

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135161.4 + phase: 0
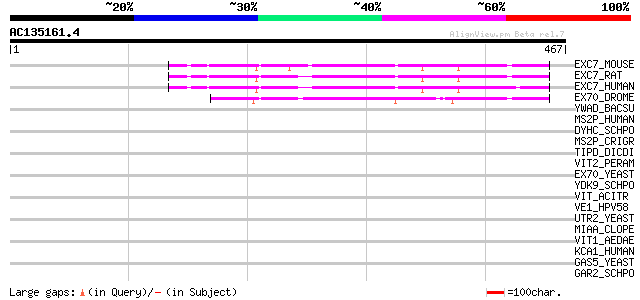
(467 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
EXC7_MOUSE (O35250) Exocyst complex component 7 (Exocyst complex... 75 5e-13
EXC7_RAT (O54922) Exocyst complex component 7 (Exocyst complex c... 69 2e-11
EXC7_HUMAN (Q9UPT5) Exocyst complex component 7 (Exocyst complex... 69 3e-11
EX70_DROME (Q9VSJ8) 70 kDa exocyst complex protein 65 4e-10
YWAD_BACSU (P25152) Hypothetical peptidase ywaD precursor (EC 3.... 38 0.049
MS2P_HUMAN (O43462) Membrane-bound transcription factor site 2 p... 37 0.083
DYHC_SCHPO (O13290) Dynein heavy chain, cytosolic (DYHC) 36 0.24
MS2P_CRIGR (O54862) Membrane-bound transcription factor site 2 p... 35 0.32
TIPD_DICDI (O15736) Protein tipD 33 1.2
VIT2_PERAM (Q9BPS0) Vitellogenin 2 precursor (Vg-2) 33 2.0
EX70_YEAST (P19658) 70 kDa exocyst complex protein 33 2.0
YDK9_SCHPO (P87115) Hypothetical UPF0202 protein C20G8.09c in ch... 32 2.7
VIT_ACITR (Q90243) Vitellogenin precursor (VTG) [Contains: Lipov... 32 2.7
VE1_HPV58 (P26543) Replication protein E1 32 2.7
UTR2_YEAST (P32623) UTR2 protein (Unknown transcript 2 protein) 32 2.7
MIAA_CLOPE (Q8XL85) tRNA delta(2)-isopentenylpyrophosphate trans... 32 2.7
VIT1_AEDAE (Q16927) Vitellogenin A1 precursor (VG) (PVG1) [Conta... 32 3.5
KCA1_HUMAN (Q12791) Calcium-activated potassium channel alpha su... 32 3.5
GAS5_YEAST (Q08193) Glycolipid anchored surface protein 5 precursor 32 3.5
GAR2_SCHPO (P41891) Protein gar2 32 3.5
>EXC7_MOUSE (O35250) Exocyst complex component 7 (Exocyst complex
component Exo70)
Length = 697
Score = 74.7 bits (182), Expect = 5e-13
Identities = 78/337 (23%), Positives = 154/337 (45%), Gaps = 29/337 (8%)
Query: 134 VIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTL 193
+I +D S +F +L L P F+ + + S +N+L ++ + L
Sbjct: 370 IIRHDFSTVLTVFPIL---RHLKQTKPEFDQVL-QGTAASTKNKLPGLITSMETIGAKAL 425
Query: 194 REFENTIRSKGPG--NAPFFGGQLHPLVRFVMNFLTWICDYRE-----ILEQVFEDHGHV 246
+F + I++ N P G +H L + FL + D++E + QV D ++
Sbjct: 426 EDFADNIKNDPDKEYNMPK-DGTVHELTSNAILFLQQLLDFQETAGAMLASQVLGDTYNI 484
Query: 247 LLEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMN 306
L+ +T S++S SS S S + +++ L+ L + ++ DP L ++L N
Sbjct: 485 PLDPR---ETSSSATSYSSEFSKRLLSTYICKVLGNLQLNLLSKSKVYEDPALSAIFLHN 541
Query: 307 SSRYIIIKTMENELGTLLGDGMLQRHSAK-LRYNFEEYIRS---SWGKVLEFLRLDNNLL 362
+ YI+ ++EL L+ + Q+ + + R + E+ I++ SW KV +++ N +
Sbjct: 542 NYNYILKSLEKSELIQLV--AVTQKTAERSYREHIEQQIQTYQRSWLKVTDYIAEKNLPV 599
