

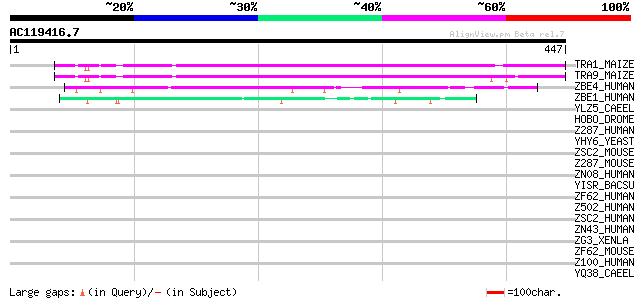
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119416.7 - phase: 0 /pseudo
(447 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TRA1_MAIZE (P08770) Putative AC transposase (ORFA) 216 1e-55
TRA9_MAIZE (P03010) Putative AC9 transposase 197 5e-50
ZBE4_HUMAN (O75132) Zinc finger BED domain containing protein 4 88 4e-17
ZBE1_HUMAN (O96006) Zinc finger BED domain containing protein 1 ... 53 2e-06
YLZ5_CAEEL (P34418) Hypothetical protein F42H10.5 in chromosome III 33 1.1
HOBO_DROME (P12258) Transposable element Hobo transposase 33 1.5
Z287_HUMAN (Q9HBT7) Zinc finger protein 287 33 1.9
YHY6_YEAST (P38873) Hypothetical 175.8 kDa Trp-Asp repeats conta... 33 1.9
ZSC2_MOUSE (Q07230) Zinc finger and SCAN domain containing prote... 32 2.5
Z287_MOUSE (Q9EQB9) Zinc finger protein 287 (Zfp-287) (Zinc fing... 32 2.5
ZN08_HUMAN (P17098) Zinc finger protein 8 (Zinc finger protein H... 32 3.3
YISR_BACSU (P40331) Putative HTH-type transcriptional regulator ... 32 3.3
ZF62_HUMAN (Q8NB50) Zinc finger protein 62 homolog (Zfp-62) 32 4.3
Z502_HUMAN (Q8TBZ5) Zinc finger protein 502 32 4.3
ZSC2_HUMAN (Q7Z7L9) Zinc finger and SCAN domain containing prote... 31 7.4
ZN43_HUMAN (P17038) Zinc finger protein 43 (Zinc protein HTF6) (... 31 7.4
ZG3_XENLA (P18718) Gastrula zinc finger protein XLCGF3.1 (Fragment) 31 7.4
ZF62_MOUSE (Q8C827) Zinc finger protein 62 homolog (Zfp-62) (ZT3) 31 7.4
Z100_HUMAN (Q8IYN0) Zinc finger protein 100 31 7.4
YQ38_CAEEL (Q09459) Hypothetical protein C09G5.8 in chromosome II 31 7.4
>TRA1_MAIZE (P08770) Putative AC transposase (ORFA)
Length = 806
Score = 216 bits (549), Expect = 1e-55
Identities = 141/428 (32%), Positives = 214/428 (49%), Gaps = 32/428 (7%)
Query: 37 QKKKPQTCSKVWRHFNRNKNLAV-------------CKY--CGKEYAANSSSHGTTNLGK 81
QK+ + S VW+HF + K + V C + C +Y A HGT+
Sbjct: 132 QKRAKKCTSDVWQHFTK-KEIEVEVDGKKYVQVWGHCNFPNCKAKYRAEGH-HGTSGFRN 189
Query: 82 HLKVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPF 141
HL+ + K Q L D N + D +L LA II+ E PF
Sbjct: 190 HLRTS-----HSLVKGQLCLKSEKDHGKDINLIEPYKYDEVVSLKKLHLA--IIMHEYPF 242
Query: 142 KHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTW 201
VE+E F F+ +P F I SRV+ + + LY +EKE L L S T D W
Sbjct: 243 NIVEHEYFVEFVKSLRPHFPIKSRVTARKYIMDLYLEEKEKLYGKLKDVQSRFSTTMDMW 302
Query: 202 TSIQNMNYMCVTAHYIDDEWVLKKKILSF-NLIADHKGETIGIALENCMKDWGI-RSICC 259
