

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137824.11 - phase: 0
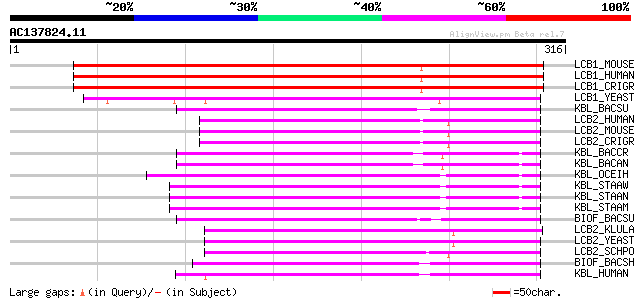
(316 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
LCB1_MOUSE (O35704) Serine palmitoyltransferase 1 (EC 2.3.1.50) ... 255 1e-67
LCB1_HUMAN (O15269) Serine palmitoyltransferase 1 (EC 2.3.1.50) ... 255 1e-67
LCB1_CRIGR (O54695) Serine palmitoyltransferase 1 (EC 2.3.1.50) ... 253 4e-67
LCB1_YEAST (P25045) Serine palmitoyltransferase 1 (EC 2.3.1.50) ... 203 5e-52
KBL_BACSU (O31777) 2-amino-3-ketobutyrate coenzyme A ligase (EC ... 130 3e-30
LCB2_HUMAN (O15270) Serine palmitoyltransferase 2 (EC 2.3.1.50) ... 128 2e-29
LCB2_MOUSE (P97363) Serine palmitoyltransferase 2 (EC 2.3.1.50) ... 127 5e-29
LCB2_CRIGR (O54694) Serine palmitoyltransferase 2 (EC 2.3.1.50) ... 127 5e-29
KBL_BACCR (Q81I05) 2-amino-3-ketobutyrate coenzyme A ligase (EC ... 125 1e-28
KBL_BACAN (Q81V80) 2-amino-3-ketobutyrate coenzyme A ligase (EC ... 125 1e-28
KBL_OCEIH (Q8EM07) 2-amino-3-ketobutyrate coenzyme A ligase (EC ... 122 9e-28
KBL_STAAW (Q8NXY3) 2-amino-3-ketobutyrate coenzyme A ligase (EC ... 122 1e-27
KBL_STAAN (P60120) 2-amino-3-ketobutyrate coenzyme A ligase (EC ... 122 1e-27
KBL_STAAM (P60121) 2-amino-3-ketobutyrate coenzyme A ligase (EC ... 122 1e-27
BIOF_BACSU (P53556) 8-amino-7-oxononanoate synthase (EC 2.3.1.47... 122 2e-27
LCB2_KLULA (P48241) Serine palmitoyltransferase 2 (EC 2.3.1.50) ... 117 4e-26
LCB2_YEAST (P40970) Serine palmitoyltransferase 2 (EC 2.3.1.50) ... 113 7e-25
LCB2_SCHPO (Q09925) Serine palmitoyltransferase 2 (EC 2.3.1.50) ... 113 7e-25
BIOF_BACSH (P22806) 8-amino-7-oxononanoate synthase (EC 2.3.1.47... 110 5e-24
KBL_HUMAN (O75600) 2-amino-3-ketobutyrate coenzyme A ligase, mit... 104 3e-22
>LCB1_MOUSE (O35704) Serine palmitoyltransferase 1 (EC 2.3.1.50)
(Long chain base biosynthesis protein 1) (LCB 1)
(Serine-palmitoyl-CoA transferase 1) (SPT 1) (SPT1)
Length = 473
Score = 255 bits (651), Expect = 1e-67
Identities = 134/274 (48%), Positives = 185/274 (66%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ L+ K+YK +R L+ KE +EL +EW P+PL+P ++ P
Sbjct: 24 HLILEGILILWIIRLVFSKTYKLQERSDLTAKEKEELIEEWQPEPLVPPVSKNHPALNYN 83
Query: 96 LESAAGPHTIV-NGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H IV NGKE VNFAS N+LGL+ + ++ + S+L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPTHNIVVNGKECVNFASFNFLGLLANPRVKAAAFSSLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG ST+ SAIPA+SK+GDII D + IQ GL SR
Sbjct: 144 VHLDLEERLAKFMKTEEAIIYSYGFSTVASAIPAYSKRGDIIFVDSAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN++ L L+ ++ + RR+IV+E LY N+G I PL E++KL
Sbjct: 204 SDIKLFKHNDVADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTICPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGISI 297
