

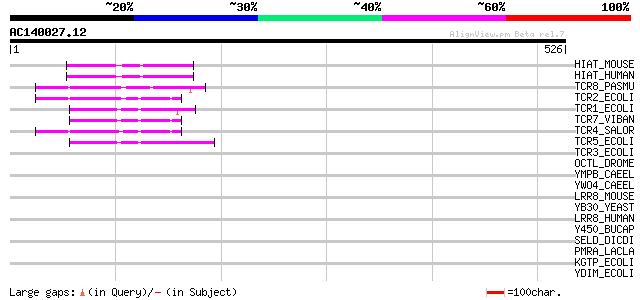
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140027.12 + phase: 0 /pseudo
(526 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
HIAT_MOUSE (P70187) Hippocampus abundant transcript 1 protein 56 3e-07
HIAT_HUMAN (Q96MC6) Hippocampus abundant transcript 1 protein (P... 54 1e-06
TCR8_PASMU (P51564) Tetracycline resistance protein, class H (TE... 52 4e-06
TCR2_ECOLI (P02980) Tetracycline resistance protein, class B (TE... 51 8e-06
TCR1_ECOLI (P02982) Tetracycline resistance protein, class A (TE... 50 1e-05
TCR7_VIBAN (P51563) Tetracycline resistance protein, class G (TE... 46 3e-04
TCR4_SALOR (P33733) Tetracycline resistance protein, class D (TE... 45 5e-04
TCR5_ECOLI (Q07282) Tetracycline resistance protein, class E (TE... 44 8e-04
TCR3_ECOLI (P02981) Tetracycline resistance protein, class C (TE... 42 0.003
OCTL_DROME (Q95R48) Organic cation transporter-like protein 41 0.007
YMPB_CAEEL (Q7Z118) Hypothetical protein B0361.11 in chromosome III 37 0.13
YWO4_CAEEL (Q10907) Probable G protein-coupled receptor AH9.4 36 0.21
LRR8_MOUSE (Q80WG5) Leucine-rich repeat-containing protein 8 pre... 35 0.37
YB30_YEAST (P38125) Hypothetical transport protein YBR180W 34 0.82
LRR8_HUMAN (Q8IWT6) Leucine-rich repeat-containing protein 8 pre... 34 1.1
Y450_BUCAP (Q8K999) Hypothetical transport protein BUsg450 33 1.8
SELD_DICDI (Q94497) Selenide,water dikinase (EC 2.7.9.3) (Seleno... 32 3.1
PMRA_LACLA (P58120) Multi-drug resistance efflux pump pmrA homolog 32 3.1
KGTP_ECOLI (P17448) Alpha-ketoglutarate permease 32 3.1
YDIM_ECOLI (P76197) Hypothetical transport protein ydiM 32 4.0
>HIAT_MOUSE (P70187) Hippocampus abundant transcript 1 protein
Length = 490
Score = 55.8 bits (133), Expect = 3e-07
Identities = 36/120 (30%), Positives = 59/120 (49%), Gaps = 7/120 (5%)
Query: 55 INGLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYY 114
+NGL Q + G+ PL+G LSD GRK LLLT+ + P L+ K + Y+
Sbjct: 73 MNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLM-----KISPWWYF 127
Query: 115 VLRTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNY 174
+ + S + + F + AYVAD+ E +R + ++ +A+ V + +L Q Y
Sbjct: 128 AVISVSGVFA--VTFSVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAIGAYLGQMY 185
>HIAT_HUMAN (Q96MC6) Hippocampus abundant transcript 1 protein
(Putative tetracycline transporter-like protein)
Length = 490
Score = 53.9 bits (128), Expect = 1e-06
Identities = 35/120 (29%), Positives = 59/120 (49%), Gaps = 7/120 (5%)
Query: 55 INGLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYY 114
+NGL Q + G+ PL+G LSD GRK LLLT+ + P L+ K + Y+
Sbjct: 73 MNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLM-----KISPWWYF 127
Query: 115 VLRTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNY 174
+ + S + + F + AYVAD+ E +R + ++ +A+ V + +L + Y