TS QN +YMCVT H+IDD+W L+K+I+ F ++ H G+ + M W I + +
Sbjct: 303 TSCQNKSYMCVTIHWIDDDWCLQKRIVGFFHVEGRHTGQRLSQTFTAIMVKWNIEKKLFA 362
Query: 260 VTVDNASANNLAINYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQR 319
+++DNASAN +A++ + ++ + + +G + H+RC HILNL+ DGL I ++++
Sbjct: 363 LSLDNASANEVAVHDIIEDLQDTDSNLVCDGAFFHVRCACHILNLVAKDGLAVIAGTIEK 422
Query: 320 IRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYR 379
I+A VKSSP + KCA + ++ DV TRWNSTYLML A Y+ R
Sbjct: 423 IKAIVLAVKSSPLQWEELMKCASECDLDKSKGISYDVSTRWNSTYLMLRDALYYKPALIR 482
Query: 380 LEHADAAFVTSLSGELGGCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQL 439
L+ +D ++ CP ++W+ + K LK F+D T SG+ + +AN F+K
Sbjct: 483 LKTSDPRRYDAI------CPKAEEWKMALTLFKCLKKFFDLTELLSGTQYSTANLFYKGF 536
Query: 440 MEIQKTLN 447
EI+ ++
Sbjct: 537 CEIKDLID 544
>TRA9_MAIZE (P03010) Putative AC9 transposase
Length = 839
Score = 197 bits (501), Expect = 5e-50
Identities = 142/448 (31%), Positives = 213/448 (46%), Gaps = 48/448 (10%)
Query: 37 QKKKPQTCSKVWRHFNRNKNLAV-------------CKY--CGKEYAANSSSHGTTNLGK 81
QK+ + S VW+HF + K + V C + C +Y A HGT+
Sbjct: 78 QKRAKKCTSDVWQHFTK-KEIEVEVDGKKYVQVWGHCNFPNCKAKYRAEGH-HGTSGFRN 135
Query: 82 HLKVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPF 141
HL+ + K Q L D N + D +L LA II+ E PF
Sbjct: 136 HLRTS-----HSLVKGQLCLKSEKDHGKDINLIEPYKYDEVVSLKKLHLA--IIMHEYPF 188
Query: 142 KHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTW 201
VE+E F F+ +P F I SRV+ + + LY +EKE L L S T D W
Sbjct: 189 NIVEHEYFVEFVKSLRPHFPIKSRVTARKYIMDLYLEEKEKLYGKLKDVQSRFSTTMDMW 248
Query: 202 TSIQNMNYMCVTAHYIDDEWVLKKKILSF-NLIADHKGETIGIALENCMKDWGI-RSICC 259
TS QN +YMCVT H+IDD+W L+K+I+ F ++ H G+ + M W I + +
Sbjct: 249 TSCQNKSYMCVTIHWIDDDWCLQKRIVGFFHVEGRHTGQRLSQTFTAIMVKWNIEKKLFA 308
Query: 260 VTVDNASANNLAINYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQR 319
+++DNASAN +A++ + ++ + + +G + H+RC HILNL+ DGL I ++++
Sbjct: 309 LSLDNASANEVAVHDIIEDLQDTDSNLVCDGAFFHVRCACHILNLVAKDGLAVIAGTIEK 368
Query: 320 IRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYR 379
I+A VKSSP + KCA + ++ DV TRWNSTYLML A Y+ R
Sbjct: 369 IKAIVLAVKSSPLQWEELMKCASECDLDKSKGISYDVSTRWNSTYLMLRDALYYKPALIR 428
Query: 380 LEHADAA---------------FVTSLSGELGGC-----PDHDDWERSRVFVKFLKTFYD 419
L+ +D + +S +G P W + K LK F+D
Sbjct: 429 LKTSDPRRYVCLNCCTCHHYKFSINQMSIIVGTMQFVLKPRSGRWHLT--LFKCLKKFFD 486
Query: 420 ATLSFSGSLHVSANSFFKQLMEIQKTLN 447
