

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139344.1 + phase: 0
(368 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
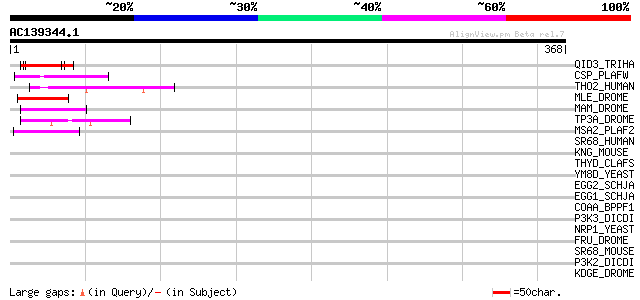
Score E
Sequences producing significant alignments: (bits) Value
QID3_TRIHA (P52755) Cell wall protein qid3 precursor 48 4e-05
CSP_PLAFW (P08307) Circumsporozoite protein precursor (CS) 45 2e-04
THO2_HUMAN (Q8NI27) THO complex subunit 2 (Tho2) 44 5e-04
MLE_DROME (P24785) Dosage compensation regulator (Male-less prot... 44 6e-04
MAM_DROME (P21519) Neurogenic protein mastermind 44 6e-04
TP3A_DROME (Q9NG98) DNA topoisomerase III alpha (EC 5.99.1.2) 44 8e-04
MSA2_PLAF2 (Q03646) Merozoite surface antigen 2 precursor (MSA-2) 44 8e-04
SR68_HUMAN (Q9UHB9) Signal recognition particle 68 kDa protein (... 43 0.001
KNG_MOUSE (O08677) Kininogen precursor [Contains: Bradykinin] 42 0.002
THYD_CLAFS (Q9UVI4) Trihydrophobin precursor (CFTH1) 42 0.002
YM8D_YEAST (P38429) Hypothetical 23.0 kDa protein in TPS3-IPP2 i... 41 0.004
EGG2_SCHJA (P19469) Eggshell protein 2A precursor 41 0.005
EGG1_SCHJA (P19470) Eggshell protein 1 precursor 41 0.005
COAA_BPPF1 (P25129) Probable coat protein A precursor 41 0.005
P3K3_DICDI (P54675) Phosphatidylinositol 3-kinase 3 (EC 2.7.1.13... 40 0.009
NRP1_YEAST (P32770) Asparagine-rich protein (ARP protein) 40 0.009
FRU_DROME (Q8IN81) Sex determination protein fruitless 40 0.009
SR68_MOUSE (Q8BMA6) Signal recognition particle 68 kDa protein (... 40 0.012
P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.13... 40 0.012
KDGE_DROME (Q09103) Eye-specific diacylglycerol kinase (EC 2.7.1... 40 0.012
>QID3_TRIHA (P52755) Cell wall protein qid3 precursor
Length = 143
Score = 47.8 bits (112), Expect = 4e-05
Identities = 21/28 (75%), Positives = 21/28 (75%), Gaps = 1/28 (3%)
Query: 8 GQGNGNGNGN-NGNGKGNGNGNGNNGNG 34
G G GNGNGN NGNG GNGNGNGN G
Sbjct: 42 GNGGGNGNGNGNGNGGGNGNGNGNTNTG 69
Score = 44.7 bits (104), Expect = 4e-04
Identities = 22/32 (68%), Positives = 22/32 (68%), Gaps = 2/32 (6%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
NGNG G NGNG GNGNG G NGNG GN G
Sbjct: 41 NGNGGG-NGNGNGNGNG-GGNGNGNGNTNTGG 70
Score = 43.5 bits (101), Expect = 8e-04
Identities = 19/28 (67%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Query: 10 GNGNGNGN-NGNGKGNGNGNGNNGNGKG 36
GNG GNGN NGNG G GNGNGN G
Sbjct: 42 GNGGGNGNGNGNGNGGGNGNGNGNTNTG 69
Score = 37.4 bits (85), Expect = 0.061
Identities = 17/26 (65%), Positives = 18/26 (68%), Gaps = 3/26 (11%)
Query: 8 GQGNGNGNGN---NGNGKGNGNGNGN 30
G GNGNGNGN NGNG GN N G+
Sbjct: 46 GNGNGNGNGNGGGNGNGNGNTNTGGS 71
>CSP_PLAFW (P08307) Circumsporozoite protein precursor (CS)
Length = 442
Score = 45.4 bits (106), Expect = 2e-04
Identities = 22/62 (35%), Positives = 29/62 (46%), Gaps = 2/62 (3%)
Query: 4 AIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEES 63
++ + N NGNN N GN N N GK DK +G + EK + PK KK +
Sbjct: 71 SLGENDDGDNDNGNNNN--GNNNNGDNGREGKDEDKRDGNNEDNEKLRKPKHKKLKQPGD 128
Query: 64 SN 65
N
Sbjct: 129 GN 130
>THO2_HUMAN (Q8NI27) THO complex subunit 2 (Tho2)
Length = 1478
Score = 44.3 bits (103), Expect = 5e-04
Identities = 34/117 (29%), Positives = 56/117 (47%), Gaps = 26/117 (22%)
Query: 14 GNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEK------------------EKAPKK 55
GN +NGN +G+ +N K NDKE+GKEK KEK ++ PK+
Sbjct: 1136 GNSSNGN-----SGSNSNKAVKENDKEKGKEKEKEKKEKTPATTPEARVLGKDGKEKPKE 1190
Query: 56 KKAPKEESSNYQQLQTLPSGQERAFCKANNTC---HFATLVCPAECKTRKPKKNKKD 109
++ K+E + + +T S +E+ K F T V AE K+ + ++ +K+
Sbjct: 1191 ERPNKDEKARETKERTPKSDKEKEKFKKEEKAKDEKFKTTVPNAESKSTQEREREKE 1247
>MLE_DROME (P24785) Dosage compensation regulator (Male-less protein)
(No action potential protein)
Length = 1293
Score = 43.9 bits (102), Expect = 6e-04
Identities = 20/35 (57%), Positives = 23/35 (65%), Gaps = 1/35 (2%)
Query: 6 AQGQGNGNGNGNNGNG-KGNGNGNGNNGNGKGNDK 39
A G GN G GNNG G + NG G GNNG G GN++
Sbjct: 1230 AGGYGNNGGYGNNGGGYRNNGGGYGNNGGGYGNNR 1264
Score = 40.4 bits (93), Expect = 0.007
Identities = 20/37 (54%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Query: 11 NGNGNGNNGNGKGN-GNGNGNNGNGKGNDKEEGKEKG 46
NG G GNNG G GN G G GNN G GN+ G G
Sbjct: 1208 NGGGYGNNGGGYGNIGGGYGNNAGGYGNNGGYGNNGG 1244
Score = 37.7 bits (86), Expect = 0.047
Identities = 18/32 (56%), Positives = 19/32 (59%), Gaps = 1/32 (3%)