Query: 363 VHPNMVGKSMKKQL-----KSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYT 417
P + + ++Q+ K FN E+CK Q +W I D +++I + + Y
Sbjct: 600 FQPGVKLRDKERQMIKERFKGFNDGLEELCKIQKVWAIPDTEQRDKIRQAQKDIVKETYG 659
Query: 418 NFIRKLHIVLKLEVKKPPDGYIEYETKDIKAILNNMF 454
F LH + K P+ YI+Y + + +++ +F
Sbjct: 660 AF---LHRYGSVPFTKNPEKYIKYRVEQVGDMIDRLF 693
>EXC7_RAT (O54922) Exocyst complex component 7 (Exocyst complex
component Exo70) (rExo70)
Length = 653
Score = 69.3 bits (168), Expect = 2e-11
Identities = 73/332 (21%), Positives = 145/332 (42%), Gaps = 32/332 (9%)
Query: 134 VIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTL 193
+I +D S +F +L L P F+ + + S +N+L ++ + L
Sbjct: 339 IIRHDFSTVLTVFPIL---RHLKQTKPEFDQVL-QGTAASTKNKLPGLITSMETIGAKAL 394
Query: 194 REFENTIRSKGPG--NAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYT 251
+F + I++ N P G +H L + FL + D++E +
Sbjct: 395 EDFADNIKNDPDKEYNMPK-DGTVHELTSNAILFLQQLLDFQETAGAMLASQ-------- 445
Query: 252 KHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYI 311
+T S++S +S S S + +++ L+ L + ++ DP L ++L N+ YI
Sbjct: 446 ---ETSSSATSYNSEFSKRLLSTYICKVLGNLQLNLLSKSKVYEDPALSAIFLHNNYNYI 502
Query: 312 IIKTMENELGTLLGDGMLQRHSAK-LRYNFEEYIRS---SWGKVLEFLRLDNNLLVHPNM 367
+ ++EL L+ + Q+ + + R + E+ I++ SW KV +++ N + P +
Sbjct: 503 LKSLEKSELIQLV--AVTQKTAERSYREHIEQQIQTYQRSWLKVTDYIAEKNLPVFQPGV 560
Query: 368 VGKSMKKQL-----KSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRK 422
+ ++Q+ K FN E+CK Q W I D +++I + Y F
Sbjct: 561 KLRDKERQMIKERFKGFNDGLEELCKIQKAWAIPDTEQRDKIRQAQKSIVKETYGAF--- 617
Query: 423 LHIVLKLEVKKPPDGYIEYETKDIKAILNNMF 454
LH + K P+ YI+Y + + +++ +F
Sbjct: 618 LHRYSSVPFTKNPEKYIKYRVEQVGDMIDRLF 649
>EXC7_HUMAN (Q9UPT5) Exocyst complex component 7 (Exocyst complex
component Exo70)
Length = 735
Score = 68.9 bits (167), Expect = 3e-11
Identities = 73/332 (21%), Positives = 145/332 (42%), Gaps = 32/332 (9%)
Query: 134 VIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNELNTVLKKLGETIVGTL 193
++ +D S +F +L L P F+ + + S +N+L ++ + L
Sbjct: 421 IVRHDFSTVLTVFPIL---RHLKQTKPEFDQVL-QGTAASTKNKLPGLITSMETIGAKAL 476
Query: 194 REFENTIRSKGPG--NAPFFGGQLHPLVRFVMNFLTWICDYREILEQVFEDHGHVLLEYT 251
+F + I++ N P G +H L + FL + D++E +
Sbjct: 477 EDFADNIKNDPDKEYNMPK-DGTVHELTSNAILFLQQLLDFQETAGAMLASQ-------- 527
Query: 252 KHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYI 311
+T S++S SS S S + +++ L+ L + ++ DP L ++L N+ YI
Sbjct: 528 ---ETSSSATSYSSEFSKRLLSTYICKVLGNLQLNLLSKSKVYEDPALSAIFLHNNYNYI 584
Query: 312 IIKTMENELGTLLGDGMLQRHSAK-LRYNFEEYIRS---SWGKVLEFLRLDNNLLVHPNM 367
+ ++EL L+ + Q+ + + R + E+ I++ SW KV +++ N + P +
