

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136449.7 - phase: 0
(471 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
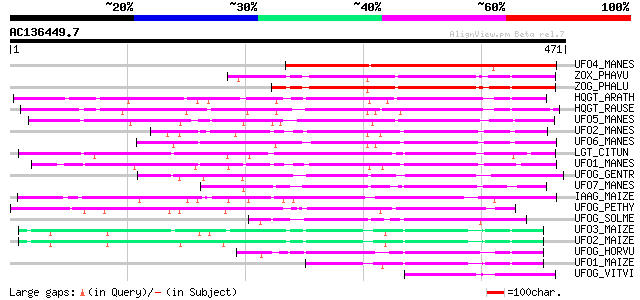
Score E
Sequences producing significant alignments: (bits) Value
UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC 2.4.1... 204 5e-52
ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40) (Ze... 177 4e-44
ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203) (... 172 2e-42
HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (E... 168 3e-41
HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.2... 165 3e-40
UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC 2.4.1... 157 6e-38
UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC 2.4.1... 146 1e-34
UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC 2.4.1... 145 3e-34
LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.21... 140 5e-33
UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC 2.4.1... 139 2e-32
UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 127 5e-29
UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC 2.4.1... 127 8e-29
IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (E... 126 1e-28
UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 124 5e-28
UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 117 5e-26
UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 104 6e-22
UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 103 1e-21
UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 102 2e-21
UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 99 3e-20
UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 72 2e-12
>UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
4) (Fragment)
Length = 241
Score = 204 bits (518), Expect = 5e-52
Identities = 105/232 (45%), Positives = 148/232 (63%), Gaps = 3/232 (1%)
Query: 235 CNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYGLEAS 294
CN+ DK ERG + ++D L WL+ +P SV+Y GS++ + QL E+ GLE++
Sbjct: 1 CNKLKLDKAERGDKASVDNTELLKWLDLWEPGSVIYACLGSISGLTSWQLAELGLGLEST 60
Query: 295 DQSFIWVVGKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQLLILEHEAVGGFMTH 354
+Q FIWV+ + S+ E W+L++ K + RGW+PQ+LIL H A+G F TH
Sbjct: 61 NQPFIWVIREG-EKSEGLEKWILEEGYEERKRKREDFWIRGWSPQVLILSHPAIGAFFTH 119
Query: 355 CGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREWGSWDEERK--ELV 412
CGWNSTLEG+ AGVP+ PL AEQF NEKL+ +VL IGV VG +W E K ++
Sbjct: 120 CGWNSTLEGISAGVPIVACPLFAEQFYNEKLVVEVLGIGVSVGVEAAVTWGLEDKCGAVM 179
Query: 413 GREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDIHALIQ 464
+E+V+ A++ +M + ++ EE RRR + I E AKR +EEGGSSY D+ LIQ
Sbjct: 180 KKEQVKKAIEIVMDKGKEGEERRRRAREIGEMAKRTIEEGGSSYLDMEMLIQ 231
>ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40)
(Zeatin O-beta-D-xylosyltransferase)
Length = 454
Score = 177 bits (450), Expect = 4e-44
Identities = 105/287 (36%), Positives = 173/287 (59%), Gaps = 25/287 (8%)
Query: 186 SQFSDRI---KQLEENSLGTLINSFYDLEPAYADYV-RNKLGKKAWLVGPVSLCNRSVED 241
+QF+D + + + + G + N+ +E Y + + R GK+ W +GP + +VE
Sbjct: 179 AQFTDFLTAQNEFRKFNNGDIYNTSRVIEGPYVELLERFNGGKEVWALGPFTPL--AVEK 236
Query: 242 KKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYGLEASDQSFIWV 301
K G C+ WL+ ++P+SV+Y+SFG+ + +Q++E+A GLE S Q FIWV
Sbjct: 237 KDSIGFS-----HPCMEWLDKQEPSSVIYVSFGTTTALRDEQIQELATGLEQSKQKFIWV 291
Query: 302 V-----GKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQLLILEHEAVGGFMTHCG 356
+ G I + S+ + + + FE R++ M GL+ R WAPQ+ IL H + GGFM+HCG
Sbjct: 292 LRDADKGDIFDGSEAKRYELPEGFEERVEGM--GLVVRDWAPQMEILSHSSTGGFMSHCG 349
Query: 357 WNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREWGSWDEERKELVGREK 416
WNS LE + GVPMATW + ++Q N L+TDVL++G+ V ++W E+RK LV
Sbjct: 350 WNSCLESLTRGVPMATWAMHSDQPRNAVLVTDVLKVGLIV--KDW----EQRKSLVSASV 403
Query: 417 VELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDIHALI 463