Query: 8 GQGN-GNGNGNNGNGKGNGNGNGNNGNGKGND 38
G GN G G GNN G GN G GNNG G N+
Sbjct: 1218 GYGNIGGGYGNNAGGYGNNGGYGNNGGGYRNN 1249
Score = 36.2 bits (82), Expect = 0.14
Identities = 18/32 (56%), Positives = 20/32 (62%), Gaps = 1/32 (3%)
Query: 8 GQGNGNGNGNNGNGKG-NGNGNGNNGNGKGND 38
G+ G G GNNG G G NG G GN G G GN+
Sbjct: 1198 GRTFGGGYGNNGGGYGNNGGGYGNIGGGYGNN 1229
Score = 35.0 bits (79), Expect = 0.30
Identities = 18/35 (51%), Positives = 19/35 (53%), Gaps = 1/35 (2%)
Query: 5 IAQGQGNGNGN-GNNGNGKGNGNGNGNNGNGKGND 38
I G GN G GNNG NG G NNG G GN+
Sbjct: 1222 IGGGYGNNAGGYGNNGGYGNNGGGYRNNGGGYGNN 1256
Score = 33.1 bits (74), Expect = 1.1
Identities = 14/31 (45%), Positives = 18/31 (57%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGND 38
G+ + G GNG+ G G GNNG G GN+
Sbjct: 1185 GRFTNSSFGRRGNGRTFGGGYGNNGGGYGNN 1215
Score = 31.6 bits (70), Expect = 3.3
Identities = 14/26 (53%), Positives = 14/26 (53%)
Query: 12 GNGNGNNGNGKGNGNGNGNNGNGKGN 37
GNG G NG G GNNG G GN
Sbjct: 1196 GNGRTFGGGYGNNGGGYGNNGGGYGN 1221
>MAM_DROME (P21519) Neurogenic protein mastermind
Length = 1596
Score = 43.9 bits (102), Expect = 6e-04
Identities = 17/44 (38%), Positives = 26/44 (58%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEK 51
G G GN NN NG G G GNGNN N G+ ++ ++ ++++
Sbjct: 360 GGGGGNSGNNNNNGGGGGGGNGNNNNNGGDHHQQQQQHQHQQQQ 403
Score = 42.4 bits (98), Expect = 0.002
Identities = 16/44 (36%), Positives = 26/44 (58%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEK 51
G GN N NNG G G GNGN NN G + +++ + +++++
Sbjct: 362 GGGNSGNNNNNGGGGGGGNGNNNNNGGDHHQQQQQHQHQQQQQQ 405
Score = 39.7 bits (91), Expect = 0.012
Identities = 15/44 (34%), Positives = 22/44 (49%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEK 51
G G G G G GN N N G G G GN+ G + +++++
Sbjct: 353 GVGGGGGGGGGGNSGNNNNNGGGGGGGNGNNNNNGGDHHQQQQQ 396
Score = 38.1 bits (87), Expect = 0.036
Identities = 16/51 (31%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Query: 8 GQGNGNGNGNNGNGKGNGNGN-GNNGNGKGNDKEEGKEKGKEKEKAPKKKK 57
G G G G GN+GN NG G G NGN N + +++ + + + ++++
Sbjct: 356 GGGGGGGGGNSGNNNNNGGGGGGGNGNNNNNGGDHHQQQQQHQHQQQQQQQ 406
Score = 36.6 bits (83), Expect = 0.10
Identities = 21/83 (25%), Positives = 28/83 (33%), Gaps = 1/83 (1%)
Query: 5 IAQGQGNGNGNGNNGNGKGNGNGNGNN-GNGKGNDKEEGKEKGKEKEKAPKKKKAPKEES 63
+ N N N NNGN N GNG+N GN N+ G + E
Sbjct: 257 LPNNNNNSNSNNNNGNANANNGGNGSNTGNNTNNNGNSTNNNGGSNNNGSENLTKFSVEI 316
Query: 64 SNYQQLQTLPSGQERAFCKANNT 86
+ T P+ + N T
Sbjct: 317 VQQLEFTTSPANSQPQQISTNVT 339
Score = 34.7 bits (78), Expect = 0.39
Identities = 17/36 (47%), Positives = 19/36 (52%), Gaps = 2/36 (5%)
Query: 4 AIAQGQGNGNGNGNNGNGKGN--GNGNGNNGNGKGN 37
A A GNG+ GNN N GN N G+N NG N
Sbjct: 273 ANANNGGNGSNTGNNTNNNGNSTNNNGGSNNNGSEN 308
Score = 33.9 bits (76), Expect = 0.67
Identities = 14/51 (27%), Positives = 22/51 (42%)
Query: 12 GNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEE 62
G G G G G GN N NNG G G G + + ++ + +++
Sbjct: 353 GVGGGGGGGGGGNSGNNNNNGGGGGGGNGNNNNNGGDHHQQQQQHQHQQQQ 403
Score = 30.8 bits (68), Expect = 5.7
Identities = 14/54 (25%), Positives = 21/54 (37%)
Query: 14 GNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSNYQ 67
G G G G G GN NN NG G G + ++++ ++ Q
Sbjct: 353 GVGGGGGGGGGGNSGNNNNNGGGGGGGNGNNNNNGGDHHQQQQQHQHQQQQQQQ 406
Score = 30.4 bits (67), Expect = 7.4
Identities = 12/26 (46%), Positives = 15/26 (57%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGN 33
G G G+ N NNG G G G GN+ +
Sbjct: 1513 GGGPGSANNNNGGGGGGAAGGGNSAS 1538
Score = 30.0 bits (66), Expect = 9.7
Identities = 12/30 (40%), Positives = 13/30 (43%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGN 37
G G G G G G+ N G G G GN
Sbjct: 1506 GPGGGGGGGGPGSANNNNGGGGGGAAGGGN 1535
>TP3A_DROME (Q9NG98) DNA topoisomerase III alpha (EC 5.99.1.2)
Length = 1250
Score = 43.5 bits (101), Expect = 8e-04
Identities = 28/85 (32%), Positives = 42/85 (48%), Gaps = 14/85 (16%)
Query: 8 GQGNGNGNGNNGNGKGNGN----GNGNNGNGKGNDKEEGKEKGKEKEKA--------PKK 55
G G G G+G G+G G G G+G G G G K GK+ G E +K+ PK
Sbjct: 801 GSGGGGGSGGWGSGTGGGGSGGWGSGTGGGGLGGGK--GKKPGGESKKSATKKPPNEPKP 858
Query: 56 KKAPKEESSNYQQLQTLPSGQERAF 80
KK + +++ ++ + SG R+F
Sbjct: 859 KKTKEPKAAPNKKTSSKSSGSIRSF 883
Score = 31.6 bits (70), Expect = 3.3
