

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136953.6 + phase: 0
(268 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
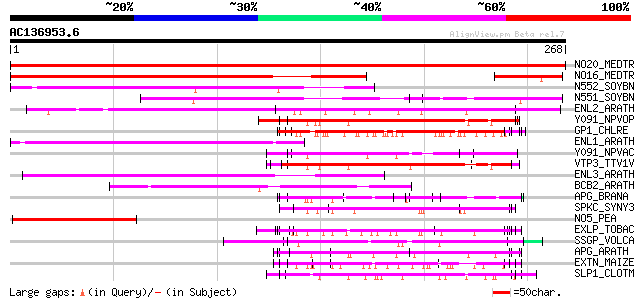
Score E
Sequences producing significant alignments: (bits) Value
NO20_MEDTR (P93329) Early nodulin 20 precursor (N-20) 553 e-157
NO16_MEDTR (P93328) Early nodulin 16 precursor (N-16) 211 2e-54
N552_SOYBN (Q02917) Early nodulin 55-2 precursor (N-55-2) (Nodul... 141 1e-33
N551_SOYBN (Q05544) Early nodulin 55-1 (N-55-1) (Fragment) 113 4e-25
ENL2_ARATH (Q9T076) Early nodulin-like protein 2 precursor (Phyt... 113 4e-25
Y091_NPVOP (O10341) Hypothetical 29.3 kDa protein (ORF92) 104 2e-22
GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor (H... 103 3e-22
ENL1_ARATH (Q9SK27) Early nodulin-like protein 1 precursor (Phyt... 96 9e-20
Y091_NPVAC (P41479) Hypothetical 24.1 kDa protein in LEF4-P33 in... 90 5e-18
VTP3_TTV1V (P19275) Viral protein TPX 90 7e-18
ENL3_ARATH (Q8LC95) Early nodulin-like protein 3 precursor (Phyt... 88 2e-17
BCB2_ARATH (O80517) Uclacyanin II precursor (Blue copper-binding... 88 2e-17
APG_BRANA (P40603) Anter-specific proline-rich protein APG (Prot... 87 3e-17
SPKC_SYNY3 (P74745) Serine/threonine-protein kinase C (EC 2.7.1.37) 85 2e-16
NO5_PEA (P25226) Early nodulin 5 precursor (N-5) 85 2e-16
EXLP_TOBAC (Q03211) Pistil-specific extensin-like protein precur... 84 5e-16
SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor ... 83 6e-16
APG_ARATH (P40602) Anter-specific proline-rich protein APG precu... 82 1e-15
EXTN_MAIZE (P14918) Extensin precursor (Proline-rich glycoprotein) 78 2e-14
SLP1_CLOTM (Q06852) Cell surface glycoprotein 1 precursor (Outer... 78 3e-14
>NO20_MEDTR (P93329) Early nodulin 20 precursor (N-20)
Length = 268
Score = 553 bits (1424), Expect = e-157
Identities = 268/268 (100%), Positives = 268/268 (100%)
Query: 1 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT 60
MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT
Sbjct: 1 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT 60
Query: 61 ITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLG 120
ITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLG
Sbjct: 61 ITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLG 120
Query: 121 LKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPR 180
LKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPR
Sbjct: 121 LKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPR 180
Query: 181 STPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPS 240
STPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPS
Sbjct: 181 STPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPS 240
Query: 241 PSSSGSKGGGAGHGFLEVSIAMMMFLIF 268
PSSSGSKGGGAGHGFLEVSIAMMMFLIF
Sbjct: 241 PSSSGSKGGGAGHGFLEVSIAMMMFLIF 268
>NO16_MEDTR (P93328) Early nodulin 16 precursor (N-16)
Length = 180
Score = 211 bits (536), Expect = 2e-54
Identities = 102/172 (59%), Positives = 125/172 (72%), Gaps = 18/172 (10%)
Query: 1 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT 60
M+SSSPILLM IFS+W+LIS+SESTDYL+GDS NSWK PLP+R A RWAS ++F VGDT
Sbjct: 1 MASSSPILLMIIFSMWLLISHSESTDYLIGDSHNSWKVPLPSRRAFARWASAHEFTVGDT 60
Query: 61 ITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLG 120
I F+Y+N+TESVHEV E DY C GEHV+H+DGNT VVL K G++HFISG KRHC++G
Sbjct: 61 ILFEYDNETESVHEVNEHDYIMCHTNGEHVEHHDGNTKVVLDKIGVYHFISGTKRHCKMG 120
Query: 121 LKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPS 172
LKLAVVV ++ + PP ++P PPSPSPSP+ S
Sbjct: 121 LKLAVVV------------------QNKHDLVLPPLITMPMPPSPSPSPNSS 154
Score = 49.3 bits (116), Expect = 1e-05
Identities = 25/36 (69%), Positives = 29/36 (80%), Gaps = 3/36 (8%)
Query: 235 PSPAPSPSSSGSKGGGAGHGF---LEVSIAMMMFLI 267
PSP+PSP+SSG+KGG AG GF L VS+ MMMFLI
Sbjct: 145 PSPSPSPNSSGNKGGAAGLGFIMWLGVSLVMMMFLI 180
>N552_SOYBN (Q02917) Early nodulin 55-2 precursor (N-55-2)
(Nodulin-315)
Length = 187
Score = 141 bits (356), Expect = 1e-33
Identities = 79/181 (43%), Positives = 108/181 (59%), Gaps = 26/181 (14%)
Query: 1 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT 60
+ ++SP L+M ++ +LIS SE+ Y+VG SE SWKFPL +L+ WA++++F +GDT
Sbjct: 5 LPNASPFLVML--AMCLLISTSEAEKYVVGGSEKSWKFPLSKPDSLSHWANSHRFKIGDT 62
Query: 61 ITFQYNNKTESVHEVEEEDYDRCGIRGE-HVDHYDGNTMVVLKKTGIHHFISGKKRHCRL 119
+ F+Y +TESVHE E DY+ C G+ H+ GNT V+L K G HFISG + HC++
Sbjct: 63 LIFKYEKRTESVHEGNETDYEGCNTVGKYHIVFNGGNTKVMLTKPGFRHFISGNQSHCQM 122
Query: 120 GLKLAVVVM----VAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSP 175
GLKLAV+V+ L SP P PSP PPS SPSPSP P
Sbjct: 123 GLKLAVLVISSNKTKKNLLSPSPSPSP-------------------PPSSLLSPSPSPLP 163
Query: 176 S 176
+
Sbjct: 164 N 164
Score = 31.2 bits (69), Expect = 2.8
Identities = 23/66 (34%), Positives = 31/66 (46%), Gaps = 19/66 (28%)
Query: 201 LSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGSKGGGAGHGFLEVSI 260
LS SPSPS PS S SPSP+P P++ G +G GF+ V +
Sbjct: 141 LSPSPSPSPPPS------------------SLLSPSPSPLPNNQGVT-SSSGAGFIGVMM 181
Query: 261 AMMMFL 266
+M+ L
Sbjct: 182 WLMLLL 187
>N551_SOYBN (Q05544) Early nodulin 55-1 (N-55-1) (Fragment)
Length = 137
Score = 113 bits (283), Expect = 4e-25
Identities = 67/137 (48%), Positives = 78/137 (56%), Gaps = 30/137 (21%)
Query: 64 QYNNKTESVHEVEEEDYDRCGIRG-EHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLGLK 122
+Y+ +TESVHEV E DY++C G EHV DGNT V+L K+G HFISG + HC++GLK
Sbjct: 1 KYDERTESVHEVNETDYEQCNTVGKEHVLFNDGNTKVMLTKSGFRHFISGNQSHCQMGLK 60
Query: 123 LAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRST 182
L VVVM PSP S PSPSPSPSPSPSPSPSP
Sbjct: 61 LMVVVMSNNTKKKLIHSPSP------------------SSPSPSPSPSPSPSPSPSP--- 99
Query: 183 PIPHPRKRSPASPSPSP 199
S +SPSPSP
Sbjct: 100 --------SLSSPSPSP 108
Score = 47.0 bits (110), Expect = 5e-05
Identities = 32/75 (42%), Positives = 45/75 (59%), Gaps = 5/75 (6%)
Query: 194 SPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSG-SKGGGAG 252
S + L SPSPS SPS +PSPS S +PS SPS SPSP+P P++ G ++ GA
Sbjct: 67 SNNTKKKLIHSPSPS-SPSPSPSPSPSP---SPSPSPSLSSPSPSPLPNNQGVTRSSGAE 122
Query: 253 HGFLEVSIAMMMFLI 267
+ + + +MM L+
Sbjct: 123 FIGVMMWLGVMMLLL 137
>ENL2_ARATH (Q9T076) Early nodulin-like protein 2 precursor
(Phytocyanin-like protein)
Length = 349
Score = 113 bits (283), Expect = 4e-25
Identities = 82/247 (33%), Positives = 121/247 (48%), Gaps = 19/247 (7%)
Query: 9 LMFIFSIWM----LISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDTITFQ 64
L F F+I + L + S + + VG S +W P + W+ +F+V DT+ F
Sbjct: 9 LSFFFTILLSLSTLFTISNARKFNVGGS-GAWVTNPPENYE--SWSGKNRFLVHDTLYFS 65
Query: 65 YNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLGLKLA 124
Y +SV EV + DYD C + DG++ + L + G +FISG + +C+ G KL
Sbjct: 66 YAKGADSVLEVNKADYDACNTKNPIKRVDDGDSEISLDRYGPFYFISGNEDNCKKGQKLN 125
Query: 125 VVVMVAPVLSSPPPP----PSPPTPRSSTPI--PHPPRRSLPSPPSPSPSPSPS-PSPSP 177
VVV+ A + S+ P P TP S TP H P+ S P P+ SP S + P +
Sbjct: 126 VVVISARIPSTAQSPHAAAPGSSTPGSMTPPGGAHSPKSSSPVSPTTSPPGSTTPPGGAH 185
Query: 178 SPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSP 237
SP+S+ P P S +P KS SP + P+P S + ++PSS+P P+P
Sbjct: 186 SPKSSSAVSPATSPPGSMAP-----KSGSPVSPTTSPPAPPKSTSPVSPSSAPMTSPPAP 240
Query: 238 APSPSSS 244
SSS
Sbjct: 241 MAPKSSS 247
Score = 63.9 bits (154), Expect = 4e-10
Identities = 50/145 (34%), Positives = 71/145 (48%), Gaps = 7/145 (4%)
Query: 129 VAPVLSSP-PPPPSPPTPRSSTPIPHPPRRSLPSPPSP-SPSPSPSPSPSPSPRSTPIPH 186
+AP SP P SPP P ST P + SPP+P +P S + PS +P ++P
Sbjct: 203 MAPKSGSPVSPTTSPPAPPKSTSPVSPSSAPMTSPPAPMAPKSSSTIPPSSAPMTSPPGS 262
Query: 187 --PRKRSPASPSP--SPSLSKSPSPSESPSLAPSPSD-SVASLAPSSSPSDESPSPAPSP 241
P+ SP S SP SPSL+ S S SPS +PS S + PS++ +P+ AP
Sbjct: 263 MAPKSSSPVSNSPTVSPSLAPGGSTSSSPSDSPSGSAMGPSGDGPSAAGDISTPAGAPGQ 322
Query: 242 SSSGSKGGGAGHGFLEVSIAMMMFL 266
S + G +S+ + +FL
Sbjct: 323 KKSSANGMTVMSITTVLSLVLTIFL 347
>Y091_NPVOP (O10341) Hypothetical 29.3 kDa protein (ORF92)
Length = 279
Score = 104 bits (260), Expect = 2e-22
Identities = 59/118 (50%), Positives = 74/118 (62%), Gaps = 9/118 (7%)
Query: 135 SPPPPPSP---PTPRSSTPIPHPPRRSLPSP---PSPSPSPSPSPSPSPSPRSTPIPHPR 188
SP P PSP PTP S TP P P PSP PSP+PSP+P+PSP+PSP TP P P
Sbjct: 78 SPTPTPSPTLSPTP-SPTPTPSPTPSPTPSPTPTPSPTPSPTPTPSPTPSPTPTPSPTPT 136
Query: 189 KRSPASPSPSPSLSKSPSPSESPSLAPSPSDS-VASLAPSSSPSDESPSPAPSPSSSG 245