+E AV++LM E+++ +E+R+R + + R+++EGG S ++ + I
Sbjct: 404 IENAVRRLM-ETKEGDEIRKRAVKLKDEIHRSMDEGGVSRMEMASFI 449
>ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203)
(Trans-zeatin O-beta-D-glucosyltransferase)
Length = 459
Score = 172 bits (436), Expect = 2e-42
Identities = 98/246 (39%), Positives = 151/246 (60%), Gaps = 21/246 (8%)
Query: 223 GKKAWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMK 282
GKK W +GP + +VE K G + C+ WL+ ++P+SV+YISFG+ + +
Sbjct: 225 GKKVWALGPFNPL--AVEKKDSIGFR-----HPCMEWLDKQEPSSVIYISFGTTTALRDE 277
Query: 283 QLKEIAYGLEASDQSFIWVV-----GKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWA 337
Q+++IA GLE S Q FIWV+ G I S+ + + FE R++ M GL+ R WA
Sbjct: 278 QIQQIATGLEQSKQKFIWVLREADKGDIFAGSEAKRYELPKGFEERVEGM--GLVVRDWA 335
Query: 338 PQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVG 397
PQL IL H + GGFM+HCGWNS LE + GVP+ATWP+ ++Q N L+T+VL++G+ V
Sbjct: 336 PQLEILSHSSTGGFMSHCGWNSCLESITMGVPIATWPMHSDQPRNAVLVTEVLKVGLVV- 394
Query: 398 SREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYD 457
++W +R LV VE V++LM E+++ +EMR+R + R+++EGG S+
Sbjct: 395 -KDWA----QRNSLVSASVVENGVRRLM-ETKEGDEMRQRAVRLKNAIHRSMDEGGVSHM 448
Query: 458 DIHALI 463
++ + I
Sbjct: 449 EMGSFI 454
>HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (EC
2.4.1.218) (Arbutin synthase)
Length = 480
Score = 168 bits (425), Expect = 3e-41
Identities = 146/485 (30%), Positives = 231/485 (47%), Gaps = 64/485 (13%)
Query: 4 ETDAVKIYFFPFVGGGHQIPMVDTA-RVFAKHGATSTIITTPSNALQFQNSITRDQKSNH 62
E+ + P G GH IP+V+ A R+ HG T T + + R +
Sbjct: 3 ESKTPHVAIIPSPGMGHLIPLVEFAKRLVHLHGLTVTFVIAGEGP---PSKAQRTVLDSL 59
Query: 63 PITIQILTTPENTEVTDTDMSAGPMIDTSILLEPLKQ----------FLVQHR-PDGIVV 111
P +I + P V TD+S+ I++ I L + F+ R P +VV
Sbjct: 60 PSSISSVFLPP---VDLTDLSSSTRIESRISLTVTRSNPELRKVFDSFVEGGRLPTALVV 116
Query: 112 DMFHRWAGDIIDELKIPRIVFNGNGCFPRCVIENTRKHVVLENLSSD----SEPFIVPGL 167
D+F A D+ E +P +F + K + E +S + +EP ++PG
Sbjct: 117 DLFGTDAFDVAVEFHVPPYIFYPTTANVLSFFLHLPK--LDETVSCEFRELTEPLMLPGC 174
Query: 168 -----PDIIEMTRSQTPIFMRNPSQFSDRIKQLEENSLGTLINSFYDLEPAYADYVRNKL 222
D ++ + + + + R K+ E G L+N+F++LEP ++
Sbjct: 175 VPVAGKDFLDPAQDRKDDAYKWLLHNTKRYKEAE----GILVNTFFELEPNAIKALQEPG 230
Query: 223 GKK--AWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVP 280
K + VGP+ V K+ KQ +E CL WL+++ SVLY+SFGS +
Sbjct: 231 LDKPPVYPVGPL------VNIGKQEAKQT--EESECLKWLDNQPLGSVLYVSFGSGGTLT 282
Query: 281 MKQLKEIAYGLEASDQSFIWVVGK---ILNSSKNEEDWVLDK-------FERRMKEMDKG 330
+QL E+A GL S+Q F+WV+ I NSS + D F R K+ +G
Sbjct: 283 CEQLNELALGLADSEQRFLWVIRSPSGIANSSYFDSHSQTDPLTFLPPGFLERTKK--RG 340
Query: 331 LIFRGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVL 390
+ WAPQ +L H + GGF+THCGWNSTLE V +G+P+ WPL AEQ +N L+++ +
Sbjct: 341 FVIPFWAPQAQVLAHPSTGGFLTHCGWNSTLESVVSGIPLIAWPLYAEQKMNAVLLSEDI 400
Query: 391 RIGVQVGSREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVE 450
R ++ + + G LV RE+V VK LM E E+ + +R ++K + E A R ++
Sbjct: 401 RAALRPRAGDDG--------LVRREEVARVVKGLM-EGEEGKGVRNKMKELKEAACRVLK 451
Query: 451 EGGSS 455
+ G+S
Sbjct: 452 DDGTS 456
>HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.218)
(Arbutin synthase)
Length = 470
Score = 165 bits (417), Expect = 3e-40
Identities = 149/485 (30%), Positives = 223/485 (45%), Gaps = 57/485 (11%)
Query: 10 IYFFPFVGGGHQIPMVDTA-RVFAKHGATSTIITTPSNALQFQNSITRDQKSNHPITIQI 68
I P G GH IP+V+ A R+ +H T I L D P +
Sbjct: 7 IAMVPTPGMGHLIPLVEFAKRLVLRHNFGVTFIIPTDGPLPKAQKSFLDAL---PAGVNY 63
Query: 69 LTTPENTEVTDTDMSAGPMIDTSILL----------EPLKQFLVQHRPDGIVVDMFHRWA 118
+ P V+ D+ A I+T I L + +K L + +VVD+F A
Sbjct: 64 VLLPP---VSFDDLPADVRIETRICLTITRSLPFVRDAVKTLLATTKLAALVVDLFGTDA 120
Query: 119 GDIIDELKIPRIVFNGNGCFPRCVIENTRK--HVVLENLSSDSEPFIVPG-LPDIIEMTR 175
D+ E K+ +F + + K +V EP +PG +P I
Sbjct: 121 FDVAIEFKVSPYIFYPTTAMCLSLFFHLPKLDQMVSCEYRDVPEPLQIPGCIP--IHGKD 178