>LCB1_HUMAN (O15269) Serine palmitoyltransferase 1 (EC 2.3.1.50)
(Long chain base biosynthesis protein 1) (LCB 1)
(Serine-palmitoyl-CoA transferase 1) (SPT 1) (SPT1)
Length = 473
Score = 255 bits (651), Expect = 1e-67
Identities = 132/274 (48%), Positives = 186/274 (67%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ LL K+YK +R L+ KE +EL +EW P+PL+P + + P
Sbjct: 24 HLILEGILILWIIRLLFSKTYKLQERSDLTVKEKEELIEEWQPEPLVPPVPKDHPALNYN 83
Query: 96 LESAAGPH-TIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H T+VNGKE +NFAS N+LGL+ + ++ + ++L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPSHKTVVNGKECINFASFNFLGLLDNPRVKAAALASLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG +T+ SAIPA+SK+GDI+ D + IQ GL SR
Sbjct: 144 VHLDLEDRLAKFMKTEEAIIYSYGFATIASAIPAYSKRGDIVFVDRAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN+M L L+ ++ + RR+IV+E LY N+G I PL E++KL
Sbjct: 204 SDIKLFKHNDMADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTICPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGINI 297
>LCB1_CRIGR (O54695) Serine palmitoyltransferase 1 (EC 2.3.1.50)
(Long chain base biosynthesis protein 1) (LCB 1)
(Serine-palmitoyl-CoA transferase 1) (SPT 1) (SPT1)
Length = 473
Score = 253 bits (646), Expect = 4e-67
Identities = 131/274 (47%), Positives = 185/274 (66%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ L+ K+YK +R L+ KE +EL +EW P+PL+P ++ P
Sbjct: 24 HLILEGILILWIIRLVFSKTYKLQERSDLTAKEKEELIEEWQPEPLVPPVSKNHPALNYN 83
Query: 96 LESAAGPHTIV-NGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H IV NGKE VNFAS N+LGL+ + ++ + ++L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPTHNIVVNGKECVNFASFNFLGLLANPRVKAAALASLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG ST+ SAIPA+SK+GDI+ D + IQ GL SR
Sbjct: 144 VHLDLEERLAKFMRTEEAIIYSYGFSTIASAIPAYSKRGDIVFVDSAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN++ L L+ ++ + RR+IV+E LY N+G + PL E++KL
Sbjct: 204 SDIKLFKHNDVADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTVCPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGISI 297
>LCB1_YEAST (P25045) Serine palmitoyltransferase 1 (EC 2.3.1.50)
(Long chain base biosynthesis protein 1) (SPT 1) (SPT1)
Length = 558
Score = 203 bits (516), Expect = 5e-52
Identities = 116/279 (41%), Positives = 164/279 (58%), Gaps = 19/279 (6%)
Query: 43 LLLVVILFLLSQ----KSYKPPKRPLSNKEIDELCDEWVPQPLI-PSLNDEMPYE----P 93
L+L I++ LS+ KS + K LS +EID L ++W P+PL+ PS DE + P
Sbjct: 63 LILYGIIYYLSKPQQKKSLQAQKPNLSPQEIDALIEDWEPEPLVDPSATDEQSWRVAKTP 122
Query: 94 PVLESAAGPHTIVNGKE-------VVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGP 146
+E H + V N AS N+L L + + + + ++ YGVG+CGP
Sbjct: 123 VTMEMPIQNHITITRNNLQEKYTNVFNLASNNFLQLSATEPVKEVVKTTIKNYGVGACGP 182
Query: 147 RGFYGTIDVHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGI 206
GFYG DVH E +++F GT S+LY S +PAF+K+GD+IVAD+ V +