SP+PSP+ + SP+PS +P+ +P+PS + S PS +P SP+P PSPS G
Sbjct: 137 PSPTPSPTPSPTPTPSPTPSPTPTPSPTPSPTPTPSPTPSPTP-PPSPTPPPSPSPLG 193
Score = 102 bits (254), Expect = 1e-21
Identities = 56/115 (48%), Positives = 74/115 (63%), Gaps = 7/115 (6%)
Query: 135 SPPPPPSP-PTPR-SSTPIPHPPRRSLPSP---PSPSPSPSPSPSPSPSPRSTPIPHPRK 189
+P P PSP PTP S TP P P PSP PSP+PSP+PSP+P+PSP +P P P
Sbjct: 64 TPTPTPSPTPTPALSPTPTPSPTLSPTPSPTPTPSPTPSPTPSPTPTPSPTPSPTPTPSP 123
Query: 190 RSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSS 244
+P+PSP+ + SP+PS +PS P+PS + S P+ SP+ SP+P PSP+ S
Sbjct: 124 TPSPTPTPSPTPTPSPTPSPTPSPTPTPSPT-PSPTPTPSPT-PSPTPTPSPTPS 176
Score = 99.0 bits (245), Expect = 1e-20
Identities = 55/122 (45%), Positives = 77/122 (63%), Gaps = 7/122 (5%)
Query: 131 PVLSSPPPPPSP---PTPRSS-TPIPHPPRRSLPSP-PSPSPSPSPSPSPSPSPRSTPIP 185
P L SP P P+P PTP S TP P P P+P PSP+P+P+ SP+P+PSP +P P
Sbjct: 32 PPLPSPTPTPTPSPTPTPTPSPTPTPTPTPTPTPTPTPSPTPTPALSPTPTPSPTLSPTP 91
Query: 186 HPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDS-VASLAPSSSPSDESPSPAPSPSSS 244
P +PSP+PS + +PSP+ SP+ PSP+ S + +P+ +PS +PSP PSP+ +
Sbjct: 92 SPTPTPSPTPSPTPSPTPTPSPTPSPTPTPSPTPSPTPTPSPTPTPS-PTPSPTPSPTPT 150
Query: 245 GS 246
S
Sbjct: 151 PS 152
Score = 98.6 bits (244), Expect = 1e-20
Identities = 52/116 (44%), Positives = 71/116 (60%), Gaps = 5/116 (4%)
Query: 135 SPPPPPSPPTPRSSTPIPHPPRRSLPSP---PSPSPSPSPSPSPSPSPRSTPIPHPRKRS 191
+P P P+P S TP P PSP P+PSP+P+PSP+PSP+P TP P P
Sbjct: 58 TPTPTPTPTPTPSPTPTPALSPTPTPSPTLSPTPSPTPTPSPTPSPTPSPTPTPSPTPSP 117
Query: 192 PASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPS-DESPSPAPSPSSSGS 246
+PSP+PS + +PSP+ +PS PSP+ S + PS +PS +PSP PSP+ + S
Sbjct: 118 TPTPSPTPSPTPTPSPTPTPSPTPSPTPS-PTPTPSPTPSPTPTPSPTPSPTPTPS 172
Score = 96.3 bits (238), Expect = 7e-20
Identities = 54/129 (41%), Positives = 79/129 (60%), Gaps = 7/129 (5%)
Query: 121 LKLAVVVMVAPVLSSPPPPPSP-PTPRSSTPIPHPPRRSLPSP-PSPSPSPSPSPSPSPS 178
L L ++ + + + PP PSP PTP S P P P P+P P+P+P+P+P+PSP+P+
Sbjct: 16 LTLWLLTSLKQNIETKPPLPSPTPTPTPS-PTPTPTPSPTPTPTPTPTPTPTPTPSPTPT 74
Query: 179 PRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDS---VASLAPSSSPSDESP 235
P +P P P +PSP+P+ S +PSP+ SP+ PSP+ S S PS +P+ SP
Sbjct: 75 PALSPTPTPSPTLSPTPSPTPTPSPTPSPTPSPTPTPSPTPSPTPTPSPTPSPTPT-PSP 133
Query: 236 SPAPSPSSS 244
+P PSP+ S
Sbjct: 134 TPTPSPTPS 142
Score = 90.9 bits (224), Expect = 3e-18
Identities = 51/115 (44%), Positives = 65/115 (56%), Gaps = 13/115 (11%)
Query: 135 SPPPPPSPPTPRSSTPIPHPPRRSLPSP---PSPSPSPSPSPSPSPSPRSTPIPHPRKRS 191
+P P PSP S TP P P PSP PSP+P+PSP+PSP+PSP TP P P
Sbjct: 100 TPSPTPSPTPTPSPTPSPTPTPSPTPSPTPTPSPTPTPSPTPSPTPSPTPTPSPTPSPTP 159
Query: 192 PASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGS 246
SP+PSP+ + SP+PS +P +P+P PS SP + P PSS G+
Sbjct: 160 TPSPTPSPTPTPSPTPSPTPPPSPTP-------PPSPSPLGD---PMYFPSSVGT 204
Score = 40.8 bits (94), Expect = 0.003
Identities = 21/58 (36%), Positives = 27/58 (46%), Gaps = 14/58 (24%)
Query: 130 APVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHP 187
+P + P P PTP + PSP+P PSP+P PSPSP P+ P
Sbjct: 156 SPTPTPSPTPSPTPTPSPT--------------PSPTPPPSPTPPPSPSPLGDPMYFP 199
>GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor
(Hydroxyproline-rich glycoprotein 1)
Length = 555
Score = 103 bits (258), Expect = 3e-22
Identities = 66/125 (52%), Positives = 73/125 (57%), Gaps = 14/125 (11%)
Query: 130 APVLSSPPPPPSPPTPRSSTPIPHPPRRSLP-SPPSPSPSPSP-----SPSPSPSPRSTP 183
+P +PPPPPSPP P P P P +P SPPSP PSP+P PSPSP P
Sbjct: 258 SPKPPAPPPPPSPPPPPPPRP-PFPANTPMPPSPPSPPPSPAPPTPPTPPSPSPPSPVPP 316
Query: 184 IPHPRKRSPASPSPSPSLSKS-----PSPSESPSLAPSPSDSVASLAPSSSPSD-ESPSP 237
P P SPA PSP+PS S PSPS SPS +PSPS S S +PS SPS SPSP
Sbjct: 317 SPAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSPSPSPSPSPS-PSPSPSPSPSPIPSPSP 375
Query: 238 APSPS 242
PSPS
Sbjct: 376 KPSPS 380
Score = 97.4 bits (241), Expect = 3e-20
Identities = 60/112 (53%), Positives = 69/112 (61%), Gaps = 7/112 (6%)
Query: 130 APVLSSPPPP-PSPPTPRSSTPIPHPPRRSLPSPPSPS-PSPSPSPSPSPSPRSTPIPHP 187
+P SP PP P+PP+P + P P PP + PSPPSP+ PSPSP PSPSP S P P P
Sbjct: 90 SPAPPSPAPPSPAPPSP--APPSPAPPSPAPPSPPSPAPPSPSPPAPPSPSPPS-PAP-P 145
Query: 188 RKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAP 239
SPA PSPSP + SPSP PS AP PS + S +P PS PSPAP
Sbjct: 146 LPPSPAPPSPSPPVPPSPSPPVPPSPAP-PSPTPPSPSPPVPPSPAPPSPAP 196
Score = 96.7 bits (239), Expect = 5e-20
Identities = 53/121 (43%), Positives = 65/121 (52%), Gaps = 3/121 (2%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P +PP PP+PP+P +P+P P PSP PSP+PSP PSP+P P +P P P
Sbjct: 294 PPSPAPPTPPTPPSPSPPSPVPPSPAPVPPSPAPPSPAPSPPPSPAP-PTPSPSPSPSPS 352
Query: 191 SPASPSPSPSLSKSPSPSESPSLAPSPSDSVASL--APSSSPSDESPSPAPSPSSSGSKG 248
SPSPSPS S SPSP SPS PSPS L A + D+ + P S+
Sbjct: 353 PSPSPSPSPSPSPSPSPIPSPSPKPSPSPVAVKLVWADDAIAFDDLNGTSTRPGSASRMV 412
Query: 249 G 249
G
Sbjct: 413 G 413
Score = 95.5 bits (236), Expect = 1e-19
Identities = 56/115 (48%), Positives = 66/115 (56%), Gaps = 6/115 (5%)
Query: 130 APVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPR- 188
AP SPP PPSP P + P+P P PSPP P PSPSP PSP+P S P P
Sbjct: 125 APPSPSPPAPPSPSPPSPAPPLPPSPAPPSPSPPVP-PSPSPPVPPSPAPPSPTPPSPSP 183
Query: 189 --KRSPASPSPSPSLSKSPS-PSESPSLAPSPS-DSVASLAPSSSPSDESPSPAP 239
SPA PSP+P + SP+ PS +P + PSP+ S S AP S PS PSP+P
Sbjct: 184 PVPPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSPPSPAPPSPSP 238
Score = 95.5 bits (236), Expect = 1e-19
Identities = 53/119 (44%), Positives = 63/119 (52%), Gaps = 7/119 (5%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIP----- 185
P +PP PPSP P + P P PP + PSP PSP+P PSP+P S P P
Sbjct: 70 PPSPAPPSPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPPSPAPPSP 129
Query: 186 -HPRKRSPASPSPSPSLSKSPS-PSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPS 242
P SP+ PSP+P L SP+ PS SP + PSPS V SP+ SPSP PS
Sbjct: 130 SPPAPPSPSPPSPAPPLPPSPAPPSPSPPVPPSPSPPVPPSPAPPSPTPPSPSPPVPPS 188
Score = 94.7 bits (234), Expect = 2e-19
Identities = 60/125 (48%), Positives = 66/125 (52%), Gaps = 17/125 (13%)
Query: 130 APVLSSPPPP--PSPPTPRSSTPIPHPPRRSLPSPPSPSP-SPSPSPSPSPSPRSTPIPH 186
AP +PP P PSP P + P P PP + PSPPSP+P SPSP PSPSP S P
Sbjct: 87 APPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPL 146
Query: 187 PRKRSPASPSPSPSLSKSPSPSESPSLAPS------------PSDSVASLAPSSSPSDES 234
P SPA PSPSP + SPSP PS AP PS + S AP PS
Sbjct: 147 PP--SPAPPSPSPPVPPSPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAP 204
Query: 235 PSPAP 239
PSPAP
Sbjct: 205 PSPAP 209
Score = 94.7 bits (234), Expect = 2e-19
Identities = 57/118 (48%), Positives = 65/118 (54%), Gaps = 7/118 (5%)
Query: 130 APVLSSPPPP--PSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHP 187
AP +PP P PSP P P P PP P+PPSP+P PSP+P PSP+P S P P
Sbjct: 49 APPSPAPPSPAPPSPAPPSPGPPSPAPPSPPSPAPPSPAP-PSPAP-PSPAPPSPAPPSP 106
Query: 188 RKRSPASPSPSPSLSKSPS-PSESPSLAPSPS--DSVASLAPSSSPSDESPSPAPSPS 242
SPA PSP+P SP+ PS SP PSPS L PS +P SP PSPS
Sbjct: 107 APPSPAPPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSPAPPSPSPPVPPSPS 164
Score = 94.4 bits (233), Expect = 3e-19
Identities = 56/125 (44%), Positives = 68/125 (53%), Gaps = 9/125 (7%)
Query: 130 APVLSSPPPP-PSPPTPR--SSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTP--- 183
+PV SP PP P+PP+P+ + P P PP P PP P+ +P P PSP P P
Sbjct: 243 SPVPPSPAPPSPAPPSPKPPAPPPPPSPPPPPPPRPPFPANTPMPPSPPSPPPSPAPPTP 302
Query: 184 --IPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSP 241
P P SP PSP+P PS +PS PSP+ S +PS SPS SPSP+PSP
Sbjct: 303 PTPPSPSPPSPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSPSPS-PSPSPSPSP 361
Query: 242 SSSGS 246
S S S
Sbjct: 362 SPSPS 366
Score = 94.4 bits (233), Expect = 3e-19
Identities = 56/127 (44%), Positives = 67/127 (52%), Gaps = 14/127 (11%)
Query: 130 APVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSP---RSTPIPH 186
AP SPP PPSP P + P P PP P PP+P P PSP P P P P +TP+P
Sbjct: 232 APPSPSPPAPPSPVPPSPAPPSPAPPS---PKPPAPPPPPSPPPPPPPRPPFPANTPMPP 288
Query: 187 PRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS-------SPSDESPSPAP 239
P SP+P P+ PSPS + PSP+ S AP S SP+ +PSP+P
Sbjct: 289 SPPSPPPSPAP-PTPPTPPSPSPPSPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTPSPSP 347