Query: 176 SQTPIFMRNPSQFSDRIKQLEENSL--GTLINSFYDLEPAYADYVRNKLGKK--AWLVGP 231
P R + + Q + L G ++N+F DLEP ++ + K + +GP
Sbjct: 179 FLDPAQDRKNDAYKCLLHQAKRYRLAEGIMVNTFNDLEPGPLKALQEEDQGKPPVYPIGP 238
Query: 232 VSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYGL 291
+ + S + +D+ CL WL+ + SVL+ISFGS V Q E+A GL
Sbjct: 239 LIRADSSSK----------VDDCECLKWLDDQPRGSVLFISFGSGGAVSHNQFIELALGL 288
Query: 292 EASDQSFIWVV----GKILNSS----KNEEDWVLDKFERRMKEMDKG--LIFRGWAPQLL 341
E S+Q F+WVV KI N++ +N+ D L E KG L+ WAPQ
Sbjct: 289 EMSEQRFLWVVRSPNDKIANATYFSIQNQND-ALAYLPEGFLERTKGRCLLVPSWAPQTE 347
Query: 342 ILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREW 401
IL H + GGF+THCGWNS LE V GVP+ WPL AEQ +N ++T+ L++ ++ + E
Sbjct: 348 ILSHGSTGGFLTHCGWNSILESVVNGVPLIAWPLYAEQKMNAVMLTEGLKVALRPKAGEN 407
Query: 402 GSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDIHA 461
G L+GR ++ AVK LM E E+ ++ R +K + + A RA+ + GSS + A
Sbjct: 408 G--------LIGRVEIANAVKGLM-EGEEGKKFRSTMKDLKDAASRALSDDGSSTKAL-A 457
Query: 462 LIQCK 466
+ CK
Sbjct: 458 ELACK 462
>UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
5)
Length = 487
Score = 157 bits (397), Expect = 6e-38
Identities = 131/480 (27%), Positives = 229/480 (47%), Gaps = 63/480 (13%)
Query: 17 GGGHQIPMVDTA-RVFAKHGATSTIITTPSNALQFQNSITRDQKSNHPITIQILTTPENT 75
G GH IP+++ R+ TI S+ + + R + P +I+ P
Sbjct: 19 GLGHLIPVLELGKRIVTLCNFDVTIFMVGSDTSAAEPQVLRSAMT--PKLCEIIQLPPPN 76
Query: 76 EVTDTDMSAGPMIDTSILLEPLKQFL------VQHRPDGIVVDMFHRWAGDIIDELKIPR 129
D A +L+ ++ ++ RP I+VD+F + ++ EL I +
Sbjct: 77 ISCLIDPEATVCTRLFVLMREIRPAFRAAVSALKFRPAAIIVDLFGTESLEVAKELGIAK 136
Query: 130 IVF-NGNGCFPRCVI------ENTRKHVVLENLSSDSEPFIVPGLPDIIEMTRSQTPIFM 182
V+ N F I + VL+ EP +PG + P+
Sbjct: 137 YVYIASNAWFLALTIYVPILDKEVEGEFVLQK-----EPMKIPGCRPV-RTEEVVDPMLD 190
Query: 183 RNPSQFSDRIKQLEE--NSLGTLINSFYDLEPAYADYVRNK--LGKKAWL----VGPVSL 234
R Q+S+ + E + G L+N++ LEP +R+ LG+ A + +GP+
Sbjct: 191 RTNQQYSEYFRLGIEIPTADGILMNTWEALEPTTFGALRDVKFLGRVAKVPVFPIGPL-- 248
Query: 235 CNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYGLEAS 294
R P L+WL+ + SV+Y+SFGS + ++Q+ E+A+GLE S
Sbjct: 249 ---------RRQAGPCGSNCELLDWLDQQPKESVVYVSFGSGGTLSLEQMIELAWGLERS 299
Query: 295 DQSFIWVVGKIL------------NSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQLLI 342
Q FIWVV + + + + + + F R++ + GL+ W+PQ+ I
Sbjct: 300 QQRFIWVVRQPTVKTGDAAFFTQGDGADDMSGYFPEGFLTRIQNV--GLVVPQWSPQIHI 357
Query: 343 LEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREWG 402
+ H +VG F++HCGWNS LE + AGVP+ WP+ AEQ +N L+T+ L + V+ +
Sbjct: 358 MSHPSVGVFLSHCGWNSVLESITAGVPIIAWPIYAEQRMNATLLTEELGVAVRPKNL--- 414
Query: 403 SWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDIHAL 462
KE+V RE++E ++++M + E+ E+R+RV+ + ++ ++A+ EGGSS++ + AL
Sbjct: 415 ----PAKEVVKREEIERMIRRIMVD-EEGSEIRKRVRELKDSGEKALNEGGSSFNYMSAL 469
>UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
2) (Fragment)
Length = 346
Score = 146 bits (368), Expect = 1e-34
Identities = 111/353 (31%), Positives = 177/353 (49%), Gaps = 42/353 (11%)
Query: 120 DIIDELKIPRIVF--NGNGCFPRCV----IENTRKHVVLENLSSDSEPFIVPGLPDIIEM 173
D+ DE IP +F +G G + I + +E SD+E IVP L +
Sbjct: 7 DLADEFGIPSYIFFASGGGFLGFMLYVQKIHDEENFNPIEFKDSDTE-LIVPSL--VNPF 63
Query: 174 TRSQTPIFMRNPSQFSD--RIKQLEENSLGTLINSFYDLEPAYADYVRNKLGKKAWLVGP 231
P + N +F I + + G ++N+F +LE + + + VGP
Sbjct: 64 PTRILPSSILNKERFGQLLAIAKKFRQAKGIIVNTFLELESRAIESFKVP---PLYHVGP 120
Query: 232 VSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYGL 291
+ D K G+ + WL+ + SV+++ FGS+ QLKEIAY L
Sbjct: 121 IL-------DVKSDGRNT---HPEIMQWLDDQPEGSVVFLCFGSMGSFSEDQLKEIAYAL 170
Query: 292 EASDQSFIWVV------GKILNSSKNEE--DWVLDKFERRMKEMDKGLIFRGWAPQLLIL 343
E S F+W + KI + + E+ D + + F R + K + GWAPQ+ +L
Sbjct: 171 ENSGHRFLWSIRRPPPPDKIASPTDYEDPRDVLPEGFLERTVAVGKVI---GWAPQVAVL 227
Query: 344 EHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREWGS 403
