

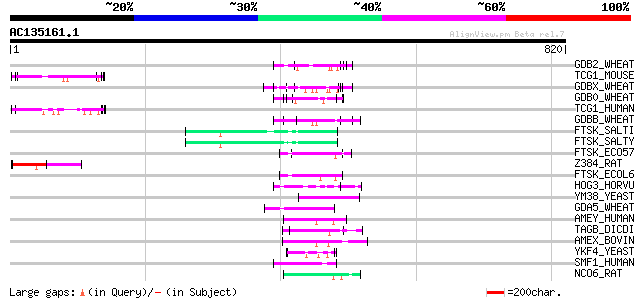
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135161.1 - phase: 0
(820 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
GDB2_WHEAT (P08453) Gamma-gliadin precursor 74 2e-12
TCG1_MOUSE (Q8CGF7) Transcription elongation regulator 1 (TATA b... 65 7e-10
GDBX_WHEAT (P21292) Gamma-gliadin precursor 64 2e-09
GDB0_WHEAT (P08079) Gamma-gliadin precursor (Fragment) 64 2e-09
TCG1_HUMAN (O14776) Transcription elongation regulator 1 (TATA b... 63 4e-09
GDBB_WHEAT (P06659) Gamma-gliadin B precursor 61 1e-08
FTSK_SALTI (Q8Z814) DNA translocase ftsK 61 1e-08
FTSK_SALTY (Q8ZQD5) DNA translocase ftsK 61 1e-08
FTSK_ECO57 (Q8X5H9) DNA translocase ftsK 58 1e-07
Z384_RAT (Q9EQJ4) Zinc finger protein 384 (Nuclear matrix transc... 57 2e-07
FTSK_ECOL6 (Q8FJC7) DNA translocase ftsK 57 2e-07
HOG3_HORVU (P80198) Gamma-hordein 3 56 3e-07
YM38_YEAST (Q03825) Hypothetical 85.0 kDa protein in HLJ1-SMP2 i... 55 6e-07
GDA5_WHEAT (P04725) Alpha/beta-gliadin A-V precursor (Prolamin) 55 7e-07
AMEY_HUMAN (Q99218) Amelogenin, Y isoform precursor 55 7e-07
TAGB_DICDI (P54683) Prestalk-specific protein tagB precursor (EC... 55 1e-06
AMEX_BOVIN (P02817) Amelogenin, class I precursor 55 1e-06
YKF4_YEAST (P35732) Hypothetical 84.0 kDa protein in NUP120-CSE4... 54 1e-06
SMF1_HUMAN (O14497) SWI/SNF-related, matrix-associated, actin-de... 54 1e-06
NCO6_RAT (Q9JLI4) Nuclear receptor coactivator 6 (Amplified in b... 54 1e-06
>GDB2_WHEAT (P08453) Gamma-gliadin precursor
Length = 327
Score = 73.9 bits (180), Expect = 2e-12
Identities = 52/117 (44%), Positives = 59/117 (49%), Gaps = 18/117 (15%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQI-PQ--YPQFPQNPSPQNVQPQNVQ 446
PQQT+P Q P QQ PQ PQ P + PQ +PQ PQ P PQ QPQ
Sbjct: 48 PQQTFPQPQQTFPHQP-----QQQVPQPQQPQQPFLQPQQPFPQQPQQPFPQTQQPQ--- 99
Query: 447 QQNFQQQPYQQYPYQQYPQQNFQQQPYQ-----QRPQQPRPP-RMPINPIPVTYAEL 497
Q F QQP Q +P Q PQQ F QQP Q Q+PQQP P + P P P +L
Sbjct: 100 -QPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQLQQPQQPFPQPQQQL 155
Score = 66.6 bits (161), Expect = 2e-10
Identities = 50/131 (38%), Positives = 59/131 (44%), Gaps = 18/131 (13%)
Query: 390 PQQTYP--AYQHIAAITPTSHPFQQTNN-HPQIPQYP----QIPQ--YPQFPQNPSPQNV 440
PQQT+P Q + PF Q PQ PQ P Q PQ +PQ PQ P PQ
Sbjct: 55 PQQTFPHQPQQQVPQPQQPQQPFLQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQ 114
Query: 441 QPQNVQQQNFQQQPYQQYPYQQYPQQNFQQ-----QPYQQRPQQPRPPRMPINPIPVTYA 495
QPQ Q F QQP Q +P Q PQQ F Q QP+ Q QQ P+ P P
Sbjct: 115 QPQ----QPFPQQPQQPFPQTQQPQQPFPQLQQPQQPFPQPQQQLPQPQQPQQSFPQQQR 170
Query: 496 ELLPGLLKKNL 506
+ L++ L
Sbjct: 171 PFIQPSLQQQL 181
Score = 62.8 bits (151), Expect = 4e-09
Identities = 49/110 (44%), Positives = 53/110 (47%), Gaps = 19/110 (17%)
Query: 391 QQTYPAYQHIAAITPTSHPFQQTNNHPQ--IPQYPQIPQYPQFPQNPSPQNVQPQNVQQQ 448
QQ P Q P S QQT PQ P PQ Q PQ PQ P +QPQ Q
Sbjct: 34 QQLVPQLQQ-----PLSQQPQQTFPQPQQTFPHQPQ-QQVPQ-PQQPQQPFLQPQ----Q 82
Query: 449 NFQQQPYQQYPYQQYPQQNFQQQPYQ-----QRPQQPRPPRMPINPIPVT 493
F QQP Q +P Q PQQ F QQP Q Q+PQQP P+ P P P T
Sbjct: 83 PFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPF-PQQPQQPFPQT 131
Score = 60.1 bits (144), Expect = 2e-08
Identities = 44/102 (43%), Positives = 49/102 (47%), Gaps = 8/102 (7%)
Query: 390 PQQTYPAYQHIAAITPTS--HPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQ 447
PQQ +P Q P PF QT PQ P +PQ PQ P FPQ PQ PQ Q
Sbjct: 88 PQQPFPQTQQPQQPFPQQPQQPFPQTQQ-PQQP-FPQQPQQP-FPQTQQPQQPFPQLQQP 144
Query: 448 QNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINP 489
Q QP QQ P Q PQQ+F P QQRP + +NP
Sbjct: 145 QQPFPQPQQQLPQPQQPQQSF---PQQQRPFIQPSLQQQLNP 183
Score = 55.5 bits (132), Expect = 6e-07
Identities = 36/84 (42%), Positives = 41/84 (47%), Gaps = 14/84 (16%)
Query: 421 QYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNF--QQQPYQQRPQ 478
Q +PQ Q P + PQ PQ QQ F QP QQ P Q PQQ F QQP+ Q+PQ
Sbjct: 33 QQQLVPQLQQ-PLSQQPQQTFPQ--PQQTFPHQPQQQVPQPQQPQQPFLQPQQPFPQQPQ 89
Query: 479 QPRP---------PRMPINPIPVT 493
QP P P+ P P P T
Sbjct: 90 QPFPQTQQPQQPFPQQPQQPFPQT 113
Score = 34.7 bits (78), Expect = 1.0
Identities = 26/58 (44%), Positives = 29/58 (49%), Gaps = 12/58 (20%)
Query: 446 QQQNFQQQ--PYQQYPYQQYPQQNFQQ-------QPYQQ--RPQQPRPPRM-PINPIP 491
Q Q QQQ P Q P Q PQQ F Q QP QQ +PQQP+ P + P P P
Sbjct: 28 QVQWLQQQLVPQLQQPLSQQPQQTFPQPQQTFPHQPQQQVPQPQQPQQPFLQPQQPFP 85
>TCG1_MOUSE (Q8CGF7) Transcription elongation regulator 1 (TATA
box-binding protein-associated factor 2S) (Transcription
factor CA150) (p144) (Formin-binding protein 28) (FBP
28)
Length = 1100
Score = 65.1 bits (157), Expect = 7e-10
Identities = 53/138 (38%), Positives = 69/138 (49%), Gaps = 18/138 (13%)
Query: 9 KTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEAS-SSWTL 67
+ A+ Q QAQAQ QAQAQA AQAQ QAQA +AQAQ+QA + QA+A + +
Sbjct: 206 QAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQA-------QAQAQAQVQAQAV 258
Query: 68 CADTPRQSAP----QRSAPWFPPFTAGEIFRPITCEAQMPTHQYTA-QVPLPAMRVTPAT 122
A TP S+P S P P + T AQ + T Q P A+ V T
Sbjct: 259 GAPTPTTSSPAPAVSTSTPTSTPSSTTATTTTATSVAQTVSTPTTQDQTPSSAVSVATPT 318
Query: 123 MTYSAPV-----IHTIPQ 135
++ SAP + T+PQ
Sbjct: 319 VSVSAPAPTATPVQTVPQ 336
Score = 63.2 bits (152), Expect = 3e-09
Identities = 48/138 (34%), Positives = 64/138 (45%), Gaps = 6/138 (4%)
Query: 3 EMAELIKTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEAS 62