Sbjct: 128 AVISVSGVFA--VTFSVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAIGAYLGRVY 185
>TCR8_PASMU (P51564) Tetracycline resistance protein, class H
(TETA(H))
Length = 400
Score = 52.0 bits (123), Expect = 4e-06
Identities = 38/166 (22%), Positives = 80/166 (47%), Gaps = 11/166 (6%)
Query: 25 IAEAMTVSVLVDVTTTALCPQQSSCSKAIYINGLQQTITGIFKMAVLPLLGQLSDEHGRK 84
+ +A+ + +++ V T L S S A + G+ + ++ P+LG+LSD++GRK
Sbjct: 13 VLDAIGIGLIMPVLPTLLNEFVSENSLATHY-GVLLALYATMQVIFAPILGRLSDKYGRK 71
Query: 85 PLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCISVAYVADVVHESK 144
P+LL ++ + + + L+A++ + ++ Y+ R + I C S ++DV
Sbjct: 72 PILLFSLLGAALDYLLMAFSTT---LWMLYIGRIIAGITGATGAVCASA--MSDVTPAKN 126
Query: 145 RVAVFSWITGLSSAAHVIANVFARFL-----PQNYIFVILNYGLLL 185
R F ++ G+ +I + L +IF +++ +LL
Sbjct: 127 RTRYFGFLGGVFGVGLIIGPMLGGLLGDISAHMPFIFAAISHSILL 172
>TCR2_ECOLI (P02980) Tetracycline resistance protein, class B
(TETA(B)) (Metal-tetracycline/H(+) antiporter)
Length = 401
Score = 50.8 bits (120), Expect = 8e-06
Identities = 38/139 (27%), Positives = 74/139 (52%), Gaps = 7/139 (5%)
Query: 25 IAEAMTVSVLVDVTTTALCPQQSSCSKAIYINGLQQTITGIFKMAVLPLLGQLSDEHGRK 84
+ +AM + +++ V T L +S A + G+ + + ++ P LG++SD GR+
Sbjct: 13 LLDAMGIGLIMPVLPTLLREFIASEDIANHF-GVLLALYALMQVIFAPWLGKMSDRFGRR 71
Query: 85 PLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCISVAYVADVVHESK 144
P+LLL++ + + + LLA++ + ++ Y+ R S I G+ ++ + +AD S+
Sbjct: 72 PVLLLSLIGASLDYLLLAFSSA---LWMLYLGRLLSGI--TGATGAVAASVIADTTSASQ 126
Query: 145 RVAVFSWITGLSSAAHVIA 163
RV F W+ G S +IA
Sbjct: 127 RVKWFGWL-GASFGLGLIA 144
>TCR1_ECOLI (P02982) Tetracycline resistance protein, class A
(TETA(A))
Length = 399
Score = 50.4 bits (119), Expect = 1e-05
Identities = 31/124 (25%), Positives = 63/124 (50%), Gaps = 9/124 (7%)
Query: 57 GLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVL 116
G+ + + + A P+LG LSD GR+P+LL++++ + + +A++A + F++ Y+
Sbjct: 46 GILLALYALMQFACAPVLGALSDRFGRRPVLLVSLAGAAVDYAIMA---TAPFLWVLYIG 102
Query: 117 RTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSS----AAHVIANVFARFLPQ 172
R + I G+ ++ AY+AD+ +R F +++ A V+ + F P
Sbjct: 103 RIVAGI--TGATGAVAGAYIADITDGDERARHFGFMSACFGFGMVAGPVLGGLMGGFSPH 160
Query: 173 NYIF 176
F
Sbjct: 161 APFF 164
>TCR7_VIBAN (P51563) Tetracycline resistance protein, class G
(TETA(G))
Length = 393
Score = 45.8 bits (107), Expect = 3e-04
Identities = 28/107 (26%), Positives = 57/107 (53%), Gaps = 6/107 (5%)
Query: 57 GLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVL 116
G ++ + ++ P+LGQLSD +GR+P+LL +++ + + + ++A S ++ Y+
Sbjct: 44 GALLSLYALMQVVFAPMLGQLSDSYGRRPVLLASLAGAAVDYTIMA---SAPVLWVLYIG 100
Query: 117 RTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIA 163
R S + G+ ++ + +AD E R F ++ G A +IA
Sbjct: 101 RLVSGV--TGATGAVAASTIADSTGEGSRARWFGYM-GACYGAGMIA 144
>TCR4_SALOR (P33733) Tetracycline resistance protein, class D
(TETA(D))
Length = 394
Score = 45.1 bits (105), Expect = 5e-04