H A+GGF++HCGWNS LE + GVP+ATWP+ AEQ N + L +GV++
Sbjct: 228 AHPAIGGFVSHCGWNSVLESLWFGVPIATWPMYAEQQFNAFEMVVELGLGVEIDM----G 283
Query: 404 WDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSY 456
+ +E +V +K+E A++KLM E+++E R++VK + E +K A+ +GGSS+
Sbjct: 284 YRKESGIIVNSDKIERAIRKLM---ENSDEKRKKVKEMREKSKMALIDGGSSF 333
>UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
6) (Fragment)
Length = 394
Score = 145 bits (365), Expect = 3e-34
Identities = 109/373 (29%), Positives = 186/373 (49%), Gaps = 34/373 (9%)
Query: 108 GIVVDMFHRWAGDIIDELKIPRIVFNGNGC------FPRCVIENTRKHVVLENLSSDSEP 161
G V+DMF D+ EL +P +F +G F +I + + + + SD+E
Sbjct: 33 GFVLDMFCTSMIDVAKELGVPYYIFFTSGAAFLGFLFYVQLIHDEQDADLTQFKDSDAE- 91
Query: 162 FIVPGLPDIIEMTRSQTPIFMRNPSQFSDRIKQLEENSLGTLINSFYDLEPAYADYVRNK 221
VP L + + + +++ RI + + G ++N+F +LE + +++
Sbjct: 92 LSVPSLANSLPARVLPASMLVKDRFYAFIRIIRGLREAKGIMVNTFMELESHALNSLKDD 151
Query: 222 LGK--KAWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARV 279
K + VGP+ + D G + + WL+ + P+SV+++ FGS+
Sbjct: 152 QSKIPPIYPVGPILKLSNQENDVGPEGSE-------IIEWLDDQPPSSVVFLCFGSMGGF 204
Query: 280 PMKQLKEIAYGLEASDQSFIWVV------GKILNSS--KNEEDWVLDKFERRMKEMDKGL 331
M Q KEIA LE S F+W + GKI S+ +N ++ + F R M K +
Sbjct: 205 DMDQAKEIACALEQSRHRFLWSLRRPPPKGKIETSTDYENLQEILPVGFSERTAGMGKVV 264
Query: 332 IFRGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLR 391
GWAPQ+ ILEH A+GGF++HCGWNS LE + VP+ATWPL AEQ N + L
Sbjct: 265 ---GWAPQVAILEHPAIGGFVSHCGWNSILESIWFSVPIATWPLYAEQQFNAFTMVTELG 321
Query: 392 IGVQVGSREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEE 451
+ V++ + +E + ++ + +E +K +M E E+R+RVK + + +++A+ +
Sbjct: 322 LAVEIKM----DYKKESEIILSADDIERGIKCVM---EHHSEIRKRVKEMSDKSRKALMD 374
Query: 452 GGSSYDDIHALIQ 464
SS + LI+
Sbjct: 375 DESSSFWLDRLIE 387
>LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.210)
(Limonoid glucosyltransferase) (Limonoid GTase) (LGTase)
Length = 511
Score = 140 bits (354), Expect = 5e-33
Identities = 123/475 (25%), Positives = 215/475 (44%), Gaps = 39/475 (8%)
Query: 8 VKIYFFPFVGGGHQIPMVDTARVFAKHGATSTIITTPSNALQFQNSITRDQKSNHPITIQ 67
V + F G GH P++ R+ A G T+ T S Q + + + P+
Sbjct: 7 VHVLLVSFPGHGHVNPLLRLGRLLASKGFFLTLTTPESFGKQMRKAGNFTYEPT-PVGDG 65
Query: 68 ILT----------TPENTEVTDTDMSAGPMIDTSILLEPLKQFLVQHRPDGIVVDM-FHR 116
+ E D M+ +I ++ + +K+ ++RP +++ F
Sbjct: 66 FIRFEFFEDGWDEDDPRREDLDQYMAQLELIGKQVIPKIIKKSAEEYRPVSCLINNPFIP 125
Query: 117 WAGDIIDELKIPRIVFNGNGCFPRCVIENTRKHVVLENLSSDSEPFIVPGLPDIIEMTRS 176
W D+ + L +P + C C L S+ EP I LP + +
Sbjct: 126 WVSDVAESLGLPSAMLWVQSC--ACFAAYYHYFHGLVPFPSEKEPEIDVQLPCMPLLKHD 183
Query: 177 QTPIFMR--NPSQFSDRIKQLEENSLGT----LINSFYDLEPAYADYVRNKLGKKAWLVG 230
+ P F+ P F R + +LG L+++FY+LE DY+ K +
Sbjct: 184 EMPSFLHPSTPYPFLRRAILGQYENLGKPFCILLDTFYELEKEIIDYMA-----KICPIK 238
Query: 231 PVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYG 290
PV ++ + + + C++WL+ K P+SV+YISFG+V + +Q++EI Y
Sbjct: 239 PVGPLFKNPKAPTLTVRDDCMKPDECIDWLDKKPPSSVVYISFGTVVYLKQEQVEEIGYA 298
Query: 291 LEASDQSFIWVVGKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQLLILEHEAVGG 350
L S SF+WV+ S + + D F ++ DKG + + W+PQ +L H +V
Sbjct: 299 LLNSGISFLWVMKPPPEDSGVKIVDLPDGFLEKVG--DKGKVVQ-WSPQEKVLAHPSVAC 355
Query: 351 FMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREWGSWDEERKE 410
F+THCGWNST+E + +GVP+ T+P +Q + + DV + G+++ E
Sbjct: 356 FVTHCGWNSTMESLASGVPVITFPQWGDQVTDAMYLCDVFKTGLRL------CRGEAENR 409
Query: 411 LVGREKVELAVKKLMA--ESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDIHALI 463
++ R++VE + + A ++ EE + K + A+ AV +GGSS +I A +
Sbjct: 410 IISRDEVEKCLLEATAGPKAVALEENALKWK---KEAEEAVADGGSSDRNIQAFV 461
>UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
1)
Length = 449
Score = 139 bits (350), Expect = 2e-32
Identities = 128/476 (26%), Positives = 212/476 (43%), Gaps = 62/476 (13%)
Query: 19 GHQIPMVDTARVFAKHGATSTIITTPSNALQFQNSITRDQKSNHPITIQILTTPENTEVT 78