Identities = 15/39 (38%), Positives = 20/39 (50%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G G G G G+G+G G+G ++G G G G G
Sbjct: 772 GGGPGPGPGGGGSGRGAGSGGWSSGPGGGGSGGGGGSGG 810
Score = 30.4 bits (67), Expect = 7.4
Identities = 14/36 (38%), Positives = 17/36 (46%)
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
+G G G G G G G G G G+G +G G G
Sbjct: 769 RGGGGGPGPGPGGGGSGRGAGSGGWSSGPGGGGSGG 804
>MSA2_PLAF2 (Q03646) Merozoite surface antigen 2 precursor (MSA-2)
Length = 347
Score = 43.5 bits (101), Expect = 8e-04
Identities = 24/46 (52%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query: 3 AAIAQGQGNGNGNG-NNGNGKGNGNGNGNN-GNGKGNDKEEGKEKG 46
A+ G GNG GNG NG G G GNG GN GNG GN G G
Sbjct: 80 ASAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNG 125
Score = 43.1 bits (100), Expect = 0.001
Identities = 24/45 (53%), Positives = 25/45 (55%), Gaps = 2/45 (4%)
Query: 3 AAIAQGQGNGNGNGNN-GNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
A + G G GNG GN GNG GNG GNG GNG GN G G
Sbjct: 78 AVASAGNGAGNGAGNGAGNGAGNGAGNG-AGNGAGNGAGNGAGNG 121
Score = 42.0 bits (97), Expect = 0.002
Identities = 23/41 (56%), Positives = 23/41 (56%), Gaps = 2/41 (4%)
Query: 8 GQGNGNGNG-NNGNGKGNGNGNGNN-GNGKGNDKEEGKEKG 46
G GNG GNG NG G G GNG GN GNG GN G G
Sbjct: 105 GAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNG 145
Score = 42.0 bits (97), Expect = 0.002
Identities = 23/41 (56%), Positives = 23/41 (56%), Gaps = 2/41 (4%)
Query: 8 GQGNGNGNG-NNGNGKGNGNGNGNN-GNGKGNDKEEGKEKG 46
G GNG GNG NG G G GNG GN GNG GN G G
Sbjct: 89 GAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNG 129
Score = 42.0 bits (97), Expect = 0.002
Identities = 23/41 (56%), Positives = 23/41 (56%), Gaps = 2/41 (4%)
Query: 8 GQGNGNGNG-NNGNGKGNGNGNGNN-GNGKGNDKEEGKEKG 46
G GNG GNG NG G G GNG GN GNG GN G G
Sbjct: 97 GAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNG 137
Score = 42.0 bits (97), Expect = 0.002
Identities = 23/41 (56%), Positives = 23/41 (56%), Gaps = 2/41 (4%)
Query: 8 GQGNGNGNG-NNGNGKGNGNGNGNN-GNGKGNDKEEGKEKG 46
G GNG GNG NG G G GNG GN GNG GN G G
Sbjct: 93 GAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNG 133
Score = 42.0 bits (97), Expect = 0.002
Identities = 23/41 (56%), Positives = 23/41 (56%), Gaps = 2/41 (4%)
Query: 8 GQGNGNGNG-NNGNGKGNGNGNGNN-GNGKGNDKEEGKEKG 46
G GNG GNG NG G G GNG GN GNG GN G G
Sbjct: 101 GAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNG 141
Score = 40.0 bits (92), Expect = 0.009
Identities = 29/93 (31%), Positives = 37/93 (39%), Gaps = 25/93 (26%)
Query: 8 GQGNGNGNG---NNGNGKGNGNGNG-------NNGNGKGNDKEEGKEKGKEKEKA----- 52
G GNG GNG GNG GNG GNG + GNG GN G E++
Sbjct: 117 GAGNGAGNGAGNGAGNGAGNGAGNGAGNGAVASAGNGAGNGAVASAGNGAVAERSSSTPA 176
Query: 53 ----------PKKKKAPKEESSNYQQLQTLPSG 75
+ + E+SN+ +T P G
Sbjct: 177 TTTTTTTTNDAEASTSTSSENSNHNNAETNPKG 209
Score = 34.3 bits (77), Expect = 0.51
Identities = 18/44 (40%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Query: 4 AIAQGQGNGNGNGNNGNGKGNGNGNGNN-GNGKGNDKEEGKEKG 46
A+A + NG G G GNG GN GNG GN G G
Sbjct: 70 AVASAGNGAVASAGNGAGNGAGNGAGNGAGNGAGNGAGNGAGNG 113
Score = 31.2 bits (69), Expect = 4.4
Identities = 20/91 (21%), Positives = 33/91 (35%), Gaps = 16/91 (17%)
Query: 2 EAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPK------- 54
EA+ + N N N N KGNG N N + + ++ + K P+
Sbjct: 188 EASTSTSSENSNHNNAETNPKGNGEVQPNQANKETQNNSNVQQDSQTKSNVPRTQDADTK 247
Query: 55 ---------KKKAPKEESSNYQQLQTLPSGQ 76
+ AP E + +LQ+ P +
Sbjct: 248 SPTAQPEQAENSAPTAEQTESPELQSAPENK 278
>SR68_HUMAN (Q9UHB9) Signal recognition particle 68 kDa protein
(SRP68)
Length = 627
Score = 42.7 bits (99), Expect = 0.001
Identities = 24/67 (35%), Positives = 36/67 (52%), Gaps = 6/67 (8%)
Query: 14 GNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKE--ESSNYQQLQT 71
G G G+G G G+G G +G G+G G E+ KE E+ KA KE +S + + LQ
Sbjct: 10 GGGGGGSGGGGGSGGGGSGGGRG----AGGEENKENERPSAGSKANKEFGDSLSLEILQI 65
Query: 72 LPSGQER 78
+ Q++
Sbjct: 66 IKESQQQ 72
>KNG_MOUSE (O08677) Kininogen precursor [Contains: Bradykinin]
Length = 661
Score = 42.4 bits (98), Expect = 0.002
Identities = 23/72 (31%), Positives = 40/72 (54%), Gaps = 4/72 (5%)
Query: 5 IAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESS 64
+ G G+G+G+G +G+G G+G+G+G +G+G G+ K K+K K+ + + SS