Sbjct: 585 LKSLEKSELIQLV--AVTQKTAERSYREHIEQQIQTYQRSWLKVTDYIAEKNLPVFQPGV 642
Query: 368 VGKSMKKQL-----KSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLLPAYTNFIRK 422
+ ++Q+ K FN E+CK Q W I D ++ I + Y F++K
Sbjct: 643 KLRDKERQIIKERFKGFNDGLEELCKIQKAWAIPDTEQRDRIRQAQKTIVKETYGAFLQK 702
Query: 423 LHIVLKLEVKKPPDGYIEYETKDIKAILNNMF 454
V K P+ YI+Y + + +++ +F
Sbjct: 703 FGSV---PFTKNPEKYIKYGVEQVGDMIDRLF 731
>EX70_DROME (Q9VSJ8) 70 kDa exocyst complex protein
Length = 693
Score = 65.1 bits (157), Expect = 4e-10
Identities = 68/301 (22%), Positives = 128/301 (41%), Gaps = 26/301 (8%)
Query: 170 YSVSLRNELNTVLKKLGETIVGTLREFENTIRSK------GPGNAPFFGGQLHPLVRFVM 223
Y + R +L VLKKL T L F + ++ + G N P +H L +
Sbjct: 399 YDPAQREQLKKVLKKLQHTGAKALEHFLDVVKGESSTNIVGQSNVPK-DATVHELTSNTI 457
Query: 224 NFLTWICDYREILEQVFEDHGHVLLEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVL 283
F+ + D+ +++ + +L T+ D + + + + ++ +++ + L
Sbjct: 458 WFIEHLYDHFDVIGSILAQD---VLYSTQLDTILMKKALPVEERNKALLAIYIKKALAEL 514
Query: 284 ESKLEAMFNIFNDPTLGYVYLMNSSRYIIIKTMENELGTL--LGDGMLQRHSAKLRYNFE 341
+ +ND +++ +N+ YI+ + L L L + + ++ +
Sbjct: 515 NLSIMNKCEQYNDQATKHLFRLNNIHYILKSLQRSNLIDLVTLAEPECEHSYMEMIRELK 574
Query: 342 EYIRSSWGKVLEFL-RLDNNLLVHPNMVGKS-------MKKQLKSFNKLFNEICKAQSLW 393
+ +W K+L + LD L P + GK +K++ +FNK F E CK Q
Sbjct: 575 ASYQKTWSKMLVGIYSLDE--LPKP-VAGKVKDKDRSVLKERFSNFNKDFEEACKIQRGI 631
Query: 394 FIMDETLKEEIIVYLGENLLPAYTNFIRKLHIVLKLEVKKPPDGYIEYETKDIKAILNNM 453
I D L+E I E++LP Y F I + K PD Y++Y +I A+L+ +
Sbjct: 632 SIPDVILREGIKRDNVEHILPIYNRF---YEIYSGVHFSKNPDKYVKYRQHEINAMLSKL 688
Query: 454 F 454
F
Sbjct: 689 F 689
>YWAD_BACSU (P25152) Hypothetical peptidase ywaD precursor (EC
3.4.11.-)
Length = 455
Score = 38.1 bits (87), Expect = 0.049
Identities = 28/104 (26%), Positives = 47/104 (44%), Gaps = 18/104 (17%)
Query: 251 TKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRY 310
T H D+VP S ++ + S +S L+M R+++ + S E F F LG L+ SS Y
Sbjct: 248 TAHYDSVPFSPGANDNGSGTSVMLEMARVLKSVPSDKEIRFIAFGAEELG---LLGSSHY 304
Query: 311 IIIKTMENELGTLLGDGMLQRHSAKLRYNFE-EYIRSSWGKVLE 353
+ D + ++ + NF + + +SW K E
Sbjct: 305 V--------------DHLSEKELKRSEVNFNLDMVGTSWEKASE 334
>MS2P_HUMAN (O43462) Membrane-bound transcription factor site 2
protease (EC 3.4.24.85) (S2P endopeptidase) (Site-2
protease) (Sterol-regulatory element-binding proteins
intramembrane protease)
Length = 519
Score = 37.4 bits (85), Expect = 0.083
Identities = 19/34 (55%), Positives = 26/34 (75%)
Query: 250 YTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVL 283
Y+ + SSSSSSSSSSSSSSSL E++++V+
Sbjct: 113 YSSSSSSSSSSSSSSSSSSSSSSSLHNEQVLQVV 146