Query: 240 SPSSSGS 246
SPS S S
Sbjct: 348 SPSPSPS 354
Score = 93.6 bits (231), Expect = 5e-19
Identities = 56/124 (45%), Positives = 68/124 (54%), Gaps = 7/124 (5%)
Query: 130 APVLSSPPPPPSPPTPRSSTPIP---HPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPH 186
+P +PP PPSP P S P+P PP P+PPSP+P PSPSP PSP
Sbjct: 138 SPPSPAPPLPPSPAPPSPSPPVPPSPSPPVPPSPAPPSPTP-PSPSPPVPPSPAPPSPAP 196
Query: 187 PRKRSPASPSPSPSLSKSPSPSESPSLAP--SPSDSVASLAPSSSPSDESPSPA-PSPSS 243
P SPA PSP+P + SP+P PS AP PS + S +P + PS PSPA PSP+
Sbjct: 197 PVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSPPSPAPPSPSPPAPPSPVPPSPAPPSPAP 256
Query: 244 SGSK 247
K
Sbjct: 257 PSPK 260
Score = 92.4 bits (228), Expect = 1e-18
Identities = 57/122 (46%), Positives = 67/122 (54%), Gaps = 8/122 (6%)
Query: 130 APVLSSPPPPPSPPTPRSSTPI---PHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPH 186
AP +PP PPSP P S P P PP + P PPSP+P PSPSP PSP P
Sbjct: 112 APPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSPAP-PSPSPPVPPSPSPPVPPS 170
Query: 187 PRKRSPASPSPSPSLSKSPS-PSESPSLAPSPSDSVASLAPSSSPSDESPS-PAPSPSSS 244
P SP PSPSP + SP+ PS +P + PSP+ S AP PS PS P+P+P S
Sbjct: 171 PAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAP--PSPAPPVPPSPAPPSPPSPAPPSP 228
Query: 245 GS 246
S
Sbjct: 229 PS 230
Score = 92.4 bits (228), Expect = 1e-18
Identities = 48/105 (45%), Positives = 60/105 (56%), Gaps = 2/105 (1%)
Query: 137 PPPPSPPTPRSSTPIPHPPRRSLPSPPSPSP-SPSPSPSPSPSPRSTPIPHPRKRSPASP 195
PP P+PP+P +P P P P+PPSP P SP+P PSP+P S P P SPA P
Sbjct: 40 PPSPAPPSPAPPSPAPPSPAPPSPAPPSPGPPSPAPPSPPSPAPPSPAPPSPAPPSPAPP 99
Query: 196 SPSPSLSKSPSPS-ESPSLAPSPSDSVASLAPSSSPSDESPSPAP 239
SP+P PSP+ SP+ PS + S +P + PS PSPAP
Sbjct: 100 SPAPPSPAPPSPAPPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAP 144
Score = 92.4 bits (228), Expect = 1e-18
Identities = 61/145 (42%), Positives = 71/145 (48%), Gaps = 29/145 (20%)
Query: 130 APVLSSPPPPPSPPTPRSSTPI--------PHPPRRSLPSPPSPSPSPSPSPSPSPSPR- 180
+P +PP PPSP P S P P PP + PSP P+P P PSP P P PR
Sbjct: 219 SPPSPAPPSPPSPAPPSPSPPAPPSPVPPSPAPPSPAPPSPKPPAPPPPPSPPPPPPPRP 278
Query: 181 ----STPIP-----------HPRKRSPASPSPSPSLSKSPSP----SESPSLAPSPSDSV 221
+TP+P P +P SPSP + SP+P PS APSP S
Sbjct: 279 PFPANTPMPPSPPSPPPSPAPPTPPTPPSPSPPSPVPPSPAPVPPSPAPPSPAPSPPPSP 338
Query: 222 ASLAPSSSPSDESPSPAPSPSSSGS 246
A PS SPS SPSP+PSPS S S
Sbjct: 339 APPTPSPSPS-PSPSPSPSPSPSPS 362
Score = 91.3 bits (225), Expect = 2e-18
Identities = 46/110 (41%), Positives = 58/110 (51%), Gaps = 1/110 (0%)
Query: 134 SSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSP-SPSPRSTPIPHPRKRSP 192
S PP P+PP+P +P P P P PPSP+P PSP+P SP+P S P P SP
Sbjct: 42 SPAPPSPAPPSPAPPSPAPPSPAPPSPGPPSPAPPSPPSPAPPSPAPPSPAPPSPAPPSP 101
Query: 193 ASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPS 242
A PSP+P PSP+ +P+P PS SP +P PSP+
Sbjct: 102 APPSPAPPSPAPPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSPA 151
Score = 91.3 bits (225), Expect = 2e-18
Identities = 56/119 (47%), Positives = 63/119 (52%), Gaps = 10/119 (8%)
Query: 131 PVLSSPPPP-PSPPTPRSSTPIPHPPRRSLPSPPSPS-PSPSPSPSPSPSPRSTPIPHPR 188
PV SP PP P+PP P S P P PP + PSPPSP+ PSPSP PSP P S P P
Sbjct: 197 PVPPSPAPPSPAPPVPPSPAP-PSPPSPAPPSPPSPAPPSPSPPAPPSPVPPSPAPPSPA 255
Query: 189 KRSPASPSPSPSLSKSPSPSESP------SLAPSPSDSVASLAPSSSPSDESPSPAPSP 241
SP P+P P S P P P + PSP S AP + P+ SPSP PSP
Sbjct: 256 PPSPKPPAPPPPPSPPPPPPPRPPFPANTPMPPSPPSPPPSPAPPTPPTPPSPSP-PSP 313
Score = 90.9 bits (224), Expect = 3e-18
Identities = 54/113 (47%), Positives = 61/113 (53%), Gaps = 4/113 (3%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPS-PSPSPRSTPIP-HPR 188
P SPP PPSP P + P P PP P+PPSP+P PSP+ PSP+P P P P
Sbjct: 160 PPSPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPS 219
Query: 189 KRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSP 241
SPA PSP SPSP PS P PS + S AP SP +P P PSP
Sbjct: 220 PPSPAPPSPPSPAPPSPSPPAPPSPVP-PSPAPPSPAP-PSPKPPAPPPPPSP 270
Score = 88.2 bits (217), Expect = 2e-17
Identities = 56/115 (48%), Positives = 64/115 (54%), Gaps = 8/115 (6%)
Query: 131 PVLSSPPPP-PSPPTPRSSTP-IPHPPRRSLPSPPSPS-PSPSPSPSPSPSPRSTPIP-H 186
PV SP PP P+PP+P P P PP + P PPSP+ PSP+P PSP+P S P P
Sbjct: 166 PVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAP 225
Query: 187 PRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSP 241
P SPA PSPSP SP P PS AP PS + S P + P SP P P P
Sbjct: 226 PSPPSPAPPSPSPPAPPSPVP---PSPAP-PSPAPPSPKPPAPPPPPSPPPPPPP 276
Score = 88.2 bits (217), Expect = 2e-17
Identities = 55/120 (45%), Positives = 67/120 (55%), Gaps = 8/120 (6%)
Query: 130 APVLSSPP--PPPSPPTPRS-STPIPHPPRRSLPSPPSPSP-SPSPSPSPSPSPRS--TP 183
AP SPP P PSPP P S + P P PP S P PPSP+P SP+P PSP+P S P
Sbjct: 151 APPSPSPPVPPSPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAPPSPAPP 210
Query: 184 IPHPRKRSPASPSPSPSLSKSPS-PSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPS 242
+P P P+ PSP+P SP+ PS SP PSP + + PS + P+P P PS
Sbjct: 211 VP-PSPAPPSPPSPAPPSPPSPAPPSPSPPAPPSPVPPSPAPPSPAPPSPKPPAPPPPPS 269
>ENL1_ARATH (Q9SK27) Early nodulin-like protein 1 precursor
(Phytocyanin-like protein)
Length = 176
Score = 95.9 bits (237), Expect = 9e-20
Identities = 55/142 (38%), Positives = 79/142 (54%), Gaps = 3/142 (2%)
Query: 1 MSSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDT 60
M+SSS L + IFS+ L S + + + VG WK P + ++ T WA +F VGD
Sbjct: 1 MASSS--LHVAIFSLIFLFSLAAANEVTVGGKSGDWKIPPSSSYSFTEWAQKARFKVGDF 58
Query: 61 ITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLG 120
I F+Y + +SV EV +E Y+ C ++ DG T V L ++G +FISG HC G
Sbjct: 59 IVFRYESGKDSVLEVTKEAYNSCNTTNPLANYTDGETKVKLDRSGPFYFISGANGHCEKG 118
Query: 121 LKLAVVVMVAPVLSSPPPPPSP 142
KL++VV ++P S P PSP
Sbjct: 119 QKLSLVV-ISPRHSVISPAPSP 139
>Y091_NPVAC (P41479) Hypothetical 24.1 kDa protein in LEF4-P33
intergenic region
Length = 224
Score = 90.1 bits (222), Expect = 5e-18
Identities = 50/110 (45%), Positives = 60/110 (54%), Gaps = 11/110 (10%)
Query: 137 PPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSP-SPSPSPSPRSTPIPHPRKRSPASP 195
PPP SPP P+P PP PSPPSP+P P+P P+P+P+P TPIP P P
Sbjct: 41 PPPTSPPI----VPLPTPPPTPTPSPPSPTPPPTPIPPTPTPTPPPTPIP-PTPTPTPPP 95
Query: 196 SPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSG 245
SP P P+P+ SP +P P S PS P +PSP PSPS G
Sbjct: 96 SPIP-----PTPTPSPPPSPIPPTPTPSPPPSPIPPTPTPSPPPSPSPLG 140
Score = 78.2 bits (191), Expect = 2e-14
Identities = 45/108 (41%), Positives = 58/108 (53%), Gaps = 8/108 (7%)
Query: 125 VVVMVAPVLSSPPPPPSP---PTPRSSTPIPHPPRRSLPSPPSPSPSPSP-SPSPSPSPR 180
+V + P + P PPSP PTP TP P PP +P P+P+P PSP P+P+PSP
Sbjct: 48 IVPLPTPPPTPTPSPPSPTPPPTPIPPTPTPTPPPTPIPPTPTPTPPPSPIPPTPTPSPP 107
Query: 181 STPIP----HPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASL 224
+PIP SP P+P+PS SPSP P PS D+ +L
Sbjct: 108 PSPIPPTPTPSPPPSPIPPTPTPSPPPSPSPLGEPMYYPSNIDTNEAL 155
Score = 78.2 bits (191), Expect = 2e-14
Identities = 41/84 (48%), Positives = 50/84 (58%), Gaps = 7/84 (8%)
Query: 135 SPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSP-SPSPSPSPRSTPIPHPRKRSPA 193
SPP P PPTP TP P PP +P P+P+P PSP P+P+PSP +PIP SP
Sbjct: 61 SPPSPTPPPTPIPPTPTPTPPPTPIPPTPTPTPPPSPIPPTPTPSPPPSPIPPTPTPSPP 120
Query: 194 SPSPSPSLSKSPSPSESPSLAPSP 217
PSP P P+P+ SP +PSP
Sbjct: 121 -PSPIP-----PTPTPSPPPSPSP 138
>VTP3_TTV1V (P19275) Viral protein TPX
Length = 474
Score = 89.7 bits (221), Expect = 7e-18
Identities = 44/125 (35%), Positives = 75/125 (59%), Gaps = 15/125 (12%)
Query: 127 VMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSP-PSPSPSPSPSPSPSPSPRSTPIP 185
V V P+ SSP P P+P + TP P P P+P P+P+P+P+P+P+P+P+P TP P
Sbjct: 268 VTVTPI-SSPSPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTP 326
Query: 186 HPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPS--------DESPSP 237
P +P+P+P+ + +P+P+ +P+ P+P+ + P+ +P+ D +PSP
Sbjct: 327 TPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPT-----PTPTPTYDITYVVFDVTPSP 381
Query: 238 APSPS 242
P+P+
Sbjct: 382 TPTPT 386
Score = 81.3 bits (199), Expect = 2e-15
Identities = 41/115 (35%), Positives = 70/115 (60%), Gaps = 10/115 (8%)
Query: 132 VLSSPPPPPSPPTPRSS---TPIPHPPRRSLPSP-PSPSPSPSPSPSPSPSPRSTPIPHP 187
VL P P TP SS TP P P P+P P+P+P+P+P+P+P+P+P TP P P
Sbjct: 259 VLPYEPDPQVTVTPISSPSPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTP 318