E+ ++ A+ Q QAQAQ QAQAQA AQAQ QAQA +AQAQ+QA + QA+A
Sbjct: 172 ELTPMLAAQAQVQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQA---QAQAQAQAQAQ 228
Query: 63 SSWTLCADTPRQSAPQRSAPWFPPFTAGEIFRPITCEAQMPTHQYTAQVPLPAMRVTPAT 122
+ A Q+ Q A A + P T P + P T AT
Sbjct: 229 AQAQAQAQAQAQAQAQAQAQAQAQVQAQAVGAP-TPTTSSPAPAVSTSTPTSTPSSTTAT 287
Query: 123 MTYSAPVIHTI--PQTEE 138
T + V T+ P T++
Sbjct: 288 TTTATSVAQTVSTPTTQD 305
Score = 50.8 bits (120), Expect = 1e-05
Identities = 41/124 (33%), Positives = 52/124 (41%), Gaps = 8/124 (6%)
Query: 9 KTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEASSSWTLC 68
+ A+ Q QAQAQ QAQ AQAQ QAQAV + +P P T SS T
Sbjct: 232 QAQAQAQAQAQAQAQAQ----AQAQVQAQAVGAPTPTTSSPAPAVSTSTPTSTPSSTTAT 287
Query: 69 ADTPRQSAPQRSAPWF---PPFTAGEIFRP-ITCEAQMPTHQYTAQVPLPAMRVTPATMT 124
T A S P P +A + P ++ A PT VP P + P +
Sbjct: 288 TTTATSVAQTVSTPTTQDQTPSSAVSVATPTVSVSAPAPTATPVQTVPQPHPQTLPPAVP 347
Query: 125 YSAP 128
+S P
Sbjct: 348 HSVP 351
Score = 46.6 bits (109), Expect = 3e-04
Identities = 42/128 (32%), Positives = 54/128 (41%), Gaps = 17/128 (13%)
Query: 12 AETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEASSSWTLCADT 71
+E AQ Q QAQA AQAQ QAQA +AQAQ+QA + QA+A + A
Sbjct: 171 SELTPMLAAQAQVQAQAQAQAQAQAQAQAQAQAQAQA-------QAQAQAQAQAQAQAQA 223
Query: 72 PRQSAPQRSAPWFPPFTAGEIFRPITCEAQMPTHQYTAQVPLPAMRVTPATMTYSAPVIH 131
Q+ Q A + +AQ Q AQV A+ T + AP +
Sbjct: 224 QAQAQAQAQA-------QAQAQAQAQAQAQA---QAQAQVQAQAVGAPTPTTSSPAPAVS 273
Query: 132 TIPQTEEP 139
T T P
Sbjct: 274 TSTPTSTP 281
>GDBX_WHEAT (P21292) Gamma-gliadin precursor
Length = 302
Score = 63.9 bits (154), Expect = 2e-09
Identities = 48/107 (44%), Positives = 56/107 (51%), Gaps = 13/107 (12%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQ--IPQYPQIPQYPQFPQNPSPQNVQPQNV-- 445
PQQTYP +Q T P QQ PQ PQ PQ+P +PQ PQ P PQ QPQ
Sbjct: 70 PQQTYP-HQPQQQFPQTQQP-QQPFPQPQQTFPQQPQLP-FPQQPQQPFPQPQQPQQPFP 126
Query: 446 QQQNFQQ---QPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINP 489
Q Q QQ QP QQ+P Q PQQ+F P QQ+P + +NP
Sbjct: 127 QSQQPQQPFPQPQQQFPQPQQPQQSF---PQQQQPAIQSFLQQQMNP 170
Score = 56.6 bits (135), Expect = 3e-07
Identities = 45/137 (32%), Positives = 58/137 (41%), Gaps = 21/137 (15%)
Query: 375 PRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQN 434
P++ + H PQQT+P Q ++P Q PQ Q PQ P FPQ
Sbjct: 48 PQRTIPQPHQTFHHQPQQTFPQPQQ-------TYPHQPQQQFPQTQQ----PQQP-FPQ- 94
Query: 435 PSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNF-----QQQPYQQRPQQPRPPRMPINP 489
PQ PQ Q F QQP Q +P Q PQQ F QQP+ Q QQ P+ P
Sbjct: 95 --PQQTFPQQPQLP-FPQQPQQPFPQPQQPQQPFPQSQQPQQPFPQPQQQFPQPQQPQQS 151
Query: 490 IPVTYAELLPGLLKKNL 506
P + L++ +
Sbjct: 152 FPQQQQPAIQSFLQQQM 168
Score = 53.9 bits (128), Expect = 2e-06
Identities = 40/102 (39%), Positives = 46/102 (44%), Gaps = 10/102 (9%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQ-YPQFPQNPSPQNVQPQNV--- 445
PQ P Q P H QT +H +PQ Q YP PQ PQ QPQ
Sbjct: 38 PQPQQPFCQQPQRTIPQPH---QTFHHQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQ 94
Query: 446 QQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRP-QQPRPPRMP 486
QQ F QQP Q P+ Q PQQ F Q Q+P Q + P+ P
Sbjct: 95 PQQTFPQQP--QLPFPQQPQQPFPQPQQPQQPFPQSQQPQQP 134
Score = 52.4 bits (124), Expect = 5e-06
Identities = 41/99 (41%), Positives = 46/99 (46%), Gaps = 20/99 (20%)
Query: 409 PFQQTNNHPQIP--QYPQ--IPQYPQ-FPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQY 463
P QQ PQ P Q PQ IPQ Q F P QP QQ + QP QQ+P Q
Sbjct: 32 PQQQPFPQPQQPFCQQPQRTIPQPHQTFHHQPQQTFPQP----QQTYPHQPQQQFPQTQQ 87
Query: 464 PQQNFQQQ----------PYQQRPQQPRP-PRMPINPIP 491
PQQ F Q P+ Q+PQQP P P+ P P P
Sbjct: 88 PQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFP 126
Score = 51.2 bits (121), Expect = 1e-05
Identities = 35/82 (42%), Positives = 40/82 (48%), Gaps = 14/82 (17%)
Query: 421 QYPQIPQYPQFPQNP---SPQNVQPQNVQQQNFQQQPYQQYPYQQ--YPQQNFQQQPYQQ 475
Q+PQ +PQ PQ P PQ PQ Q F QP Q +P Q YP Q QQ P Q
Sbjct: 30 QWPQQQPFPQ-PQQPFCQQPQRTIPQ--PHQTFHHQPQQTFPQPQQTYPHQPQQQFPQTQ 86
Query: 476 RPQQPRP------PRMPINPIP 491
+PQQP P P+ P P P
Sbjct: 87 QPQQPFPQPQQTFPQQPQLPFP 108
Score = 34.3 bits (77), Expect = 1.4
Identities = 29/75 (38%), Positives = 34/75 (44%), Gaps = 9/75 (12%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQP--QNVQQ 447
PQQ +P Q P S QQ PQ Q+PQ PQ PQ PQ QP Q+ Q
Sbjct: 111 PQQPFPQPQQPQQPFPQSQQPQQPFPQPQ-QQFPQ----PQQPQQSFPQQQQPAIQSFLQ 165
Query: 448 QNFQQQPYQQYPYQQ 462
Q Q P + + QQ
Sbjct: 166 Q--QMNPCKNFLLQQ 178
>GDB0_WHEAT (P08079) Gamma-gliadin precursor (Fragment)
Length = 251
Score = 63.5 bits (153), Expect = 2e-09
Identities = 47/106 (44%), Positives = 53/106 (49%), Gaps = 23/106 (21%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQ---------YPQIPQ--YPQFPQNPSPQ 438
PQQT+P Q P QQ PQ PQ +PQ PQ YPQ PQ P PQ
Sbjct: 48 PQQTFPQPQQTFPHQP-----QQQFPQPQQPQQQFLQPQQPFPQQPQQPYPQQPQQPFPQ 102
Query: 439 NVQPQNVQQQNFQQQPYQQY--PYQQYPQQNFQQQPYQQRPQQPRP 482
QPQ + Q+ QQP QQ+ P QQ+PQ QQP Q PQQ P
Sbjct: 103 TQQPQQLFPQS--QQPQQQFSQPQQQFPQ---PQQPQQSFPQQQPP 143
Score = 59.3 bits (142), Expect = 4e-08
Identities = 37/79 (46%), Positives = 42/79 (52%), Gaps = 8/79 (10%)
Query: 418 QIPQYPQIPQYPQ-FPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNF--QQQPYQ 474
Q PQ +PQ Q F Q P QP QQ F QP QQ+P Q PQQ F QQP+
Sbjct: 30 QWPQQQPVPQPHQPFSQQPQQTFPQP----QQTFPHQPQQQFPQPQQPQQQFLQPQQPFP 85
Query: 475 QRPQQPRPPRMPINPIPVT 493