T SG+ + +AN F+K EI+ ++
Sbjct: 487 LTELLSGTQYSTANLFYKGFCEIKDLID 514
>ZBE4_HUMAN (O75132) Zinc finger BED domain containing protein 4
Length = 1171
Score = 88.2 bits (217), Expect = 4e-17
Identities = 98/416 (23%), Positives = 178/416 (42%), Gaps = 63/416 (15%)
Query: 45 SKVWRHFN---RNKNLAVCKYCGKEYAANS--SSHGTTNLGKHLKVCLKNPYRVVDKK-- 97
SK+W HF+ + VC +CG+ + ++ GT+ L +HL+ N + +
Sbjct: 561 SKLWNHFSICSADSTKVVCLHCGRTISRGKKPTNLGTSCLLRHLQRFHSNVLKTEVSETA 620
Query: 98 ----------QKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKHVENE 147
+ T G S DD N + +++ T L +A+MI +D P+ V+N
Sbjct: 621 RPSSPDTRVPRGTELSGASSFDDTNEKFYDSHPVAKKITSL-IAEMIALDLQPYSFVDNV 679
Query: 148 GFKMFMGEAQPKFKIPSRVSVARDCL-RLYFDEKETLKSVL-SANNQMVSLTTDTWTSIQ 205
GF + +P++ +P+ +R + +Y + K+ + S L A + ++ T+ W S Q
Sbjct: 680 GFNRLLEYLKPQYSLPAPSYFSRTAIPGMYDNVKQIIMSHLKEAESGVIHFTSGIWMSNQ 739
Query: 206 NMNYMCVTAHYIDDEWVLKKK--------ILSFNLI-ADHKGETIGIALENCMKDW---- 252
Y+ +TAH++ E + + +L + + D+ G +I LE + W
Sbjct: 740 TREYLTLTAHWVSFESPARPRCDDHHCSALLDVSQVDCDYSGNSIQKQLECWWEAWVTST 799
Query: 253 GIRSICCVTVDNASANNLAINYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKD 312
G++ VT DNAS G+TL GE+ ++C +H +NLIV + +K
Sbjct: 800 GLQVGITVT-DNASI----------------GKTLNEGEHSSVQCFSHTVNLIVSEAIKS 842
Query: 313 ---IDASVQRIRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNI 369
+ + R C+ V SP + + ++ DV ++W++++ ML
Sbjct: 843 QRMVQNLLSLARKICERVHRSPKAKEKLAELQREYALPQHHLIQ-DVPSKWSTSFHML-- 899
Query: 370 AEKYEHVFYRLEHADAAFVTSLSGELGGCPDHDDWERSRVFVKFLKTFYDATLSFS 425
E + + + V EL C D WE + + LK F A+ S
Sbjct: 900 ----ERLIEQKRAINEMSVECNFRELISC---DQWEVMQSVCRALKPFEAASREMS 948
Score = 31.6 bits (70), Expect = 4.3
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 10/56 (17%)
Query: 34 VSKQKKKPQTCSKVWRHF---NRNKNLAVCKYCGKEYA--ANSSSHGTTNLGKHLK 84
+S +KK P W+HF R+ A+C YC KE++ N T+ L +H++
Sbjct: 112 MSSRKKSP-----AWKHFFISPRDSTKAICMYCVKEFSRGKNEKDLSTSCLMRHVR 162
>ZBE1_HUMAN (O96006) Zinc finger BED domain containing protein 1
(dREF homolog) (Putative Ac-like transposable element)
Length = 694
Score = 52.8 bits (125), Expect = 2e-06
Identities = 81/369 (21%), Positives = 146/369 (38%), Gaps = 50/369 (13%)
Query: 41 PQTCSKVWRHFNRNKNLAVCK------YCGKEYAANSSSHGTTNLGKHLK------VC-- 86
P+ SKVW++F + N C YC A + S T+NL HL+ C
Sbjct: 19 PRAKSKVWKYFGFDTNAEGCILQWKKIYCRICMAQIAYSGNTSNLSYHLEKNHPEEFCEF 78