Query: 188 RKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPS 242
+P+P+P+ + +P+P+ +P+ P+P+ + P+ +P+ +P+P P+P+
Sbjct: 319 TPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPT-----PTPTPT-PTPTPTPTPT 367
Score = 78.6 bits (192), Expect = 2e-14
Identities = 39/119 (32%), Positives = 71/119 (58%), Gaps = 7/119 (5%)
Query: 135 SPPPPPSP-PTPRSSTPIPHPPRRSLPSP---PSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
+P P P+P PTP + I + PSP P+P+P+P+P+P+P+P+P TP P P
Sbjct: 353 TPTPTPTPTPTPTPTYDITYVVFDVTPSPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPT 412
Query: 191 SPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPS---DESPSPAPSPSSSGS 246
+P+P+P+ + +P+P+ +P+ + + + PS +P+ +P+P P+P+S+ S
Sbjct: 413 PTPTPTPTPTPTPTPTPTPTPTPTYDITYVIFDVTPSPTPTPTPTPTPTPTPTPTSTTS 471
Score = 72.4 bits (176), Expect = 1e-12
Identities = 36/107 (33%), Positives = 60/107 (55%), Gaps = 12/107 (11%)
Query: 125 VVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSP-PSPSPSPSPSPSPSPSPRSTP 183
+ +V V SP P P+P + TP P P P+P P+P+P+P+P+P+P+P+P TP
Sbjct: 370 ITYVVFDVTPSPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTPTP 429
Query: 184 IPHPRKRSP-------ASPSPSPSLSKSPSPSESPSLAPSPSDSVAS 223
P P +PSP+P+ P+P+ +P+ P+P+ + +S
Sbjct: 430 TPTPTPTYDITYVIFDVTPSPTPT----PTPTPTPTPTPTPTSTTSS 472
>ENL3_ARATH (Q8LC95) Early nodulin-like protein 3 precursor
(Phytocyanin-like protein)
Length = 186
Score = 88.2 bits (217), Expect = 2e-17
Identities = 52/175 (29%), Positives = 89/175 (50%), Gaps = 19/175 (10%)
Query: 7 ILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDTITFQYN 66
++ F ++L + S + +VG +SWK P +L +WA + +F VGDT+ ++Y+
Sbjct: 6 LVATFFLIFFLLTNLVCSKEIIVGGKTSSWKIPSSPSESLNKWAESLRFRVGDTLVWKYD 65
Query: 67 NKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHHFISGKKRHCRLGLKLAVV 126
+ +SV +V ++ Y C ++ +G+T V L+++G + FISG K +C G KL +V
Sbjct: 66 EEKDSVLQVTKDAYINCNTTNPAANYSNGDTKVKLERSGPYFFISGSKSNCVEGEKLHIV 125
Query: 127 VMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRS 181
VM S+ H S PSP+PSP+ +P+ +P S
Sbjct: 126 VM-------------------SSRGGHTGGFFTGSSPSPAPSPALLGAPTVAPAS 161
>BCB2_ARATH (O80517) Uclacyanin II precursor (Blue copper-binding
protein II) (BCB II) (Phytocyanin 2)
Length = 202
Score = 88.2 bits (217), Expect = 2e-17
Identities = 57/148 (38%), Positives = 78/148 (52%), Gaps = 13/148 (8%)
Query: 49 WASNYQFIVGDTITFQYNNKTESVHEVEEEDYDRCGIRGEHVDHYDGNTMVVLKKTGIHH 108
WA+ F VGD + F+Y + + +V V++ YD C +H DG+T + LK GI++
Sbjct: 45 WATGKTFRVGDILEFKYGS-SHTVDVVDKAGYDGCDASSSTENHSDGDTKIDLKTVGINY 103
Query: 109 FISGKKRHCRL--GLKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPS 166
FI HCR G+KLAV V++ PP+ PTP SSTP S PS SP+
Sbjct: 104 FICSTPGHCRTNGGMKLAV-----NVVAGSAGPPATPTPPSSTPGTPTTPESPPSGGSPT 158
Query: 167 PSPSPSPSPSPSPRSTPIPHPRKRSPAS 194
P+ +P+P ST P P K S AS
Sbjct: 159 PT-----TPTPGAGSTSPPPPPKASGAS 181
Score = 38.9 bits (89), Expect = 0.013
Identities = 22/71 (30%), Positives = 36/71 (49%), Gaps = 6/71 (8%)
Query: 192 PASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGSKGGGA 251
PA+P+P S +P+ ESP SP+ + + +P S SP P P +SG+ G
Sbjct: 132 PATPTPPSSTPGTPTTPESPPSGGSPTPT------TPTPGAGSTSPPPPPKASGASKGVM 185
Query: 252 GHGFLEVSIAM 262
+ + VS+ +
Sbjct: 186 SYVLVGVSMVL 196
>APG_BRANA (P40603) Anter-specific proline-rich protein APG (Protein
CEX) (Fragment)
Length = 449
Score = 87.4 bits (215), Expect = 3e-17
Identities = 51/125 (40%), Positives = 59/125 (46%), Gaps = 12/125 (9%)
Query: 131 PVLSSPPPPPS-----PPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIP 185
P P PPP PP P S P PP+ PP+P+PSP P P P P+ P P
Sbjct: 1 PPKPQPKPPPKPQPKPPPAPTPSPCPPQPPKPQPKPPPAPTPSPCPPQPPKPQPKPPPAP 60
Query: 186 HPRKR---SPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPS 242
P + SP+ P P PS + P P SPS PSP APS PS P PAPSP
Sbjct: 61 GPSPKPGPSPSPPKPPPSPAPKPVPPPSPSPKPSPPKPP---APSPKPSPPKP-PAPSPP 116
Query: 243 SSGSK 247
+K
Sbjct: 117 KPQNK 121
Score = 67.4 bits (163), Expect = 3e-11
Identities = 37/79 (46%), Positives = 42/79 (52%), Gaps = 7/79 (8%)
Query: 130 APVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRK 189
+P PP P P P P P P PSPP P PSP+P P P PSP +P P P K
Sbjct: 43 SPCPPQPPKPQPKPPP---APGPSPKPGPSPSPPKPPPSPAPKPVPPPSP--SPKPSPPK 97
Query: 190 RSPASPSPSPSLSKSPSPS 208
P +PSP PS K P+PS
Sbjct: 98 --PPAPSPKPSPPKPPAPS 114
Score = 61.6 bits (148), Expect = 2e-09
Identities = 31/75 (41%), Positives = 38/75 (50%), Gaps = 2/75 (2%)
Query: 131 PVLSSPPPPPSP-PTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRK 189
P P PPP+P P+P+ P P PP+ P P P PSPSP PSP P P P
Sbjct: 49 PPKPQPKPPPAPGPSPKPG-PSPSPPKPPPSPAPKPVPPPSPSPKPSPPKPPAPSPKPSP 107
Query: 190 RSPASPSPSPSLSKS 204
P +PSP +K+
Sbjct: 108 PKPPAPSPPKPQNKT 122
Score = 56.6 bits (135), Expect = 6e-08
Identities = 30/87 (34%), Positives = 33/87 (37%), Gaps = 20/87 (22%)
Query: 162 PPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSV 221
PP P P P P P P P P TP P P P P P P +P+PS P P P
Sbjct: 1 PPKPQPKPPPKPQPKPPPAPTPSPCP----PQPPKPQPKPPPAPTPSPCPPQPPKPQ--- 53
Query: 222 ASLAPSSSPSDESPSPAPSPSSSGSKG 248
P P P+P S G
Sbjct: 54 -------------PKPPPAPGPSPKPG 67
Score = 53.9 bits (128), Expect = 4e-07
Identities = 26/65 (40%), Positives = 35/65 (53%), Gaps = 8/65 (12%)
Query: 130 APVLSSPPPPPSPPTPRSSTPIPHP---PRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPH 186
+P P PP PP + P+P P P+ S P PP+PSP PSP P+PSP P
Sbjct: 63 SPKPGPSPSPPKPPPSPAPKPVPPPSPSPKPSPPKPPAPSPKPSPPKPPAPSP-----PK 117
Query: 187 PRKRS 191
P+ ++
Sbjct: 118 PQNKT 122
Score = 53.1 bits (126), Expect = 7e-07
Identities = 27/59 (45%), Positives = 31/59 (51%), Gaps = 4/59 (6%)
Query: 135 SPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPA 193
SPP PP P P+ P+P P PSPP P P+PSP PSP P +P K PA
Sbjct: 71 SPPKPPPSPAPK---PVPPPSPSPKPSPPKP-PAPSPKPSPPKPPAPSPPKPQNKTIPA 125
Score = 49.7 bits (117), Expect = 8e-06
Identities = 28/59 (47%), Positives = 33/59 (55%), Gaps = 6/59 (10%)
Query: 131 PVLSSPPPPPSPPT----PRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIP 185
P S P PPPSP P S +P P PP+ PS P PSP P+PSP P P++ IP
Sbjct: 68 PSPSPPKPPPSPAPKPVPPPSPSPKPSPPKPPAPS-PKPSPPKPPAPSP-PKPQNKTIP 124
Score = 37.0 bits (84), Expect = 0.050
Identities = 18/44 (40%), Positives = 22/44 (49%), Gaps = 2/44 (4%)
Query: 129 VAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPS 172
V P SP P PP P + +P P PP+ PSPP P P+
Sbjct: 84 VPPPSPSPKPS--PPKPPAPSPKPSPPKPPAPSPPKPQNKTIPA 125
>SPKC_SYNY3 (P74745) Serine/threonine-protein kinase C (EC 2.7.1.37)
Length = 535
Score = 85.1 bits (209), Expect = 2e-16
Identities = 47/116 (40%), Positives = 62/116 (52%), Gaps = 11/116 (9%)
Query: 138 PPPSPPTPRSSTPIPHP---PRRSLPSPPSP-----SPSPSPSPSPSPSPRSTPIPHPRK 189
PPP+ P TPIP P PR +L PSP +PSP P+PSPSPSP +T P
Sbjct: 373 PPPAVEEPTEETPIPLPSLEPRPNLFETPSPIPTPATPSPEPTPSPSPSPETTSSPTEDT 432
Query: 190 RSPASPSPS---PSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPS 242
+P P PS P+ P PS SP+++P PS +++ + SPSP P P+
Sbjct: 433 ITPMEPEPSLDEPAPIPEPKPSPSPTISPQPSPTISIPVTPAPVPKPSPSPTPKPT 488
Score = 77.4 bits (189), Expect = 3e-14
Identities = 44/117 (37%), Positives = 62/117 (52%), Gaps = 6/117 (5%)
Query: 131 PVLSSPPPPPSP-PTPRS-STPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPR 188
P SP P PSP P+P + S+P P P P+P P P PSPSP +P P P
Sbjct: 407 PATPSPEPTPSPSPSPETTSSPTEDTITPMEPEPSLDEPAPIPEPKPSPSPTISPQPSPT 466
Query: 189 KRSPASPS--PSPSLSKSPSPSESPSLAPS--PSDSVASLAPSSSPSDESPSPAPSP 241
P +P+ P PS S +P P+ P ++P+ PS++V + P +PS E+ P+P
Sbjct: 467 ISIPVTPAPVPKPSPSPTPKPTVPPQISPTPQPSNTVPVIPPPENPSAETEPNLPAP 523
Score = 59.3 bits (142), Expect = 9e-09
Identities = 34/90 (37%), Positives = 43/90 (47%), Gaps = 8/90 (8%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHP-PRRSLPSPPSP--SPSPSPSPSPSPSPRSTPIPHP 187
P L P P P P S T P P P S+P P+P PSPSP+P P+ P+ +P P P
Sbjct: 440 PSLDEPAPIPEPKPSPSPTISPQPSPTISIPVTPAPVPKPSPSPTPKPTVPPQISPTPQP 499
Query: 188 RKRSPASPSPSPSLSKSPSPSESPSLAPSP 217
P P P ++PS P+L P
Sbjct: 500 SNTVPVIPPP-----ENPSAETEPNLPAPP 524
Score = 50.4 bits (119), Expect = 4e-06
Identities = 32/94 (34%), Positives = 44/94 (46%), Gaps = 14/94 (14%)
Query: 155 PRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSP---SPSLSKSPSPSESP 211