Q+PQQP P+ P P P T
Sbjct: 86 QQPQQPY-PQQPQQPFPQT 103
Score = 53.5 bits (127), Expect = 2e-06
Identities = 37/81 (45%), Positives = 42/81 (51%), Gaps = 8/81 (9%)
Query: 405 PTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQ-QQPYQQYPYQQY 463
P P Q + PQ +PQ Q FP P Q QPQ QQQ Q QQP+ Q P Q Y
Sbjct: 36 PVPQPHQPFSQQPQ-QTFPQPQQ--TFPHQPQQQFPQPQQPQQQFLQPQQPFPQQPQQPY 92
Query: 464 PQQNFQQQPY--QQRPQQPRP 482
PQQ QQP+ Q+PQQ P
Sbjct: 93 PQQ--PQQPFPQTQQPQQLFP 111
Score = 53.5 bits (127), Expect = 2e-06
Identities = 46/102 (45%), Positives = 50/102 (48%), Gaps = 21/102 (20%)
Query: 405 PTSHPFQQTNNHPQ--IPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQ- 461
P S QQT PQ P PQ Q+PQ PQ P Q +QPQ Q F QQP Q YP Q
Sbjct: 43 PFSQQPQQTFPQPQQTFPHQPQ-QQFPQ-PQQPQQQFLQPQ----QPFPQQPQQPYPQQP 96
Query: 462 -------QYPQQNF--QQQPYQQ--RPQQPRP-PRMPINPIP 491
Q PQQ F QQP QQ +PQQ P P+ P P
Sbjct: 97 QQPFPQTQQPQQLFPQSQQPQQQFSQPQQQFPQPQQPQQSFP 138
Score = 47.4 bits (111), Expect = 2e-04
Identities = 36/76 (47%), Positives = 40/76 (52%), Gaps = 8/76 (10%)
Query: 409 PFQQTNNHPQIPQYPQIPQ--YPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQ 466
P QQ P P + Q PQ +PQ PQ P Q Q Q Q QQQ Q P Q +PQQ
Sbjct: 32 PQQQPVPQPHQP-FSQQPQQTFPQ-PQQTFPHQPQQQFPQPQQPQQQFLQ--PQQPFPQQ 87
Query: 467 NFQQQPYQQRPQQPRP 482
QQPY Q+PQQP P
Sbjct: 88 --PQQPYPQQPQQPFP 101
>TCG1_HUMAN (O14776) Transcription elongation regulator 1 (TATA
box-binding protein-associated factor 2S) (Transcription
factor CA150)
Length = 1098
Score = 62.8 bits (151), Expect = 4e-09
Identities = 45/133 (33%), Positives = 63/133 (46%), Gaps = 6/133 (4%)
Query: 9 KTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPP-PIRTQAEASSSWTL 67
+ A+ Q QAQAQ QAQAQA AQAQ QAQA +AQAQ+QA + QA+A + T
Sbjct: 202 QAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQVQAQVQAQVQAQAVGASTP 261
Query: 68 CADTPRQSAPQRSAPWFPPFTAGEIFRPITCEAQMPTHQYTAQVPLPAMRVTPATMTYSA 127
+P + ++ P T + + T Q P A+ V T++ S
Sbjct: 262 TTSSPAPAVSTSTSSSTPSSTTSTTTTATSVAQTVSTPTTQDQTPSSAVSVATPTVSVST 321
Query: 128 PV-----IHTIPQ 135
P + T+PQ
Sbjct: 322 PARTATPVQTVPQ 334
Score = 58.2 bits (139), Expect = 9e-08
Identities = 47/137 (34%), Positives = 64/137 (46%), Gaps = 29/137 (21%)
Query: 3 EMAELIKTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEAS 62
E+ ++ A+ Q QAQAQ QAQAQA AQAQ QAQA +AQAQ+QA + QA+A
Sbjct: 172 ELTPMLAAQAQVQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQA-------QAQAQAQ 224
Query: 63 SSWTLCADTPRQSAPQRSAPWFPPFTAGEIFRPITCEAQMPTHQYTAQVPLPAMRVTPAT 122
+ A Q+ Q A +AQ+ Q AQV A+ + T
Sbjct: 225 AQ----AQAQAQAQAQAQA-----------------QAQVQA-QVQAQVQAQAVGASTPT 262
Query: 123 MTYSAPVIHTIPQTEEP 139
+ AP + T + P
Sbjct: 263 TSSPAPAVSTSTSSSTP 279
Score = 56.2 bits (134), Expect = 3e-07
Identities = 45/137 (32%), Positives = 55/137 (39%), Gaps = 8/137 (5%)
Query: 9 KTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEASSSWTLC 68
+ A+ Q QAQAQ QAQ QA QAQ QAQAV + + +P P T + SS T
Sbjct: 226 QAQAQAQAQAQAQAQAQVQAQVQAQVQAQAVGASTPTTSSPAPAVSTSTSSSTPSSTTST 285
Query: 69 ADTPRQSAPQRSAPWFPPFTAGEIFRPITCEAQMPTHQYTA----QVPLPAMRVTPATMT 124
T A S P T T + T TA VP P + P +
Sbjct: 286 TTTATSVAQTVSTPTTQDQTPSSAVSVATPTVSVSTPARTATPVQTVPQPHPQTLPPAVP 345
Query: 125 YSAPVIHTIPQTEEPIF 141
+S P P T P F
Sbjct: 346 HSVPQ----PTTAIPAF 358
Score = 50.8 bits (120), Expect = 1e-05
Identities = 52/183 (28%), Positives = 67/183 (36%), Gaps = 54/183 (29%)
Query: 9 KTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQA--------PPPPPPIRTQAE 60
+ A+ Q QAQAQ QAQAQA AQAQ Q QA +AQ Q+QA P P + T
Sbjct: 216 QAQAQAQAQAQAQAQAQAQAQAQAQAQVQAQVQAQVQAQAVGASTPTTSSPAPAVSTSTS 275
Query: 61 ASS-----SWTLCAD------------------------------------TPRQSAPQR 79
+S+ S T A TP Q+ PQ
Sbjct: 276 SSTPSSTTSTTTTATSVAQTVSTPTTQDQTPSSAVSVATPTVSVSTPARTATPVQTVPQP 335
Query: 80 SAPWFPPFTAGEIFRPITC-EAQMPTHQYTAQVPLPAMRV----TPATMTYSAPVIHTIP 134
PP + +P T A P +VPLP M + S P + T+
Sbjct: 336 HPQTLPPAVPHSVPQPTTAIPAFPPVMVPPFRVPLPGMPIPLPGVAMMQIVSCPYVKTVA 395
Query: 135 QTE 137
T+
Sbjct: 396 TTK 398
>GDBB_WHEAT (P06659) Gamma-gliadin B precursor
Length = 291
Score = 61.2 bits (147), Expect = 1e-08
Identities = 46/117 (39%), Positives = 53/117 (44%), Gaps = 17/117 (14%)
Query: 405 PTSHPFQQTNNHPQIPQYPQ--IPQYPQ-FPQNPSPQNVQPQNVQQQNFQ---------Q 452
P PF Q H Q PQ PQ Q FP P Q QPQ QQQ Q Q
Sbjct: 32 PQQQPFLQP--HQPFSQQPQQIFPQPQQTFPHQPQQQFPQPQQPQQQFLQPRQPFPQQPQ 89
Query: 453 QPYQQYPYQQYPQQNFQQQPYQQ--RPQQPRP-PRMPINPIPVTYAELLPGLLKKNL 506
QPY Q P Q +PQ QQP+ Q +PQQP P P+ P P L+ L++ L
Sbjct: 90 QPYPQQPQQPFPQTQQPQQPFPQSKQPQQPFPQPQQPQQSFPQQQPSLIQQSLQQQL 146
Score = 61.2 bits (147), Expect = 1e-08
Identities = 40/100 (40%), Positives = 48/100 (48%), Gaps = 6/100 (6%)
Query: 424 QIPQYPQFPQNPSPQNVQPQNV---QQQNFQQQPYQQYPYQQYPQQNFQQ--QPYQQRPQ 478
Q PQ F Q P + QPQ + QQ F QP QQ+P Q PQQ F Q QP+ Q+PQ
Sbjct: 30 QWPQQQPFLQPHQPFSQQPQQIFPQPQQTFPHQPQQQFPQPQQPQQQFLQPRQPFPQQPQ 89
Query: 479 QPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEK 518
QP P+ P P P T P K Q P P++
Sbjct: 90 QPY-PQQPQQPFPQTQQPQQPFPQSKQPQQPFPQPQQPQQ 128
Score = 52.4 bits (124), Expect = 5e-06
Identities = 37/100 (37%), Positives = 44/100 (44%), Gaps = 6/100 (6%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQN 449
PQQT+P P Q PQ PQ P YPQ PQ P PQ QPQ Q+
Sbjct: 55 PQQTFPHQPQQQFPQPQQPQQQFLQPRQPFPQQPQQP-YPQQPQQPFPQTQQPQQPFPQS 113
Query: 450 FQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINP 489