Sbjct: 493 VGHGHGHGHGHG-HGHGHGHGHGHG-HGHGHGHGKHTNKDKNSVKQTTQRTESL--ASSS 548
Query: 65 NYQQLQTLPSGQ 76
Y T G+
Sbjct: 549 EYSTTSTQMQGR 560
>THYD_CLAFS (Q9UVI4) Trihydrophobin precursor (CFTH1)
Length = 394
Score = 42.0 bits (97), Expect = 0.002
Identities = 18/46 (39%), Positives = 20/46 (43%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAP 53
G GN GNNG G NG N GN GN+ G G + P
Sbjct: 160 GNNGGNNGGNNGGNNGGNNGGNNGGNNGGNNGGNGGNGGNGYQACP 205
Score = 40.4 bits (93), Expect = 0.007
Identities = 20/45 (44%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query: 2 EAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
+A + Q NG GNG N G G GN GN GN GN G G
Sbjct: 109 QAVLCQDSINGGGNGGNNGGNG-GNNGGNGGNNGGNTDYPGGNGG 152
Score = 39.7 bits (91), Expect = 0.012
Identities = 22/49 (44%), Positives = 23/49 (46%), Gaps = 4/49 (8%)
Query: 2 EAAIAQGQGNGNGNGNNGNGKGNGNGNGNN-GNGK---GNDKEEGKEKG 46
+ AI G GNG N GNG GN GNN GNG GN G G
Sbjct: 267 QPAIGGGNPGGNGGNNGGNGGNGGNNGGNNGGNGDYPGGNGGNNGGSNG 315
Score = 38.9 bits (89), Expect = 0.021
Identities = 15/31 (48%), Positives = 16/31 (51%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGND 38
G GN GNNG G NG N GN GN+
Sbjct: 152 GNNGGNNGGNNGGNNGGNNGGNNGGNNGGNN 182
Score = 38.9 bits (89), Expect = 0.021
Identities = 15/31 (48%), Positives = 16/31 (51%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGND 38
G GN GNNG G NG N GN GN+
Sbjct: 156 GNNGGNNGGNNGGNNGGNNGGNNGGNNGGNN 186
Score = 38.1 bits (87), Expect = 0.036
Identities = 15/31 (48%), Positives = 16/31 (51%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGND 38
G GN GNNG G NG N GN GN+
Sbjct: 148 GGNGGNNGGNNGGNNGGNNGGNNGGNNGGNN 178
Score = 38.1 bits (87), Expect = 0.036
Identities = 16/39 (41%), Positives = 16/39 (41%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G NG N GN GN GN NG N G G
Sbjct: 149 GNGGNNGGNNGGNNGGNNGGNNGGNNGGNNGGNNGGNNG 187
Score = 37.7 bits (86), Expect = 0.047
Identities = 19/45 (42%), Positives = 19/45 (42%), Gaps = 8/45 (17%)
Query: 10 GNGNGNGNNGNGKGNGNGNGNN--------GNGKGNDKEEGKEKG 46
G GNG N GNG NG GNN GNG N G G
Sbjct: 119 GGGNGGNNGGNGGNNGGNGGNNGGNTDYPGGNGGNNGGNNGGNNG 163
Score = 35.8 bits (81), Expect = 0.18
Identities = 15/37 (40%), Positives = 15/37 (40%)
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G NG N GN GN GN NG N G G
Sbjct: 155 GGNNGGNNGGNNGGNNGGNNGGNNGGNNGGNNGGNNG 191
Score = 35.4 bits (80), Expect = 0.23
Identities = 14/25 (56%), Positives = 15/25 (60%)
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGNG 34
GNG+ G NG G NG GN GNG
Sbjct: 298 GNGDYPGGNGGNNGGSNGGGNGGNG 322
Score = 35.4 bits (80), Expect = 0.23
Identities = 17/39 (43%), Positives = 18/39 (45%), Gaps = 8/39 (20%)
Query: 8 GQGNGNGNGNNGNGKGN--------GNGNGNNGNGKGND 38
G GNG N GNG N GNG N GN GN+
Sbjct: 124 GNNGGNGGNNGGNGGNNGGNTDYPGGNGGNNGGNNGGNN 162
Score = 35.4 bits (80), Expect = 0.23
Identities = 18/41 (43%), Positives = 18/41 (43%), Gaps = 2/41 (4%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNG--NNGNGKGNDKEEGKEKG 46
G NG GN GN GN GNG GNG N G G
Sbjct: 279 GGNNGGNGGNGGNNGGNNGGNGDYPGGNGGNNGGSNGGGNG 319
Score = 35.0 bits (79), Expect = 0.30
Identities = 18/41 (43%), Positives = 19/41 (45%), Gaps = 9/41 (21%)
Query: 8 GQGNGNGNGNNG---------NGKGNGNGNGNNGNGKGNDK 39
G NG NG NG NG NG GNG NG G+ K
Sbjct: 289 GGNNGGNNGGNGDYPGGNGGNNGGSNGGGNGGNGGNGGSFK 329
Score = 35.0 bits (79), Expect = 0.30
Identities = 17/42 (40%), Positives = 17/42 (40%), Gaps = 5/42 (11%)
Query: 10 GNGNGNGNN-----GNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
GNG NG N GNG NG NG N G G G
Sbjct: 135 GNGGNNGGNTDYPGGNGGNNGGNNGGNNGGNNGGNNGGNNGG 176
Score = 35.0 bits (79), Expect = 0.30
Identities = 16/36 (44%), Positives = 17/36 (46%), Gaps = 5/36 (13%)
Query: 8 GQGNGNGNGNNGN-----GKGNGNGNGNNGNGKGND 38
G GNG N GN G G NG N GN GN+
Sbjct: 131 GNNGGNGGNNGGNTDYPGGNGGNNGGNNGGNNGGNN 166
Score = 34.7 bits (78), Expect = 0.39
Identities = 16/39 (41%), Positives = 19/39 (48%), Gaps = 6/39 (15%)
Query: 10 GNGNGNGNNGNGKGN------GNGNGNNGNGKGNDKEEG 42
GNG NG N G G+ GN G+NG G G + G
Sbjct: 287 GNGGNNGGNNGGNGDYPGGNGGNNGGSNGGGNGGNGGNG 325
Score = 33.9 bits (76), Expect = 0.67
Identities = 15/36 (41%), Positives = 16/36 (43%)
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEK 45
GN GNG+ G G NG N G GN G K
Sbjct: 294 GNNGGNGDYPGGNGGNNGGSNGGGNGGNGGNGGSFK 329
