

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148819.7 - phase: 0
(343 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
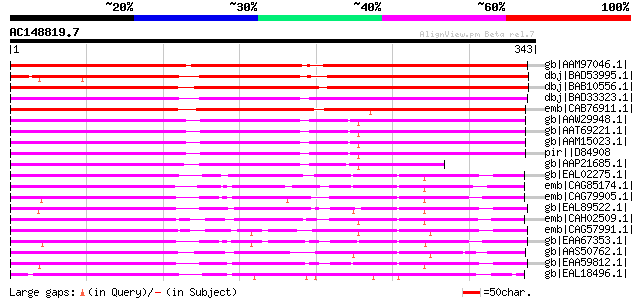
gb|AAM97046.1| putative protein [Arabidopsis thaliana] gi|884381... 458 e-128
dbj|BAD53995.1| calcineurin-like phosphoesterase-like [Oryza sat... 398 e-109
dbj|BAB10556.1| unnamed protein product [Arabidopsis thaliana] g... 326 5e-88
dbj|BAD33323.1| PTS protein-like [Oryza sativa (japonica cultiva... 320 5e-86
emb|CAB76911.1| putative PTS protein [Cicer arietinum] 309 7e-83
gb|AAW29948.1| putative purple acid phosphatase [Arabidopsis tha... 294 3e-78
gb|AAT69221.1| hypothetical protein At2g46880 [Arabidopsis thali... 294 3e-78
gb|AAM15023.1| hypothetical protein [Arabidopsis thaliana] gi|20... 294 3e-78
pir||D84908 probable phosphoesterase (EC 3.1.-.-) At2g46880 - Ar... 294 3e-78
gb|AAP21685.1| hypothetical protein [Arabidopsis thaliana] gi|42... 233 8e-60
gb|EAL02275.1| hypothetical protein CaO19.8463 [Candida albicans... 193 5e-48
emb|CAG85174.1| unnamed protein product [Debaryomyces hansenii C... 191 3e-47
emb|CAG79905.1| unnamed protein product [Yarrowia lipolytica CLI... 176 9e-43
gb|EAL89522.1| phosphoesterase, putative [Aspergillus fumigatus ... 167 4e-40
emb|CAH02509.1| unnamed protein product [Kluyveromyces lactis NR... 166 9e-40
emb|CAG57991.1| unnamed protein product [Candida glabrata CBS138... 162 1e-38
gb|EAA67353.1| hypothetical protein FG01846.1 [Gibberella zeae P... 159 9e-38
gb|AAS50762.1| ABL009Wp [Ashbya gossypii ATCC 10895] gi|45185221... 157 6e-37
gb|EAA59812.1| hypothetical protein AN3604.2 [Aspergillus nidula... 156 1e-36
gb|EAL18496.1| hypothetical protein CNBJ1380 [Cryptococcus neofo... 151 2e-35
>gb|AAM97046.1| putative protein [Arabidopsis thaliana] gi|8843816|dbj|BAA97364.1|
unnamed protein product [Arabidopsis thaliana]
gi|15242042|ref|NP_200524.1| calcineurin-like
phosphoesterase family protein [Arabidopsis thaliana]
gi|58618187|gb|AAW80660.1| putative purple acid
phosphatase [Arabidopsis thaliana]
gi|25083815|gb|AAN72121.1| putative protein [Arabidopsis
thaliana]
Length = 397
Score = 458 bits (1179), Expect = e-128
Identities = 221/339 (65%), Positives = 265/339 (77%), Gaps = 15/339 (4%)
Query: 1 MHFGNG-ITKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAE 59
MHFG G IT+CRDVL SEFE+CSDLNTT FL+R+I+ E PD IAFTGDNIFG S+ DAAE
Sbjct: 66 MHFGMGMITRCRDVLDSEFEYCSDLNTTRFLRRMIESERPDLIAFTGDNIFGSSTTDAAE 125