+QP Q +P Q PQQ P QQ + + +NP
Sbjct: 114 --KQPQQPFPQPQQPQQ---SFPQQQPSLIQQSLQQQLNP 148
Score = 38.5 bits (88), Expect = 0.073
Identities = 38/109 (34%), Positives = 45/109 (40%), Gaps = 23/109 (21%)
Query: 374 FPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYP----QIPQYP 429
FP + +Q+ PQ P Q + P P Q +PQ PQ P Q PQ P
Sbjct: 59 FPHQPQQQF-------PQPQQPQQQFLQPRQP--FPQQPQQPYPQQPQQPFPQTQQPQQP 109
Query: 430 ----QFPQNPSPQNVQPQNVQQQNFQQQP---YQQYPYQQYPQQNFQQQ 471
+ PQ P P QPQ QQ QQQP Q Q P +NF Q
Sbjct: 110 FPQSKQPQQPFP---QPQQPQQSFPQQQPSLIQQSLQQQLNPCKNFLLQ 155
>FTSK_SALTI (Q8Z814) DNA translocase ftsK
Length = 1343
Score = 61.2 bits (147), Expect = 1e-08
Identities = 67/239 (28%), Positives = 95/239 (39%), Gaps = 29/239 (12%)
Query: 261 EAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIE------EKEM 314
EA QY +T + D + ++ + R+ +T + + E E E+
Sbjct: 606 EAERNQYETGAQLTDEEIDAMHQDELARQFAQSQQHRYGETYQHDTQQAEDDDTAAEAEL 665
Query: 315 TKLFLKTLNHFYY-KKMVGSTPKSFAEM--VGMGVQLEEGVREGRLVKNTTPASGTKKTG 371
+ F + Y ++ G+ P S ++ M V ++EG E P S T
Sbjct: 666 ARQFAASQQQRYSGEQPAGAQPFSLDDLDFSPMKVLVDEGPHEPLFTPGVMPES-TPVQQ 724
Query: 372 NHFPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQY--P 429
P+ + Q+ P P YQ + +QQ PQ P PQ PQY P
Sbjct: 725 PVAPQPQYQQ--------PVAPQPQYQQPQQPVASQPQYQQ----PQQPVAPQ-PQYQQP 771
Query: 430 QFPQNPSPQNVQPQN--VQQQNFQQ--QPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPR 484
Q P P PQ QPQ Q +QQ QP P Q PQQ QP Q+PQQP P+
Sbjct: 772 QQPVAPQPQYQQPQQPVAPQPQYQQPQQPVAPQPQYQQPQQPVAPQPQYQQPQQPTAPQ 830
Score = 41.6 bits (96), Expect = 0.009
Identities = 26/74 (35%), Positives = 32/74 (43%), Gaps = 8/74 (10%)
Query: 426 PQYPQFPQNPSPQNVQPQN-VQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQP---- 480
P P F P++ Q V Q QQP P Q PQQ QP Q+PQQP
Sbjct: 706 PHEPLFTPGVMPESTPVQQPVAPQPQYQQPVAPQPQYQQPQQPVASQPQYQQPQQPVAPQ 765
Query: 481 ---RPPRMPINPIP 491
+ P+ P+ P P
Sbjct: 766 PQYQQPQQPVAPQP 779
Score = 31.6 bits (70), Expect = 8.9
Identities = 28/105 (26%), Positives = 41/105 (38%), Gaps = 9/105 (8%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQ---NVQ 446
P+ P Q+ S P+QQ PQY P + +PQ QPQ
Sbjct: 375 PEGYQPHPQYAQPQEAQSAPWQQPVPVASAPQYAATPATAAEYDSLAPQETQPQWQAPDA 434
Query: 447 QQNFQQQPYQQYPYQQYPQQNFQQQPYQQRP----QQPRPPRMPI 487
+Q++Q +P P P +QP P ++ RP R P+
Sbjct: 435 EQHWQPEPIAAEPSHMPPP--VIEQPVATEPEPGIEETRPARPPL 477
>FTSK_SALTY (Q8ZQD5) DNA translocase ftsK
Length = 1351
Score = 60.8 bits (146), Expect = 1e-08
Identities = 69/239 (28%), Positives = 95/239 (38%), Gaps = 24/239 (10%)
Query: 261 EAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIE------EKEM 314
EA QY +T + D + ++ + R+ +T + + E E E+
Sbjct: 609 EAERNQYETGAQLTDEEIDAMHQDELARQFAQSQQHRYGETYQHDTQQAEDDDTAAEAEL 668
Query: 315 TKLFLKTLNHFYY-KKMVGSTPKSFAEM--VGMGVQLEEGVREGRLVKNTTPASGTKKTG 371
+ F + Y ++ G+ P S ++ M V ++EG E P S T
Sbjct: 669 ARQFAASQQQRYSGEQPAGAQPFSLDDLDFSPMKVLVDEGPHEPLFTPGVMPES-TPVQQ 727
Query: 372 NHFPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQY--P 429
P+ + Q PQ Y Q A P +QQ PQ P PQ PQY P
Sbjct: 728 PVAPQPQPQYQQPQQPVAPQPQYQQPQQPVAPQPQ---YQQ----PQQPVAPQ-PQYQQP 779
Query: 430 QFPQNPSPQNVQPQN--VQQQNFQQ--QPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPR 484
Q P P PQ QPQ Q +QQ QP P Q PQQ QP Q+PQQP P+
Sbjct: 780 QQPVAPQPQYQQPQQPVAPQPQYQQPQQPVAPQPQYQQPQQPVAPQPQYQQPQQPTAPQ 838
Score = 43.1 bits (100), Expect = 0.003
Identities = 25/64 (39%), Positives = 29/64 (45%), Gaps = 7/64 (10%)
Query: 435 PSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQP-------RPPRMPI 487
P Q V PQ Q QQP P Q PQQ QP Q+PQQP + P+ P+
Sbjct: 724 PVQQPVAPQPQPQYQQPQQPVAPQPQYQQPQQPVAPQPQYQQPQQPVAPQPQYQQPQQPV 783
Query: 488 NPIP 491
P P
Sbjct: 784 APQP 787
Score = 32.3 bits (72), Expect = 5.2
Identities = 30/111 (27%), Positives = 39/111 (35%), Gaps = 5/111 (4%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQN 449
P+ P Q+ S P+QQ PQY P + +PQ QPQ +
Sbjct: 375 PEGYQPHPQYAQPQEAQSAPWQQPVPVASAPQYAATPATAAEYDSLAPQETQPQWQPEPT 434
Query: 450 FQQQP-YQQYPYQQYPQQ---NFQQQPYQQRPQQPRPPRMPINPIPVTYAE 496
Q P YQ P P +QP P+ P P P+ Y E
Sbjct: 435 HQPTPVYQPEPIAAEPSHMPPPVIEQPVATEPEPDTEETRPARP-PLYYFE 484
>FTSK_ECO57 (Q8X5H9) DNA translocase ftsK
Length = 1342
Score = 57.8 bits (138), Expect = 1e-07
Identities = 50/119 (42%), Positives = 55/119 (46%), Gaps = 18/119 (15%)
Query: 399 HIAAITPTSHPFQQTNNHPQIPQYPQIP-QYPQFPQNPSPQNVQPQN-VQQQNFQQQPYQ 456
H TP P QQ PQ P PQ Q PQ P P PQ QPQ V Q QQP Q
Sbjct: 728 HEPLFTPIVEPVQQ----PQQPVAPQQQYQQPQQPVAPQPQYQQPQQQVAPQPQYQQPQQ 783
Query: 457 QY-PYQQY--PQQNFQQQPYQQRPQQP-------RPPRMPINPIPVTYAELLPGLLKKN 505
P QQY PQQ QP Q+PQQP + P+ P+ P P LL LL +N
Sbjct: 784 PVAPQQQYQQPQQPVAPQPQYQQPQQPVAPQPQYQQPQQPVAPQP--QDTLLHPLLMRN 840
Score = 46.2 bits (108), Expect = 3e-04
Identities = 31/82 (37%), Positives = 37/82 (44%), Gaps = 13/82 (15%)
Query: 417 PQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQR 476
P P + I + Q PQ P V PQ QQ QQP P Q PQQ QP Q+
Sbjct: 727 PHEPLFTPIVEPVQQPQQP----VAPQQQYQQ--PQQPVAPQPQYQQPQQQVAPQPQYQQ 780
Query: 477 PQQP-------RPPRMPINPIP 491
PQQP + P+ P+ P P
Sbjct: 781 PQQPVAPQQQYQQPQQPVAPQP 802
Score = 42.7 bits (99), Expect = 0.004
Identities = 42/158 (26%), Positives = 57/158 (35%), Gaps = 16/158 (10%)
Query: 375 PRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNH--PQIPQYPQIPQYPQFP 432
