDLCIQDTDNYIWKNFTAVSCLPKDWTSG 541
Query: 620 F-NPAFSYYVYYCYANLYTLNKLRE---------------------SKGMTTIKFRPHAG 657
NP S Y+YY Y NL LN +R+ S+ T+ F H
Sbjct: 542 GDNPTISQYMYYVYVNLTKLNHIRQALHQNTFTLRSSCSPTSMNRTSQFSNTLNFTEHT- 600
Query: 658 EAGDIDHLAAT--FLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSN---------- 705
EA + L A FL A N+ + P L YL+YL+QI + ++PL++
Sbjct: 601 EAILNNFLLACGGFLNAENLWNA-----PPSLVYLFYLSQIPMVVAPLNSIVDSKPTMLQ 655
Query: 706 ----NSLFLD----YHRNPLPVFFLRGLNVSLSTDDPLQIH-LTKEPLVEEYSIAASVWK 756
L L+ Y +NP FF G +SLS++ L + TKEP++EEYS+AAS+++
Sbjct: 656 EQAPTGLVLEPSKPYKKNPFMKFFEMGFKISLSSESILYNNSYTKEPIIEEYSVAASIYR 715
Query: 757 LSSCDLCEIARNSVYQSGFSHALKSHWIG----------------KEYYKRGPNGNDIHR 800
L S DLCE+ RNSV SGFS LK+ W+G ++Y PN + H
Sbjct: 716 LHSADLCELLRNSVITSGFSSTLKNKWLGVSLASHDYFVENTGFVDKWYDCKPNTSLEH- 774
Query: 801 TNVPHIRLEFRDT 813
NVP IR ++R +
Sbjct: 775 -NVPIIRRQYRSS 786
>YJH0_YEAST (P40361) Hypothetical 104.3 kDa protein in SMC3-MRPL8
intergenic region
Length = 888
Score = 205 bits (521), Expect = 4e-52
Identities = 181/644 (28%), Positives = 282/644 (43%), Gaps = 137/644 (21%)
Query: 295 YFEMQDG----VIHVYPNKNSNEELFP------------VADATTFFT---DLHQILRVI 335
YF+++D + H+ +N+ L P + D TF D I+ +I
Sbjct: 233 YFDLEDYKKQYIYHLSNQENTQNPLSPYSSKEESLEEEFLTDVPTFQEFRDDFAYIIELI 292
Query: 336 AAGNIRTLCHHRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMN 395
+ + RL+ L KF L LN+ +E LA K+ P+RDFYN RKVD + S C++
Sbjct: 293 QSHKFNEVSRKRLSYLLDKFELFQYLNSKKEILANKNVPYRDFYNSRKVDRDLSLSGCIS 352
Query: 396 QKHLLRFIKSKLRKEPDEVVIFRD---GTYLTLREVFE-----------------SLDLT 435
Q+ L +I K+ EP+ +V ++D L+LR++F+ +
Sbjct: 353 QRQLSEYIWEKINLEPERIV-YQDPETSRKLSLRDIFQFGCSSNDQPIAIGLKLIDDEFL 411
Query: 436 GYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGRFLGEL-TKQV 494
+ N+ L+D H + + +++ L ++FL+ DN I+G +L E+ K V
Sbjct: 412 DWYRNIYLIDYHLTPNKVAKLVGKEMRF-----YLLAKVFLEFDNFIEGEYLAEIFIKYV 466
Query: 495 FSDLEASKYQMAEYRIS--IYGRKQSEWDQLASWIVNNDLYSENVVWLIQLPRLYNIYKD 552
LE SKYQ+A+ ++ Y + + + + W++ L S N+ W IQ+ R++
Sbjct: 467 IHILEKSKYQLAQVSVNFQFYSSGEDWYKKFSQWLLRWKLVSYNIRWNIQIARIFPKLFK 526
Query: 553 MGIVTSFQNMLDNIFIPLF-----EVTVDPDSHPQ---LHVFLKQVVGLDLVDDESKPE- 603
+V++FQ LD IF PLF ++ +D + L FL V +DLV ES
Sbjct: 527 ENVVSNFQEFLDLIFNPLFTLEKEQLPIDSSVNTDIIGLQFFLSNVCSMDLVIKESDEYY 586
Query: 604 -RRPTKHMPTPAQWT-NVFNPAFSYYVYYCYANLYTLNKLRESKGMTTIKFRPHAGE--- 658
+ T P WT NP ++Y+YY Y +L +N LR TI R +
Sbjct: 587 WKEFTDMNCKPKFWTAQGDNPTVAHYMYYIYKSLAKVNFLRSQNLQNTITLRNYCSPLSS 646
Query: 659 -----------AGDIDHLAATFLTAHNIAHGINLRKSP------VLQYLYYLAQIGLAMS 701