Query: 60 SMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSAK 119
S+ +A GPA+E G+PWAA+LGNHD ESTLNR ELM+ +SLMD+SVSQINP + T K
Sbjct: 126 SLLEAIGPAIEYGIPWAAVLGNHDHESTLNRLELMTFLSLMDFSVSQINPLVEDET---K 182
Query: 120 GHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIRTYDWIKDSQLHW 179
G M IDGFGNY +RVYGAPGS++ANS+V +LFF DSGDR + QG RTY WIK+SQL W
Sbjct: 183 GDTMRLIDGFGNYRVRVYGAPGSVLANSTVFDLFFFDSGDREIVQGKRTYGWIKESQLRW 242
Query: 180 LRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVGQFQEGVACS 239
L+ S + +Q +H + PPALAFFHIPI EVR+L+Y +GQFQEGVACS
Sbjct: 243 LQDTSIQGHSQR---IH--------VNPPALAFFHIPILEVRELWYTPFIGQFQEGVACS 291
Query: 240 RVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPRRARI 299
V S VLQTFVSMG+VKA F+GHDH NDFCG L G+WFCYGGGFGYH YG+ W RRAR+
Sbjct: 292 IVQSGVLQTFVSMGNVKAAFMGHDHVNDFCGTLKGVWFCYGGGFGYHAYGRPNWHRRARV 351
Query: 300 ILAELQKGKESWTSVQKIMTWKRLDDEKMSKIDEQILWD 338
I A+L KG+++W ++ I TWKRLDDE +SKIDEQ+LW+
Sbjct: 352 IEAKLGKGRDTWEGIKLIKTWKRLDDEYLSKIDEQVLWE 390
>dbj|BAD53995.1| calcineurin-like phosphoesterase-like [Oryza sativa (japonica
cultivar-group)]
Length = 409
Score = 398 bits (1023), Expect = e-109
Identities = 212/370 (57%), Positives = 248/370 (66%), Gaps = 58/370 (15%)
Query: 1 MHFGNGI-TKCRDVLASEF--EFCSDLNTTLFLKRVIQDETPDFIAFTG----------- 46
MHFGNG T+CRDV A E CSDLNTT FL+RVI+ E PD IAFT
Sbjct: 64 MHFGNGAATRCRDV-APEVGGARCSDLNTTRFLRRVIEAERPDLIAFTEGLGGISRQITC 122
Query: 47 -----------------DNIFGPSSHDAAESMFKAFGPAMESGLPWAAILGNHDQESTLN 89
DNIFG S+ DAAES+ KA PA+E +PWAAILGNHDQEST+
Sbjct: 123 CLPMFAIVEKYLRQSTRDNIFGGSASDAAESLLKAISPAIEYKVPWAAILGNHDQESTMT 182
Query: 90 REELMSLISLMDYSVSQINPSADSLTNSAKGHKMSKIDGFGNYNLRVYGAPGSMMANSSV 149
REELM +SLMDYSVSQ+NP S + GFGNY++ ++G GS N+S+
Sbjct: 183 REELMVFMSLMDYSVSQVNPPG------------SLVHGFGNYHVSIHGPFGSEFVNTSL 230
Query: 150 LNLFFLDSGDRVVYQGIRTYDWIKDSQLHWLRHVSQEPQAQEQDPLHSTDHVTSPITPPA 209
LNL+FLDSGDR V G++TY WIK+SQL WLR SQE Q LH+ PA
Sbjct: 231 LNLYFLDSGDREVVNGVKTYGWIKESQLAWLRATSQELQQN----LHA----------PA 276
Query: 210 LAFFHIPIPEVRQLFYKQIVGQFQEGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFC 269
AFFHIPIPEVR L+Y GQ+QEGVACS VNS VL T SMGDVKAVF+GHDH NDFC
Sbjct: 277 FAFFHIPIPEVRGLWYTGFKGQYQEGVACSTVNSGVLGTLTSMGDVKAVFLGHDHLNDFC 336
Query: 270 GNLDGIWFCYGGGFGYHGYGKAGWPRRARIILAELQKGKESWTSVQKIMTWKRLDDEKMS 329
G+L+GIWFCYGGGFGYH YG+ WPRRAR+I EL+KG++S V+ I TWK LDDEK++
Sbjct: 337 GDLNGIWFCYGGGFGYHAYGRPHWPRRARVIHTELKKGQKSLVEVESIHTWKLLDDEKLT 396
Query: 330 KIDEQILWDH 339