Identities = 37/139 (26%), Positives = 71/139 (50%), Gaps = 7/139 (5%)
Query: 25 IAEAMTVSVLVDVTTTALCPQQSSCSKAIYINGLQQTITGIFKMAVLPLLGQLSDEHGRK 84
+ +AM + +++ V + L A + G+ + + ++ PLLG+ SD+ GR+
Sbjct: 13 LLDAMGIGLIMPVLPSLLREYLPEADVANHY-GILLALYAVMQVCFAPLLGRWSDKLGRR 71
Query: 85 PLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCISVAYVADVVHESK 144
P+LLL+++ + + LLA + ++ Y+ R S I G+ ++ + VAD S+
Sbjct: 72 PVLLLSLAGAAFDYTLLALS---NVLWMLYLGRIISGI--TGATGAVAASVVADSTAVSE 126
Query: 145 RVAVFSWITGLSSAAHVIA 163
R A F + G + A +IA
Sbjct: 127 RTAWFGRL-GAAFGAGLIA 144
Score = 34.3 bits (77), Expect = 0.82
Identities = 14/39 (35%), Positives = 24/39 (60%)
Query: 441 PATYGIISKASSSTNQGKAQTFIAGANSISGLLSPIVMS 479
PA GIIS +S+ NQGK Q + +++G+ P++ +
Sbjct: 314 PALQGIISAGASAANQGKLQGVLVSLTNLTGVAGPLLFA 352
>TCR5_ECOLI (Q07282) Tetracycline resistance protein, class E
(TETA(E))
Length = 405
Score = 44.3 bits (103), Expect = 8e-04
Identities = 32/138 (23%), Positives = 62/138 (44%), Gaps = 5/138 (3%)
Query: 57 GLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVL 116
G+ + + ++ PLLG+ SD GR+P+LLL++ + + +AL+A + V+ Y+
Sbjct: 44 GVLLALYAMMQVIFAPLLGRWSDRIGRRPVLLLSLLGATLDYALMA---TASVVWVLYLG 100
Query: 117 RTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYIF 176
R + I G+ ++ + +ADV E R F + + V F Q +
Sbjct: 101 RLIAGI--TGATGAVAASTIADVTPEESRTHWFGMMGACFGGGMIAGPVIGGFAGQLSVQ 158
Query: 177 VILNYGLLLSGFNNTIDL 194
+ ++G + L
Sbjct: 159 APFMFAAAINGLAFLVSL 176
>TCR3_ECOLI (P02981) Tetracycline resistance protein, class C
(TETA(C))
Length = 396
Score = 42.4 bits (98), Expect = 0.003
Identities = 29/109 (26%), Positives = 52/109 (47%), Gaps = 9/109 (8%)
Query: 57 GLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVL 116
G+ + + + P+LG LSD GR+P+LL ++ + I +A++A +YA ++
Sbjct: 46 GVLLALYALMQFLCAPVLGALSDRFGRRPVLLASLLGATIDYAIMATTPVLWILYAGRIV 105
Query: 117 RTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANV 165
+ G+ ++ AY+AD+ R F GL SA + V
Sbjct: 106 AGIT-----GATGAVAGAYIADITDGEDRARHF----GLMSACFGVGMV 145
>OCTL_DROME (Q95R48) Organic cation transporter-like protein
Length = 567
Score = 41.2 bits (95), Expect = 0.007
Identities = 29/113 (25%), Positives = 55/113 (48%), Gaps = 7/113 (6%)
Query: 61 TITGIFKMAVLP---LLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLR 117
T +F + VL + GQLSD++GRKP+L ++ ++ F +LA + F Y + L
Sbjct: 132 TSDSLFMLGVLLGSIVFGQLSDKYGRKPILFASLVIQVL-FGVLAGVAPEYFTYTFARLM 190
Query: 118 TFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFL 170
+ + +F ++ ++V KR+ ++ S ++ VFA F+
Sbjct: 191 VGA---TTSGVFLVAYVVAMEMVGPDKRLYAGIFVMMFFSVGFMLTAVFAYFV 240
>YMPB_CAEEL (Q7Z118) Hypothetical protein B0361.11 in chromosome III
Length = 534
Score = 37.0 bits (84), Expect = 0.13
Identities = 44/142 (30%), Positives = 71/142 (49%), Gaps = 18/142 (12%)
Query: 32 SVLVDVT-----TTALCPQQSSCSKAIYIN-GLQQTITGIFKMAVLPLLGQLSDEHGRKP 85