GH + V+TA++ + +I L F NS+ + N+ + QI ++
Sbjct: 2 GHLVSAVETAKLLLSRCHSLSI-----TVLIFNNSVVTSKVHNY-VDSQIASSSNRLRFI 55
Query: 79 DTDMSAGPMIDTSILLEPLKQFLVQH--------------RPDGIVVDMFHRWAGDIIDE 124
+ S L+E K + + R G +VDMF D+ +E
Sbjct: 56 YLPRDETGISSFSSLIEKQKPHVKESVMKITEFGSSVESPRLVGFIVDMFCTAMIDVANE 115
Query: 125 LKIPRIVFNGNGCFPRCVIENTRKHVVLENLS------SDSEPFIVPGLPDIIEMTRSQT 178
+P +F +G + + +K EN + SD E VPGL + T
Sbjct: 116 FGVPSYIFYTSGAAFLNFMLHVQKIHDEENFNPTEFNASDGE-LQVPGLVNSFPSKAMPT 174
Query: 179 PIFMRN--PSQFSDRIKQLEENSLGTLINSFYDLEPAYADYVRNKLGKKAWLVGPVSLCN 236
I + P + + E G +IN+F++LE + ++ + VGP+
Sbjct: 175 AILSKQWFPPLLENTRRYGEAK--GVIINTFFELESHAIESFKDP---PIYPVGPIL--- 226
Query: 237 RSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYGLEASDQ 296
D + G+ Q + WL+ + P+SV+++ FGS Q+KEIA LE S
Sbjct: 227 ----DVRSNGRNTN---QEIMQWLDDQPPSSVVFLCFGSNGSFSKDQVKEIACALEDSGH 279
Query: 297 SFIWVVGK-----ILNSSKNEEDW---VLDKFERRMKEMDKGLIFRGWAPQLLILEHEAV 348
F+W + L S + ED + + F R ++K + GWAPQ+ +L H A
Sbjct: 280 RFLWSLADHRAPGFLESPSDYEDLQEVLPEGFLERTSGIEKVI---GWAPQVAVLAHPAT 336
Query: 349 GGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREWGSWDEER 408
GG ++H GWNS LE + GVP+ATWP+ AEQ N + L + V++ + +
Sbjct: 337 GGLVSHSGWNSILESIWFGVPVATWPMYAEQQFNAFQMVIELGLAVEIKM----DYRNDS 392
Query: 409 KELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDIHALIQ 464
E+V +++E ++ LM D R++VK + E ++ A+ EGGSSY + LI+
Sbjct: 393 GEIVKCDQIERGIRCLMKHDSD---RRKKVKEMSEKSRGALMEGGSSYCWLDNLIK 445
>UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 453
Score = 127 bits (320), Expect = 5e-29
Identities = 100/371 (26%), Positives = 175/371 (46%), Gaps = 41/371 (11%)
Query: 109 IVVDMFHRWAGDIIDELKIPRI-VFNGNGCFPRCV---IENTRKHVVLENLSSDSEPFI- 163
++ D F +A D +++ +P I V+ C C+ + R +++ +E I
Sbjct: 114 LLTDAFLWFAADFSEKIGVPWIPVWTAASC-SLCLHVYTDEIRSRFAEFDIAEKAEKTID 172
Query: 164 -VPGLPDIIEMTRSQTPIFMRNPSQFSDRIKQLE---ENSLGTLINSFYDLEPAYADYVR 219
+PGL I + I + S F+ + + + +NSF +++P +++R
Sbjct: 173 FIPGLSAISFSDLPEELIMEDSQSIFALTLHNMGLKLHKATAVAVNSFEEIDPIITNHLR 232
Query: 220 NKLGKKAWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARV 279
+ +GP+ + S+ ++ CL WL ++K +SV+Y+SFG+V
Sbjct: 233 STNQLNILNIGPLQTLSSSIPP----------EDNECLKWLQTQKESSVVYLSFGTVINP 282
Query: 280 PMKQLKEIAYGLEASDQSFIWVVGKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQ 339
P ++ +A LE+ F+W + + K+ + +D+ K + WAPQ
Sbjct: 283 PPNEMAALASTLESRKIPFLWSLRD--EARKHLPENFIDRTSTFGKIVS-------WAPQ 333
Query: 340 LLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSR 399
L +LE+ A+G F+THCGWNSTLE + VP+ P +Q +N +++ DV +IGV V
Sbjct: 334 LHVLENPAIGVFVTHCGWNSTLESIFCRVPVIGRPFFGDQKVNARMVEDVWKIGVGV--- 390
Query: 400 EWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDI 459
K V E V +L+ S+ +EMR+ V + E AK AV+ GSS +
Sbjct: 391 ---------KGGVFTEDETTRVLELVLFSDKGKEMRQNVGRLKEKAKDAVKANGSSTRNF 441
Query: 460 HALIQCKTSLD 470
+L+ LD
Sbjct: 442 ESLLAAFNKLD 452
>UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
7) (Fragment)
Length = 287
Score = 127 bits (318), Expect = 8e-29
Identities = 93/296 (31%), Positives = 142/296 (47%), Gaps = 34/296 (11%)
Query: 163 IVPGLPDIIEMTRSQTPIFMRNPSQFSDRIKQLEE---NSLGTLINSFYDLEPAYADYVR 219
++PG+ I + +F S FS + + + L+NSF +L+P +
Sbjct: 4 LIPGMSKIQIRDLPEGVLFGNLESLFSQMLHNMGRMLPRAAAVLMNSFEELDPTIVSDLN 63
Query: 220 NKLGKKAWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARV 279
+K +GP +L + P D C+ WL+ +KP SV YISFGSVA
Sbjct: 64 SKFNN-ILCIGPFNLVSPP---------PPVPDTYGCMAWLDKQKPASVAYISFGSVATP 113
Query: 280 PMKQLKEIAYGLEASDQSFIWVVGKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQ 339
P +L +A LEAS F+W + +S + + LD+ + G++ WAPQ
Sbjct: 114 PPHELVALAEALEASKVPFLWSLKD--HSKVHLPNGFLDRTKSH------GIVL-SWAPQ 164
Query: 340 LLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSR 399
+ ILEH A+G F+THCGWNS LE + GVPM P +Q +N +++ DV IG+ +