Score = 33.5 bits (75), Expect = 0.88
Identities = 16/42 (38%), Positives = 16/42 (38%), Gaps = 5/42 (11%)
Query: 10 GNGNGNGNNGNGK-----GNGNGNGNNGNGKGNDKEEGKEKG 46
GN GNG N G GNG NG N G G G
Sbjct: 131 GNNGGNGGNNGGNTDYPGGNGGNNGGNNGGNNGGNNGGNNGG 172
>YM8D_YEAST (P38429) Hypothetical 23.0 kDa protein in TPS3-IPP2
intergenic region (ORF2)
Length = 201
Score = 41.2 bits (95), Expect = 0.004
Identities = 16/46 (34%), Positives = 23/46 (49%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKK 56
N NG+ NN N N N N NN N G+ GK++ A +++
Sbjct: 33 NNNGSNNNNNNNNNNNNNSNNSNNNNGPTSSGRTNGKQRLTAAQQQ 78
>EGG2_SCHJA (P19469) Eggshell protein 2A precursor
Length = 207
Score = 40.8 bits (94), Expect = 0.005
Identities = 21/47 (44%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGN-NGNGKGNDKEEGKEKGKEKEKAP 53
G+G G G G GKG GNG GN G G N +G GK AP
Sbjct: 158 GKGGNGGGGGKGGGKGGGNGEGNGKGGGGKNGGGKGGNGGKGGSYAP 204
Score = 40.8 bits (94), Expect = 0.005
Identities = 18/39 (46%), Positives = 19/39 (48%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G NG NG+ GKG G G G GKG EG KG
Sbjct: 145 GGSNGRRNGHGKGGKGGNGGGGGKGGGKGGGNGEGNGKG 183
Score = 39.3 bits (90), Expect = 0.016
Identities = 21/46 (45%), Positives = 26/46 (55%), Gaps = 2/46 (4%)
Query: 6 AQGQGNGNGNGNNG-NGKGNGNGNGNNG-NGKGNDKEEGKEKGKEK 49
+ G+ NG+G G G NG G G G G G NG+GN K G + G K
Sbjct: 147 SNGRRNGHGKGGKGGNGGGGGKGGGKGGGNGEGNGKGGGGKNGGGK 192
Score = 38.1 bits (87), Expect = 0.036
Identities = 19/30 (63%), Positives = 20/30 (66%), Gaps = 2/30 (6%)
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
+G GNG GNG G GK NG G G NG GKG
Sbjct: 172 KGGGNGEGNGKGGGGK-NGGGKGGNG-GKG 199
Score = 36.6 bits (83), Expect = 0.10
Identities = 19/36 (52%), Positives = 22/36 (60%), Gaps = 4/36 (11%)
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
+G G G GNG GNGKG G G NG GKG + +G
Sbjct: 168 KGGGKGGGNG-EGNGKG---GGGKNGGGKGGNGGKG 199
Score = 34.3 bits (77), Expect = 0.51
Identities = 19/39 (48%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query: 12 GNGNGN-NGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEK 49
G NG NG+GKG GNG G GKG K G +G K
Sbjct: 145 GGSNGRRNGHGKGGKGGNG-GGGGKGGGKGGGNGEGNGK 182
Score = 31.6 bits (70), Expect = 3.3
Identities = 17/40 (42%), Positives = 19/40 (47%), Gaps = 1/40 (2%)
Query: 8 GQGNGNGNGNNGNGKGNGNG-NGNNGNGKGNDKEEGKEKG 46
G GN NG+ NG+G G GNG G K GK G
Sbjct: 136 GDSYGNDYYGGSNGRRNGHGKGGKGGNGGGGGKGGGKGGG 175
Score = 30.0 bits (66), Expect = 9.7
Identities = 14/29 (48%), Positives = 15/29 (51%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G GNG GNG G G G G + GKG
Sbjct: 101 GGGNGGGNGGGGGCNGGGCGGVPDFYGKG 129
>EGG1_SCHJA (P19470) Eggshell protein 1 precursor
Length = 212
Score = 40.8 bits (94), Expect = 0.005
Identities = 21/47 (44%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGN-NGNGKGNDKEEGKEKGKEKEKAP 53
G+G G G G GKG GNG GN G G N +G GK AP
Sbjct: 163 GKGGNGGGGGKGGGKGGGNGKGNGKGGGGKNGGGKGGNGGKGGSYAP 209
Score = 40.8 bits (94), Expect = 0.005
Identities = 22/46 (47%), Positives = 26/46 (55%), Gaps = 2/46 (4%)
Query: 6 AQGQGNGNGNGNNG-NGKGNGNGNGNNG-NGKGNDKEEGKEKGKEK 49
+ G+ NG+G G G NG G G G G G NGKGN K G + G K
Sbjct: 152 SNGRKNGHGKGGKGGNGGGGGKGGGKGGGNGKGNGKGGGGKNGGGK 197
Score = 40.0 bits (92), Expect = 0.009
Identities = 17/39 (43%), Positives = 19/39 (48%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G NG NG+ GKG G G G GKG +G KG
Sbjct: 150 GDSNGRKNGHGKGGKGGNGGGGGKGGGKGGGNGKGNGKG 188
Score = 38.1 bits (87), Expect = 0.036
Identities = 19/30 (63%), Positives = 20/30 (66%), Gaps = 2/30 (6%)
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
+G GNG GNG G GK NG G G NG GKG
Sbjct: 177 KGGGNGKGNGKGGGGK-NGGGKGGNG-GKG 204
Score = 36.6 bits (83), Expect = 0.10
Identities = 19/36 (52%), Positives = 22/36 (60%), Gaps = 4/36 (11%)
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
+G G G GNG GNGKG G G NG GKG + +G
Sbjct: 173 KGGGKGGGNG-KGNGKG---GGGKNGGGKGGNGGKG 204
Score = 36.2 bits (82), Expect = 0.14
Identities = 20/39 (51%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Query: 12 GNGNGN-NGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEK 49
G+ NG NG+GKG GNG G GKG K G KG K
Sbjct: 150 GDSNGRKNGHGKGGKGGNG-GGGGKGGGKGGGNGKGNGK 187
Score = 34.7 bits (78), Expect = 0.39