Sbjct: 183 AGFYGNQDVHYTLEYDLAQFFGTQGSVLYGQDFCAAPSVLPAFTKRGDVIVADDQVSLPV 242
Query: 207 QNGLYLSRSTVVYFKHNNMDSLRETLENITSIYKRTK---NLRRYIVIEALYQNSGQIAP 263
QN L LSRSTV YF HN+M+SL L +T K K R++IV E ++ NSG +AP
Sbjct: 243 QNALQLSRSTVYYFNHNDMNSLECLLNELTEQEKLEKLPAIPRKFIVTEGIFHNSGDLAP 302
Query: 264 LDEIIKLKEKYCFRILLDESNSFGVLGSSGRGLTEHYGV 302
L E+ KLK KY FR+ +DE+ S GVLG++GRGL+EH+ +
Sbjct: 303 LPELTKLKNKYKFRLFVDETFSIGVLGATGRGLSEHFNM 341
>KBL_BACSU (O31777) 2-amino-3-ketobutyrate coenzyme A ligase (EC
2.3.1.29) (AKB ligase) (Glycine C-acetyltransferase)
Length = 392
Score = 130 bits (328), Expect = 3e-30
Identities = 67/208 (32%), Positives = 120/208 (57%), Gaps = 8/208 (3%)
Query: 96 LESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDV 155
LES GP VN ++V+ +S NYLG H +L+++ A+++YG G+ R GT +
Sbjct: 27 LESMQGPSVTVNHQKVIQLSSNNYLGFTSHPRLINAAQEAVQQYGAGTGSVRTIAGTFTM 86
Query: 156 HLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRS 215
H + E +++ F T ++++ G +T + + K DI+++DE H I +G+ L+++
Sbjct: 87 HQELEKKLAAFKKTEAALVFQSGFTTNQGVLSSILSKEDIVISDELNHASIIDGIRLTKA 146
Query: 216 TVVYFKHNNMDSLRETLENITSIYKRTKNLR-RYIVIEALYQNSGQIAPLDEIIKLKEKY 274
++H NM L L +++ N R R IV + ++ G IAPL +I++L EKY
Sbjct: 147 DKKVYQHVNMSDLERVL-------RKSMNYRMRLIVTDGVFSMDGNIAPLPDIVELAEKY 199
Query: 275 CFRILLDESNSFGVLGSSGRGLTEHYGV 302
+++D++++ GVLG +GRG H+G+
Sbjct: 200 DAFVMVDDAHASGVLGENGRGTVNHFGL 227
>LCB2_HUMAN (O15270) Serine palmitoyltransferase 2 (EC 2.3.1.50)
(Long chain base biosynthesis protein 2) (LCB 2)
(Serine-palmitoyl-CoA transferase 2) (SPT 2)
Length = 562
Score = 128 bits (322), Expect = 2e-29
Identities = 73/199 (36%), Positives = 113/199 (56%), Gaps = 6/199 (3%)
Query: 109 KEVVNFASANYLGLIGHQ-KLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFL 167
K V+N S NYLG + ++ + LE+YG G C R G +D H + E +++FL
Sbjct: 168 KGVINMGSYNYLGFARNTGSCQEAAAKVLEEYGAGVCSTRQEIGNLDKHEELEELVARFL 227
Query: 168 GTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDS 227
G ++ Y G +T IPA KG +I++DE H + G LS +T+ FKHNNM S
Sbjct: 228 GVEAAMAYGMGFATNSMNIPALVGKGCLILSDELNHASLVLGARLSGATIRIFKHNNMQS 287
Query: 228 LRETLENITSIYKRTKNLRRY----IVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDES 283
L + L++ +Y + + R + I++E +Y G I L E+I LK+KY + LDE+
Sbjct: 288 LEKLLKD-AIVYGQPRTRRPWKKILILVEGIYSMEGSIVRLPEVIALKKKYKAYLYLDEA 346
Query: 284 NSFGVLGSSGRGLTEHYGV 302
+S G LG +GRG+ E++G+
Sbjct: 347 HSIGALGPTGRGVVEYFGL 365
>LCB2_MOUSE (P97363) Serine palmitoyltransferase 2 (EC 2.3.1.50)
(Long chain base biosynthesis protein 2) (LCB 2)
(Serine-palmitoyl-CoA transferase 2) (SPT 2)
Length = 560
Score = 127 bits (318), Expect = 5e-29
Identities = 72/199 (36%), Positives = 113/199 (56%), Gaps = 6/199 (3%)
Query: 109 KEVVNFASANYLGLIGHQ-KLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFL 167