P+ E + G PQQ+ A + P P Q + P Q Q P Y P
Sbjct: 366 PQTGEPVIAPAPEGYPQQSQYAQPAVQYNEPLQQPVQPQQPYYAPAAEQPAQQPYYAPAP 425
Query: 433 QNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRP--QQPRP----PRMP 486
+ P N QQ F P Y + QQP Q P QQP+P P +
Sbjct: 426 EQPVAGNAWQAEEQQSTFA-------PQSTYQTEQTYQQPAAQEPLYQQPQPVEQQPVVE 478
Query: 487 INPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYR 524
P+ P L V+ + A E+L +WY+
Sbjct: 479 PEPVVEETKPARPPLYYFEEVEEKRARE-REQLAAWYQ 515
Score = 38.5 bits (88), Expect = 0.073
Identities = 56/210 (26%), Positives = 76/210 (35%), Gaps = 15/210 (7%)
Query: 285 QGDKETFKEYAQRWRDTAAQVSPRIEEKEMTKLFLKTLNHFYY-KKMVGSTPKSFA--EM 341
Q ++ + E Q A+ + E E+ + F +T Y ++ G+ P S E
Sbjct: 657 QTQQQRYGEQYQHDVPVNAEDADAAAEAELARQFAQTQQQRYSGEQPAGANPFSLDDFEF 716
Query: 342 VGMGVQLEEGVREGRLVKNTTPASGTKKTGNHFPRKKEQEVGMVTHGGPQQTYPAYQHIA 401
M L++G E P ++ P+++ Q+ PQ Y Q
Sbjct: 717 SPMKALLDDGPHEPLFTPIVEPVQQPQQPVA--PQQQYQQPQQPV--APQPQYQQPQQQV 772
Query: 402 AITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQ 461
A P QQ Q Q PQ P PQ PQ PQ Q V Q QQP Q P
Sbjct: 773 APQPQYQQPQQPVAPQQQYQQPQQPVAPQ-PQYQQPQ----QPVAPQPQYQQP--QQPVA 825
Query: 462 QYPQQNFQQQPYQQRPQQPRPPRMPINPIP 491
PQ P R RP P P+P
Sbjct: 826 PQPQDTL-LHPLLMRNGDSRPLHKPTTPLP 854
>Z384_RAT (Q9EQJ4) Zinc finger protein 384 (Nuclear matrix
transcription factor 4) (Cas-associated zinc finger
protein)
Length = 579
Score = 57.4 bits (137), Expect = 2e-07
Identities = 28/50 (56%), Positives = 33/50 (66%)
Query: 5 AELIKTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPP 54
A + + A+ Q QAQAQ QAQAQA AQAQ QA ++ Q Q Q PPP PP
Sbjct: 472 AAVAQAQAQAQAQAQAQAQAQAQAQAQAQAQASQASQQQQQQQPPPPQPP 521
Score = 55.1 bits (131), Expect = 7e-07
Identities = 38/107 (35%), Positives = 49/107 (45%), Gaps = 3/107 (2%)
Query: 3 EMAELIKTMAETQTQAQAQIQAQAQALAQAQTQAQA---VTEAQAQSQAPPPPPPIRTQA 59
+ A +A+ Q QAQAQ QAQAQA AQAQ QAQA Q Q Q PPPP P Q+
Sbjct: 466 QAAAAAAAVAQAQAQAQAQAQAQAQAQAQAQAQAQAQASQASQQQQQQQPPPPQPPHFQS 525
Query: 60 EASSSWTLCADTPRQSAPQRSAPWFPPFTAGEIFRPITCEAQMPTHQ 106
++ Q+ P + + P+ E + I T Q
Sbjct: 526 PGAAPQGGGGGDSNQNPPPQCSFDLTPYKPAEHHKDICLTVTTSTIQ 572
>FTSK_ECOL6 (Q8FJC7) DNA translocase ftsK
Length = 1347
Score = 57.0 bits (136), Expect = 2e-07
Identities = 47/123 (38%), Positives = 50/123 (40%), Gaps = 34/123 (27%)
Query: 399 HIAAITPTSHPFQQTNNHPQIPQYPQIP-QYPQFPQNPSPQNVQPQN--VQQQNFQQQPY 455
H TP P QQ PQ P PQ Q PQ P P PQ QPQ Q +QQ Y
Sbjct: 741 HEPLFTPIVEPVQQ----PQQPVAPQQQYQQPQQPVAPQPQYQQPQQPVAPQPQYQQPQY 796
Query: 456 QQ-----YPYQQY--PQQNFQQQPYQQRPQQP--------------------RPPRMPIN 488
QQ P QQY PQQ QQP Q+PQQP RP P
Sbjct: 797 QQPQQPVAPQQQYQQPQQPVTQQPQYQQPQQPVVPQPQDTLLHPLLMRNGDSRPLHKPTT 856
Query: 489 PIP 491
P+P
Sbjct: 857 PLP 859
Score = 42.0 bits (97), Expect = 0.007
Identities = 27/68 (39%), Positives = 31/68 (44%), Gaps = 1/68 (1%)
Query: 417 PQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQR 476
P P + I + Q PQ P Q Q QQ Q YQQ PQ +QQ YQQ
Sbjct: 740 PHEPLFTPIVEPVQQPQQPVAPQQQYQQPQQPVAPQPQYQQPQQPVAPQPQYQQPQYQQ- 798
Query: 477 PQQPRPPR 484
PQQP P+
Sbjct: 799 PQQPVAPQ 806
Score = 40.8 bits (94), Expect = 0.015
Identities = 39/136 (28%), Positives = 51/136 (36%), Gaps = 11/136 (8%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQN 449
PQQ Y A P QQ P Q Q P Y P+ P N QQ
Sbjct: 403 PQQPYYA-------PAAEQPVQQPYYAPAAEQPVQQPYYAPAPEQPVAGNAWQAEEQQST 455
Query: 450 FQQQPYQQYPYQQYPQQNFQQQPYQQRPQQ-PRPPRMPINPIPVTYAELLPGLLKKNLVQ 508
F P Y +Q QQ Q+P Q+PQ + P + P+ P L V+
Sbjct: 456 F--APQSTYQTEQTYQQPAAQEPLYQQPQPVEQQPVVEPEPVVEETKPTRPPLYYFEEVE 513
Query: 509 TRTAPPIPEKLPSWYR 524
+ A E+L +WY+
Sbjct: 514 EKRARE-REQLAAWYQ 528
Score = 36.2 bits (82), Expect = 0.36
Identities = 29/92 (31%), Positives = 36/92 (38%), Gaps = 15/92 (16%)
Query: 389 GPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQ 448
GPQ P + A P +P Q P + QY + P Q VQPQ
Sbjct: 365 GPQTGEP----VIAPAPEGYPHQSQYAQPAV-------QYNE----PLQQPVQPQQPYYA 409
Query: 449 NFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQP 480
+QP QQ Y +Q QQ Y P+QP
Sbjct: 410 PAAEQPVQQPYYAPAAEQPVQQPYYAPAPEQP 441
>HOG3_HORVU (P80198) Gamma-hordein 3
Length = 289
Score = 56.2 bits (134), Expect = 3e-07
Identities = 48/133 (36%), Positives = 60/133 (45%), Gaps = 16/133 (12%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYP-QFPQNPSPQNVQPQNVQQQ 448
P+Q YP Q + P PF PQ PQ P Q+P Q PQ PQ + P QQQ
Sbjct: 39 PEQPYPQQQPL----PQQQPF------PQQPQLPHQHQFPQQLPQQQFPQQM-PLQPQQQ 87
Query: 449 NFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPI-NPIPVTYAELLPGLLKKNLV 507
QQ P Q Q+PQQ Q Q QQP P + P+ P + L K+ L+
Sbjct: 88 FPQQMPLQPQQQPQFPQQKPFGQYQQPLTQQPYPQQQPLAQQQPSIEEQHQLNLCKEFLL 147
Query: 508 QTRTAPPIPEKLP 520
Q T + EK+P
Sbjct: 148 QQCT---LDEKVP 157
Score = 53.9 bits (128), Expect = 2e-06
Identities = 39/100 (39%), Positives = 45/100 (45%), Gaps = 21/100 (21%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQN 449
PQQ +P +Q + P PF Q PQ +PQ FPQ QPQ Q
Sbjct: 18 PQQLFPQWQPL----PQQPPFLQQEPEQPYPQQQPLPQQQPFPQ-------QPQLPHQHQ 66
Query: 450 FQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINP 489
F QQ P QQ+PQQ QP QQ PQQ MP+ P
Sbjct: 67 FPQQ----LPQQQFPQQ-MPLQPQQQFPQQ-----MPLQP 96
Score = 40.8 bits (94), Expect = 0.015
Identities = 39/126 (30%), Positives = 54/126 (41%), Gaps = 27/126 (21%)
Query: 403 ITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQ 462