++ L L + +G L+ P ++QYL+YL QI + +
Sbjct: 647 RTSQFGVDLYFTDQVESLVCNLL----LCNGGLLQVEPLWDTATMIQYLFYLFQIPILAA 702
Query: 702 PLSNNSL-------FL--------------------------------DYHRNPLPVFFL 722
PLS+ SL FL Y NP F
Sbjct: 703 PLSSVSLLNSQKSTFLKNKNVLLEHDYLKDQETAKINPSRDITVGEQRSYETNPFMKMFK 762
Query: 723 RGLNVSLSTDDPL-QIHLTKEPLVEEYSIAASVWKLSSCDLCEIARNSVYQSGFSHALKS 781
GL +SLS+ L T EPL+EEYS+AAS++ L+ DLCE++R SV SG+ K+
Sbjct: 763 MGLKISLSSKSILYNSSYTLEPLIEEYSVAASIYLLNPTDLCELSRTSVLSSGYEGWYKA 822
Query: 782 HWIG-----KEYYKRGPNGND---------IHRTNVPHIRLEFR 811
HWIG Y++ G D + NVP IR +R
Sbjct: 823 HWIGVGVKKAPYFEENVGGIDNWYDTAKDTSIKHNVPMIRRRYR 866
>AMD2_RAT (Q02356) AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase
isoform L) (Fragment)
Length = 88
Score = 140 bits (352), Expect = 2e-32
Identities = 66/88 (75%), Positives = 73/88 (82%)
Query: 654 PHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNNSLFLDYH 713
PH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLSNNSLFL YH
Sbjct: 1 PHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLSNNSLFLSYH 60
Query: 714 RNPLPVFFLRGLNVSLSTDDPLQIHLTK 741
RNPLP + RGL VSLSTDDPLQ H TK
Sbjct: 61 RNPLPEYLSRGLMVSLSTDDPLQFHFTK 88
>ADD_SHEON (Q8E8D4) Adenosine deaminase (EC 3.5.4.4) (Adenosine
aminohydrolase)
Length = 331
Score = 51.6 bits (122), Expect = 8e-06
Identities = 40/128 (31%), Positives = 56/128 (43%), Gaps = 7/128 (5%)
Query: 655 HAGEAGDIDHL--AATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSN--NSLFL 710
HAGEA + A L A I HG+N P L +IG+ P SN S
Sbjct: 197 HAGEAAGSQSMWQAIQELGATRIGHGVNAIHDPKLMEYLAKHRIGIESCPTSNLHTSTVS 256
Query: 711 DYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCEIARNSV 770
Y +P F G+ + L+TDDP ++ + EY IA LS +L ++ RN V
Sbjct: 257 SYAEHPFRTFMDAGVLIGLNTDDP---GVSAIDIKHEYRIAKFELGLSDAELAQVQRNGV 313
Query: 771 YQSGFSHA 778
+ S +
Sbjct: 314 EMAFLSES 321
>ADD_CLOAB (Q97EV1) Adenosine deaminase (EC 3.5.4.4) (Adenosine
aminohydrolase)
Length = 334
Score = 48.9 bits (115), Expect = 5e-05
Identities = 42/142 (29%), Positives = 69/142 (48%), Gaps = 9/142 (6%)
Query: 650 IKFRPHAGEAGDIDHLAATF--LTAHNIAHGINLRKSP-VLQYLYYLAQIGLAMSPLSN- 705
IK HAGE G +++ + L A I HGI KS +LQY+ Q+ L M P SN
Sbjct: 195 IKITIHAGETGIAENILKSIKLLHADRIGHGIFAYKSEEILQYVIE-NQVPLEMCPKSNV 253
Query: 706 -NSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCE 764
+Y +P +F G+ V+L+TD+ ++ LV+EY A+++ ++
Sbjct: 254 DTKAVKNYKNHPFKKYFDLGVKVTLNTDNRT---VSNVSLVDEYLNLANIFDFGIEEIKT 310
Query: 765 IARNSVYQSGFSHALKSHWIGK 786
+ RN + S + K + + K
Sbjct: 311 VIRNGISASFATEEFKVNLLKK 332
>ADD_ECOL6 (Q8FH99) Adenosine deaminase (EC 3.5.4.4) (Adenosine