P ++ P +P P PS P P+ TP SP P +PS +PSPS SP
Sbjct: 373 PPPAVEEPTEETPIPLPSLEPRPNLFETP----------SPIPTPATPSPEPTPSPSPSP 422
Query: 212 SLAPSPS-DSVASLAPSSSPSDESPSPAPSPSSS 244
SP+ D++ + P S + +P P P PS S
Sbjct: 423 ETTSSPTEDTITPMEPEPSLDEPAPIPEPKPSPS 456
Score = 42.0 bits (97), Expect = 0.002
Identities = 24/67 (35%), Positives = 36/67 (52%), Gaps = 9/67 (13%)
Query: 180 RSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAP 239
R+ P P + + +P P PSL P+ E+PS P+P+ PS P+ PSP+P
Sbjct: 370 RNNPPPAVEEPTEETPIPLPSLEPRPNLFETPSPIPTPA------TPSPEPT---PSPSP 420
Query: 240 SPSSSGS 246
SP ++ S
Sbjct: 421 SPETTSS 427
Score = 41.2 bits (95), Expect = 0.003
Identities = 23/75 (30%), Positives = 31/75 (40%), Gaps = 1/75 (1%)
Query: 135 SPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPAS 194
SP P P+ P + P+P P P P P P SP+P PS + P P
Sbjct: 460 SPQPSPTISIPVTPAPVPKPSPSPTPKPTVP-PQISPTPQPSNTVPVIPPPENPSAETEP 518
Query: 195 PSPSPSLSKSPSPSE 209
P+P + + P E
Sbjct: 519 NLPAPPVGEKPIDPE 533
Score = 31.2 bits (69), Expect = 2.8
Identities = 23/70 (32%), Positives = 32/70 (44%), Gaps = 14/70 (20%)
Query: 123 LAVVVMVAPVLS-SPPPPPSP-------PTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPS 174
+++ V APV SP P P P PTP+ S +P +P P +PS P+
Sbjct: 467 ISIPVTPAPVPKPSPSPTPKPTVPPQISPTPQPSNTVP-----VIPPPENPSAETEPN-L 520
Query: 175 PSPSPRSTPI 184
P+P PI
Sbjct: 521 PAPPVGEKPI 530
>NO5_PEA (P25226) Early nodulin 5 precursor (N-5)
Length = 135
Score = 85.1 bits (209), Expect = 2e-16
Identities = 39/60 (65%), Positives = 46/60 (76%)
Query: 2 SSSSPILLMFIFSIWMLISYSESTDYLVGDSENSWKFPLPTRHALTRWASNYQFIVGDTI 61
SSSSPI LM IFS+W+L SYSEST+YLV DSENSWK P+R AL RW + +Q + DTI
Sbjct: 3 SSSSPIFLMIIFSMWLLFSYSESTEYLVRDSENSWKVNFPSRDALNRWVTRHQLTIHDTI 62
Score = 39.3 bits (90), Expect = 0.010
Identities = 22/38 (57%), Positives = 27/38 (70%), Gaps = 6/38 (15%)
Query: 234 SPSPAPSPSSSGSKGGGAGHGF---LEVSIAMMMFLIF 268
SPSPAP+PS SG+ AGHGF L S+ M+MFLI+
Sbjct: 100 SPSPAPAPSLSGA---AAGHGFIVWLGASLPMLMFLIW 134
>EXLP_TOBAC (Q03211) Pistil-specific extensin-like protein precursor
(PELP)
Length = 426
Score = 83.6 bits (205), Expect = 5e-16
Identities = 49/121 (40%), Positives = 63/121 (51%), Gaps = 9/121 (7%)
Query: 130 APVLSSPPPPPSP----PTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIP 185
+P++ PPPPPSP P +S+ P PP PSPP P P P +PSPSP+ + P P
Sbjct: 143 SPLVKPPPPPPSPCKPSPPDQSAKQPPQPPPAKQPSPP-PPPPPVKAPSPSPAKQPPPPP 201
Query: 186 HPRKRSPASPSPSPSLSKSPSP---SESPSLAPSPSDSVAS-LAPSSSPSDESPSPAPSP 241
P K SP+ P + P P +SP L P P + + PS SP+ E P AP P
Sbjct: 202 PPVKAPSPSPATQPPTKQPPPPPRAKKSPLLPPPPPVAYPPVMTPSPSPAAEPPIIAPFP 261
Query: 242 S 242
S
Sbjct: 262 S 262
Score = 68.2 bits (165), Expect = 2e-11
Identities = 45/121 (37%), Positives = 53/121 (43%), Gaps = 21/121 (17%)
Query: 129 VAPVLSSPP---PPPSPPTPRSSTPIPHPPRRSLPSPPSPSPS--PSPSPSPSPSPRSTP 183
V P+ PP P PS P+P P P PP PSPP S P P P+ PSP P
Sbjct: 125 VPPIGGGPPVNQPKPSSPSPLVKPP-PPPPSPCKPSPPDQSAKQPPQPPPAKQPSPPPPP 183
Query: 184 IPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSS 243
P +PSPS +K P P P APSPS P++ P + P P P
Sbjct: 184 --------PPVKAPSPSPAKQPPPPPPPVKAPSPS-------PATQPPTKQPPPPPRAKK 228
Query: 244 S 244
S
Sbjct: 229 S 229
Score = 66.6 bits (161), Expect = 6e-11
Identities = 43/129 (33%), Positives = 56/129 (43%), Gaps = 7/129 (5%)
Query: 120 GLKLAVVVMVAPVLSS-PPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPS 178
G KL + P++ + P PP P + P P P L PP P PSP P S
Sbjct: 106 GSKLPDFAGLLPLIPNLPDVPPIGGGPPVNQPKPSSPS-PLVKPPPPPPSPCKPSPPDQS 164
Query: 179 PRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPA 238
+ P P P K+ P P P + SPSP++ P P P APS SP+ + P+
Sbjct: 165 AKQPPQPPPAKQPSPPPPPPPVKAPSPSPAKQPPPPPPPVK-----APSPSPATQPPTKQ 219
Query: 239 PSPSSSGSK 247
P P K
Sbjct: 220 PPPPPRAKK 228
Score = 65.9 bits (159), Expect = 1e-10
Identities = 41/113 (36%), Positives = 53/113 (46%), Gaps = 16/113 (14%)
Query: 136 PPPPPSPPTPRSSTPIPHP---PRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSP 192
PPP P P P+ P P + P PP P +PSPSP+ P + P P K+SP
Sbjct: 171 PPPAKQPSPPPPPPPVKAPSPSPAKQPPPPPPPVKAPSPSPATQPPTKQPPPPPRAKKSP 230
Query: 193 ASPSP-----SPSLSKSPSP-SESPSLAPSPSDSVASLAPSSSPSDESPSPAP 239
P P P ++ SPSP +E P +AP PS P ++P PAP
Sbjct: 231 LLPPPPPVAYPPVMTPSPSPAAEPPIIAPFPS-------PPANPPLIPRRPAP 276
Score = 64.3 bits (155), Expect = 3e-10
Identities = 51/142 (35%), Positives = 55/142 (37%), Gaps = 42/142 (29%)
Query: 138 PPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRST-------------PI 184
P P P+P PP LPSPP PSPSP P PSPSP P ST P+
Sbjct: 46 PLPDIPSPFDGPTFVLPPPSPLPSPPPPSPSP-PPPSPSPPPPSTIPLIPPFTGGFLPPL 104
Query: 185 P-------------------------HPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSD 219
P P P SPSP L K P P SP PSP D
Sbjct: 105 PGSKLPDFAGLLPLIPNLPDVPPIGGGPPVNQPKPSSPSP-LVKPPPPPPSP-CKPSPPD 162
Query: 220 SVASLAPSSSPSDESPSPAPSP 241
A P P+ + PSP P P
Sbjct: 163 QSAKQPPQPPPA-KQPSPPPPP 183
Score = 62.8 bits (151), Expect = 9e-10
Identities = 49/150 (32%), Positives = 58/150 (38%), Gaps = 41/150 (27%)
Query: 130 APVLSSPPPPPSPPTPRSSTP---------------IPHPPRRSLPS------------- 161
+P+ S PPP PSPP P S P +P P LP
Sbjct: 65 SPLPSPPPPSPSPPPPSPSPPPPSTIPLIPPFTGGFLPPLPGSKLPDFAGLLPLIPNLPD 124
Query: 162 -PPSPSPSPSPSPSPS-PSPRSTPIPHPRKRSPASPS---------PSPSLSKSPSPSES 210
PP P P PS PSP P P P SP PS P P +K PSP
Sbjct: 125 VPPIGGGPPVNQPKPSSPSPLVKPPPPP--PSPCKPSPPDQSAKQPPQPPPAKQPSPPPP 182
Query: 211 PSLAPSPSDSVASLAPSSSPSDESPSPAPS 240
P +PS S A P P ++PSP+P+
Sbjct: 183 PPPVKAPSPSPAKQPPPPPPPVKAPSPSPA 212
Score = 53.9 bits (128), Expect = 4e-07
Identities = 30/78 (38%), Positives = 39/78 (49%), Gaps = 6/78 (7%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSP---SPSPSPRSTPIPHP 187
P PPP PP + S +P PP + P +PSPSP+ P +P PSP + P P
Sbjct: 215 PPTKQPPP---PPRAKKSPLLPPPPPVAYPPVMTPSPSPAAEPPIIAPFPSPPANPPLIP 271
Query: 188 RKRSPASPSPSPSLSKSP 205
R+ +P P P L K P
Sbjct: 272 RRPAPPVVKPLPPLGKPP 289
Score = 33.5 bits (75), Expect = 0.56
Identities = 19/53 (35%), Positives = 26/53 (48%), Gaps = 4/53 (7%)
Query: 192 PASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSS 244
P + P P + PSP + P+ P + S P SPS PSP+P P S+
Sbjct: 41 PPAEIPLPDI---PSPFDGPTFVLPPPSPLPS-PPPPSPSPPPPSPSPPPPST 89
>SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor
(SSG 185)
Length = 485
Score = 83.2 bits (204), Expect = 6e-16
Identities = 54/144 (37%), Positives = 63/144 (43%), Gaps = 8/144 (5%)
Query: 104 TGIHHFISGKKRHCRLGLKLAVVVMVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPP 163
T + + K C GL V + P ++ P PPSP SS P PP PSPP
Sbjct: 195 TASYSVFNSDKDCCPTGLSGPNVNPIGPAPNNSPLPPSPQPTASSRPPSPPPSPRPPSPP 254
Query: 164 SPSPSPSPSPSPSPSPRSTPIPH-------PRKRSPASPSPSPSLSKSPSPSESPSLAPS 216
PSPSP P P P P P P P P P P PSPS + P PS SP + P
Sbjct: 255 PPSPSPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPSPSPPRKP-PSPSPPVPPP 313
Query: 217 PSDSVASLAPSSSPSDESPSPAPS 240
PS A + P E S +PS
Sbjct: 314 PSPPSVLPAATGFPFCECVSRSPS 337
Score = 76.6 bits (187), Expect = 6e-14
Identities = 51/119 (42%), Positives = 58/119 (47%), Gaps = 8/119 (6%)
Query: 135 SPPP---PPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPR-KR 190
SPPP PPSPP P S P P PP P PP PSP P P P P P P P P P R
Sbjct: 243 SPPPSPRPPSPPPPSPSPPPPPPPPPPPPPPPPPSPPPPPPP-PPPPPPPPPPPSPSPPR 301
Query: 191 SPASPSPSPSLSKSPS-PSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGSKG 248
P PSPSP + PS PS P+ P S +PSS P + + + + SG G
Sbjct: 302 KP--PSPSPPVPPPPSPPSVLPAATGFPFCECVSRSPSSYPWRVTVANVSAVTISGGAG 358
Score = 67.4 bits (163), Expect = 3e-11
Identities = 44/125 (35%), Positives = 47/125 (37%), Gaps = 6/125 (4%)
Query: 133 LSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSP 192
LS P P P P +S P PP P P + S PSP PSP P P P P P
Sbjct: 212 LSGPNVNPIGPAPNNS---PLPPS---PQPTASSRPPSPPPSPRPPSPPPPSPSPPPPPP 265
Query: 193 ASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGSKGGGAG 252
P P P SP P P P P S +P P SP P PS G
Sbjct: 266 PPPPPPPPPPPSPPPPPPPPPPPPPPPPPPSPSPPRKPPSPSPPVPPPPSPPSVLPAATG 325
Query: 253 HGFLE 257
F E
Sbjct: 326 FPFCE 330
>APG_ARATH (P40602) Anter-specific proline-rich protein APG
precursor
Length = 534
Score = 82.4 bits (202), Expect = 1e-15
Identities = 41/117 (35%), Positives = 51/117 (43%), Gaps = 16/117 (13%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P SP P PSPP P+P P P P P P+P P P P+P P P+P P