IT T+ F + + PQ PQ+ PQ QP +QQ+ Q P QQ QQ
Sbjct: 1 ITTTTMQFNPSGLELERPQQ-LFPQWQPLPQ-------QPPFLQQEPEQPYPQQQPLPQQ 52
Query: 463 YPQQNFQQQPY--------QQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPP 514
P F QQP QQ PQQ P +MP+ P + + +P +Q + P
Sbjct: 53 QP---FPQQPQLPHQHQFPQQLPQQQFPQQMPLQP-QQQFPQQMP-------LQPQQQPQ 101
Query: 515 IPEKLP 520
P++ P
Sbjct: 102 FPQQKP 107
>YM38_YEAST (Q03825) Hypothetical 85.0 kDa protein in HLJ1-SMP2
intergenic region
Length = 758
Score = 55.5 bits (132), Expect = 6e-07
Identities = 31/90 (34%), Positives = 43/90 (47%)
Query: 427 QYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMP 486
Q P PQ S Q +Q Q Q+ QQ QQ QQ QQ QQQ QQ+ QQ + +
Sbjct: 269 QSPAQPQQSSQQQIQQPQHQPQHQPQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHQQQQ 328
Query: 487 INPIPVTYAELLPGLLKKNLVQTRTAPPIP 516
P P+ +++P + +N T P +P
Sbjct: 329 QTPYPIVNPQMVPHIPSENSHSTGLMPSVP 358
Score = 38.5 bits (88), Expect = 0.073
Identities = 29/91 (31%), Positives = 37/91 (39%), Gaps = 6/91 (6%)
Query: 388 GGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQY-----PQIPQYPQFPQNPSPQNVQP 442
G T ++H + + P Q + Q PQ+ PQ Q Q Q Q Q
Sbjct: 253 GQQYATINLHKHFNDLQSPAQPQQSSQQQIQQPQHQPQHQPQQQQQQQQQQQQQQQQQQQ 312
Query: 443 QNVQQQNFQQQPYQQYPYQQYPQQNFQQQPY 473
Q QQQ QQQ +QQ YP N Q P+
Sbjct: 313 QQQQQQQ-QQQQHQQQQQTPYPIVNPQMVPH 342
>GDA5_WHEAT (P04725) Alpha/beta-gliadin A-V precursor (Prolamin)
Length = 319
Score = 55.1 bits (131), Expect = 7e-07
Identities = 39/103 (37%), Positives = 46/103 (43%), Gaps = 5/103 (4%)
Query: 377 KKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPS 436
+++Q G PQQ YP Q P+ P+ Q PQ +P YPQ PQ+
Sbjct: 46 QQQQFPGQQQQFPPQQPYPQPQPF----PSQQPYLQLQPFPQPQPFPPQLPYPQ-PQSFP 100
Query: 437 PQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQ 479
PQ PQ Q QQP Q QQ QQ QQQ QQ QQ
Sbjct: 101 PQQPYPQQQPQYLQPQQPISQQQAQQQQQQQQQQQQQQQILQQ 143
Score = 43.5 bits (101), Expect = 0.002
Identities = 37/77 (48%), Positives = 41/77 (53%), Gaps = 11/77 (14%)
Query: 432 PQNPSPQNVQPQN--VQQQNF---QQQPYQQYPYQQYPQQNFQQQPY---QQRPQ-QPRP 482
PQNPS Q Q Q VQQQ F QQQ Q PY Q PQ QQPY Q PQ QP P
Sbjct: 30 PQNPSQQQPQEQVPLVQQQQFPGQQQQFPPQQPYPQ-PQPFPSQQPYLQLQPFPQPQPFP 88
Query: 483 PRMPINPIPVTYAELLP 499
P++P P P ++ P
Sbjct: 89 PQLPY-PQPQSFPPQQP 104
Score = 43.1 bits (100), Expect = 0.003
Identities = 37/97 (38%), Positives = 46/97 (47%), Gaps = 9/97 (9%)
Query: 400 IAAITPTSHPFQQTNNHPQIPQYPQIP-QYPQFP-QNPSPQNVQPQNVQQQNFQQQPYQQ 457
+ + P + QQ + Q Q P Q QFP Q P PQ QP QQ Q QP+ Q
Sbjct: 25 VPQLQPQNPSQQQPQEQVPLVQQQQFPGQQQQFPPQQPYPQP-QPFPSQQPYLQLQPFPQ 83
Query: 458 Y----PYQQYPQ-QNFQ-QQPYQQRPQQPRPPRMPIN 488
P YPQ Q+F QQPY Q+ Q P+ PI+
Sbjct: 84 PQPFPPQLPYPQPQSFPPQQPYPQQQPQYLQPQQPIS 120
Score = 39.7 bits (91), Expect = 0.033
Identities = 39/133 (29%), Positives = 50/133 (37%), Gaps = 21/133 (15%)
Query: 391 QQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQN---------PSPQNVQ 441
Q P+ Q P Q Q P PQ FP P PQ
Sbjct: 29 QPQNPSQQQPQEQVPLVQQQQFPGQQQQFPPQQPYPQPQPFPSQQPYLQLQPFPQPQPFP 88
Query: 442 PQ--NVQQQNF-QQQPY-QQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINPIPVTYAEL 497
PQ Q Q+F QQPY QQ P PQQ QQ QQ+ QQ + + ++
Sbjct: 89 PQLPYPQPQSFPPQQPYPQQQPQYLQPQQPISQQQAQQQQQQQQQQQQ--------QQQI 140
Query: 498 LPGLLKKNLVQTR 510
L +L++ L+ R
Sbjct: 141 LQQILQQQLIPCR 153
Score = 39.7 bits (91), Expect = 0.033
Identities = 34/92 (36%), Positives = 38/92 (40%), Gaps = 7/92 (7%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYP-QFPQNPSPQNVQPQNVQQQ 448
P Q P Q + + P QQ PQ P YPQ +P Q P QPQ Q
Sbjct: 33 PSQQQPQEQ-VPLVQQQQFPGQQQQFPPQQP-YPQPQPFPSQQPYLQLQPFPQPQPFPPQ 90
Query: 449 NFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQP 480
QP P Q YP QQQP +PQQP
Sbjct: 91 LPYPQPQSFPPQQPYP----QQQPQYLQPQQP 118
Score = 37.7 bits (86), Expect = 0.12
Identities = 26/74 (35%), Positives = 30/74 (40%), Gaps = 3/74 (4%)
Query: 416 HPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQ 475
H Q Q + Q Q Q Q Q Q QQQ Q Q P QQYP QP Q
Sbjct: 208 HQQQQQQQEQKQQLQQQQQQQQQLQQQQQQQQQQPSSQVSFQQPQQQYPSSQVSFQPSQL 267
Query: 476 RPQ---QPRPPRMP 486
PQ +P ++P
Sbjct: 268 NPQAQGSVQPQQLP 281
Score = 36.6 bits (83), Expect = 0.28
Identities = 28/68 (41%), Positives = 31/68 (45%), Gaps = 14/68 (20%)
Query: 435 PSPQNVQPQNVQQQNFQQQPYQQYPYQQYP---QQNFQQQPYQQRPQQPRPPRMPINPIP 491
P PQ +QPQN P QQ P +Q P QQ F Q Q PQQP P P P
Sbjct: 24 PVPQ-LQPQN---------PSQQQPQEQVPLVQQQQFPGQQQQFPPQQPYPQPQPF-PSQ 72
Query: 492 VTYAELLP 499
Y +L P
Sbjct: 73 QPYLQLQP 80
Score = 36.6 bits (83), Expect = 0.28
Identities = 30/92 (32%), Positives = 36/92 (38%), Gaps = 3/92 (3%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQN 449
P+Q+ H A H QQ + Q Q Q Q Q QP + Q +
Sbjct: 190 PEQSQCQAIHNVAHAIIMHQQQQQQQEQKQQLQQQQQQQQQLQQQQQQQQQQPSS--QVS 247
Query: 450 FQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPR 481
F QQP QQYP Q Q Q P Q QP+
Sbjct: 248 F-QQPQQQYPSSQVSFQPSQLNPQAQGSVQPQ 278
Score = 35.4 bits (80), Expect = 0.61
Identities = 25/59 (42%), Positives = 31/59 (52%), Gaps = 3/59 (5%)
Query: 424 QIPQYPQFPQNPSPQNVQPQNV--QQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQP 480
Q+ Q P+ Q + NV + QQQ QQ+ QQ QQ QQ QQQ QQ+ QQP
Sbjct: 185 QLLQIPEQSQCQAIHNVAHAIIMHQQQQQQQEQKQQLQQQQQQQQQLQQQQ-QQQQQQP 242
>AMEY_HUMAN (Q99218) Amelogenin, Y isoform precursor
Length = 192
Score = 55.1 bits (131), Expect = 7e-07