K V+N S NYLG + ++ + L++YG G C R G +D H + E +++FL
Sbjct: 166 KGVINMGSYNYLGFARNTGSCQEAAAEVLKEYGAGVCSTRQEIGNLDKHEELEKLVARFL 225
Query: 168 GTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDS 227
G ++ Y G +T IPA KG +I++DE H + G LS +T+ FKHNNM S
Sbjct: 226 GVEAAMTYGMGFATNSMNIPALVGKGCLILSDELNHASLVLGARLSGATIRIFKHNNMQS 285
Query: 228 LRETLENITSIYKRTKNLRRY----IVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDES 283
L + L++ +Y + + R + I++E +Y G I L E+I LK+KY + LDE+
Sbjct: 286 LEKLLKD-AIVYGQPRTRRPWKKILILVEGIYSMEGSIVRLPEVIALKKKYKAYLYLDEA 344
Query: 284 NSFGVLGSSGRGLTEHYGV 302
+S G LG SGRG+ +++G+
Sbjct: 345 HSIGALGPSGRGVVDYFGL 363
>LCB2_CRIGR (O54694) Serine palmitoyltransferase 2 (EC 2.3.1.50)
(Long chain base biosynthesis protein 2) (LCB 2)
(Serine-palmitoyl-CoA transferase 2) (SPT 2)
Length = 560
Score = 127 bits (318), Expect = 5e-29
Identities = 72/199 (36%), Positives = 113/199 (56%), Gaps = 6/199 (3%)
Query: 109 KEVVNFASANYLGLIGHQ-KLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFL 167
K V+N S NYLG + ++ + L++YG G C R G +D H + E +++FL
Sbjct: 166 KGVINMGSYNYLGFARNTGSCQEAAAEVLKEYGAGVCSTRQEIGNLDKHEELEKLVARFL 225
Query: 168 GTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDS 227
G ++ Y G +T IPA KG +I++DE H + G LS +T+ FKHNNM S
Sbjct: 226 GVEAAMTYGMGFATNSMNIPALVGKGCLILSDELNHASLVLGARLSGATIRIFKHNNMQS 285
Query: 228 LRETLENITSIYKRTKNLRRY----IVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDES 283
L + L++ +Y + + R + I++E +Y G I L E+I LK+KY + LDE+
Sbjct: 286 LEKLLKD-AIVYGQPRTRRPWKKILILVEGIYSMEGSIVRLPEVIALKKKYKAYLYLDEA 344
Query: 284 NSFGVLGSSGRGLTEHYGV 302
+S G LG SGRG+ +++G+
Sbjct: 345 HSIGALGPSGRGVVDYFGL 363
>KBL_BACCR (Q81I05) 2-amino-3-ketobutyrate coenzyme A ligase (EC
2.3.1.29) (AKB ligase) (Glycine C-acetyltransferase)
Length = 396
Score = 125 bits (314), Expect = 1e-28
Identities = 70/209 (33%), Positives = 118/209 (55%), Gaps = 8/209 (3%)
Query: 96 LESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDV 155
LES+ GP + GKE +N +S NYLGL +L ++ A+ KYGVG+ R GT+D+
Sbjct: 29 LESSNGPIITIGGKEYINLSSNNYLGLATDSRLQEAAIGAIHKYGVGAGAVRTINGTLDL 88
Query: 156 HLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRS 215
H+ E I+KF T +I Y G + +AI A K D I++DE H I +G LS++
Sbjct: 89 HIKLEETIAKFKHTEAAIAYQSGFNCNMAAISAVMDKNDAILSDELNHASIIDGSRLSKA 148
Query: 216 TVVYFKHNNMDSLRETLENITSIYKRTKNL--RRYIVIEALYQNSGQIAPLDEIIKLKEK 273
++ +KH++M+ LR+ +I + L + ++ + ++ G +A L EI+++ E+
Sbjct: 149 KIIVYKHSDMEDLRQ-----KAIAAKESGLYNKLMVITDGVFSMDGDVAKLPEIVEIAEE 203
Query: 274 YCFRILLDESNSFGVLGSSGRGLTEHYGV 302
+D+++ GVLG G G +H+G+
Sbjct: 204 LDLMTYVDDAHGSGVLG-KGAGTVKHFGL 231
>KBL_BACAN (Q81V80) 2-amino-3-ketobutyrate coenzyme A ligase (EC
2.3.1.29) (AKB ligase) (Glycine C-acetyltransferase)
Length = 396
Score = 125 bits (314), Expect = 1e-28