Identities = 16/35 (45%), Positives = 18/35 (50%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
G GNG GNG G G G G G + GKG + G
Sbjct: 106 GGGNGGGNGGGGGCNGGGCGGGPDFYGKGYEDSYG 140
Score = 33.1 bits (74), Expect = 1.1
Identities = 18/43 (41%), Positives = 21/43 (47%), Gaps = 1/43 (2%)
Query: 8 GQGNGNGNGNNGNGKGNGNG-NGNNGNGKGNDKEEGKEKGKEK 49
G GN + NG+ NG+G G GNG G K GK G K
Sbjct: 141 GDSYGNDYYGDSNGRKNGHGKGGKGGNGGGGGKGGGKGGGNGK 183
>COAA_BPPF1 (P25129) Probable coat protein A precursor
Length = 437
Score = 40.8 bits (94), Expect = 0.005
Identities = 20/61 (32%), Positives = 31/61 (50%), Gaps = 3/61 (4%)
Query: 2 EAAIAQGQGNGNGNGN---NGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKA 58
+ G G+G G+GN +G+G G+G+G G +GNG + +E G E K+
Sbjct: 285 DGGTGNGDGSGGGDGNGAGDGSGDGDGSGTGGDGNGTCDPAKENCSTGPEGPGGELKEPT 344
Query: 59 P 59
P
Sbjct: 345 P 345
Score = 40.4 bits (93), Expect = 0.007
Identities = 20/40 (50%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Query: 8 GQGNGNGNGNNGNGKGNGNGN-GNNGNGKGNDKEEGKEKG 46
G GN NG GN+G G GNG+G+ G +GNG G+ +G G
Sbjct: 275 GGGNNNGGGNDG-GTGNGDGSGGGDGNGAGDGSGDGDGSG 313
Score = 39.3 bits (90), Expect = 0.016
Identities = 20/43 (46%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query: 8 GQGNGNGNG---NNGNGKGNGNGNGN-NGNGKGNDKEEGKEKG 46
G G G+GNG NNG G G GNG+ +G G GN +G G
Sbjct: 267 GDGGGDGNGGGNNNGGGNDGGTGNGDGSGGGDGNGAGDGSGDG 309
Score = 37.0 bits (84), Expect = 0.079
Identities = 19/40 (47%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query: 8 GQGNGNGN-GNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G NG GN G GNG G+G G+G NG G G+ +G G
Sbjct: 277 GNNNGGGNDGGTGNGDGSGGGDG-NGAGDGSGDGDGSGTG 315
Score = 35.8 bits (81), Expect = 0.18
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 3/38 (7%)
Query: 12 GNGNGNNGNGKGNGNGNGNN---GNGKGNDKEEGKEKG 46
G G +GNG GN NG GN+ GNG G+ +G G
Sbjct: 266 GGDGGGDGNGGGNNNGGGNDGGTGNGDGSGGGDGNGAG 303
>P3K3_DICDI (P54675) Phosphatidylinositol 3-kinase 3 (EC 2.7.1.137)
(PI3-kinase) (PtdIns-3-kinase) (PI3K) (Fragment)
Length = 1585
Score = 40.0 bits (92), Expect = 0.009
Identities = 19/57 (33%), Positives = 21/57 (36%)
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEES 63
+ N N N NN N N N N NN N N+ EE K EES
Sbjct: 346 ENNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNEELINNNNNNNNDENYKIEETEES 402
Score = 38.5 bits (88), Expect = 0.027
Identities = 29/99 (29%), Positives = 39/99 (39%), Gaps = 24/99 (24%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGNDKE----------------EGKEKG------KE 48
N N N NN N N N N NN N N++E E E+ KE
Sbjct: 351 NNNNNNNNNNNNNNNNNNNNNNNNNNNNEELINNNNNNNNDENYKIEETEESLKELLEKE 410
Query: 49 KEKAPKKKKAPKE--ESSNYQQLQTLPSGQERAFCKANN 85
K + +++K KE E N ++ L G C A+N
Sbjct: 411 KLENEEREKILKERNEIDNLKKKNHLSKGYFMRACNASN 449
Score = 38.1 bits (87), Expect = 0.036
Identities = 19/69 (27%), Positives = 26/69 (37%)
Query: 2 EAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKE 61
++ I N N N NN N N N N NN N N+ E + K +
Sbjct: 340 QSDIFNENNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNEELINNNNNNNNDENYKIEET 399
Query: 62 ESSNYQQLQ 70
E S + L+
Sbjct: 400 EESLKELLE 408
Score = 37.4 bits (85), Expect = 0.061
Identities = 14/26 (53%), Positives = 15/26 (56%)
Query: 13 NGNGNNGNGKGNGNGNGNNGNGKGND 38
N N NN NG GN N N NN N N+
Sbjct: 200 NNNNNNNNGNGNNNSNNNNSNSNNNN 225
Score = 37.4 bits (85), Expect = 0.061
Identities = 14/28 (50%), Positives = 15/28 (53%)
Query: 15 NGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
N NN N GNGN N NN N N+ G
Sbjct: 200 NNNNNNNNGNGNNNSNNNNSNSNNNNNG 227
Score = 37.4 bits (85), Expect = 0.061
Identities = 14/27 (51%), Positives = 15/27 (54%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGN 37
N N N NNGNG N N N +N N N
Sbjct: 200 NNNNNNNNGNGNNNSNNNNSNSNNNNN 226
Score = 35.8 bits (81), Expect = 0.18
Identities = 17/42 (40%), Positives = 18/42 (42%), Gaps = 11/42 (26%)
Query: 8 GQGNGNGNGNNGNGKGNGNG-----------NGNNGNGKGND 38
G GN N N NN N N NG NGNN N N+
Sbjct: 208 GNGNNNSNNNNSNSNNNNNGISPSSSPPSHLNGNNNNNNSNN 249
Score = 34.7 bits (78), Expect = 0.39
Identities = 13/27 (48%), Positives = 13/27 (48%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGN 37
N N N NN N N N N NN N N
Sbjct: 58 NNNNNNNNNNNNNNNNNNNNNNNNNNN 84
Score = 33.9 bits (76), Expect = 0.67