Query: 87 -------LKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDEL 139
++ + K K + +D + D +++ A +I + L
Sbjct: 79 VKSNTEQMREAFATAFSKLKPESSQQPGQDALAVKAGHGYDSKKQQELTAAVLGLICEGL 138
Query: 140 -PFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCL-RLYFDEKETLKSVLSANNQMVSLT 197
P V+ FK+ + A P++++PSR ++ + Y +E + L A ++
Sbjct: 139 YPASIVDEPTFKVLLKTADPRYELPSRKYISTKAIPEKYGAVREVILKEL-AEATWCGIS 197
Query: 198 TDTWTS-IQNMNYMCVTAHYI-----DDEWVLKKKILSFNLIADHKGETIGIALENCMKD 251
TD W S QN Y+ + AH++ + + + + +F + ++ ETI L +
Sbjct: 198 TDMWRSENQNRAYVTLAAHFLGLGAPNCLSMGSRCLKTFEVPEENTAETITRVLYEVFIE 257
Query: 252 WGIRSICCVTVDNASANNLAINYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDG-- 309
WGI +A NY G + +L + +HM C H N +
Sbjct: 258 WGI---------SAKVFGATTNY---GKDIVKACSLLD-VAVHMPCLGHTFNAGIQQAFQ 304
Query: 310 LKDIDASVQRIRAACKFVKSSPSRLATF--KKCADSVGVCSKAMVTLDVVTRWNSTYLML 367
L + A + R R ++ + S + K+ +V C M+ + V+ W ST ML
Sbjct: 305 LPKLGALLSRCRKLVEYFQQSAVAMYMLYEKQKQQNVAHC---MLVSNRVSWWGSTLAML 361
Query: 368 NIAEKYEHV 376
++ + V
Sbjct: 362 QRLKEQQFV 370
>YLZ5_CAEEL (P34418) Hypothetical protein F42H10.5 in chromosome III
Length = 810
Score = 33.5 bits (75), Expect = 1.1
Identities = 35/148 (23%), Positives = 60/148 (39%), Gaps = 4/148 (2%)
Query: 126 TRLALAKMIIIDELPFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKS 185
T+ ALA + + F+ V+N +K AQPKF IP+ ++LKS
Sbjct: 288 TKFALA--LATSHVDFEVVQNPLWKEVFEMAQPKFSIPTESQYEHIVNSTSHKLIQSLKS 345
Query: 186 VLSANNQMVSL--TTDTWTSIQNMNYMCVTAHYIDDEWVLKKKILSFNLIADHKGETIGI 243
LSA+ ++ L T I + + + + +L+F I ++ E +
Sbjct: 346 QLSASKKLNLLLDITKITADISRVTVSVALTGGAGNSYETQVILLAFRNINGNQSEDLTA 405
Query: 244 ALENCMKDWGIRSICCVTVDNASANNLA 271
E ++D+ I V + N LA
Sbjct: 406 VFEKVLQDYNISPSSINRVICSGLNELA 433
>HOBO_DROME (P12258) Transposable element Hobo transposase
Length = 644
Score = 33.1 bits (74), Expect = 1.5
Identities = 61/291 (20%), Positives = 110/291 (36%), Gaps = 61/291 (20%)
Query: 122 SQEKTRLALAKM---IIIDELPFKHVENEGFK-----------MFMGEAQPKFKIPSRVS 167
S+ ++A+ K ++ D PF V GFK ++ + +P +
Sbjct: 135 SENDKKVAIEKCTQWVVQDCRPFSAVTGAGFKNLVKFFLQIGAIYGEQVDVDDLLPDPTT 194
Query: 168 VARDCLRLYFDEKETLKSVL--SANNQMVSLTTDTWTS-IQNMNYMCVTAHYIDDEWVLK 224
++R +++ + S + + ++ S T D WT N++ +T HY + E+ L
Sbjct: 195 LSRKAKSDAEEKRSLISSEIKKAVDSGRASATVDMWTDQYVQRNFLGITFHY-EKEFKLC 253
Query: 225 KKILSFNLIADHKG--ETIGIALENCMKDWGIRSICCVTVDNASANNLAINYLNRGMRVW 282
IL + K E I + ++ ++ + +I V N+ +