Identities = 70/209 (33%), Positives = 118/209 (55%), Gaps = 8/209 (3%)
Query: 96 LESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDV 155
LES+ GP + GKE +N +S NYLGL +L ++ A+ KYGVG+ R GT+D+
Sbjct: 29 LESSNGPIITIGGKEYINLSSNNYLGLATDSRLQEAAIGAIHKYGVGAGAVRTINGTLDL 88
Query: 156 HLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRS 215
H+ E I+KF T +I Y G + +AI A K D I++DE H I +G LS++
Sbjct: 89 HIKLEETIAKFKHTEAAIAYQSGFNCNMAAISAVMDKNDAILSDELNHASIIDGSRLSKA 148
Query: 216 TVVYFKHNNMDSLRETLENITSIYKRTKNL--RRYIVIEALYQNSGQIAPLDEIIKLKEK 273
++ +KH++M+ LR+ +I + L + ++ + ++ G +A L EI+++ E+
Sbjct: 149 KIIVYKHSDMEDLRQ-----KAIAAKESGLYNKLMVITDGVFSMDGDVAKLPEIVEIAEE 203
Query: 274 YCFRILLDESNSFGVLGSSGRGLTEHYGV 302
+D+++ GVLG G G +H+G+
Sbjct: 204 LDLMTYVDDAHGSGVLG-KGAGTVKHFGL 231
>KBL_OCEIH (Q8EM07) 2-amino-3-ketobutyrate coenzyme A ligase (EC
2.3.1.29) (AKB ligase) (Glycine C-acetyltransferase)
Length = 396
Score = 122 bits (307), Expect = 9e-28
Identities = 68/225 (30%), Positives = 124/225 (54%), Gaps = 5/225 (2%)
Query: 79 QPLIPSLNDEMPY-EPPVLESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALE 137
Q I L D Y E ++ A GP + GK+++N +S NYLGL + ++ + A++
Sbjct: 11 QENISDLKDRGLYNEIDTVQGANGPEITIEGKKLINLSSNNYLGLATNNEMKEIAKKAID 70
Query: 138 KYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIV 197
+GVG+ R GT+D+H++ E +I++F GT +I Y G + +AI A K D I+
Sbjct: 71 SHGVGAGAVRTINGTLDLHIELEKKIAQFKGTEAAIAYQSGFNCNMAAISAVMDKNDAIL 130
Query: 198 ADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALYQN 257
+D H I +G LS++ ++ F+H++M+ LR+ + + K + ++ + ++
Sbjct: 131 SDSLNHASIIDGCRLSKAKIIRFEHSDMNDLRQKAKEAVESGQYNKIM---VITDGVFSM 187
Query: 258 SGQIAPLDEIIKLKEKYCFRILLDESNSFGVLGSSGRGLTEHYGV 302
G IA L EI+ + E++ +D+++ GV G G G +H+G+
Sbjct: 188 DGDIAKLPEIVDIAEEFDLITYVDDAHGSGVTG-DGAGTVKHFGL 231
>KBL_STAAW (Q8NXY3) 2-amino-3-ketobutyrate coenzyme A ligase (EC
2.3.1.29) (AKB ligase) (Glycine C-acetyltransferase)
Length = 395
Score = 122 bits (306), Expect = 1e-27
Identities = 66/211 (31%), Positives = 116/211 (54%), Gaps = 4/211 (1%)
Query: 92 EPPVLESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYG 151
E +E A GP +NGK +N +S NYLGL ++ L + +A++ +GVG+ R G
Sbjct: 24 EIDTIEGANGPEIKINGKSYINLSSNNYLGLATNEDLKSAAKAAIDTHGVGAGAVRTING 83
Query: 152 TIDVHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLY 211
T+D+H + E ++KF GT +I Y G + +AI A K D I++DE H I +G
Sbjct: 84 TLDLHDELEETLAKFKGTEAAIAYQSGFNCNMAAISAVMNKNDAILSDELNHASIIDGCR 143
Query: 212 LSRSTVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLK 271
LS++ ++ H++MD LR + + K + + + ++ G +A L EI+++
Sbjct: 144 LSKAKIIRVNHSDMDDLRAKAKEAVESGQYNKVM---YITDGVFSMDGDVAKLPEIVEIA 200
Query: 272 EKYCFRILLDESNSFGVLGSSGRGLTEHYGV 302
E++ +D+++ GV+G G G +H+G+
Sbjct: 201 EEFGLLTYVDDAHGSGVMG-KGAGTVKHFGL 230