+ LV+VT T L SCS + GL TI I + +P + L+D +GRKP
Sbjct: 118 NTLVNVTNQKASTNLLVDFDLSCSHWFFQEFGL--TIFTIGAVIAVPFMSMLADRYGRKP 175
Query: 86 LLLLTISTSIIPF-ALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCISVAYVADVVHESK 144
++ ++T+I+ F A +A + S F + +LR F I S +SVA VA + S+
Sbjct: 176 II---VTTAILAFLANMAASFSPNFA-IFLILRAF---IGACSDSYLSVASVATCEYLSE 228
Query: 145 RVAVFSWITGLSSAAHVIANVF 166
+ +WIT + + A + V+
Sbjct: 229 KAR--AWITVVYNVAWSLGMVW 248
>YWO4_CAEEL (Q10907) Probable G protein-coupled receptor AH9.4
Length = 364
Score = 36.2 bits (82), Expect = 0.21
Identities = 24/98 (24%), Positives = 45/98 (45%), Gaps = 9/98 (9%)
Query: 349 LPILNPLVGEKVILCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQ----IYMFIWLA 404
LP+ VG+ + LC+ L+ +I + G +S I F I LI ++ W
Sbjct: 47 LPLAAMSVGDSITLCALLMQAIFHITPKGEVPTVVLSSICKFGIFLIHSTSAFSVWCWFF 106
Query: 405 ENFITYISIMKVIEFNPLWQVP-----YLGGSFGIIYI 437
+ + YI++ ++ +W+ P +L G+ G+ I
Sbjct: 107 LSVLRYIAVFHPFKYRTIWRQPRNALKFLAGAVGMFQI 144
>LRR8_MOUSE (Q80WG5) Leucine-rich repeat-containing protein 8
precursor
Length = 810
Score = 35.4 bits (80), Expect = 0.37
Identities = 22/79 (27%), Positives = 39/79 (48%), Gaps = 3/79 (3%)
Query: 108 EFVYAYYVLRTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSS--AAHVIANV 165
+ VY Y+ +T +I I C +V YV ++ + +TG + AH +A +
Sbjct: 258 DIVYRLYMRQTIIKVIKFALIICYTVYYVHNIKFDVDCTVDIESLTGYRTYRCAHPLATL 317
Query: 166 FARFLPQNYIFVILNYGLL 184
F + L YI +++ YGL+
Sbjct: 318 F-KILASFYISLVIFYGLI 335
>YB30_YEAST (P38125) Hypothetical transport protein YBR180W
Length = 572
Score = 34.3 bits (77), Expect = 0.82
Identities = 32/101 (31%), Positives = 45/101 (43%), Gaps = 7/101 (6%)
Query: 52 AIYINGLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVY 111
A IN +F + L G L+D GRK L ++++S +I LLA +
Sbjct: 145 ATTINATVSVFMAVFSVGPL-FWGALADFGGRKFLYMVSLSLMLIVNILLA--AVPVNIA 201
Query: 112 AYYVLRTFSHIISQGSIFCISVAYVADVV---HESKRVAVF 149
A +VLR F S S+ + V DVV H K +A F
Sbjct: 202 ALFVLRIFQAFAS-SSVISLGAGTVTDVVPPKHRGKAIAYF 241
>LRR8_HUMAN (Q8IWT6) Leucine-rich repeat-containing protein 8
precursor
Length = 810
Score = 33.9 bits (76), Expect = 1.1
Identities = 22/79 (27%), Positives = 39/79 (48%), Gaps = 3/79 (3%)
Query: 108 EFVYAYYVLRTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSS--AAHVIANV 165
+ VY Y+ +T +I I C +V YV ++ + +TG + AH +A +
Sbjct: 258 DIVYRLYMRQTIIKVIKFILIICYTVYYVHNIKFDVDCTVDIESLTGYRTYRCAHPLATL 317
Query: 166 FARFLPQNYIFVILNYGLL 184
F + L YI +++ YGL+
Sbjct: 318 F-KILASFYISLVIFYGLI 335
>Y450_BUCAP (Q8K999) Hypothetical transport protein BUsg450
Length = 391
Score = 33.1 bits (74), Expect = 1.8
Identities = 28/123 (22%), Positives = 57/123 (45%), Gaps = 6/123 (4%)
Query: 54 YINGLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAY 113
++ GL I G ++ G LSD GRK +++ + I ++ S ++
Sbjct: 48 FLIGLAVGIYGATQIVFQIPFGILSDRFGRKQIIIFGLFIFFIGSLIVV---STNSIFGL 104