KIDEQ+LW H
Sbjct: 397 KIDEQVLWRH 406
>dbj|BAB10556.1| unnamed protein product [Arabidopsis thaliana]
gi|50198940|gb|AAT70473.1| At5g63140 [Arabidopsis
thaliana] gi|15242654|ref|NP_201119.1| calcineurin-like
phosphoesterase family protein [Arabidopsis thaliana]
gi|48525337|gb|AAT44970.1| At5g63140 [Arabidopsis
thaliana] gi|58618189|gb|AAW80661.1| putative purple
acid phosphatase [Arabidopsis thaliana]
Length = 389
Score = 326 bits (836), Expect = 5e-88
Identities = 166/342 (48%), Positives = 217/342 (62%), Gaps = 18/342 (5%)
Query: 1 MHFGNGI-TKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAE 59
MHF NG T+C++VL S+ CSDLNTT+F+ RVI E PD I FTGDNIFG DA +
Sbjct: 55 MHFANGAKTQCQNVLPSQRAHCSDLNTTIFMSRVIAAEKPDLIVFTGDNIFGFDVKDALK 114
Query: 60 SMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSAK 119
S+ AF PA+ S +PW AILGNHDQEST R+++M+ I + ++SQ+NP
Sbjct: 115 SINAAFAPAIASKIPWVAILGNHDQESTFTRQQVMNHIVKLPNTLSQVNPP--------- 165
Query: 120 GHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIRTYDWIKDSQLHW 179
IDGFGNYNL+++GA S + N SVLNL+FLDSGD + YDWIK SQ W
Sbjct: 166 -EAAHYIDGFGNYNLQIHGAADSKLQNKSVLNLYFLDSGDYSSVPYMEGYDWIKTSQQFW 224
Query: 180 LRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIV-GQFQEGVAC 238
S+ + + + + + P LA+FHIP+PE K G QEG +
Sbjct: 225 FDRTSKRLKREYNAKPNPQEGIA-----PGLAYFHIPLPEFLSFDSKNATKGVRQEGTSA 279
Query: 239 SRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPRRAR 298
+ NS T ++ GDVK+VF+GHDH NDFCG L G+ CYGGGFGYH YGKAGW RRAR
Sbjct: 280 ASTNSGFFTTLIARGDVKSVFVGHDHVNDFCGELKGLNLCYGGGFGYHAYGKAGWERRAR 339
Query: 299 IILAEL-QKGKESWTSVQKIMTWKRLDDEKMSKIDEQILWDH 339
+++ +L +K K W +V+ I TWKRLDD+ +S ID Q+LW++
Sbjct: 340 VVVVDLNKKRKGKWGAVKSIKTWKRLDDKHLSVIDSQVLWNN 381
>dbj|BAD33323.1| PTS protein-like [Oryza sativa (japonica cultivar-group)]
gi|52075952|dbj|BAD46032.1| PTS protein-like [Oryza
sativa (japonica cultivar-group)]
Length = 398
Score = 320 bits (819), Expect = 5e-86
Identities = 161/340 (47%), Positives = 205/340 (59%), Gaps = 19/340 (5%)
Query: 1 MHFGNGI-TKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAE 59
MH+ +G T C DVL SE CSDLNTT FL R+ +DE PD + FTGDNI+G + DAA+
Sbjct: 60 MHYADGRRTGCLDVLPSEAAGCSDLNTTAFLYRLFRDEDPDLVVFTGDNIYGFDATDAAK 119
Query: 60 SMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSAK 119
SM A PA+ LPWAA++GNHDQE TL+RE +M + M ++S+ NP
Sbjct: 120 SMDAAIAPAINMNLPWAAVIGNHDQEGTLSREGVMRHLVGMKNTLSRFNPEG-------- 171
Query: 120 GHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIRTYDWIKDSQLHW 179
+IDG+GNYNL V G G+++AN SVLNL+FLDSGD I Y WIK SQ W
Sbjct: 172 ----IEIDGYGNYNLEVGGVEGTLLANKSVLNLYFLDSGDYSTVPSIGGYGWIKASQQFW 227
Query: 180 LRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVGQFQEGVACS 239
+ S Q + + + P L +FHIP+PE G QEG++
Sbjct: 228 FQQTSSNLQTK-----YMKEEPKQKAAAPGLVYFHIPLPEFSSFTSSNFTGVKQEGISSP 282
Query: 240 RVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPRRARI 299
+NS + V GDVKA FIGHDH NDFCG L+GI CY GGFGYH YGKAGW RRAR+
Sbjct: 283 SINSGFFASMVEAGDVKAAFIGHDHVNDFCGKLNGIQLCYAGGFGYHAYGKAGWSRRARV 342
Query: 300 ILAELQKGK-ESWTSVQKIMTWKRLDDEKMSKIDEQILWD 338
+ +L+K W V+ I TWKRLDD ++ ID ++LW+
Sbjct: 343 VSVQLEKSDGGEWRGVKSIKTWKRLDDPHLTTIDSEVLWN 382
>emb|CAB76911.1| putative PTS protein [Cicer arietinum]
Length = 405
Score = 309 bits (792), Expect = 7e-83
Identities = 165/342 (48%), Positives = 211/342 (61%), Gaps = 22/342 (6%)
Query: 1 MHFGNGI-TKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAE 59
MH+ +G T C DVL S+ C+DLNTT F++R I E P+ I FTGDNIFG S D+A+
Sbjct: 65 MHYADGKNTLCLDVLPSQNASCTDLNTTAFIQRTILAEKPNLIVFTGDNIFGFDSSDSAK 124
Query: 60 SMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSAK 119
SM AF PA+ S +PW A+LGNHDQE +L+RE +M I M ++S++NP
Sbjct: 125 SMDAAFAPAIASNIPWVAVLGNHDQEGSLSREGVMKYIVGMKNTLSKLNPP--------- 175
Query: 120 GHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIRTYDWIKDSQLHW 179
++ IDGFGNYNL V G G++ N SVLNL+FLDSGD I YDWIK SQ W
Sbjct: 176 --EVHIIDGFGNYNLEVGGVQGTVFENKSVLNLYFLDSGDYSKVPAIFGYDWIKPSQQLW 233
Query: 180 LRHVSQE-PQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVGQFQE--GV 236
+S + +A + P+ + P LA+FHIP+PE G E G+
Sbjct: 234 FERMSAKLRKAYIKGPVPQKE------AAPGLAYFHIPLPEYASFDSSNFTGVKMEPDGI 287
Query: 237 ACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPRR 296
+ + VNS T V GDVKAVF GHDH NDFCG L I CY GGFGYH YGKAGW RR
Sbjct: 288 SSASVNSGFFTTLVEAGDVKAVFTGHDHLNDFCGKLMDIQLCYAGGFGYHAYGKAGWSRR 347
Query: 297 ARIILAELQK-GKESWTSVQKIMTWKRLDDEKMSKIDEQILW 337
AR+++A L+K K SW V+ I +WKRLDD+ ++ ID ++LW
Sbjct: 348 ARVVVASLEKTDKGSWGDVKSIKSWKRLDDQHLTGIDGEVLW 389
>gb|AAW29948.1| putative purple acid phosphatase [Arabidopsis thaliana]
Length = 387
Score = 294 bits (752), Expect = 3e-78
Identities = 160/343 (46%), Positives = 202/343 (58%), Gaps = 21/343 (6%)
Query: 1 MHFGNGI-TKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGP-SSHDAA 58
MH+G G T+C DV +EF +CSDLNTT FL+R I E PD I F+GDN++G + D A
Sbjct: 54 MHYGFGKETQCSDVSPAEFPYCSDLNTTSFLQRTIASEEPDLIVFSGDNVYGLCETSDVA 113