>KBL_STAAN (P60120) 2-amino-3-ketobutyrate coenzyme A ligase (EC
2.3.1.29) (AKB ligase) (Glycine C-acetyltransferase)
Length = 395
Score = 122 bits (306), Expect = 1e-27
Identities = 66/211 (31%), Positives = 116/211 (54%), Gaps = 4/211 (1%)
Query: 92 EPPVLESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYG 151
E +E A GP +NGK +N +S NYLGL ++ L + +A++ +GVG+ R G
Sbjct: 24 EIDTIEGANGPEIKINGKSYINLSSNNYLGLATNEDLKSAAKAAIDTHGVGAGAVRTING 83
Query: 152 TIDVHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLY 211
T+D+H + E ++KF GT +I Y G + +AI A K D I++DE H I +G
Sbjct: 84 TLDLHDELEETLAKFKGTEAAIAYQSGFNCNMAAISAVMNKNDAILSDELNHASIIDGCR 143
Query: 212 LSRSTVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLK 271
LS++ ++ H++MD LR + + K + + + ++ G +A L EI+++
Sbjct: 144 LSKAKIIRVNHSDMDDLRAKAKEAVESGQYNKVM---YITDGVFSMDGDVAKLPEIVEIA 200
Query: 272 EKYCFRILLDESNSFGVLGSSGRGLTEHYGV 302
E++ +D+++ GV+G G G +H+G+
Sbjct: 201 EEFGLLTYVDDAHGSGVMG-KGAGTVKHFGL 230
>KBL_STAAM (P60121) 2-amino-3-ketobutyrate coenzyme A ligase (EC
2.3.1.29) (AKB ligase) (Glycine C-acetyltransferase)
Length = 395
Score = 122 bits (306), Expect = 1e-27
Identities = 66/211 (31%), Positives = 116/211 (54%), Gaps = 4/211 (1%)
Query: 92 EPPVLESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYG 151
E +E A GP +NGK +N +S NYLGL ++ L + +A++ +GVG+ R G
Sbjct: 24 EIDTIEGANGPEIKINGKSYINLSSNNYLGLATNEDLKSAAKAAIDTHGVGAGAVRTING 83
Query: 152 TIDVHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLY 211
T+D+H + E ++KF GT +I Y G + +AI A K D I++DE H I +G
Sbjct: 84 TLDLHDELEETLAKFKGTEAAIAYQSGFNCNMAAISAVMNKNDAILSDELNHASIIDGCR 143
Query: 212 LSRSTVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLK 271
LS++ ++ H++MD LR + + K + + + ++ G +A L EI+++
Sbjct: 144 LSKAKIIRVNHSDMDDLRAKAKEAVESGQYNKVM---YITDGVFSMDGDVAKLPEIVEIA 200
Query: 272 EKYCFRILLDESNSFGVLGSSGRGLTEHYGV 302
E++ +D+++ GV+G G G +H+G+
Sbjct: 201 EEFGLLTYVDDAHGSGVMG-KGAGTVKHFGL 230
>BIOF_BACSU (P53556) 8-amino-7-oxononanoate synthase (EC 2.3.1.47)
(AONS) (8-amino-7-ketopelargonate synthase)
(7-keto-8-amino-pelargonic acid synthetase) (7-KAP
synthetase) (L-alanine--pimelyl CoA ligase)
Length = 389
Score = 122 bits (305), Expect = 2e-27
Identities = 68/207 (32%), Positives = 122/207 (58%), Gaps = 6/207 (2%)
Query: 96 LESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDV 155
++ A P ++G+ ++S NYLGL ++L+D+ +AL+++G GS G R G
Sbjct: 26 MDGAPVPERNIDGENQTVWSSNNYLGLASDRRLIDAAQTALQQFGTGSSGSRLTTGNSVW 85
Query: 156 HLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRS 215
H E +I+ F T ++L+S G + + +K D+I++D+ H + +G LS++
Sbjct: 86 HEKLEKKIASFKLTEAALLFSSGYLANVGVLSSLPEKEDVILSDQLNHASMIDGCRLSKA 145
Query: 216 TVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKYC 275
V ++H +M+ L L N T Y+ RR+IV + ++ G IAPLD+II L ++Y