Sbjct: 254 DMILGLKSMNFQKSTAENILMKIKGLFSEFNVENIDNVKFVTDRGANIK--------KAL 305
Query: 283 NGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSPSRLATFKKCAD 342
G T N C +H+L+ + ++ S K VKS C
Sbjct: 306 EGNTRLN-------CSSHLLSNV-------LEKSFNEANELKKIVKS----------CKK 341
Query: 343 SVGVCSKAMV--TLDVV------TRWNSTY-LMLNIAEKYEHVFYRLEHAD 384
V C K+ + TL+ TRWNS Y +M +I + + V L AD
Sbjct: 342 IVKYCKKSNLQHTLETTLKSACPTRWNSNYKMMKSILDNWRSVDKILGEAD 392
>Z287_HUMAN (Q9HBT7) Zinc finger protein 287
Length = 754
Score = 32.7 bits (73), Expect = 1.9
Identities = 18/62 (29%), Positives = 30/62 (48%), Gaps = 14/62 (22%)
Query: 39 KKPQTCSKVWRHFNRNKNLA------------VCKYCGKEYAANSS--SHGTTNLGKHLK 84
KKP C++ W+ F+++ L C CGK +A +S+ H TT+ G+
Sbjct: 526 KKPYKCNECWKVFSQSTYLIRHQRIHSGEKCYKCNECGKAFAHSSTLIQHQTTHTGEKSY 585
Query: 85 VC 86
+C
Sbjct: 586 IC 587
>YHY6_YEAST (P38873) Hypothetical 175.8 kDa Trp-Asp repeats containing
protein in GND1-IKI1 intergenic region
Length = 1541
Score = 32.7 bits (73), Expect = 1.9
Identities = 21/78 (26%), Positives = 33/78 (41%)
Query: 121 FSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEK 180
F +++ K + P + V+N FK + PK P R S+A+ L F E
Sbjct: 1011 FGFNSSQVQFVKSSLRSFSPNERVDNNAFKKEQQQHDPKISHPMRTSLAKLFQSLGFSES 1070
Query: 181 ETLKSVLSANNQMVSLTT 198
+ S+N M S T+
Sbjct: 1071 NSDSDTQSSNTSMKSHTS 1088
>ZSC2_MOUSE (Q07230) Zinc finger and SCAN domain containing protein
2 (Zinc finger protein 29) (Zfp-29)
Length = 614
Score = 32.3 bits (72), Expect = 2.5
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 14/63 (22%)
Query: 39 KKPQTCSKVWRHFNRNKNLAV------------CKYCGKEYAANSS--SHGTTNLGKHLK 84
+KP C + ++F+R+ NLA C CGK ++ +SS +H T+ G+
Sbjct: 415 EKPYQCGECGKNFSRSSNLATHRRTHLVEKPYKCGLCGKSFSQSSSLIAHQGTHTGEKPY 474
Query: 85 VCL 87
CL
Sbjct: 475 ECL 477
>Z287_MOUSE (Q9EQB9) Zinc finger protein 287 (Zfp-287) (Zinc finger
protein SKAT-2)
Length = 759
Score = 32.3 bits (72), Expect = 2.5
Identities = 18/62 (29%), Positives = 30/62 (48%), Gaps = 14/62 (22%)
Query: 39 KKPQTCSKVWRHFNRNKNLA------------VCKYCGKEYAANSS--SHGTTNLGKHLK 84
KKP C++ W+ F+++ L C CGK +A +S+ H TT+ G+
Sbjct: 531 KKPYKCNECWKVFSQSTYLIRHQRIHSGEKCYKCTACGKAFAHSSTLIQHQTTHTGEKSY 590
Query: 85 VC 86
+C
Sbjct: 591 IC 592
>ZN08_HUMAN (P17098) Zinc finger protein 8 (Zinc finger protein
HF.18)
Length = 575
Score = 32.0 bits (71), Expect = 3.3
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 12/50 (24%)