Identities = 13/29 (44%), Positives = 14/29 (47%)
Query: 5 IAQGQGNGNGNGNNGNGKGNGNGNGNNGN 33
I + N N NGN N N N N NN N
Sbjct: 197 IKKNNNNNNNNGNGNNNSNNNNSNSNNNN 225
Score = 32.7 bits (73), Expect = 1.5
Identities = 14/39 (35%), Positives = 16/39 (40%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEK 49
N N N NN N N N N NN N + KE+
Sbjct: 59 NNNNNNNNNNNNNNNNNNNNNNNNNNVIIPSASTENKEE 97
Score = 32.7 bits (73), Expect = 1.5
Identities = 12/26 (46%), Positives = 13/26 (49%)
Query: 13 NGNGNNGNGKGNGNGNGNNGNGKGND 38
N N NN N N N N NN N N+
Sbjct: 58 NNNNNNNNNNNNNNNNNNNNNNNNNN 83
Score = 32.7 bits (73), Expect = 1.5
Identities = 12/26 (46%), Positives = 13/26 (49%)
Query: 13 NGNGNNGNGKGNGNGNGNNGNGKGND 38
N N NN N N N N NN N N+
Sbjct: 59 NNNNNNNNNNNNNNNNNNNNNNNNNN 84
Score = 32.7 bits (73), Expect = 1.5
Identities = 12/29 (41%), Positives = 13/29 (44%)
Query: 5 IAQGQGNGNGNGNNGNGKGNGNGNGNNGN 33
+ N N N NN N N N N NN N
Sbjct: 56 LINNNNNNNNNNNNNNNNNNNNNNNNNNN 84
Score = 31.6 bits (70), Expect = 3.3
Identities = 15/41 (36%), Positives = 17/41 (40%)
Query: 5 IAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEK 45
I N N N NN N N N N NN + E KE+
Sbjct: 57 INNNNNNNNNNNNNNNNNNNNNNNNNNNVIIPSASTENKEE 97
Score = 30.8 bits (68), Expect = 5.7
Identities = 11/26 (42%), Positives = 13/26 (49%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKG 36
N N N NGN N N + +N N G
Sbjct: 202 NNNNNNGNGNNNSNNNNSNSNNNNNG 227
Score = 30.4 bits (67), Expect = 7.4
Identities = 12/39 (30%), Positives = 14/39 (35%)
Query: 15 NGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAP 53
N N N NGNGNN + N G +P
Sbjct: 196 NIKKNNNNNNNNGNGNNNSNNNNSNSNNNNNGISPSSSP 234
>NRP1_YEAST (P32770) Asparagine-rich protein (ARP protein)
Length = 719
Score = 40.0 bits (92), Expect = 0.009
Identities = 16/28 (57%), Positives = 18/28 (64%)
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGNGKGN 37
GNGNGNGNN N N N N NN + G+
Sbjct: 516 GNGNGNGNNSNNNNNHNNNHNNNHHNGS 543
Score = 38.5 bits (88), Expect = 0.027
Identities = 16/30 (53%), Positives = 18/30 (59%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGN 37
G GNGNGN +N N N N N N+ NG N
Sbjct: 516 GNGNGNGNNSNNNNNHNNNHNNNHHNGSIN 545
Score = 35.4 bits (80), Expect = 0.23
Identities = 14/27 (51%), Positives = 15/27 (54%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGN 37
NGNGNGN N N N N N+ N N
Sbjct: 515 NGNGNGNGNNSNNNNNHNNNHNNNHHN 541
Score = 34.3 bits (77), Expect = 0.51
Identities = 14/30 (46%), Positives = 17/30 (56%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGN 37
G GNGN + NN N N N N +NG+ N
Sbjct: 518 GNGNGNNSNNNNNHNNNHNNNHHNGSINSN 547
Score = 33.1 bits (74), Expect = 1.1
Identities = 13/23 (56%), Positives = 14/23 (60%)
Query: 15 NGNNGNGKGNGNGNGNNGNGKGN 37
N NGNG GNGN + NN N N
Sbjct: 512 NNINGNGNGNGNNSNNNNNHNNN 534
Score = 33.1 bits (74), Expect = 1.1
Identities = 13/20 (65%), Positives = 13/20 (65%)
Query: 18 NGNGKGNGNGNGNNGNGKGN 37
N N GNGNGNGNN N N
Sbjct: 511 NNNINGNGNGNGNNSNNNNN 530
Score = 33.1 bits (74), Expect = 1.1
Identities = 13/22 (59%), Positives = 15/22 (68%)
Query: 17 NNGNGKGNGNGNGNNGNGKGND 38
NN NG GNGNGN +N N N+
Sbjct: 512 NNINGNGNGNGNNSNNNNNHNN 533
Score = 32.3 bits (72), Expect = 2.0
Identities = 15/28 (53%), Positives = 17/28 (60%), Gaps = 1/28 (3%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGND 38
N N NGN GNG GN + N NN N N+
Sbjct: 511 NNNINGN-GNGNGNNSNNNNNHNNNHNN 537
Score = 30.8 bits (68), Expect = 5.7
Identities = 14/27 (51%), Positives = 16/27 (58%), Gaps = 1/27 (3%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGN 37
N NGNG NGNG + N N +N N N
Sbjct: 513 NINGNG-NGNGNNSNNNNNHNNNHNNN 538
>FRU_DROME (Q8IN81) Sex determination protein fruitless
Length = 955
Score = 40.0 bits (92), Expect = 0.009
Identities = 18/83 (21%), Positives = 39/83 (46%), Gaps = 1/83 (1%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEE-SSNY 66
G N N N NN + N N + N ++E +E+ +E+++ + P E+ SS+
Sbjct: 399 GNNNNNNNNNNNSSSNNNNSSSNRERNNSGERERERERERERDRDRELSTTPVEQLSSSK 458
Query: 67 QQLQTLPSGQERAFCKANNTCHF 89
++ + S + + ++ H+
Sbjct: 459 RRRKNSSSNCDNSLSSSHQDRHY 481
Score = 35.8 bits (81), Expect = 0.18
Identities = 14/75 (18%), Positives = 33/75 (43%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSNYQQLQ 70
N + G N N N N N ++ N + E G+ + + ++++ ++ + ++