Sbjct: 146 DTVVYRHIDMNDLENKL-NETQRYQ-----RRFIVTDGVFSMDGTIAPLDQIISLAKRYH 199
Query: 276 FRILLDESNSFGVLGSSGRGLTEHYGV 302
+++D++++ GVLG SG+G +E++GV
Sbjct: 200 AFVVVDDAHATGVLGDSGQGTSEYFGV 226
>LCB2_KLULA (P48241) Serine palmitoyltransferase 2 (EC 2.3.1.50)
(Long chain base biosynthesis protein 2) (SPT 2)
Length = 562
Score = 117 bits (293), Expect = 4e-26
Identities = 68/196 (34%), Positives = 113/196 (56%), Gaps = 4/196 (2%)
Query: 112 VNFASANYLGLIGHQ-KLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTP 170
+N +S NYLG + + + A +KYGV S GPR GT D+H+ E +++F+G
Sbjct: 157 LNLSSYNYLGFAQSEGQCTTAALEATDKYGVYSGGPRTRIGTTDLHVMTEKYVAQFVGKE 216
Query: 171 DSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRE 230
D+IL+S G T + +F ++++D H I+ G+ LS + V FKHN+M +L +
Sbjct: 217 DAILFSMGYGTNANFFNSFLDSKCLVISDSLNHTSIRTGVRLSGAAVKTFKHNDMRALEK 276
Query: 231 TL-ENITSIYKRTKNLRRYIVI--EALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFG 287
+ E I +T + I+I E LY G +A L ++++LK+KY + +DE++S G
Sbjct: 277 LIREQIVQGQSKTHRPWKKIIICVEGLYSMEGTMANLPKLVELKKKYKCYLFVDEAHSIG 336
Query: 288 VLGSSGRGLTEHYGVP 303
+G SGRG+ + +G+P
Sbjct: 337 AMGPSGRGVCDFFGIP 352
>LCB2_YEAST (P40970) Serine palmitoyltransferase 2 (EC 2.3.1.50)
(Long chain base biosynthesis protein 2) (SPT 2)
Length = 561
Score = 113 bits (282), Expect = 7e-25
Identities = 64/195 (32%), Positives = 112/195 (56%), Gaps = 4/195 (2%)
Query: 112 VNFASANYLGLIGHQ-KLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTP 170
+N +S NYLG + + D+ +++KY + S GPR GT D+H+ E +++F+G
Sbjct: 158 MNLSSYNYLGFAQSKGQCTDAALESVDKYSIQSGGPRAQIGTTDLHIKAEKLVARFIGKE 217
Query: 171 DSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRE 230
D++++S G T + AF K ++++DE H I+ G+ LS + V FKH +M L +
Sbjct: 218 DALVFSMGYGTNANLFNAFLDKKCLVISDELNHTSIRTGVRLSGAAVRTFKHGDMVGLEK 277
Query: 231 TL-ENITSIYKRTKNLRRYIVI--EALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFG 287
+ E I +T + I+I E L+ G + L ++++LK+KY + +DE++S G
Sbjct: 278 LIREQIVLGQPKTNRPWKKILICAEGLFSMEGTLCNLPKLVELKKKYKCYLFIDEAHSIG 337
Query: 288 VLGSSGRGLTEHYGV 302
+G +GRG+ E +GV
Sbjct: 338 AMGPTGRGVCEIFGV 352
>LCB2_SCHPO (Q09925) Serine palmitoyltransferase 2 (EC 2.3.1.50)
(Long chain base biosynthesis protein 2) (SPT 2)
Length = 603
Score = 113 bits (282), Expect = 7e-25
Identities = 60/196 (30%), Positives = 102/196 (51%), Gaps = 6/196 (3%)
Query: 112 VNFASANYLGLI-GHQKLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTP 170
+N +S NYLG H A++KYG+ +C GT +H + E + F+G P
Sbjct: 190 LNVSSYNYLGFAQSHGPCATKVEEAMQKYGLSTCSSNAICGTYGLHKEVEELTANFVGKP 249
Query: 171 DSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRE 230
++++S G ST + G +I++DE H I+ G LS + + +KHN+M L
Sbjct: 250 AALVFSQGFSTNATVFSTLMCPGSLIISDELNHTSIRFGARLSGANIRVYKHNDMTDLER 309
Query: 231 TLENITSIYKRTKNLRRY----IVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSF 286