Query: 35 SKQKKKPQTCSKVWRHFNRNKNLAV------------CKYCGKEYAANSS 72
S+ + KP C+ + FN N +L V CK CGK ++ NSS
Sbjct: 250 SQVQDKPYKCTDCGKSFNHNAHLTVHKRIHTGERPYMCKECGKAFSQNSS 299
>YISR_BACSU (P40331) Putative HTH-type transcriptional regulator
yisR
Length = 287
Score = 32.0 bits (71), Expect = 3.3
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 8/70 (11%)
Query: 117 KLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLY 176
KL+D+S E + L L K I+ +EL H++ E F++ P K AR L+ +
Sbjct: 141 KLIDYSAENSDLPLRKQILFEEL-MLHLQKEAFQI------PSAKERVAWEAAR-YLQEH 192
Query: 177 FDEKETLKSV 186
+ EK T+K +
Sbjct: 193 YKEKTTIKDL 202
>ZF62_HUMAN (Q8NB50) Zinc finger protein 62 homolog (Zfp-62)
Length = 694
Score = 31.6 bits (70), Expect = 4.3
Identities = 19/63 (30%), Positives = 29/63 (45%), Gaps = 14/63 (22%)
Query: 38 KKKPQTCSKVWRHFNRNKNLAV------------CKYCGKEYAANSS--SHGTTNLGKHL 83
++KP C + + F N +L V C CGK Y ++SS +H +T+ GK
Sbjct: 435 REKPYECDRCEKVFRNNSSLKVHKRIHTGERPYECDVCGKAYISHSSLINHKSTHPGKTP 494
Query: 84 KVC 86
C
Sbjct: 495 HTC 497
>Z502_HUMAN (Q8TBZ5) Zinc finger protein 502
Length = 544
Score = 31.6 bits (70), Expect = 4.3
Identities = 20/62 (32%), Positives = 29/62 (46%), Gaps = 14/62 (22%)
Query: 39 KKPQTCSKVWRHFNRNKNLAV------------CKYCGKEYAANSS--SHGTTNLGKHLK 84
+KP C++ + FN+N L CK CGK +A +SS H T+ G+ L
Sbjct: 432 EKPYKCNECGKGFNQNTCLTQHMRIHTGEKPYKCKECGKAFAHSSSLTEHHRTHTGEKLY 491
Query: 85 VC 86
C
Sbjct: 492 KC 493
>ZSC2_HUMAN (Q7Z7L9) Zinc finger and SCAN domain containing protein
2 (Zinc finger protein 29 homolog) (Zfp-29)
Length = 613
Score = 30.8 bits (68), Expect = 7.4
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 14/63 (22%)
Query: 39 KKPQTCSKVWRHFNRNKNLAV------------CKYCGKEYAANSS--SHGTTNLGKHLK 84
+KP CS+ + F+R+ NLA C CGK ++ +SS +H + G+
Sbjct: 414 EKPYQCSECGKSFSRSSNLATHRRTHMVEKPYKCGVCGKSFSQSSSLIAHQGMHTGEKPY 473
Query: 85 VCL 87
CL
Sbjct: 474 ECL 476
>ZN43_HUMAN (P17038) Zinc finger protein 43 (Zinc protein HTF6)
(Zinc finger protein KOX27)
Length = 803
Score = 30.8 bits (68), Expect = 7.4
Identities = 23/66 (34%), Positives = 29/66 (43%), Gaps = 17/66 (25%)
Query: 39 KKPQTCSKVWRHFNRNKNLA------------VCKYCGKEYAANSSSHGTTNLGKHLKVC 86
+KP C K + FNR NL C+ CGK A N SSH T+ H K
Sbjct: 697 EKPYKCEKCGKAFNRPSNLIEHKKIHTGEQPYKCEECGK--AFNYSSHLNTHKRIHTK-- 752
Query: 87 LKNPYR 92
+ PY+
Sbjct: 753 -EQPYK 757