Query: 114 YVLRTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQN 173
+ R I G+I +S+A ++D++ + R+ S I + + +I+ V A + +N
Sbjct: 105 IIGRA---IQGSGAISGVSMALLSDLIRKENRIKSISIIGVSFAVSFLISVVSAPIIAEN 161
Query: 174 YIF 176
+ F
Sbjct: 162 FGF 164
>SELD_DICDI (Q94497) Selenide,water dikinase (EC 2.7.9.3)
(Selenophosphate synthetase) (Selenium donor protein)
(Fragment)
Length = 370
Score = 32.3 bits (72), Expect = 3.1
Identities = 17/53 (32%), Positives = 28/53 (52%)
Query: 394 LIQIYMFIWLAENFITYISIMKVIEFNPLWQVPYLGGSFGIIYILEKPATYGI 446
LI++Y++I+L +I +I +K F PL PY G +L ++GI
Sbjct: 78 LIKLYIYIYLIYIYIFFIIFLKKKFFFPLVDDPYFQGKIACANVLSDLYSFGI 130
>PMRA_LACLA (P58120) Multi-drug resistance efflux pump pmrA homolog
Length = 405
Score = 32.3 bits (72), Expect = 3.1
Identities = 17/53 (32%), Positives = 30/53 (56%), Gaps = 5/53 (9%)
Query: 56 NGLQQTITGIFKMAVLPLLGQLSDEHGRKPL-----LLLTISTSIIPFALLAW 103
+GL ++ + V P+ G+L+DEHGRK + +++T++ I FA W
Sbjct: 52 SGLAFSLPALASGLVAPIWGRLADEHGRKVMMVRASIVMTLTMGGIAFAPNVW 104
Score = 31.6 bits (70), Expect = 5.3
Identities = 17/60 (28%), Positives = 34/60 (56%)
Query: 47 SSCSKAIYINGLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQS 106
+S + ++I+GL + G+ M LG+L D++G L+L+ + + I + +A+ QS
Sbjct: 247 TSTNNLMFISGLIVSAVGLSAMLSSSFLGRLGDKYGSHRLILIGLVFTFIIYLPMAFVQS 306
>KGTP_ECOLI (P17448) Alpha-ketoglutarate permease
Length = 432
Score = 32.3 bits (72), Expect = 3.1
Identities = 22/87 (25%), Positives = 43/87 (49%), Gaps = 6/87 (6%)
Query: 73 LLGQLSDEHGRKPLLLLTIST----SIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSI 128
L G+++D+HGRK +LL++ S++ L + + A +L +S G
Sbjct: 82 LFGRIADKHGRKKSMLLSVCMMCFGSLVIACLPGYETIGTWAPALLLLARLFQGLSVGGE 141
Query: 129 FCISVAYVADVVHESKR--VAVFSWIT 153
+ S Y+++V E ++ A F ++T
Sbjct: 142 YGTSATYMSEVAVEGRKGFYASFQYVT 168
>YDIM_ECOLI (P76197) Hypothetical transport protein ydiM
Length = 404
Score = 32.0 bits (71), Expect = 4.0
Identities = 22/79 (27%), Positives = 41/79 (51%), Gaps = 6/79 (7%)
Query: 12 PLFHLLLPLSIHWIAEAMTVSVL-VDVTTTALCPQQSSCSKAIYINGLQQTITGIFKMAV 70
P F L L +++ M V ++ +++ + Q ++ +I I+ L GI +++V
Sbjct: 4 PYFPTALGLYFNYLVHGMGVLLMSLNMASLETLWQTNAAGVSIVISSL-----GIGRLSV 58
Query: 71 LPLLGQLSDEHGRKPLLLL 89
L G LSD GR+P ++L
Sbjct: 59 LLFAGLLSDRFGRRPFIML 77
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.339 0.147 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,927,679
Number of Sequences: 164201
Number of extensions: 2145205
Number of successful extensions: 8441
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 8409
Number of HSP's gapped (non-prelim): 49
length of query: 526
length of database: 59,974,054
effective HSP length: 115
effective length of query: 411
effective length of database: 41,090,939
effective search space: 16888375929
effective search space used: 16888375929
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC140027.12