Sbjct: 100 PPAPSPSPCPSPPPKPQPKPVPPPACPPTPPKPQPKPAPPPEPKPAPPPAPKPVPCPSPP 159
Query: 191 SPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGSK 247
P +P+P P P P +P+ P+P SP PAPSP +K
Sbjct: 160 KPPAPTPKPVPPHGPPPKPAPAPTPAP----------------SPKPAPSPPKPENK 200
Score = 82.0 bits (201), Expect = 1e-15
Identities = 47/132 (35%), Positives = 64/132 (47%), Gaps = 15/132 (11%)
Query: 130 APVLSSPPPPPSPPT-PRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIP--- 185
+P +PP P S P P +P P PP + P PP P+PSPSP PSP P P+ P+P
Sbjct: 66 SPKPVAPPGPSSKPVAPPGPSPCPSPPPKPQPKPP-PAPSPSPCPSPPPKPQPKPVPPPA 124
Query: 186 --------HPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDS-VASLAPSSSPSDE-SP 235
P+ P P P+P + P P SP P+P+ V P P+ +P
Sbjct: 125 CPPTPPKPQPKPAPPPEPKPAPPPAPKPVPCPSPPKPPAPTPKPVPPHGPPPKPAPAPTP 184
Query: 236 SPAPSPSSSGSK 247
+P+P P+ S K
Sbjct: 185 APSPKPAPSPPK 196
Score = 77.8 bits (190), Expect = 3e-14
Identities = 46/131 (35%), Positives = 57/131 (43%), Gaps = 13/131 (9%)
Query: 128 MVAPVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPP-SPSPSPSPSPSPSPSPRSTPIPH 186
M P P P +PP P SS P+ P PSPP P P P P+PSPSP P P P
Sbjct: 58 MNPPTPDPSPKPVAPPGP-SSKPVAPPGPSPCPSPPPKPQPKPPPAPSPSPCPSPPPKPQ 116
Query: 187 PRKRSPAS-----------PSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESP 235
P+ P + P+P P +P P+ P PSP A P P
Sbjct: 117 PKPVPPPACPPTPPKPQPKPAPPPEPKPAPPPAPKPVPCPSPPKPPAPTPKPVPPHGPPP 176
Query: 236 SPAPSPSSSGS 246
PAP+P+ + S
Sbjct: 177 KPAPAPTPAPS 187
Score = 71.2 bits (173), Expect = 2e-12
Identities = 41/111 (36%), Positives = 46/111 (40%), Gaps = 4/111 (3%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P+ P P P P P + P P P P PS P P PSP PSP P P P
Sbjct: 46 PLWPRPYPQPWPMNPPTPDPSPKPVA---PPGPSSKPVAPPGPSPCPSPPPKPQPKPPPA 102
Query: 191 SPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSP 241
SP PSP P P P+ P+P AP P +P PAP P
Sbjct: 103 PSPSPCPSPPPKPQPKPVPPPACPPTPPKPQPKPAPPPEPK-PAPPPAPKP 152
Score = 58.5 bits (140), Expect = 2e-08
Identities = 36/91 (39%), Positives = 40/91 (43%), Gaps = 10/91 (10%)
Query: 156 RRSLPSPPSPSPSPSPSPS----PSPSPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESP 211
RR P P P P P P P P PSP+ P P + A P PSP S P P P
Sbjct: 40 RRLWPWPLWPRPYPQPWPMNPPTPDPSPKPVAPPGPSSKPVAPPGPSPCPSPPPKPQPKP 99
Query: 212 SLAPSPSDSVASLAPSSSPSDESPSPAPSPS 242
APSPS PS P + P P P P+
Sbjct: 100 PPAPSPSP-----CPSPPPKPQ-PKPVPPPA 124
Score = 55.5 bits (132), Expect = 1e-07
Identities = 25/65 (38%), Positives = 33/65 (50%), Gaps = 7/65 (10%)
Query: 136 PPPPPSPPTPRSSTPIPHPPRRSLPSP-------PSPSPSPSPSPSPSPSPRSTPIPHPR 188
P P P+PP P P PP+ P+P P P P+P+P+P+PSP P +P
Sbjct: 140 PEPKPAPPPAPKPVPCPSPPKPPAPTPKPVPPHGPPPKPAPAPTPAPSPKPAPSPPKPEN 199
Query: 189 KRSPA 193
K PA
Sbjct: 200 KTIPA 204
Score = 31.2 bits (69), Expect = 2.8
Identities = 18/58 (31%), Positives = 21/58 (36%), Gaps = 14/58 (24%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPR--------------RSLPSPPSPSPSPSPSPS 174
P P P PSPP P + TP P PP P+P P P P+
Sbjct: 147 PPAPKPVPCPSPPKPPAPTPKPVPPHGPPPKPAPAPTPAPSPKPAPSPPKPENKTIPA 204
>EXTN_MAIZE (P14918) Extensin precursor (Proline-rich glycoprotein)
Length = 267
Score = 78.2 bits (191), Expect = 2e-14
Identities = 40/108 (37%), Positives = 58/108 (53%), Gaps = 10/108 (9%)
Query: 135 SPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPAS 194
SP PP PTP + TP P PP P+P P+ +PSP P P+P+ TP + P +
Sbjct: 98 SPKPPTPKPTPPTYTPSPKPPATKPPTPKPTPPTYTPSPKP-PTPKPTPPTYTPSPKPPT 156
Query: 195 PSPS-PSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSP 241
P P+ P+ + SP P P+ P+P P+ +PS + P+P P+P
Sbjct: 157 PKPTPPTYTPSPKPPTHPTPKPTP--------PTYTPSPKPPTPKPTP 196
Score = 76.6 bits (187), Expect = 6e-14
Identities = 46/131 (35%), Positives = 66/131 (50%), Gaps = 18/131 (13%)
Query: 128 MVAPVLSSPPPPPSP-PTPRSSTPIPHPPRRS----LPSPPSPSPSPSPSPSPSPSPRS- 181
+ P + P PP+P PTP + TP P PP P+PP+ +PSP P P+P P+P +
Sbjct: 16 LTPPTYTPSPKPPTPKPTPPTYTPSPKPPASKPPTPKPTPPTYTPSPKP-PTPKPTPPTY 74
Query: 182 TPIPHPRKRSPASPSPSPSL----SKSPSPSES-PSLAPSPSDSVAS------LAPSSSP 230
TP P P P +P P+P K P+P + P+ PSP P+ +P
Sbjct: 75 TPSPKPPATKPPTPKPTPPTYTPSPKPPTPKPTPPTYTPSPKPPATKPPTPKPTPPTYTP 134
Query: 231 SDESPSPAPSP 241
S + P+P P+P
Sbjct: 135 SPKPPTPKPTP 145
Score = 76.3 bits (186), Expect = 7e-14
Identities = 41/109 (37%), Positives = 58/109 (52%), Gaps = 10/109 (9%)
Query: 135 SPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPAS 194
SP PP PTP + TP P PP P+P P+ +PSP P P+P+ TP + +
Sbjct: 61 SPKPPTPKPTPPTYTPSPKPPATKPPTPKPTPPTYTPSPKP-PTPKPTPPTY-------T 112
Query: 195 PSPSPSLSKSPSPSES-PSLAPSPSDSVAS-LAPSSSPSDESPSPAPSP 241
PSP P +K P+P + P+ PSP P+ +PS + P+P P+P
Sbjct: 113 PSPKPPATKPPTPKPTPPTYTPSPKPPTPKPTPPTYTPSPKPPTPKPTP 161
Score = 75.5 bits (184), Expect = 1e-13
Identities = 44/118 (37%), Positives = 63/118 (53%), Gaps = 9/118 (7%)
Query: 131 PVLSSPPPPPSPPT----PRSSTPIPHPPRRS-LPSPPSPSPSPSPSPSPSPSPRSTPIP 185
P P P P+PPT P+ TP P PP + P PP+P P+P P+ +PSP P + P P
Sbjct: 118 PATKPPTPKPTPPTYTPSPKPPTPKPTPPTYTPSPKPPTPKPTP-PTYTPSPKPPTHPTP 176
Query: 186 HPRKRSPASPSPSPSLSKSPSPSESPS-LAPSPSDSVASLAPS-SSPSDESPSPAPSP 241
P + +PSP P K P+ +PS P+P + + PS P+ + P+P P+P
Sbjct: 177 KPTPPT-YTPSPKPPTPKPTPPTYTPSPKPPTPKPTPPTYTPSPKPPATKPPTPKPTP 233
Score = 75.1 bits (183), Expect = 2e-13
Identities = 42/114 (36%), Positives = 56/114 (48%), Gaps = 9/114 (7%)
Query: 135 SPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPS-PSPSPSPRS-TPIPHPRKRSP 192
SP PP PTP + TP P PP P P P+ +PSP P+P P+P + TP P P P
Sbjct: 151 SPKPPTPKPTPPTYTPSPKPPTHPTPKPTPPTYTPSPKPPTPKPTPPTYTPSPKPPTPKP 210
Query: 193 A----SPSPSPSLSKSPSPSESP---SLAPSPSDSVASLAPSSSPSDESPSPAP 239
+PSP P +K P+P +P + P P + + P +PSP P
Sbjct: 211 TPPTYTPSPKPPATKPPTPKPTPPTYTPTPKPPATKPPTYTPTPPVSHTPSPPP 264
Score = 70.5 bits (171), Expect = 4e-12
Identities = 43/119 (36%), Positives = 60/119 (50%), Gaps = 10/119 (8%)
Query: 131 PVLSSPPPPPSPPTP-----RSSTPIPHPPRRS-LPSPPSPSPSPSPSPSPSPSPRSTPI 184
P P PP P+P + TP P PP + P PP+P P+P P+ +PSP P +T
Sbjct: 64 PPTPKPTPPTYTPSPKPPATKPPTPKPTPPTYTPSPKPPTPKPTP-PTYTPSPKPPATKP 122
Query: 185 PHPRKRSPA-SPSPSPSLSKSPSPSESPS-LAPSPSDSVASLAPSSSPSDESPSPAPSP 241
P P+ P +PSP P K P+ +PS P+P + + PS P P+P P+P
Sbjct: 123 PTPKPTPPTYTPSPKPPTPKPTPPTYTPSPKPPTPKPTPPTYTPSPKPPTH-PTPKPTP 180
Score = 70.5 bits (171), Expect = 4e-12
Identities = 41/115 (35%), Positives = 58/115 (49%), Gaps = 8/115 (6%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P P PP P+P+ TP P PP + PSP P P+P P+P P TP P P
Sbjct: 138 PPTPKPTPPTYTPSPKPPTPKPTPPTYT----PSPKPPTHPTPKPTP-PTYTPSPKPPTP 192
Query: 191 SPASPSPSPSLSKSPSPSES-PSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSS 244
P P+ +PS K P+P + P+ PSP A+ P+ P+ + +P P P ++
Sbjct: 193 KPTPPTYTPS-PKPPTPKPTPPTYTPSPKPP-ATKPPTPKPTPPTYTPTPKPPAT 245
Score = 69.7 bits (169), Expect = 7e-12
Identities = 43/116 (37%), Positives = 65/116 (55%), Gaps = 19/116 (16%)
Query: 135 SPP---PPPSPPTPRSSTPIPHPPRRS-LPSPPSPSPSP---SPSPSPSPSPRSTPIPH- 186
+PP P P PP + TP P PP + P PP+P P+P +PSP P P+P+ TP +
Sbjct: 107 TPPTYTPSPKPPATKPPTPKPTPPTYTPSPKPPTPKPTPPTYTPSPKP-PTPKPTPPTYT 165
Query: 187 PRKRSPASPSPSPS-LSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSP 241
P + P P+P P+ + +PSP + P+ P+P P+ +PS + P+P P+P
Sbjct: 166 PSPKPPTHPTPKPTPPTYTPSP-KPPTPKPTP--------PTYTPSPKPPTPKPTP 212
Score = 59.3 bits (142), Expect = 9e-09
Identities = 36/111 (32%), Positives = 53/111 (47%), Gaps = 14/111 (12%)
Query: 147 SSTPIPHPPRRSLPSPPSPSPSP---SPSPSP----SPSPRSTPIPHPRKRSPASPSPSP 199
S TP + P P PP+P P+P +PSP P P+P+ TP + P +P P+P
Sbjct: 15 SLTPPTYTPS---PKPPTPKPTPPTYTPSPKPPASKPPTPKPTPPTYTPSPKPPTPKPTP 71
Query: 200 SLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSP---APSPSSSGSK 247
+ +PSP + P+P + + PS P P+P PSP +K
Sbjct: 72 P-TYTPSPKPPATKPPTPKPTPPTYTPSPKPPTPKPTPPTYTPSPKPPATK 121
Score = 58.9 bits (141), Expect = 1e-08
Identities = 33/80 (41%), Positives = 42/80 (52%), Gaps = 5/80 (6%)
Query: 131 PVLSSPPPPPSPPT--PRSSTPIPHP-PRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHP 187
P P P P+PPT P P P P P PSP P+ P P+P P+P P TP P P
Sbjct: 185 PSPKPPTPKPTPPTYTPSPKPPTPKPTPPTYTPSPKPPATKP-PTPKPTP-PTYTPTPKP 242
Query: 188 RKRSPASPSPSPSLSKSPSP 207