>ZG3_XENLA (P18718) Gastrula zinc finger protein XLCGF3.1 (Fragment)
Length = 196
Score = 30.8 bits (68), Expect = 7.4
Identities = 31/126 (24%), Positives = 55/126 (43%), Gaps = 16/126 (12%)
Query: 39 KKPQTCSKVWRHFNRNKNLAV------------CKYCGKEYAANSS--SHGTTNLGKHLK 84
+KP TC++ + F+RN L + C CGK ++ N + H T+ G+
Sbjct: 31 EKPFTCTECGKGFSRNDYLKIHQTIHTGEKPFTCIECGKGFSINRTLKLHYMTHTGEKPF 90
Query: 85 VCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKHV 144
C + K+ + M + + P + + FS K +L++ + + E PF
Sbjct: 91 TCTECSKGFSTKRDLEIHQTMHTGEKPLTCTECSKGFS-TKHKLSIHQRVHTGEKPFTCT 149
Query: 145 E-NEGF 149
E N+GF
Sbjct: 150 ECNKGF 155
>ZF62_MOUSE (Q8C827) Zinc finger protein 62 homolog (Zfp-62) (ZT3)
Length = 914
Score = 30.8 bits (68), Expect = 7.4
Identities = 19/63 (30%), Positives = 29/63 (45%), Gaps = 14/63 (22%)
Query: 38 KKKPQTCSKVWRHFNRNKNLAV------------CKYCGKEYAANSS--SHGTTNLGKHL 83
++KP C + + F N +L V C CGK Y ++SS +H +T+ GK
Sbjct: 631 REKPFECDRCEKVFRNNSSLKVHKRIHTGEKPYECDICGKAYISHSSLINHKSTHPGKTS 690
Query: 84 KVC 86
C
Sbjct: 691 YTC 693
>Z100_HUMAN (Q8IYN0) Zinc finger protein 100
Length = 542
Score = 30.8 bits (68), Expect = 7.4
Identities = 22/69 (31%), Positives = 31/69 (44%), Gaps = 17/69 (24%)
Query: 39 KKPQTCSKVWRHFNRNKNLAV------------CKYCGKEYAANSSSHGTTNLGKHLKVC 86
+KP C + + FNR+ +L C+ CGK A N SSH TT+ H V
Sbjct: 258 EKPYKCEECGKAFNRSSHLTTHKIIHTGEKPYRCEECGK--AFNRSSHLTTHKRIHTGV- 314
Query: 87 LKNPYRVVD 95
PY+ +
Sbjct: 315 --KPYKCTE 321
>YQ38_CAEEL (Q09459) Hypothetical protein C09G5.8 in chromosome II
Length = 1531
Score = 30.8 bits (68), Expect = 7.4
Identities = 17/46 (36%), Positives = 27/46 (57%), Gaps = 2/46 (4%)
Query: 13 QGKDGEVTEAGAVT--EVGAATAVSKQKKKPQTCSKVWRHFNRNKN 56
+GKD E+ E ++ E G +T V ++KKKP+ S+ H +KN
Sbjct: 401 EGKDSELEEMSEMSDDESGRSTPVIEEKKKPRRKSRKSSHQEPSKN 446
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.133 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,391,707
Number of Sequences: 164201
Number of extensions: 2026995
Number of successful extensions: 5301
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 5268
Number of HSP's gapped (non-prelim): 47
length of query: 447
length of database: 59,974,054
effective HSP length: 113
effective length of query: 334
effective length of database: 41,419,341
effective search space: 13834059894
effective search space used: 13834059894
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC119416.7