>DYHC_SCHPO (O13290) Dynein heavy chain, cytosolic (DYHC)
Length = 4196
Score = 35.8 bits (81), Expect = 0.24
Identities = 42/172 (24%), Positives = 71/172 (40%), Gaps = 11/172 (6%)
Query: 61 LNSDNLTIKDVNMEDLGLRIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISD--ISFT 118
L DN T+ N E + L+ ++K F A + T + S I D +S T
Sbjct: 2264 LLDDNKTLTLSNGERIALQ--PYVKIFFEADSVASLTRATISRCGLICISNIDDNILSST 2321
Query: 119 DVCREFTIRLLNFPNVIANDQSNTTLLFRMLDMYETLHDLIPNFESLFCDQYSVSLRNEL 178
D FT N+P +ND+ +T + D E + +LI + C ++SV L++ +
Sbjct: 2322 DKMLSFTSGATNYPLGSSNDEFSTVFSKVLTD--EVMMNLISS-----CYKFSVDLQHIM 2374
Query: 179 NTVLKKLGETIVGTLREFENTIRSKGPGNAPFFGGQLHPLVRFVMNFLTWIC 230
N ++ T L + + RS + F + L + + L W C
Sbjct: 2375 NFTKQRFFTTFYSLLDQTKLFTRSSNITESLSFKELCNYLKKKICYILAWCC 2426
>MS2P_CRIGR (O54862) Membrane-bound transcription factor site 2
protease (EC 3.4.24.85) (S2P endopeptidase) (Site-2
protease) (Sterol-regulatory element-binding proteins
intramembrane protease)
Length = 510
Score = 35.4 bits (80), Expect = 0.32
Identities = 18/30 (60%), Positives = 24/30 (80%)
Query: 254 DDTVPSSSSSSSSSSSSSSSLQMERIMEVL 283
D SSSSSSSSSSSSSSS+ E++++V+
Sbjct: 108 DSPSSSSSSSSSSSSSSSSSIHNEQVLQVV 137
>TIPD_DICDI (O15736) Protein tipD
Length = 612
Score = 33.5 bits (75), Expect = 1.2
Identities = 22/68 (32%), Positives = 36/68 (52%), Gaps = 5/68 (7%)
Query: 258 PSSSSSSSSSSSSSSSLQMERIMEVLESKLEAMFNIFNDPTLGYVYLMNSSRYIII---- 313
PS SS SSSSSSSS ++ + +E + +F + D T Y +++ I++
Sbjct: 105 PSGSSKMDSSSSSSSSNRVSGMGSTIEEMEQKLFKLQEDLTNSYKRNADNASSILLLNDK 164
Query: 314 -KTMENEL 320
K ++NEL
Sbjct: 165 NKDLQNEL 172
>VIT2_PERAM (Q9BPS0) Vitellogenin 2 precursor (Vg-2)
Length = 1876
Score = 32.7 bits (73), Expect = 2.0
Identities = 22/49 (44%), Positives = 27/49 (54%), Gaps = 4/49 (8%)
Query: 229 ICDYREILEQVFEDHGHV----LLEYTKHDDTVPSSSSSSSSSSSSSSS 273
IC ++ +H H + Y DD+ SSSSSSSSSSSSSSS
Sbjct: 1709 ICVREDVQLVNLTNHRHAEKSGIRPYDIDDDSSSSSSSSSSSSSSSSSS 1757
>EX70_YEAST (P19658) 70 kDa exocyst complex protein
Length = 623
Score = 32.7 bits (73), Expect = 2.0
Identities = 35/162 (21%), Positives = 74/162 (45%), Gaps = 15/162 (9%)
Query: 299 LGYVYLMNSSRYIIIKTME-NELGTLL-GDGMLQRHSAKLRYNFEEYIRSSWGKVLEFLR 356
+G+ LMN + ++ + +E +EL +L G+G + K RY Y+ S W + L
Sbjct: 472 IGFFILMNLT--LVEQIVEKSELNLMLAGEGHSRLERLKKRYI--SYMVSDWRDLTANL- 526
Query: 357 LDNNLLVHPNMVGKS---MKKQLKSFNKLFNEICKAQSLWFIMDETLKEEIIVYLGENLL 413
+D+ + K +K++ + FN+ F ++ + + D +LK + + ++
Sbjct: 527 MDSVFIDSSGKKSKDKEQIKEKFRKFNEGFEDLVSKTKQYKLSDPSLKVTLKSEIISLVM 586
Query: 414 PAYTNFIRKLHIVLKLEVKKPPDGYIEYETKDIKAILNNMFK 455