Query: 59 ESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSA 118
+SM AF PA+ESG+PW AILGNHDQES + RE +M I + S+SQ+NP L
Sbjct: 114 KSMDMAFAPAIESGIPWVAILGNHDQESDMTRETMMKYIMKLPNSLSQVNPPDAWLY--- 170
Query: 119 KGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIR-TYDWIKDSQL 177
+IDGFGNYNL++ G GS + S+LNL+ LD G G YDW+K SQ
Sbjct: 171 ------QIDGFGNYNLQIEGPFGSPLFFKSILNLYLLDGGSYTKLDGFGYKYDWVKTSQQ 224
Query: 178 HWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYK--QIVGQFQEG 235
+W H S+ + + H T P L + HIP+PE LF K ++ G QE
Sbjct: 225 NWYEHTSKWLEME-----HKRWPFPQNSTAPGLVYLHIPMPEFA-LFNKSTEMTGVRQES 278
Query: 236 VACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPR 295
+NS V G+VK VF GHDH NDFC L GI CY GG GYHGYG+ GW R
Sbjct: 279 TCSPPINSGFFTKLVERGEVKGVFSGHDHVNDFCAELHGINLCYAGGAGYHGYGQVGWAR 338
Query: 296 RARIILAELQKGKES-WTSVQKIMTWKRLDDEKMSKIDEQILW 337
R R++ A+L+K W +V I TWKRLDD+ S ID Q+LW
Sbjct: 339 RVRVVEAQLEKTMYGRWGAVDTIKTWKRLDDKNHSLIDTQLLW 381
>gb|AAT69221.1| hypothetical protein At2g46880 [Arabidopsis thaliana]
gi|30267807|gb|AAP21684.1| hypothetical protein
[Arabidopsis thaliana] gi|42569990|ref|NP_182211.2|
calcineurin-like phosphoesterase family protein
[Arabidopsis thaliana]
Length = 401
Score = 294 bits (752), Expect = 3e-78
Identities = 160/343 (46%), Positives = 202/343 (58%), Gaps = 21/343 (6%)
Query: 1 MHFGNGI-TKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGP-SSHDAA 58
MH+G G T+C DV +EF +CSDLNTT FL+R I E PD I F+GDN++G + D A
Sbjct: 54 MHYGFGKETQCSDVSPAEFPYCSDLNTTSFLQRTIASEKPDLIVFSGDNVYGLCETSDVA 113
Query: 59 ESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSA 118
+SM AF PA+ESG+PW AILGNHDQES + RE +M I + S+SQ+NP L
Sbjct: 114 KSMDMAFAPAIESGIPWVAILGNHDQESDMTRETMMKYIMKLPNSLSQVNPPDAWLY--- 170
Query: 119 KGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIR-TYDWIKDSQL 177
+IDGFGNYNL++ G GS + S+LNL+ LD G G YDW+K SQ
Sbjct: 171 ------QIDGFGNYNLQIEGPFGSPLFFKSILNLYLLDGGSYTKLDGFGYKYDWVKTSQQ 224
Query: 178 HWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYK--QIVGQFQEG 235
+W H S+ + + H T P L + HIP+PE LF K ++ G QE
Sbjct: 225 NWYEHTSKWLEME-----HKRWPFPQNSTAPGLVYLHIPMPEFA-LFNKSTEMTGVRQES 278
Query: 236 VACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPR 295
+NS V G+VK VF GHDH NDFC L GI CY GG GYHGYG+ GW R
Sbjct: 279 TCSPPINSGFFTKLVERGEVKGVFSGHDHVNDFCAELHGINLCYAGGAGYHGYGQVGWAR 338
Query: 296 RARIILAELQKGKES-WTSVQKIMTWKRLDDEKMSKIDEQILW 337
R R++ A+L+K W +V I TWKRLDD+ S ID Q+LW