Sbjct: 165 VEILEHAALGVFVTHCGWNSILESIVGGVPMICRPFFGDQRLNGRMVEDVWEIGLLMD-- 222
Query: 400 EWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSS 455
G + + G ++ L K ++MR +K + E AK A E GSS
Sbjct: 223 --GGVLTKNGAIDGLNQILLQGK--------GKKMRENIKRLKELAKGATEPKGSS 268
>IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (EC
2.4.1.121) (IAA-Glu synthetase) ((Uridine
5'-diphosphate-glucose:indol-3-ylacetyl)-beta-D-glucosyl
transferase)
Length = 471
Score = 126 bits (316), Expect = 1e-28
Identities = 128/493 (25%), Positives = 202/493 (40%), Gaps = 67/493 (13%)
Query: 7 AVKIYFFPFVGGGHQIPMVDTARVFAKHGATSTIITTPSNALQFQNSITRDQKSNHPITI 66
A + PF G GH PMV A+ A G +T++TT Q + D HP +
Sbjct: 2 APHVLVVPFPGQGHMNPMVQFAKRLASKGVATTLVTTRF----IQRTADVDA---HPAMV 54
Query: 67 QILTTPENTEVTDTDMSAGPMIDTSILLEPLK-QFLVQHRPDG------IVVDMFHRWAG 119
+ ++ + + ++ LV+ R +V D + W
Sbjct: 55 EAISDGHDEGGFASAAGVAEYLEKQAAAASASLASLVEARASSADAFTCVVYDSYEDWVL 114
Query: 120 DIIDELKIPRIVFNGNGCFPRCVIENTRKH--VVLENLSSD-----------SEPFIVPG 166
+ + +P + F+ C V + + V ++D SE F+ G
Sbjct: 115 PVARRMGLPAVPFSTQSCAVSAVYYHFSQGRLAVPPGAAADGSDGGAGAAALSEAFL--G 172
Query: 167 LPDIIEMTRSQTPIFMRN----PSQFSDRIKQLEENSLG--TLINSFYDLEPAYADYVRN 220
LP EM RS+ P F+ + P+ IKQ L NSF +LE +
Sbjct: 173 LP---EMERSELPSFVFDHGPYPTIAMQAIKQFAHAGKDDWVLFNSFEELETEVLAGLTK 229
Query: 221 KLGKKAWLVGP---VSLCNRSV--EDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGS 275
L KA +GP + R+ + G E +C WL++K SV Y+SFGS
Sbjct: 230 YL--KARAIGPCVPLPTAGRTAGANGRITYGANLVKPEDACTKWLDTKPDRSVAYVSFGS 287
Query: 276 VARVPMKQLKEIAYGLEASDQSFIWVVGKILNSSKNEEDWVLDKFERRMKEMDKGLIFRG 335
+A + Q +E+A GL A+ + F+WVV + ++ + ++ +
Sbjct: 288 LASLGNAQKEELARGLLAAGKPFLWVV-------RASDEHQVPRYLLAEATATGAAMVVP 340
Query: 336 WAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQ 395
W PQL +L H AVG F+THCGWNSTLE + GVPM L +Q N + +
Sbjct: 341 WCPQLDVLAHPAVGCFVTHCGWNSTLEALSFGVPMVAMALWTDQPTNARNV--------- 391
Query: 396 VGSREWGSWDEERKE----LVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEE 451
WG+ R++ + R +VE V+ +M E R+ + A+ AV
Sbjct: 392 --ELAWGAGVRARRDAGAGVFLRGEVERCVRAVMDGGEAASAARKAAGEWRDRARAAVAP 449
Query: 452 GGSSYDDIHALIQ 464
GGSS ++ +Q
Sbjct: 450 GGSSDRNLDEFVQ 462
>UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Anthocyanin rhamnosyl transferase)
Length = 473
Score = 124 bits (311), Expect = 5e-28
Identities = 116/446 (26%), Positives = 193/446 (43%), Gaps = 38/446 (8%)
Query: 1 MNHETDAVKIYFFPFVGGGHQIPMVDTARVFAKHGATSTIITTPSNALQFQNSITRDQKS 60
M H DA+ + FPF GH P V A + +G + T NA + + S+ +
Sbjct: 5 MKHSNDALHVVMFPFFAFGHISPFVQLANKLSSYGVKVSFFTASGNASRVK-SMLNSAPT 63
Query: 61 NH--PITI-QILTTPENTEVTD--TDMSAGPMIDTSILLEPLKQFLVQH-RPDGIVVDMF 114
H P+T+ + P E T T SA + L++P + L+ H +P ++ D
Sbjct: 64 THIVPLTLPHVEGLPPGAESTAELTPASAELLKVALDLMQPQIKTLLSHLKPHFVLFDFA 123
Query: 115 HRWAGDIIDELKIPRIVFNG----NGCFPRCV--IENTRKHVVLENLSSDSEPFIVPGLP 168
W + + L I + ++ + F C + +K+ LE++ F +
Sbjct: 124 QEWLPKMANGLGIKTVYYSVVVALSTAFLTCPARVLEPKKYPSLEDMKKPPLGFPQTSVT 183
Query: 169 DIIEM-TRSQTPIF--MRNPSQFSDRIKQLEENSLGTLINSFYDLEPAYADYVRNKLGKK 225
+ R +F N DRI+ L + +E Y YV + K
Sbjct: 184 SVRTFEARDFLYVFKSFHNGPTLYDRIQSGLRGCSAILAKTCSQMEGPYIKYVEAQFNKP 243
Query: 226 AWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLK 285
+L+GPV V D GK E+ WLN + +V+Y SFGS + Q+K
Sbjct: 244 VFLIGPV------VPDPPS-GKL----EEKWATWLNKFEGGTVIYCSFGSETFLTDDQVK 292
Query: 286 EIAYGLEASDQSFIWVVGKILNSSKNEE--DWVLDKFERRMKEMDKGLIFRGWAPQLLIL 343
E+A GLE + F V+ N + E + + F R+K DKG+I GW Q IL
Sbjct: 293 ELALGLEQTGLPFFLVLNFPANVDVSAELNRALPEGFLERVK--DKGIIHSGWVQQQNIL 350
Query: 344 EHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREWGS 403
H +VG ++ H G++S +E + + P +Q +N KL++ + GV++ R+
Sbjct: 351 AHSSVGCYVCHAGFSSVIEALVNDCQVVMLPQKGDQILNAKLVSGDMEAGVEINRRDEDG 410
Query: 404 WDEERKELVGREKVELAVKKLMAESE 429
+ G+E ++ AV+K+M + E