aminohydrolase)
Length = 333
Score = 43.9 bits (102), Expect = 0.002
Identities = 37/124 (29%), Positives = 55/124 (43%), Gaps = 11/124 (8%)
Query: 639 NKLRESKGMTTIKFRPHAGEAGDIDHL--AATFLTAHNIAHGINLRKSPVLQYLYYLAQI 696
N+ R++ T+ HAGEA + + A L A I HG+ + L QI
Sbjct: 185 NRARDAGWHITV----HAGEAAGPESIWQAIRELGAERIGHGVKAIEDRALMDFLAEQQI 240
Query: 697 GLAMSPLSN--NSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASV 754
G+ SN S D +PL F G+ S++TDDP + ++ EY++AA
Sbjct: 241 GIESCLTSNIQTSTVADLAAHPLKTFLEHGIRASINTDDP---GVQGVDIIHEYTVAAPA 297
Query: 755 WKLS 758
LS
Sbjct: 298 AGLS 301
>ADD1_STRCO (O86737) Probable adenosine deaminase 1 (EC 3.5.4.4)
(Adenosine aminohydrolase 1)
Length = 387
Score = 43.1 bits (100), Expect = 0.003
Identities = 42/128 (32%), Positives = 62/128 (47%), Gaps = 15/128 (11%)
Query: 654 PHAGEAGDIDHL--AATFLTAHNIAHGINLRKSPVLQYLYYLAQ--IGLAMSPLSNNSL- 708
PHAGE + A L A I HG + + P L L +LA+ I L + P SN +
Sbjct: 243 PHAGETTGPQTVWEALIDLRAERIGHGTSSAQDPKL--LAHLAERRIPLEVCPTSNIATR 300
Query: 709 ---FLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCEI 765
LD H P+ F G+ V++++DDP + L EY++AA + L L ++
Sbjct: 301 AVRTLDEH--PIKEFVRAGVPVTINSDDP---PMFGTDLNNEYAVAARLLGLDERGLADL 355
Query: 766 ARNSVYQS 773
A+N V S
Sbjct: 356 AKNGVEAS 363
>ADD_SHIFL (Q83RC0) Adenosine deaminase (EC 3.5.4.4) (Adenosine
aminohydrolase)
Length = 333
Score = 42.4 bits (98), Expect = 0.005
Identities = 36/124 (29%), Positives = 55/124 (44%), Gaps = 11/124 (8%)
Query: 639 NKLRESKGMTTIKFRPHAGEAGDIDHL--AATFLTAHNIAHGINLRKSPVLQYLYYLAQI 696
N+ R++ T+ HAGEA + + A L A I HG+ + L QI
Sbjct: 185 NRARDAGWHITV----HAGEAAGPESIWQAIRELGAERIGHGVKAIEDRALMDFLAEQQI 240
Query: 697 GLAMSPLSN--NSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASV 754
G+ SN S + +PL F G+ S++TDDP + ++ EY++AA
Sbjct: 241 GIESCLTSNIQTSTVAELAAHPLKTFLEHGIRASINTDDP---GVQGVDIIHEYTVAAPA 297
Query: 755 WKLS 758
LS
Sbjct: 298 AGLS 301
>ADD_ECOLI (P22333) Adenosine deaminase (EC 3.5.4.4) (Adenosine
aminohydrolase)
Length = 333
Score = 42.4 bits (98), Expect = 0.005
Identities = 36/124 (29%), Positives = 55/124 (44%), Gaps = 11/124 (8%)
Query: 639 NKLRESKGMTTIKFRPHAGEAGDIDHL--AATFLTAHNIAHGINLRKSPVLQYLYYLAQI 696
N+ R++ T+ HAGEA + + A L A I HG+ + L QI
Sbjct: 185 NRARDAGWHITV----HAGEAAGPESIWQAIRELGAERIGHGVKAIEDRALMDFLAEQQI 240
Query: 697 GLAMSPLSN--NSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASV 754
G+ SN S + +PL F G+ S++TDDP + ++ EY++AA
Sbjct: 241 GIESCLTSNIQTSTVAELAAHPLKTFLEHGIRASINTDDP---GVQGVDIIHEYTVAAPA 297
Query: 755 WKLS 758
LS
Sbjct: 298 AGLS 301