P + +P+P +S +PSP
Sbjct: 243 PATKPPTYTPTPPVSHTPSP 262
Score = 56.2 bits (134), Expect = 8e-08
Identities = 34/98 (34%), Positives = 50/98 (50%), Gaps = 13/98 (13%)
Query: 156 RRSLPSPPSPSPSPSPSPSPSPSPRS-TPIPHPRKRSPASPSPSPSL----SKSPSPSES 210
RR +PP+ +PSP P P+P P+P + TP P P P +P P+P K P+P +
Sbjct: 12 RRYSLTPPTYTPSPKP-PTPKPTPPTYTPSPKPPASKPPTPKPTPPTYTPSPKPPTPKPT 70
Query: 211 -PSLAPSPSDSVAS------LAPSSSPSDESPSPAPSP 241
P+ PSP P+ +PS + P+P P+P
Sbjct: 71 PPTYTPSPKPPATKPPTPKPTPPTYTPSPKPPTPKPTP 108
Score = 40.8 bits (94), Expect = 0.003
Identities = 21/49 (42%), Positives = 27/49 (54%), Gaps = 5/49 (10%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSP 179
P + PP P PTP + TP P PP PP+ +P+P S +PSP P
Sbjct: 221 PPATKPPTP--KPTPPTYTPTPKPP---ATKPPTYTPTPPVSHTPSPPP 264
Score = 32.3 bits (72), Expect = 1.2
Identities = 15/34 (44%), Positives = 17/34 (49%), Gaps = 3/34 (8%)
Query: 135 SPP---PPPSPPTPRSSTPIPHPPRRSLPSPPSP 165
+PP P P PP + T P PP PSPP P
Sbjct: 232 TPPTYTPTPKPPATKPPTYTPTPPVSHTPSPPPP 265
>SLP1_CLOTM (Q06852) Cell surface glycoprotein 1 precursor (Outer
layer protein B) (S-layer protein 1)
Length = 1664
Score = 77.8 bits (190), Expect = 3e-14
Identities = 41/124 (33%), Positives = 57/124 (45%), Gaps = 4/124 (3%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S P P +P P P+ +PS P+PS TP P
Sbjct: 1291 PTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 1349
Query: 191 SPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESPSPAPSPSSSGSKGGG 250
+PS P+ S P+PS+ P+ + +P + + P+ +P S +P SG GGG
Sbjct: 1350 DEPTPSDEPTPSDEPTPSDEPTPSETPEEPTPTTTPTPTP---STTPTSGSGGSGGSGGG 1406
Query: 251 AGHG 254
G G
Sbjct: 1407 GGGG 1410
Score = 77.4 bits (189), Expect = 3e-14
Identities = 50/139 (35%), Positives = 66/139 (46%), Gaps = 21/139 (15%)
Query: 134 SSPPPPPSPPTPRSS-TPIPHPPRRSLPSP---PSPSPSPSPSPSPS-PSPRSTPIPHPR 188
S P P PTP TP P P+P P+PS P+PS +P P P TP P
Sbjct: 1276 SDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPT 1335
Query: 189 KRSPASPSPSPSLSKSPSPSE--SPSLAPSPSDSVASLAPSSSPSDESPSPAPSPS---- 242
+PS P+ S P+PS+ +PS P+PSD PS +P + +P+ P+P+
Sbjct: 1336 PSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPT---PSETPEEPTPTTTPTPTPSTT 1392
Query: 243 -------SSGSKGGGAGHG 254
S GS GGG G G
Sbjct: 1393 PTSGSGGSGGSGGGGGGGG 1411
Score = 73.9 bits (180), Expect = 4e-13
Identities = 41/115 (35%), Positives = 58/115 (49%), Gaps = 4/115 (3%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S P P +P P P+ +PS P+PS TP P
Sbjct: 911 PTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 969
Query: 191 SPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS--SPSDE-SPSPAPSPS 242
+PS P+ S P+PS+ P+ + +P + + + PS +PSDE +PS P+PS
Sbjct: 970 DEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 1024
Score = 73.9 bits (180), Expect = 4e-13
Identities = 41/115 (35%), Positives = 58/115 (49%), Gaps = 4/115 (3%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S P P +P P P+ +PS P+PS TP P
Sbjct: 966 PTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 1024
Query: 191 SPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS--SPSDE-SPSPAPSPS 242
+PS P+ S P+PS+ P+ + +P + + + PS +PSDE +PS P+PS
Sbjct: 1025 DEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 1079
Score = 73.9 bits (180), Expect = 4e-13
Identities = 41/115 (35%), Positives = 58/115 (49%), Gaps = 4/115 (3%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S P P +P P P+ +PS P+PS TP P
Sbjct: 1236 PTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 1294
Query: 191 SPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS--SPSDE-SPSPAPSPS 242
+PS P+ S P+PS+ P+ + +P + + + PS +PSDE +PS P+PS
Sbjct: 1295 DEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 1349
Score = 73.2 bits (178), Expect = 6e-13
Identities = 44/118 (37%), Positives = 62/118 (52%), Gaps = 6/118 (5%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S TP P + P+PS P+PS P+PS TP P
Sbjct: 978 PTPSDEPTPSDEPTP-SETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPS 1036
Query: 191 SPASPSPSPSLSKSPS---PSESPSLAPSPSDSVASLAPSSSPSDE-SPSPAPSPSSS 244
+PS P+ S++P P+++PS P+PSD + + +PSDE +PS P+PS +
Sbjct: 1037 DEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE-PTPSDEPTPSDEPTPSDEPTPSET 1093
Score = 73.2 bits (178), Expect = 6e-13
Identities = 45/120 (37%), Positives = 62/120 (51%), Gaps = 8/120 (6%)
Query: 131 PVLSSPPPPPSPPTPRSS-TPIPHPPRRSLPSP---PSPSPSPS-PSPSPSPSPRSTPIP 185
P S P P PTP TP P P+P P+PS +P P P+ +PS TP
Sbjct: 954 PTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSD 1013
Query: 186 HPRKRSPASPSPSPSLSKSPSPSE--SPSLAPSPSDSVASLAPSSSPSDE-SPSPAPSPS 242
P +PS P+ S P+PS+ +PS P+PS++ P+ +PSDE +PS P+PS
Sbjct: 1014 EPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPS 1073
Score = 72.0 bits (175), Expect = 1e-12
Identities = 44/126 (34%), Positives = 65/126 (50%), Gaps = 8/126 (6%)
Query: 125 VVVMVAPVLSSPPPPPSPPTPRSS-TPIPHPPRRSLPSP---PSPSPSPSPSPSPS-PSP 179
VV+ AP+ ++ P TP TP P P+P P+PS P+PS +P P P
Sbjct: 758 VVIQPAPIKAASDEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIP 817
Query: 180 RSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS--SPSDE-SPS 236
TP P +PS P+ S P+PS+ P+ + +P + + + PS +PSDE +PS
Sbjct: 818 TDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPS 877
Query: 237 PAPSPS 242
P+PS
Sbjct: 878 DEPTPS 883
Score = 72.0 bits (175), Expect = 1e-12
Identities = 43/117 (36%), Positives = 60/117 (50%), Gaps = 8/117 (6%)
Query: 134 SSPPPPPSPPTPRSS-TPIPHPPRRSLPSP---PSPSPSPSPSPSPS-PSPRSTPIPHPR 188
S P P PTP TP P P+P P+PS P+PS +P P P TP P
Sbjct: 1006 SDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPT 1065
Query: 189 KRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS--SPSDE-SPSPAPSPS 242
+PS P+ S P+PS+ P+ + +P + + + PS +PSDE +PS P+PS
Sbjct: 1066 PSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 1122
Score = 71.2 bits (173), Expect = 2e-12
Identities = 45/133 (33%), Positives = 63/133 (46%), Gaps = 21/133 (15%)
Query: 131 PVLSSPPPPPSPPTPRSS--------TPIPHPPRRSLPSP---PSPSPSPSPSPSPSPS- 178
P S P P PTP + TP P P+P P+PS P+PS P+PS
Sbjct: 1119 PTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSE 1178
Query: 179 ------PRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS--SP 230
P TP P +PS P+ S P+PS+ P+ + +P + + + PS +P
Sbjct: 1179 TPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTP 1238
Query: 231 SDE-SPSPAPSPS 242
SDE +PS P+PS
Sbjct: 1239 SDEPTPSDEPTPS 1251
Score = 71.2 bits (173), Expect = 2e-12
Identities = 45/133 (33%), Positives = 63/133 (46%), Gaps = 21/133 (15%)
Query: 131 PVLSSPPPPPSPPTPRSS--------TPIPHPPRRSLPSP---PSPSPSPSPSPSPSPS- 178
P S P P PTP + TP P P+P P+PS P+PS P+PS
Sbjct: 837 PTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSE 896
Query: 179 ------PRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS--SP 230
P TP P +PS P+ S P+PS+ P+ + +P + + + PS +P
Sbjct: 897 TPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTP 956
Query: 231 SDE-SPSPAPSPS 242
SDE +PS P+PS
Sbjct: 957 SDEPTPSDEPTPS 969
Score = 71.2 bits (173), Expect = 2e-12
Identities = 45/133 (33%), Positives = 63/133 (46%), Gaps = 21/133 (15%)
Query: 131 PVLSSPPPPPSPPTPRSS--------TPIPHPPRRSLPSP---PSPSPSPSPSPSPSPS- 178
P S P P PTP + TP P P+P P+PS P+PS P+PS
Sbjct: 1076 PTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSE 1135
Query: 179 ------PRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS--SP 230
P TP P +PS P+ S P+PS+ P+ + +P + + + PS +P
Sbjct: 1136 TPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTP 1195
Query: 231 SDE-SPSPAPSPS 242
SDE +PS P+PS
Sbjct: 1196 SDEPTPSDEPTPS 1208
Score = 71.2 bits (173), Expect = 2e-12
Identities = 45/118 (38%), Positives = 62/118 (52%), Gaps = 6/118 (5%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPR-K 189
P S P P PTP S TP P + P+PS P+PS P+PS TP P
Sbjct: 1205 PTPSDEPTPSDEPTP-SETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPS 1263
Query: 190 RSPASPSPSPSLSKSPSPSE--SPSLAPSPSDSVASLAPSSSPSDE-SPSPAPSPSSS 244
+P P P+ + S P+PS+ +PS P+PSD + + +PSDE +PS P+PS +
Sbjct: 1264 ETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDE-PTPSDEPTPSDEPTPSDEPTPSET 1320
Score = 71.2 bits (173), Expect = 2e-12
Identities = 45/133 (33%), Positives = 63/133 (46%), Gaps = 21/133 (15%)