Sbjct: 411 Y-------FGKEDIKEAVEKVMVDVE 429
>UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 433
Score = 117 bits (294), Expect = 5e-26
Identities = 79/247 (31%), Positives = 124/247 (49%), Gaps = 32/247 (12%)
Query: 203 LINSFYDLE--PAYADYVRNKLGKKAWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWL 260
++NSF +L+ P ++ L +K + +GP+ L + +DE C+ WL
Sbjct: 205 VLNSFQELDRDPLINKDLQKNL-QKVFNIGPLVLQSSR-----------KLDESGCIQWL 252
Query: 261 NSKKPNSVLYISFGSVARVPMKQLKEIAYGLEASDQSFIWVVGKILNSSKNEEDWVLDKF 320
+ +K SV+Y+SFG+V +P ++ IA LE FIW + N KN L+
Sbjct: 253 DKQKEKSVVYLSFGTVTTLPPNEIGSIAEALETKKTPFIWSLRN--NGVKNLPKGFLE-- 308
Query: 321 ERRMKEMDKGLIFRGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQF 380
R KE K + WAPQL IL H++VG F+THCGWNS LEG+ GVPM P +Q
Sbjct: 309 --RTKEFGKIV---SWAPQLEILAHKSVGVFVTHCGWNSILEGISFGVPMICRPFFGDQK 363
Query: 381 INEKLITDVLRIGVQVGS---------REWGSWDEERKELVGREKVELAVKKLMAESEDT 431
+N +++ V IG+Q+ ++ E K + RE VE +K +
Sbjct: 364 LNSRMVESVWEIGLQIEGGIFTKSGIISALDTFFNEEKGKILRENVEGLKEKALEAVNQM 423
Query: 432 EEMRRRV 438
E+++++
Sbjct: 424 MEVQQKI 430
>UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-W22 allele)
Length = 471
Score = 104 bits (259), Expect = 6e-22
Identities = 123/475 (25%), Positives = 188/475 (38%), Gaps = 62/475 (13%)
Query: 8 VKIYFFPFVGGGHQIPMVDTARVFAK----HGATSTIITTPSNALQFQNSITRDQKSNHP 63
V + FPF H ++ AR A GAT + ++T S+ Q + + + P
Sbjct: 14 VAVVAFPF--SSHAAVLLSIARALAAAAAPSGATLSFLSTASSLAQLRKASSASAGHGLP 71
Query: 64 ITIQILTTPENTEVTDTD----------MSAGPMIDTSILLEPLKQFLVQHRPDGIVVDM 113
++ + P+ + M A LE + R +V D
Sbjct: 72 GNLRFVEVPDGAPAAEESVPVPRQMQLFMEAAEAGGVKAWLEAARAAAGGARVTCVVGDA 131
Query: 114 FHRWAGDIIDELKIPRI-VFNGNGCFPRCVIENTRKHVVLENLSSDS-----EPFIV-PG 166
F A D P + V+ C ++ + R + E++ + EP I PG
Sbjct: 132 FVWPAADAAASAGAPWVPVWTAASC---ALLAHIRTDALREDVGDQAANRVDEPLISHPG 188
Query: 167 LP-----DIIEMTRSQTPIFMRNPSQFSDRIKQ-LEENSLGTLINSFYDLEPAYADYVRN 220
L D+ + S ++ N R+ Q L ++ +N+F L+P
Sbjct: 189 LASYRVRDLPDGVVSGDFNYVIN--LLVHRMGQCLPRSAAAVALNTFPGLDPPDVTAALA 246
Query: 221 KLGKKAWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVP 280
++ GP L ED + D CL WL + V Y+SFG+VA
Sbjct: 247 EILPNCVPFGPYHLL--LAEDDADTAAPA--DPHGCLAWLGRQPARGVAYVSFGTVACPR 302
Query: 281 MKQLKEIAYGLEASDQSFIWVVGKILNSSKNEEDWVL--DKFERRMKEMDKGLIFRGWAP 338
+L+E+A GLEAS F+W S E+ W L F R GL+ WAP
Sbjct: 303 PDELRELAAGLEASGAPFLW--------SLREDSWTLLPPGFLDRAAGTGSGLVVP-WAP 353
Query: 339 QLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGS 398
Q+ +L H +VG F+TH GW S LEGV +GVPMA P +Q +N + + V
Sbjct: 354 QVAVLRHPSVGAFVTHAGWASVLEGVSSGVPMACRPFFGDQRMNARSVAHV--------- 404
Query: 399 REWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGG 453
WG + + + V AV++L+ E+ MR R K + A GG
Sbjct: 405 --WG-FGAAFEGAMTSAGVAAAVEELL-RGEEGARMRARAKVLQALVAEAFGPGG 455
>UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-Mc2 allele)
Length = 471
Score = 103 bits (257), Expect = 1e-21
Identities = 123/470 (26%), Positives = 183/470 (38%), Gaps = 52/470 (11%)
Query: 8 VKIYFFPFVGGGHQIPMVDTARVFAK----HGATSTIITTPSNALQFQNSITRDQKSNHP 63
V + FPF H ++ AR A GAT + ++T S+ Q + + + P
Sbjct: 14 VAVVAFPF--SSHAAVLLSIARALAAAAAPSGATLSFLSTASSLAQLRKASSASAGHGLP 71
Query: 64 ITIQILTTPENTEVTDTD----------MSAGPMIDTSILLEPLKQFLVQHRPDGIVVDM 113
++ + P+ + M A LE + R +V D
Sbjct: 72 GNLRFVEVPDGAPAAEETVPVPRQMQLFMEAAEAGGVKAWLEAARAAAGGARVTCVVGDA 131
Query: 114 FHRWAGDIIDELKIPRI-VFNGNGCFPRCVI--ENTRKHVVLENLSSDSEPFIV-PGLPD 169
F A D P + V+ C I ++ R+ V + + EP I PGL
Sbjct: 132 FVWPAADAAASAGAPWVPVWTAASCALLAHIRTDSLREDVGDQAANRVDEPLISHPGLAS 191
Query: 170 IIEMTRSQTPI---FMRNPSQFSDRIKQ-LEENSLGTLINSFYDLEPAYADYVRNKLGKK 225
+ F S R+ Q L ++ +N+F L+P ++
Sbjct: 192 YRVRDLPDGVVSGDFNYVISLLVHRMGQCLPRSAAAVALNTFPGLDPPDVTAALAEILPN 251
Query: 226 AWLVGPVSLCNRSVEDKKERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLK 285
GP L ED + D CL WL + V Y+SFG+VA +L+