Sbjct: 339 RVRVVEAQLEKTMYGRWGAVDTIKTWKRLDDKNHSLIDTQLLW 381
>gb|AAM15023.1| hypothetical protein [Arabidopsis thaliana]
gi|20197136|gb|AAC34232.2| hypothetical protein
[Arabidopsis thaliana] gi|7485755|pir||T02689
hypothetical protein F19D11.16 - Arabidopsis thaliana
Length = 437
Score = 294 bits (752), Expect = 3e-78
Identities = 160/343 (46%), Positives = 202/343 (58%), Gaps = 21/343 (6%)
Query: 1 MHFGNGI-TKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGP-SSHDAA 58
MH+G G T+C DV +EF +CSDLNTT FL+R I E PD I F+GDN++G + D A
Sbjct: 54 MHYGFGKETQCSDVSPAEFPYCSDLNTTSFLQRTIASEKPDLIVFSGDNVYGLCETSDVA 113
Query: 59 ESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSA 118
+SM AF PA+ESG+PW AILGNHDQES + RE +M I + S+SQ+NP L
Sbjct: 114 KSMDMAFAPAIESGIPWVAILGNHDQESDMTRETMMKYIMKLPNSLSQVNPPDAWLY--- 170
Query: 119 KGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIR-TYDWIKDSQL 177
+IDGFGNYNL++ G GS + S+LNL+ LD G G YDW+K SQ
Sbjct: 171 ------QIDGFGNYNLQIEGPFGSPLFFKSILNLYLLDGGSYTKLDGFGYKYDWVKTSQQ 224
Query: 178 HWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYK--QIVGQFQEG 235
+W H S+ + + H T P L + HIP+PE LF K ++ G QE
Sbjct: 225 NWYEHTSKWLEME-----HKRWPFPQNSTAPGLVYLHIPMPEFA-LFNKSTEMTGVRQES 278
Query: 236 VACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPR 295
+NS V G+VK VF GHDH NDFC L GI CY GG GYHGYG+ GW R
Sbjct: 279 TCSPPINSGFFTKLVERGEVKGVFSGHDHVNDFCAELHGINLCYAGGAGYHGYGQVGWAR 338
Query: 296 RARIILAELQKGKES-WTSVQKIMTWKRLDDEKMSKIDEQILW 337
R R++ A+L+K W +V I TWKRLDD+ S ID Q+LW
Sbjct: 339 RVRVVEAQLEKTMYGRWGAVDTIKTWKRLDDKNHSLIDTQLLW 381
>pir||D84908 probable phosphoesterase (EC 3.1.-.-) At2g46880 - Arabidopsis
thaliana
Length = 387
Score = 294 bits (752), Expect = 3e-78
Identities = 160/343 (46%), Positives = 202/343 (58%), Gaps = 21/343 (6%)
Query: 1 MHFGNGI-TKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGP-SSHDAA 58
MH+G G T+C DV +EF +CSDLNTT FL+R I E PD I F+GDN++G + D A
Sbjct: 54 MHYGFGKETQCSDVSPAEFPYCSDLNTTSFLQRTIASEKPDLIVFSGDNVYGLCETSDVA 113
Query: 59 ESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSA 118
+SM AF PA+ESG+PW AILGNHDQES + RE +M I + S+SQ+NP L
Sbjct: 114 KSMDMAFAPAIESGIPWVAILGNHDQESDMTRETMMKYIMKLPNSLSQVNPPDAWLY--- 170
Query: 119 KGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIR-TYDWIKDSQL 177
+IDGFGNYNL++ G GS + S+LNL+ LD G G YDW+K SQ
Sbjct: 171 ------QIDGFGNYNLQIEGPFGSPLFFKSILNLYLLDGGSYTKLDGFGYKYDWVKTSQQ 224