>ADD_ECO57 (Q8X661) Adenosine deaminase (EC 3.5.4.4) (Adenosine
aminohydrolase)
Length = 333
Score = 42.4 bits (98), Expect = 0.005
Identities = 36/124 (29%), Positives = 55/124 (44%), Gaps = 11/124 (8%)
Query: 639 NKLRESKGMTTIKFRPHAGEAGDIDHL--AATFLTAHNIAHGINLRKSPVLQYLYYLAQI 696
N+ R++ T+ HAGEA + + A L A I HG+ + L QI
Sbjct: 185 NRARDAGWHITV----HAGEAAGPESIWQAIRELGAERIGHGVKAIEDRALMDFLAEQQI 240
Query: 697 GLAMSPLSN--NSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASV 754
G+ SN S + +PL F G+ S++TDDP + ++ EY++AA
Sbjct: 241 GIESCLTSNIQTSTVAELAAHPLKTFLEHGIRASINTDDP---GVQGVDIIHEYTVAAPA 297
Query: 755 WKLS 758
LS
Sbjct: 298 AGLS 301
>ADD_STRVG (P53984) Adenosine deaminase (EC 3.5.4.4) (Adenosine
aminohydrolase)
Length = 339
Score = 42.0 bits (97), Expect = 0.007
Identities = 39/122 (31%), Positives = 52/122 (41%), Gaps = 9/122 (7%)
Query: 654 PHAGEAGDIDHLAATF--LTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNNSLFLD 711
PH GE + L A I HG+ + P L QI + P SN +L +
Sbjct: 199 PHGGELTGPSSVRDCLDDLHASRIGHGVRAAEDPRLLKRLADRQITCEVCPASNVALGV- 257
Query: 712 YHRN---PLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCEIARN 768
Y R PL F G+ ++L DDPL L L +Y IA + +L E+AR
Sbjct: 258 YERPEDVPLRTLFEAGVPMALGADDPL---LFGSRLAAQYEIARRHHAFTDTELAELARQ 314
Query: 769 SV 770
SV
Sbjct: 315 SV 316
>ADD_ENTFA (Q839J4) Adenosine deaminase (EC 3.5.4.4) (Adenosine
aminohydrolase)
Length = 337
Score = 40.8 bits (94), Expect = 0.015
Identities = 37/138 (26%), Positives = 62/138 (44%), Gaps = 6/138 (4%)
Query: 636 YTLNKLRESKGMTTIKFRPHAGEAGDIDHLA-ATFLTAHNIAHGINLRKSPVLQYLYYLA 694
YT + +I HAGE G ++A A L A I HGI L+ +P L
Sbjct: 180 YTFEDVLALANQLSIPLTLHAGECGCGKNVADAVTLGATRIGHGIALKDTPEYLALLKEK 239
Query: 695 QIGLAMSPLSN--NSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAA 752
++ L M P SN P F GL V ++TD+ ++ L +E+ A
Sbjct: 240 KVLLEMCPTSNFQTGTVKTLAEYPFQQFIEAGLAVCINTDNRT---VSDTTLTKEFMKLA 296
Query: 753 SVWKLSSCDLCEIARNSV 770
+ ++LS ++ ++ +N++
Sbjct: 297 TWYQLSYDEMKQLTKNAL 314
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 103,293,672
Number of Sequences: 164201
Number of extensions: 4855864
Number of successful extensions: 15300
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 14873
Number of HSP's gapped (non-prelim): 294
length of query: 814
length of database: 59,974,054
effective HSP length: 119
effective length of query: 695
effective length of database: 40,434,135
effective search space: 28101723825
effective search space used: 28101723825
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Medicago: description of AC140721.5