Sbjct: 252 CVPFGPYHLL--LAEDDADTAAPA--DPHGCLAWLGRQPARGVAYVSFGTVACPRPDELR 307
Query: 286 EIAYGLEASDQSFIWVVGKILNSSKNEEDWVL--DKFERRMKEMDKGLIFRGWAPQLLIL 343
E+A GLEAS F+W S E+ W L F R GL+ WAPQ+ +L
Sbjct: 308 ELAAGLEASAAPFLW--------SLREDSWTLLPPGFLDRAAGTGSGLVVP-WAPQVAVL 358
Query: 344 EHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSREWGS 403
H +VG F+TH GW S LEGV +GVPMA P +Q +N + + V WG
Sbjct: 359 RHPSVGAFVTHAGWASVLEGVSSGVPMACRPFFGDQRMNARSVAHV-----------WG- 406
Query: 404 WDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGG 453
+ + + V AV++L+ E+ MR R K + A GG
Sbjct: 407 FGAAFEGAMTSAGVAAAVEELL-RGEEGAGMRARAKELQALVAEAFGPGG 455
>UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1)
Length = 455
Score = 102 bits (254), Expect = 2e-21
Identities = 81/266 (30%), Positives = 121/266 (45%), Gaps = 37/266 (13%)
Query: 193 KQLEENSLGTLINSFYDLEP-----AYADYVRNKLGKKAWLVGPVSLCNRSVEDKKERGK 247
++L + + +N+F L+P A A + N L +GP L E + +
Sbjct: 206 QRLPKAATAVALNTFPGLDPPDLIAALAAELPNCLP-----LGPYHLLP-GAEPTADTNE 259
Query: 248 QPTIDEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYGLEASDQSFIWVVGKILN 307
P D CL WL+ + SV Y+SFG+ A +L+E+A GLEAS F+W + ++
Sbjct: 260 APA-DPHGCLAWLDRRPARSVAYVSFGTNATARPDELQELAAGLEASGAPFLWSLRGVVA 318
Query: 308 SSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAG 367
++ R E GL+ WAPQ+ +L H AVG F+TH GW S +EGV +G
Sbjct: 319 AAP-----------RGFLERAPGLVVP-WAPQVGVLRHAAVGAFVTHAGWASVMEGVSSG 366
Query: 368 VPMATWPLSAEQFINEKLITDVLRIGVQVGSREWGSWDEERKELVGREKVELAVKKLMAE 427
VPMA P +Q +N + + V G + R V AV L+
Sbjct: 367 VPMACRPFFGDQTMNARSVASVWGFGTAFDGP------------MTRGAVANAVATLL-R 413
Query: 428 SEDTEEMRRRVKSIFENAKRAVEEGG 453
ED E MR + + + +A E G
Sbjct: 414 GEDGERMRAKAQELQAMVGKAFEPDG 439
>UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-McC allele)
Length = 471
Score = 98.6 bits (244), Expect = 3e-20
Identities = 68/204 (33%), Positives = 96/204 (46%), Gaps = 24/204 (11%)
Query: 252 DEQSCLNWLNSKKPNSVLYISFGSVARVPMKQLKEIAYGLEASDQSFIWVVGKILNSSKN 311
D CL WL + V Y+SFG+VA +L+E+A GLE S F+W S
Sbjct: 274 DPHGCLAWLGRQPARGVAYVSFGTVACPRPDELRELAAGLEDSGAPFLW--------SLR 325
Query: 312 EEDW--VLDKFERRMKEMDKGLIFRGWAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVP 369
E+ W + F R GL+ WAPQ+ +L H +VG F+TH GW S LEG+ +GVP
Sbjct: 326 EDSWPHLPPGFLDRAAGTGSGLVVP-WAPQVAVLRHPSVGAFVTHAGWASVLEGLSSGVP 384
Query: 370 MATWPLSAEQFINEKLITDVLRIGVQVGSREWGSWDEERKELVGREKVELAVKKLMAESE 429
MA P +Q +N + + V WG + + + V AV++L+ E
Sbjct: 385 MACRPFFGDQRMNARSVAHV-----------WG-FGAAFEGAMTSAGVATAVEELL-RGE 431
Query: 430 DTEEMRRRVKSIFENAKRAVEEGG 453
+ MR R K + A GG
Sbjct: 432 EGARMRARAKELQALVAEAFGPGG 455
>UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Fragment)
Length = 154
Score = 72.4 bits (176), Expect = 2e-12
Identities = 42/128 (32%), Positives = 66/128 (50%), Gaps = 12/128 (9%)
Query: 336 WAPQLLILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQ 395
WAPQ +L G F+THCGWNS E V GVP+ P +Q +N +++ DVL IGV+
Sbjct: 30 WAPQAEVLALRQFGAFVTHCGWNSLWESVAGGVPLICRPFFGDQRLNGRMVEDVLEIGVR 89
Query: 396 VGSREWGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSS 455
+ E G V + ++ + E +++R ++++ E A RAV GSS
Sbjct: 90 I---EGG---------VFTKSGLMSCFDQILSQEKGKKLRENLRALRETADRAVGPKGSS 137
Query: 456 YDDIHALI 463
++ L+
Sbjct: 138 TENFKTLV 145
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 56,538,122
Number of Sequences: 164201
Number of extensions: 2421432
Number of successful extensions: 7378
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 7219
Number of HSP's gapped (non-prelim): 115
length of query: 471
length of database: 59,974,054
effective HSP length: 114
effective length of query: 357
effective length of database: 41,255,140
effective search space: 14728084980
effective search space used: 14728084980
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC136449.7