L + S + + R Y +VIE LY G L ++++LK +Y F + +DE++S
Sbjct: 310 VLREVIS-QGQPRTHRPYSKILVVIEGLYSMEGNFCDLPKVVELKNRYKFYLFIDEAHSI 368
Query: 287 GVLGSSGRGLTEHYGV 302
G +G G G+ +++G+
Sbjct: 369 GAIGPRGGGICDYFGI 384
>BIOF_BACSH (P22806) 8-amino-7-oxononanoate synthase (EC 2.3.1.47)
(AONS) (8-amino-7-ketopelargonate synthase)
(7-keto-8-amino-pelargonic acid synthetase) (7-KAP
synthetase) (L-alanine--pimelyl CoA ligase)
Length = 389
Score = 110 bits (275), Expect = 5e-24
Identities = 61/198 (30%), Positives = 107/198 (53%), Gaps = 6/198 (3%)
Query: 105 IVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRIS 164
++NGK+ + F+S NYLGL +L + + KYG G+ G R G D+H E I+
Sbjct: 33 VMNGKKFLLFSSNNYLGLATDSRLKKKATEGISKYGTGAGGSRLTTGNFDIHEQLESEIA 92
Query: 165 KFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNN 224
F T +I++S G I + K GD I +D H I +G LS++ + ++H +
Sbjct: 93 DFKKTEAAIVFSSGYLANVGVISSVMKAGDTIFSDAWNHASIIDGCRLSKAKTIVYEHAD 152
Query: 225 MDSLRETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESN 284
M L L + + ++IV + ++ G IAPL +I++L ++Y I++D+++
Sbjct: 153 MVDLERKLR------QSHGDGLKFIVTDGVFSMDGDIAPLPKIVELAKEYKAYIMIDDAH 206
Query: 285 SFGVLGSSGRGLTEHYGV 302
+ GVLG+ G G +++G+
Sbjct: 207 ATGVLGNDGCGTADYFGL 224
>KBL_HUMAN (O75600) 2-amino-3-ketobutyrate coenzyme A ligase,
mitochondrial precursor (EC 2.3.1.29) (AKB ligase)
(Glycine acetyltransferase)
Length = 419
Score = 104 bits (259), Expect = 3e-22
Identities = 65/210 (30%), Positives = 104/210 (48%), Gaps = 8/210 (3%)
Query: 95 VLESAAGPHTIVNGKE--VVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGT 152
V+ S GPH V+G ++NF + NYLGL H +++ + ALE++G G R GT
Sbjct: 49 VITSRQGPHIRVDGVSGGILNFCANNYLGLSSHPEVIQAGLQALEEFGAGLSSVRFICGT 108
Query: 153 IDVHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYL 212
+H + E +I++F D+ILY A D +++DE H I +G+ L
Sbjct: 109 QSIHKNLEAKIARFHQREDAILYPSCYDANAGLFEALLTPEDAVLSDELNHASIIDGIRL 168
Query: 213 SRSTVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLKE 272
++ ++H +M L L+ + K+ R + + + G IAPL EI L
Sbjct: 169 CKAHKYRYRHLDMADLEAKLQ------EAQKHRLRLVATDGAFSMDGDIAPLQEICCLAS 222
Query: 273 KYCFRILLDESNSFGVLGSSGRGLTEHYGV 302
+Y + +DE ++ G LG +GRG E GV
Sbjct: 223 RYGALVFMDECHATGFLGPTGRGTDELLGV 252
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,607,748
Number of Sequences: 164201
Number of extensions: 1693757
Number of successful extensions: 4185
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 4016
Number of HSP's gapped (non-prelim): 77
length of query: 316
length of database: 59,974,054
effective HSP length: 110
effective length of query: 206
effective length of database: 41,911,944
effective search space: 8633860464
effective search space used: 8633860464
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC137824.11