Query: 178 HWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYK--QIVGQFQEG 235
+W H S+ + + H T P L + HIP+PE LF K ++ G QE
Sbjct: 225 NWYEHTSKWLEME-----HKRWPFPQNSTAPGLVYLHIPMPEFA-LFNKSTEMTGVRQES 278
Query: 236 VACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPR 295
+NS V G+VK VF GHDH NDFC L GI CY GG GYHGYG+ GW R
Sbjct: 279 TCSPPINSGFFTKLVERGEVKGVFSGHDHVNDFCAELHGINLCYAGGAGYHGYGQVGWAR 338
Query: 296 RARIILAELQKGKES-WTSVQKIMTWKRLDDEKMSKIDEQILW 337
R R++ A+L+K W +V I TWKRLDD+ S ID Q+LW
Sbjct: 339 RVRVVEAQLEKTMYGRWGAVDTIKTWKRLDDKNHSLIDTQLLW 381
>gb|AAP21685.1| hypothetical protein [Arabidopsis thaliana]
gi|42571261|ref|NP_973704.1| calcineurin-like
phosphoesterase family protein [Arabidopsis thaliana]
Length = 327
Score = 233 bits (593), Expect = 8e-60
Identities = 131/289 (45%), Positives = 165/289 (56%), Gaps = 20/289 (6%)
Query: 1 MHFGNGI-TKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGP-SSHDAA 58
MH+G G T+C DV +EF +CSDLNTT FL+R I E PD I F+GDN++G + D A
Sbjct: 54 MHYGFGKETQCSDVSPAEFPYCSDLNTTSFLQRTIASEKPDLIVFSGDNVYGLCETSDVA 113
Query: 59 ESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSA 118
+SM AF PA+ESG+PW AILGNHDQES + RE +M I + S+SQ+NP L
Sbjct: 114 KSMDMAFAPAIESGIPWVAILGNHDQESDMTRETMMKYIMKLPNSLSQVNPPDAWL---- 169
Query: 119 KGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGI-RTYDWIKDSQL 177
+IDGFGNYNL++ G GS + S+LNL+ LD G G YDW+K SQ
Sbjct: 170 -----YQIDGFGNYNLQIEGPFGSPLFFKSILNLYLLDGGSYTKLDGFGYKYDWVKTSQQ 224
Query: 178 HWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYK--QIVGQFQEG 235
+W H S+ + + H T P L + HIP+PE LF K ++ G QE
Sbjct: 225 NWYEHTSKWLEME-----HKRWPFPQNSTAPGLVYLHIPMPEF-ALFNKSTEMTGVRQES 278
Query: 236 VACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFG 284
+NS V G+VK VF GHDH NDFC L GI CY GG G
Sbjct: 279 TCSPPINSGFFTKLVERGEVKGVFSGHDHVNDFCAELHGINLCYAGGAG 327
>gb|EAL02275.1| hypothetical protein CaO19.8463 [Candida albicans SC5314]
gi|46442861|gb|EAL02147.1| hypothetical protein
CaO19.843 [Candida albicans SC5314]
Length = 728
Score = 193 bits (491), Expect = 5e-48
Identities = 126/349 (36%), Positives = 181/349 (51%), Gaps = 47/349 (13%)
Query: 1 MHFGNGITKCRDVLA-SEFEFC-SDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAA 58
+HF G KC D S + C +D T F+ +V+ E PD + TGD IFG +S D+
Sbjct: 405 LHFSTGYGKCLDPQPPSSAKGCKADSRTLEFINKVLDLEKPDMVVLTGDQIFGDASPDSE 464
Query: 59 ESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSA 118
S FKA P +E +P+A +GNHD E +L REE+M L + M YSV+ + P++