Identities = 35/102 (34%), Positives = 47/102 (45%), Gaps = 10/102 (9%)
Query: 405 PTSHPFQQTNNHPQIP-QYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQ---QQPYQQYPY 460
P +H Q ++ P +P Q P++ Q P P Q++ P Q N QQP+Q P
Sbjct: 75 PLTHTLQSHHHIPVVPAQQPRVRQQALMPV-PGQQSMTPTQHHQPNLPLPAQQPFQPQPV 133
Query: 461 QQYPQQNFQQQPYQQR-----PQQPRPPRMPINPIPVTYAEL 497
Q P Q Q QP Q PQ P PP P+ P+P +L
Sbjct: 134 QPQPHQPMQPQPPVQPMQPLLPQPPLPPMFPLRPLPPILPDL 175
>TAGB_DICDI (P54683) Prestalk-specific protein tagB precursor (EC
3.4.21.-)
Length = 1905
Score = 54.7 bits (130), Expect = 1e-06
Identities = 38/108 (35%), Positives = 47/108 (43%), Gaps = 13/108 (12%)
Query: 414 NNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPY 473
N+ P P Q Q P P PQ Q Q QQQ QQ+ QQ QQ QQ QQQ
Sbjct: 1789 NDQPTPPPQEQQEQKNDQPPPPPPQEQQEQQEQQQQQQQEQQQQQQQQQQQQQQQQQQQQ 1848
Query: 474 QQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPS 521
QQ+ QQ + + P P Y ++ P PP+P + PS
Sbjct: 1849 QQQQQQQQQQQQNDQP-PNDYDQVPP------------PPPLPSESPS 1883
Score = 50.4 bits (119), Expect = 2e-05
Identities = 37/94 (39%), Positives = 40/94 (42%), Gaps = 4/94 (4%)
Query: 404 TPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQY 463
TP Q+ N P PQ Q Q Q Q Q Q QQQ QQQ QQ QQ
Sbjct: 1793 TPPPQEQQEQKNDQPPPPPPQEQQEQQEQQQQQQQEQQQQQQQQQQQQQQQQQQQQQQQQ 1852
Query: 464 PQQNFQQ---QPYQQRPQQPRPPRMPI-NPIPVT 493
QQ QQ QP Q P PP +P +P P T
Sbjct: 1853 QQQQQQQQNDQPPNDYDQVPPPPPLPSESPSPPT 1886
Score = 34.7 bits (78), Expect = 1.0
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 7/54 (12%)
Query: 13 ETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPP-------PPPPIRTQA 59
E Q Q Q Q Q Q Q Q Q Q Q + Q Q+ PP PPPP+ +++
Sbjct: 1828 EQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQNDQPPNDYDQVPPPPPLPSES 1881
Score = 33.9 bits (76), Expect = 1.8
Identities = 21/71 (29%), Positives = 28/71 (38%)
Query: 409 PFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNF 468
P Q NN+ + P+ Q P QQ+ QP P +Q QQ
Sbjct: 1762 PLQNNNNNKNNNNNNNNNEPSSSSTPPNDQPTPPPQEQQEQKNDQPPPPPPQEQQEQQEQ 1821
Query: 469 QQQPYQQRPQQ 479
QQQ Q++ QQ
Sbjct: 1822 QQQQQQEQQQQ 1832
Score = 32.3 bits (72), Expect = 5.2
Identities = 26/67 (38%), Positives = 28/67 (40%), Gaps = 5/67 (7%)
Query: 414 NNHPQIPQYPQIPQYPQFPQNPSPQ-NVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQP 472
NN P P Q PQ Q N QP Q Q+Q QQ Q QQ QQQ
Sbjct: 1778 NNEPSSSSTPPNDQPTPPPQEQQEQKNDQPPPPPPQEQQEQQEQQ----QQQQQEQQQQQ 1833
Query: 473 YQQRPQQ 479
QQ+ QQ
Sbjct: 1834 QQQQQQQ 1840
>AMEX_BOVIN (P02817) Amelogenin, class I precursor
Length = 213
Score = 54.7 bits (130), Expect = 1e-06
Identities = 44/142 (30%), Positives = 60/142 (41%), Gaps = 25/142 (17%)
Query: 404 TPTSHPFQQTNNHPQIP-QYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQ---QQPYQQYP 459
TP +H Q ++ P +P Q P +PQ P P P ++ P Q N QQP+Q
Sbjct: 74 TPQNHALQPHHHIPMVPAQQPVVPQQPMMPV-PGQHSMTPTQHHQPNLPLPAQQPFQPQS 132
Query: 460 YQQYPQQNFQ----------QQPYQQ-RPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQ 508
Q P Q Q QP Q +P QP+PP PI P+P P +
Sbjct: 133 IQPQPHQPLQPHQPLQPMQPMQPLQPLQPLQPQPPVHPIQPLP-------PQPPLPPIFP 185
Query: 509 TRTAPPIPEKLP--SWYRLDQT 528
+ PP+ LP +W D+T
Sbjct: 186 MQPLPPMLPDLPLEAWPATDKT 207
Score = 44.7 bits (104), Expect = 0.001
Identities = 42/134 (31%), Positives = 53/134 (39%), Gaps = 19/134 (14%)
Query: 357 LVKNTTPASGTKKTGNHFPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNH 416
+V TP + + +H P Q+ + PQQ ++TPT H
Sbjct: 69 VVSQQTPQNHALQPHHHIPMVPAQQPVV-----PQQPMMPVPGQHSMTPTQH-------- 115
Query: 417 PQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQR 476
P P Q P PQ+ PQ QP Q QP Q P Q P Q Q QP
Sbjct: 116 -HQPNLPLPAQQPFQPQSIQPQPHQPLQPHQPLQPMQPMQ--PLQ--PLQPLQPQP-PVH 169
Query: 477 PQQPRPPRMPINPI 490
P QP PP+ P+ PI
Sbjct: 170 PIQPLPPQPPLPPI 183
>YKF4_YEAST (P35732) Hypothetical 84.0 kDa protein in NUP120-CSE4
intergenic region
Length = 738
Score = 54.3 bits (129), Expect = 1e-06
Identities = 40/101 (39%), Positives = 43/101 (41%), Gaps = 29/101 (28%)
Query: 411 QQTNNHPQIPQYPQIPQYPQFPQNPS----PQNVQPQNVQQQNFQQQP------------ 454
QQ + PQ PQ PQ PQ PQ PQ PQ Q Q QQQ QQQP
Sbjct: 384 QQQSQQPQQPQQPQQPQQPQQPQQQQQPQQPQQPQQQLQQQQQQQQQPVQAQAQAQEEQL 443
Query: 455 ----YQQYPYQQYPQQNF---------QQQPYQQRPQQPRP 482
Y Q QQY QQ QQQ QQ+ Q P+P
Sbjct: 444 SQNYYTQQQQQQYAQQQHQLQQQYLSQQQQYAQQQQQHPQP 484
Score = 52.4 bits (124), Expect = 5e-06
Identities = 35/72 (48%), Positives = 36/72 (49%), Gaps = 13/72 (18%)
Query: 409 PFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNF 468
P Q N PQ Q Q PQ PQ PQ P QPQ QQQ QQP Q PQQ
Sbjct: 373 PEDQANTVPQPQQQSQQPQQPQQPQQPQ----QPQQPQQQQQPQQP-------QQPQQQL 421
Query: 469 QQQPYQQRPQQP 480
QQQ QQ+ QQP
Sbjct: 422 QQQ--QQQQQQP 431
Score = 42.4 bits (98), Expect = 0.005
Identities = 40/111 (36%), Positives = 55/111 (49%), Gaps = 15/111 (13%)
Query: 387 HGGPQQTYPAYQHIAAITPT-SHPFQQTNNHPQIPQYPQIPQYP-QFPQNPSPQNVQPQN 444
H PQQ P + + P +H +QQ+ + Q PQY QYP Q P+N + Q QN
Sbjct: 586 HAQPQQQQP---YGGSFMPYYAHFYQQSFPYGQ-PQYGVAGQYPYQLPKN-NYNYYQTQN 640
Query: 445 VQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQ-QPRPP-----RMPINP 489
Q+Q Q Q+ +Q+ QQQ QQ+PQ QP+P P+NP
Sbjct: 641 GQEQQSPNQGVAQHSEDSQQKQSQQQQ--QQQPQGQPQPEVQMQNGQPVNP 689
Score = 41.2 bits (95), Expect = 0.011
Identities = 29/65 (44%), Positives = 33/65 (50%), Gaps = 4/65 (6%)
Query: 423 PQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRP 482