Query: 131 PVLSSPPPPPSPPTPRSS--------TPIPHPPRRSLPSP---PSPSPSPSPSPSPSPS- 178
P S P P PTP + TP P P+P P+PS P+PS P+PS
Sbjct: 794 PTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSE 853
Query: 179 ------PRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS--SP 230
P TP P +PS P+ S P+PS+ P+ + +P + + + PS +P
Sbjct: 854 TPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTP 913
Query: 231 SDE-SPSPAPSPS 242
SDE +PS P+PS
Sbjct: 914 SDEPTPSDEPTPS 926
Score = 71.2 bits (173), Expect = 2e-12
Identities = 45/133 (33%), Positives = 63/133 (46%), Gaps = 21/133 (15%)
Query: 131 PVLSSPPPPPSPPTPRSS--------TPIPHPPRRSLPSP---PSPSPSPSPSPSPSPS- 178
P S P P PTP + TP P P+P P+PS P+PS P+PS
Sbjct: 1033 PTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSE 1092
Query: 179 ------PRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS--SP 230
P TP P +PS P+ S P+PS+ P+ + +P + + + PS +P
Sbjct: 1093 TPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTP 1152
Query: 231 SDE-SPSPAPSPS 242
SDE +PS P+PS
Sbjct: 1153 SDEPTPSDEPTPS 1165
Score = 71.2 bits (173), Expect = 2e-12
Identities = 45/133 (33%), Positives = 63/133 (46%), Gaps = 21/133 (15%)
Query: 131 PVLSSPPPPPSPPTPRSS--------TPIPHPPRRSLPSP---PSPSPSPSPSPSPSPS- 178
P S P P PTP + TP P P+P P+PS P+PS P+PS
Sbjct: 1162 PTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSE 1221
Query: 179 ------PRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSS--SP 230
P TP P +PS P+ S P+PS+ P+ + +P + + + PS +P
Sbjct: 1222 TPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTP 1281
Query: 231 SDE-SPSPAPSPS 242
SDE +PS P+PS
Sbjct: 1282 SDEPTPSDEPTPS 1294
Score = 70.5 bits (171), Expect = 4e-12
Identities = 42/118 (35%), Positives = 60/118 (50%), Gaps = 6/118 (5%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S P P +P P P+ +PS P+PS TP P
Sbjct: 1150 PTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 1208
Query: 191 SPASPSPSPSLSKSPS---PSESPSLAPSPSDSVASLAPSSSPSDE-SPSPAPSPSSS 244
+PS P+ S++P P+++PS P+PSD + + +PSDE +PS P+PS +
Sbjct: 1209 DEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE-PTPSDEPTPSDEPTPSDEPTPSET 1265
Score = 70.5 bits (171), Expect = 4e-12
Identities = 42/118 (35%), Positives = 60/118 (50%), Gaps = 6/118 (5%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S P P +P P P+ +PS P+PS TP P
Sbjct: 1064 PTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 1122
Query: 191 SPASPSPSPSLSKSPS---PSESPSLAPSPSDSVASLAPSSSPSDE-SPSPAPSPSSS 244
+PS P+ S++P P+++PS P+PSD + + +PSDE +PS P+PS +
Sbjct: 1123 DEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE-PTPSDEPTPSDEPTPSDEPTPSET 1179
Score = 70.5 bits (171), Expect = 4e-12
Identities = 42/118 (35%), Positives = 60/118 (50%), Gaps = 6/118 (5%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S P P +P P P+ +PS P+PS TP P
Sbjct: 1107 PTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 1165
Query: 191 SPASPSPSPSLSKSPS---PSESPSLAPSPSDSVASLAPSSSPSDE-SPSPAPSPSSS 244
+PS P+ S++P P+++PS P+PSD + + +PSDE +PS P+PS +
Sbjct: 1166 DEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE-PTPSDEPTPSDEPTPSDEPTPSET 1222
Score = 70.5 bits (171), Expect = 4e-12
Identities = 42/118 (35%), Positives = 60/118 (50%), Gaps = 6/118 (5%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S P P +P P P+ +PS P+PS TP P
Sbjct: 782 PTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 840
Query: 191 SPASPSPSPSLSKSPS---PSESPSLAPSPSDSVASLAPSSSPSDE-SPSPAPSPSSS 244
+PS P+ S++P P+++PS P+PSD + + +PSDE +PS P+PS +
Sbjct: 841 DEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE-PTPSDEPTPSDEPTPSDEPTPSET 897
Score = 70.5 bits (171), Expect = 4e-12
Identities = 42/118 (35%), Positives = 60/118 (50%), Gaps = 6/118 (5%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S P P +P P P+ +PS P+PS TP P
Sbjct: 825 PTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 883
Query: 191 SPASPSPSPSLSKSPS---PSESPSLAPSPSDSVASLAPSSSPSDE-SPSPAPSPSSS 244
+PS P+ S++P P+++PS P+PSD + + +PSDE +PS P+PS +
Sbjct: 884 DEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE-PTPSDEPTPSDEPTPSDEPTPSET 940
Score = 70.1 bits (170), Expect = 5e-12
Identities = 42/116 (36%), Positives = 59/116 (50%), Gaps = 6/116 (5%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S P P +P P P+ +PS P+PS TP P
Sbjct: 1193 PTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 1251
Query: 191 SPASPSPSPSLSKSPS---PSESPSLAPSPSDSVASLAPSSSPSDE-SPSPAPSPS 242
+PS P+ S++P P+++PS P+PSD + + +PSDE +PS P+PS
Sbjct: 1252 DEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE-PTPSDEPTPSDEPTPSDEPTPS 1306
Score = 70.1 bits (170), Expect = 5e-12
Identities = 42/116 (36%), Positives = 59/116 (50%), Gaps = 6/116 (5%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKR 190
P S P P PTP S P P +P P P+ +PS P+PS TP P
Sbjct: 868 PTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPS 926
Query: 191 SPASPSPSPSLSKSPS---PSESPSLAPSPSDSVASLAPSSSPSDE-SPSPAPSPS 242
+PS P+ S++P P+++PS P+PSD + + +PSDE +PS P+PS
Sbjct: 927 DEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE-PTPSDEPTPSDEPTPSDEPTPS 981
Score = 69.7 bits (169), Expect = 7e-12
Identities = 46/129 (35%), Positives = 64/129 (48%), Gaps = 16/129 (12%)
Query: 131 PVLSSPPPPPSPPTPRSS--------TPIPHPPRRSLPSP---PSPSPSPSPSPSPSPSP 179
P S P P PTP + TP P P+P P+PS P+PS P+PS
Sbjct: 923 PTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSD 982
Query: 180 RSTPIPHPR-KRSPASPSPSPSLSKSPSPSE--SPSLAPSPSDSVASLAPSSSPSDE-SP 235
TP P +P P P+ + S P+PS+ +PS P+PSD + + +PSDE +P
Sbjct: 983 EPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDE-PTPSDEPTPSDEPTP 1041
Query: 236 SPAPSPSSS 244
S P+PS +
Sbjct: 1042 SDEPTPSET 1050
Score = 68.6 bits (166), Expect = 2e-11
Identities = 48/139 (34%), Positives = 64/139 (45%), Gaps = 27/139 (19%)
Query: 131 PVLSSPPPPPSPPTPRSS--------TPIPHPPRRSLPSP---PSPSPSPSPSPSPSPS- 178
P S P P PTP + TP P P+P P+PS P+PS P+PS
Sbjct: 880 PTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSE 939
Query: 179 ------PRSTPIPHPRKRSPASPSPSPSLSKSPSPSE--SPSLAPSPSD------SVASL 224
P TP P +PS P+ S P+PS+ +PS P+PSD +
Sbjct: 940 TPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEP 999
Query: 225 APSSSPSDE-SPSPAPSPS 242
P+ +PSDE +PS P+PS
Sbjct: 1000 IPTDTPSDEPTPSDEPTPS 1018
Score = 53.1 bits (126), Expect = 7e-07
Identities = 39/122 (31%), Positives = 57/122 (45%), Gaps = 10/122 (8%)
Query: 134 SSPPPPPSPPTPRSS-TPIPHPPRRSLPSP---PSPSPSPSPSPSP-SPSPRSTPIPHPR 188
S P P PTP TP P P+P P+PS P+PS +P P+P +TP P P
Sbjct: 1331 SDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPTPTTTPTPTP- 1389
Query: 189 KRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSD---ESPSPAPSPSSSG 245
+P S S S ++ SP+ + S P+S+P+ E P+P+ P + G
Sbjct: 1390 STTPTSGSGGSGGSGGGGGGGGGTVPTSPTPTPTS-KPTSTPAPTEIEEPTPSDVPGAIG 1448
Query: 246 SK 247
+
Sbjct: 1449 GE 1450
Score = 38.1 bits (87), Expect = 0.023
Identities = 20/51 (39%), Positives = 32/51 (62%), Gaps = 2/51 (3%)
Query: 195 PSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDE-SPSPAPSPSSS 244
P+P + S P P+++PS P+PSD + + +PSDE +PS P+PS +
Sbjct: 762 PAPIKAASDEPIPTDTPSDEPTPSDE-PTPSDEPTPSDEPTPSDEPTPSET 811
Score = 34.7 bits (78), Expect = 0.25
Identities = 19/71 (26%), Positives = 30/71 (41%), Gaps = 16/71 (22%)
Query: 131 PVLSSPPPPPSPPTPRSSTPIPHPPRRS----------------LPSPPSPSPSPSPSPS 174
P S P P+P T + TP P S +P+ P+P+P+ P+ +
Sbjct: 1370 PTPSETPEEPTPTTTPTPTPSTTPTSGSGGSGGSGGGGGGGGGTVPTSPTPTPTSKPTST 1429
Query: 175 PSPSPRSTPIP 185
P+P+ P P
Sbjct: 1430 PAPTEIEEPTP 1440
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.311 0.129 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,496,403
Number of Sequences: 164201
Number of extensions: 2489168
Number of successful extensions: 102633
Number of sequences better than 10.0: 3967
Number of HSP's better than 10.0 without gapping: 1977
Number of HSP's successfully gapped in prelim test: 2084
Number of HSP's that attempted gapping in prelim test: 22692
Number of HSP's gapped (non-prelim): 29687
length of query: 268
length of database: 59,974,054
effective HSP length: 108
effective length of query: 160
effective length of database: 42,240,346
effective search space: 6758455360
effective search space used: 6758455360
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC136953.6