P Y F + + K P +I+Y ++ +LN + +
Sbjct: 587 PMYERFYSRYK-----DSFKNPRKHIKYTPDELTTVLNQLVR 623
>YDK9_SCHPO (P87115) Hypothetical UPF0202 protein C20G8.09c in
chromosome I
Length = 1033
Score = 32.3 bits (72), Expect = 2.7
Identities = 33/123 (26%), Positives = 50/123 (39%), Gaps = 20/123 (16%)
Query: 79 RIKRWIKAFKVAFKILFPTERQLCDIVFFEFSAISDISFTDVCREFTIRLLNFPNVIAND 138
R W+ AF F F + L F EF+AI+ +S D C T ++N + + N+
Sbjct: 757 RDSEWLGAFAQNFYRRFLS---LLGYQFREFAAITALSVLDACNNGTKYVVNSTSKLTNE 813
Query: 139 QSNTTLLFRMLDM------------YETLHDLIPNFESLFCD---QYSVSLRNELNTVLK 183
+ N +F D+ Y + DL+P L+ SV L +VL
Sbjct: 814 EINN--VFESYDLKRLESYSNNLLDYHVIVDLLPKLAHLYFSGKFPDSVKLSPVQQSVLL 871
Query: 184 KLG 186
LG
Sbjct: 872 ALG 874
>VIT_ACITR (Q90243) Vitellogenin precursor (VTG) [Contains:
Lipovitellin I (LVI); Phosvitin (PV); Lipovitellin II
(LVII)] (Fragment)
Length = 1677
Score = 32.3 bits (72), Expect = 2.7
Identities = 18/26 (69%), Positives = 21/26 (80%)
Query: 259 SSSSSSSSSSSSSSSLQMERIMEVLE 284
SSSSSSSSSSSSSSS Q R+ + +E
Sbjct: 1104 SSSSSSSSSSSSSSSSQQSRMEKRME 1129
Score = 32.0 bits (71), Expect = 3.5
Identities = 16/22 (72%), Positives = 18/22 (81%)
Query: 252 KHDDTVPSSSSSSSSSSSSSSS 273
KH +PSSSSSSSSSSS S+S
Sbjct: 1162 KHHKQLPSSSSSSSSSSSGSNS 1183
>VE1_HPV58 (P26543) Replication protein E1
Length = 644
Score = 32.3 bits (72), Expect = 2.7
Identities = 40/174 (22%), Positives = 78/174 (43%), Gaps = 21/174 (12%)
Query: 6 SLEGTLMLEALSLETVNNLQETVKLMLNSGFNKECLIV--YSSCRRECLEECLVK----- 58
S++ T EA + + N+QE V N C + +++C +E+C+ +
Sbjct: 54 SVQSTTQAEAEAARALFNVQEGV-----DDINAVCALKRKFAACSESAVEDCVDRAANVC 108
Query: 59 ---QFLNSD--NLTIKDVNMEDLGLRIKRWIKAFKVAFKILFPT-ERQLCDIVFFEFSAI 112
++ N + + K + +ED G ++ ++A ++ + L D A
Sbjct: 109 VSWKYKNKECTHRKRKIIELEDSGYGNTE-VETEQMAHQVESQNGDADLNDSESSGVGAS 167
Query: 113 SDISF-TDVCREFTIRLLNFPNVIANDQSNTTLLFRMLDMY-ETLHDLIPNFES 164
SD+S TDV T+ L N N++ N + TLL++ + Y + +L+ F+S
Sbjct: 168 SDVSSETDVDSCNTVPLQNISNILHNSNTKATLLYKFKEAYGVSFMELVRPFKS 221
>UTR2_YEAST (P32623) UTR2 protein (Unknown transcript 2 protein)
Length = 347
Score = 32.3 bits (72), Expect = 2.7
Identities = 18/32 (56%), Positives = 21/32 (65%)
Query: 242 DHGHVLLEYTKHDDTVPSSSSSSSSSSSSSSS 273
DH + +K T SSSSSSSSSSSSSS+
Sbjct: 253 DHSSSTKKSSKTSSTASSSSSSSSSSSSSSST 284
>MIAA_CLOPE (Q8XL85) tRNA delta(2)-isopentenylpyrophosphate
transferase (EC 2.5.1.8) (IPP transferase)
(Isopentenyl-diphosphate:tRNA isopentenyltransferase)
(IPTase) (IPPT)
Length = 310
Score = 32.3 bits (72), Expect = 2.7
Identities = 23/101 (22%), Positives = 46/101 (44%), Gaps = 1/101 (0%)