P Q PQ P Q+ QPQ QQ QQP QQ QQ PQQ QQP QQ QQ +
Sbjct: 372 PPEDQANTVPQ-PQQQSQQPQQPQQPQQPQQP-QQPQQQQQPQQ--PQQPQQQLQQQQQQ 427
Query: 483 PRMPI 487
+ P+
Sbjct: 428 QQQPV 432
Score = 32.7 bits (73), Expect = 4.0
Identities = 32/86 (37%), Positives = 35/86 (40%), Gaps = 8/86 (9%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQ-NVQPQNVQQQ 448
P+ Y YQ S P Q H + Q Q Q Q PQ VQ QN Q
Sbjct: 629 PKNNYNYYQTQNGQEQQS-PNQGVAQHSEDSQQKQSQQQQQQQPQGQPQPEVQMQNGQPV 687
Query: 449 NFQQQPYQQYPYQQYPQQNFQQQPYQ 474
N P QQ +QQY Q FQQQ Q
Sbjct: 688 N----PQQQMQFQQYYQ--FQQQQQQ 707
Score = 32.7 bits (73), Expect = 4.0
Identities = 20/43 (46%), Positives = 23/43 (52%), Gaps = 4/43 (9%)
Query: 447 QQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINP 489
Q N QP QQ Q PQQ QQP Q+PQQP+ + P P
Sbjct: 376 QANTVPQPQQQSQQPQQPQQ--PQQP--QQPQQPQQQQQPQQP 414
Score = 32.3 bits (72), Expect = 5.2
Identities = 31/108 (28%), Positives = 34/108 (30%), Gaps = 45/108 (41%)
Query: 412 QTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQ---------------------------- 443
Q PQ PQ PQ Q PQ PQ P Q Q Q
Sbjct: 394 QQPQQPQQPQQPQQQQQPQQPQQPQQQLQQQQQQQQQPVQAQAQAQEEQLSQNYYTQQQQ 453
Query: 444 ---------------NVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQR 476
+ QQQ QQQ QQ+P Q Q QQ P Q+
Sbjct: 454 QQYAQQQHQLQQQYLSQQQQYAQQQ--QQHPQPQSQQPQSQQSPQSQK 499
>SMF1_HUMAN (O14497) SWI/SNF-related, matrix-associated,
actin-dependent regulator of chromatin subfamily F
member 1 (SWI-SNF complex protein p270) (B120)
Length = 1902
Score = 54.3 bits (129), Expect = 1e-06
Identities = 42/97 (43%), Positives = 46/97 (47%), Gaps = 8/97 (8%)
Query: 390 PQQTYPAY-QHIAAITPTSHPFQQTNNHPQIPQYPQI-PQYPQFP-QNPSPQNVQPQNVQ 446
PQQ P Y Q + TP + P Q Q PQ P Y Q P Q P Q+ P Q
Sbjct: 95 PQQQQPPYSQQPPSQTPHAQPSYQQQPQSQPPQLQSSQPPYSQQPSQPPHQQSPAPYPSQ 154
Query: 447 QQNFQQQPYQQYPYQQYPQQNFQQQPY-QQRPQQPRP 482
Q QQ P Q PY Q PQ Q PY QQ+PQQP P
Sbjct: 155 QSTTQQHPQSQPPYSQ-PQ---AQSPYQQQQPQQPAP 187
Score = 44.3 bits (103), Expect = 0.001
Identities = 38/129 (29%), Positives = 45/129 (34%), Gaps = 13/129 (10%)
Query: 363 PASGTKKTGNHFPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQY 422
P G G + + Q M G Q + I P Q
Sbjct: 28 PQQGHGYPGQPYGSQTPQRYPMTVQGRAQSAMGGLSYTQQIPPYGQ-----QGPSGYGQQ 82
Query: 423 PQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQ-----NFQQQPYQQRP 477
Q P Y Q Q+P PQ QP QQ Q P+ Q YQQ PQ Q PY Q+P
Sbjct: 83 GQTPYYNQ--QSPHPQQQQPPYSQQPP-SQTPHAQPSYQQQPQSQPPQLQSSQPPYSQQP 139
Query: 478 QQPRPPRMP 486
QP + P
Sbjct: 140 SQPPHQQSP 148
Score = 35.8 bits (81), Expect = 0.47
Identities = 34/123 (27%), Positives = 49/123 (39%), Gaps = 13/123 (10%)
Query: 361 TTPASGTKKTGNHFPRKKEQEVGMVTHGGPQQTYPAY--QHIAAITPTSHPF--QQTNNH 416
+ P SG + P++++Q+ QQ + +Y Q TP+ PF QQT +
Sbjct: 929 SNPDSGMYSPSRYPPQQQQQQ---------QQRHDSYGNQFSTQGTPSGSPFPSQQTTMY 979
Query: 417 PQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQR 476
Q Q + P + + +V Q QP QQ PQ QQQ Q
Sbjct: 980 QQQQQNYKRPMDGTYGPPAKRHEGEMYSVPYSTGQGQPQQQQLPPAQPQPASQQQAAQPS 1039
Query: 477 PQQ 479
PQQ
Sbjct: 1040 PQQ 1042
Score = 33.5 bits (75), Expect = 2.3
Identities = 26/82 (31%), Positives = 33/82 (39%), Gaps = 5/82 (6%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQN 449
P Q QH + P S P Q+ Q PQ P Q P PQ+ Q QQ
Sbjct: 152 PSQQSTTQQHPQSQPPYSQPQAQSPYQQQQPQQPAPSTLSQQAAYPQPQSQQS---QQTA 208
Query: 450 FQQQPYQQYPYQQYPQQNFQQQ 471
+ QQ + P Q+ Q +F Q
Sbjct: 209 YSQQRFP--PPQELSQDSFGSQ 228
>NCO6_RAT (Q9JLI4) Nuclear receptor coactivator 6 (Amplified in
breast cancer-3 protein) (Cancer-amplified
transcriptional coactivator ASC-2) (Activating signal
cointegrator-2) (ASC-2) (Peroxisome
proliferator-activated receptor-interacting protein)
(PPAR
Length = 418
Score = 54.3 bits (129), Expect = 1e-06
Identities = 41/135 (30%), Positives = 54/135 (39%), Gaps = 23/135 (17%)
Query: 405 PTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYP 464
P +P Q P PQ+ Q+ P P Q QPQ QQQ Q P P Q
Sbjct: 257 PPGYPTQPVEQRPLQQMPPQLMQHVAPPPQPPQQQPQPQLPQQQPQPQPPPPSQPQSQQQ 316
Query: 465 QQNFQQQPYQQR-----------------PQQPRPPRMPINP----IPVTYAELLPGLLK 503
QQ QQQ QQ Q PPR P+NP +P+ + +P ++
Sbjct: 317 QQQQQQQQQQQMMMMLMMQQDPKSIRLPVSQNVHPPRGPLNPDSQRVPMQQSGNVPVMV- 375
Query: 504 KNLVQTRTAPPIPEK 518
+L + PP P+K
Sbjct: 376 -SLQGPASVPPSPDK 389
Score = 38.5 bits (88), Expect = 0.073
Identities = 25/66 (37%), Positives = 29/66 (43%), Gaps = 7/66 (10%)
Query: 426 PQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRP-----QQP 480
P+ P F P QP V+Q+ QQ P Q + P Q QQQP Q P QP
Sbjct: 248 PKLPDFSNRPPGYPTQP--VEQRPLQQMPPQLMQHVAPPPQPPQQQPQPQLPQQQPQPQP 305
Query: 481 RPPRMP 486
PP P
Sbjct: 306 PPPSQP 311
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 103,042,216
Number of Sequences: 164201
Number of extensions: 4900705
Number of successful extensions: 36318
Number of sequences better than 10.0: 542
Number of HSP's better than 10.0 without gapping: 279
Number of HSP's successfully gapped in prelim test: 289
Number of HSP's that attempted gapping in prelim test: 22491
Number of HSP's gapped (non-prelim): 4655
length of query: 820
length of database: 59,974,054
effective HSP length: 119
effective length of query: 701
effective length of database: 40,434,135
effective search space: 28344328635
effective search space used: 28344328635
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC135161.1