Sbjct: 393 NSSSTGGNNNNNNNNNNNSSSNNNNSSSNRERNNSGERERERERERERDRDRELSTTPVE 452
Query: 71 TLPSGQERAFCKANN 85
L S + R ++N
Sbjct: 453 QLSSSKRRRKNSSSN 467
Score = 34.7 bits (78), Expect = 0.39
Identities = 14/66 (21%), Positives = 28/66 (42%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSNYQQLQ 70
+ N + ++ G N N N NN N N+ + + ++++ + E ++L
Sbjct: 388 DNNNSNSSSTGGNNNNNNNNNNNSSSNNNNSSSNRERNNSGERERERERERERDRDRELS 447
Query: 71 TLPSGQ 76
T P Q
Sbjct: 448 TTPVEQ 453
>SR68_MOUSE (Q8BMA6) Signal recognition particle 68 kDa protein
(SRP68)
Length = 625
Score = 39.7 bits (91), Expect = 0.012
Identities = 24/75 (32%), Positives = 37/75 (49%), Gaps = 8/75 (10%)
Query: 6 AQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKE--ES 63
A+ Q G G+G G+G G G G G G + G ++ KE E+ KA KE +S
Sbjct: 3 AEKQIPGGGSGGGGSGSGGG------GGGSGGGRSAGGDENKENERPSAGSKANKEFGDS 56
Query: 64 SNYQQLQTLPSGQER 78
+ + LQ + Q++
Sbjct: 57 LSLEILQIIKESQQQ 71
>P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.137)
(PI3-kinase) (PtdIns-3-kinase) (PI3K)
Length = 1858
Score = 39.7 bits (91), Expect = 0.012
Identities = 15/28 (53%), Positives = 16/28 (56%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGND 38
N N N NN N N N N NNGN GN+
Sbjct: 1022 NNNNNNNNNNNNNNNNNNNNNGNNNGNN 1049
Score = 39.3 bits (90), Expect = 0.016
Identities = 22/68 (32%), Positives = 25/68 (36%), Gaps = 19/68 (27%)
Query: 11 NGNGNGNNGNGKGNGNGNGNN-------------------GNGKGNDKEEGKEKGKEKEK 51
N N N NN N NGN NGNN G+G GN E+ G +
Sbjct: 1029 NNNNNNNNNNNNNNGNNNGNNSNNNSNSNISRGSIDSEGNGSGSGNGSEQPTLIGVQNFS 1088
Query: 52 APKKKKAP 59
P K P
Sbjct: 1089 LPNNSKLP 1096
Score = 38.5 bits (88), Expect = 0.027
Identities = 22/76 (28%), Positives = 26/76 (33%), Gaps = 2/76 (2%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSNYQQLQ 70
N N N NN N N N N NN NG N + + S N +
Sbjct: 1020 NNNNNNNNNNNNNNNNNNNNNNNGNNNGNNSNNNSNSNISRGSIDSEGNGSGSGNGSEQP 1079
Query: 71 TLPSGQERAFCKANNT 86
TL Q F NN+
Sbjct: 1080 TLIGVQN--FSLPNNS 1093
Score = 37.7 bits (86), Expect = 0.047
Identities = 14/33 (42%), Positives = 16/33 (48%)
Query: 6 AQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGND 38
+ N N N NN N N N N NN N GN+
Sbjct: 1013 SSNNNNNNNNNNNNNNNNNNNNNNNNNNNNGNN 1045
Score = 36.6 bits (83), Expect = 0.10
Identities = 13/40 (32%), Positives = 18/40 (44%)
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
+ + + + N NN N N N N NN N N+ G G
Sbjct: 1008 ENKDSSSNNNNNNNNNNNNNNNNNNNNNNNNNNNNGNNNG 1047
Score = 36.2 bits (82), Expect = 0.14
Identities = 19/80 (23%), Positives = 24/80 (29%)
Query: 6 AQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSN 65
+ N N N NN N N N N NN N N+ + SS
Sbjct: 184 SNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTTSTTTTTTSILISSSP 243
Query: 66 YQQLQTLPSGQERAFCKANN 85
+ S + F NN
Sbjct: 244 PPSSSSSSSSNDEQFNNNNN 263
Score = 34.7 bits (78), Expect = 0.39
Identities = 13/33 (39%), Positives = 15/33 (45%)
Query: 6 AQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGND 38
+ N N N NN N N N N NN N N+
Sbjct: 1014 SNNNNNNNNNNNNNNNNNNNNNNNNNNNNGNNN 1046
>KDGE_DROME (Q09103) Eye-specific diacylglycerol kinase (EC
2.7.1.107) (Retinal degeneration A protein) (Diglyceride
kinase 2) (DGK 2) (DAG kinase 2)
Length = 1454
Score = 39.7 bits (91), Expect = 0.012
Identities = 18/53 (33%), Positives = 30/53 (55%), Gaps = 1/53 (1%)
Query: 2 EAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPK 54
+++I N +G+ G G G G G G G GK + + ++KGKE++K P+
Sbjct: 744 KSSIRVSNKNNAASGSGGGGAGGGAGGGGGG-GKSKKQTQRRQKGKEEKKEPR 795
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.133 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,439,196
Number of Sequences: 164201
Number of extensions: 2517825
Number of successful extensions: 31564
Number of sequences better than 10.0: 562
Number of HSP's better than 10.0 without gapping: 375
Number of HSP's successfully gapped in prelim test: 206
Number of HSP's that attempted gapping in prelim test: 16757
Number of HSP's gapped (non-prelim): 6742
length of query: 368
length of database: 59,974,054
effective HSP length: 112
effective length of query: 256
effective length of database: 41,583,542
effective search space: 10645386752
effective search space used: 10645386752
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC139344.1