Query: 231 DYREILEQVFEDHGHVLL-EYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEA 289
+YRE LE++ +HG+ L E K D +S ++ +L+ ++ S +A
Sbjct: 122 EYREELEKIANEHGNEYLHEMLKDIDLESYNSIHFNNRKRVIRALETYKLTGKPFSSFKA 181
Query: 290 MFNIFNDPTLGYVYLMNSSRYIIIKTMENELGTLLGDGMLQ 330
+I+ P Y Y++N R + + + + G+L+
Sbjct: 182 KNSIYETPYNIYYYVLNMDRAKLYDRINKRVDIMFEKGLLE 222
>VIT1_AEDAE (Q16927) Vitellogenin A1 precursor (VG) (PVG1) [Contains:
Vitellin light chain (VL); Vitellin heavy chain (VH)]
Length = 2148
Score = 32.0 bits (71), Expect = 3.5
Identities = 18/37 (48%), Positives = 22/37 (58%)
Query: 241 EDHGHVLLEYTKHDDTVPSSSSSSSSSSSSSSSLQME 277
+++ H Y H+ SSSSSSSSS SSSSS E
Sbjct: 1990 QEYYHPRYRYYNHNVEESSSSSSSSSSDSSSSSSSSE 2026
>KCA1_HUMAN (Q12791) Calcium-activated potassium channel alpha
subunit 1 (Calcium-activated potassium channel,
subfamily M, alpha subunit 1) (Maxi K channel) (MaxiK)
(BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1)
(Slowpoke homolog) (Slo homolog) (Slo-al
Length = 1236
Score = 32.0 bits (71), Expect = 3.5
Identities = 18/35 (51%), Positives = 24/35 (68%)
Query: 256 TVPSSSSSSSSSSSSSSSLQMERIMEVLESKLEAM 290
++ +SSSSSSSSSSSSSS V E K++A+
Sbjct: 35 SLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDAL 69
>GAS5_YEAST (Q08193) Glycolipid anchored surface protein 5 precursor
Length = 484
Score = 32.0 bits (71), Expect = 3.5
Identities = 17/20 (85%), Positives = 18/20 (90%)
Query: 254 DDTVPSSSSSSSSSSSSSSS 273
DDT SSSSSSSSSSS+SSS
Sbjct: 402 DDTSSSSSSSSSSSSSASSS 421
Score = 30.8 bits (68), Expect = 7.8
Identities = 16/22 (72%), Positives = 18/22 (81%)
Query: 252 KHDDTVPSSSSSSSSSSSSSSS 273
+ DD SSSSSSSSSSSS+SS
Sbjct: 399 EEDDDTSSSSSSSSSSSSSASS 420
>GAR2_SCHPO (P41891) Protein gar2
Length = 500
Score = 32.0 bits (71), Expect = 3.5
Identities = 20/48 (41%), Positives = 25/48 (51%)
Query: 241 EDHGHVLLEYTKHDDTVPSSSSSSSSSSSSSSSLQMERIMEVLESKLE 288
E+ V +E K + SS SSSS S S SSS + E EV+E E
Sbjct: 149 EEEAVVKIEEKKESSSDSSSESSSSESESESSSSESEEEEEVVEKTEE 196
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,974,895
Number of Sequences: 164201
Number of extensions: 2229104
Number of successful extensions: 12629
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 11978
Number of HSP's gapped (non-prelim): 285
length of query: 467
length of database: 59,974,054
effective HSP length: 114
effective length of query: 353
effective length of database: 41,255,140
effective search space: 14563064420
effective search space used: 14563064420
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 68 (30.8 bits)
Medicago: description of AC135161.4