Sbjct: 465 SSAFKALNPFVERKIPFAITVGNHDDEGSLKREEIMGLYADMPYSVAAMGPAS------- 517
Query: 119 KGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIRT-YDWIKDSQL 177
IDGFGNY + V G + ++ L+L+F+DS + YDWIK++QL
Sbjct: 518 -------IDGFGNYVVTVQG----KSSKATALSLYFVDSHAYSKTPKVTPGYDWIKENQL 566
Query: 178 HWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVGQFQEGVA 237
+L+ ++ Q + + S P A+AFFHIP+PE R L + +G+ +EGV
Sbjct: 567 IYLKQEAESIQNSVE------KYRKSNKIPLAMAFFHIPLPEFRNL-NQPFIGENREGVT 619
Query: 238 CSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFC---------GNLDGIWFCYGGGFGYHGY 288
R NS Q +G V +GHDH ND+C + +W C+GGG G GY
Sbjct: 620 APRYNSGARQVLSEIG-VSVASVGHDHCNDYCLQDTQQSSSPGDNKMWLCFGGGAGLGGY 678
Query: 289 -GKAGWPRRARIILAELQKGKESWTSVQKIMTWKRLDDEKMSKIDEQIL 336
G G+ RR R+ + KG +I TWKR +D + IDEQ+L
Sbjct: 679 GGYNGYIRRMRVYELDTSKG--------EIKTWKRTEDNPGNIIDEQVL 719
>emb|CAG85174.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50412907|ref|XP_457179.1| unnamed protein product
[Debaryomyces hansenii]
Length = 763
Score = 191 bits (485), Expect = 3e-47
Identities = 121/347 (34%), Positives = 181/347 (51%), Gaps = 48/347 (13%)
Query: 1 MHFGNGITKCRDVLASEFEF-C-SDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAA 58
+HF G+ KCRD +E + C +D T FL++V+ E PD + TGD IFG + D+
Sbjct: 444 LHFSTGVGKCRDPSPAETKSGCQADSRTLKFLEKVLDLEKPDLVVLTGDQIFGDEAKDSE 503
Query: 59 ESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSA 118
++FKA P ++ G+P+A +GNHD E +L+R E+MSL + + YS++ +
Sbjct: 504 TALFKALNPFIKRGIPFAVTMGNHDDEGSLSRTEIMSLSANLPYSLASLG---------- 553
Query: 119 KGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIRT-YDWIKDSQL 177
++ G GNY L + G P S ++ + LFFLD+ + + YDW+K+SQL
Sbjct: 554 ----ADEVAGVGNYALTIEG-PSS---RNTAMTLFFLDTHKYSLNPKVTPGYDWLKESQL 605
Query: 178 HWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVGQFQEGVA 237
WL + A Q + + H+ ++AFFHIP+PE R L + +VG+ +EG+
Sbjct: 606 KWL----EREAASLQKSIAAYTHIHL-----SMAFFHIPLPEYRNL-DQPMVGEKKEGIT 655
Query: 238 CSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFC-------GNLDGIWFCYGGGFGYHGY-G 289
R NS T +G V +GHDH ND+C N + +W CYGGG G GY G
Sbjct: 656 APRYNSGARSTLGKLG-VSVASVGHDHCNDYCLQDATNNENENALWLCYGGGSGEGGYGG 714
Query: 290 KAGWPRRARIILAELQKGKESWTSVQKIMTWKRLDDEKMSKIDEQIL 336
G+ RR R+ + TS +I +WKR + E D Q L
Sbjct: 715 YGGYIRRMRVFDID--------TSAGEIKSWKRKESEPNVDFDHQTL 753
>emb|CAG79905.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50553790|ref|XP_504306.1| hypothetical protein
[Yarrowia lipolytica]
Length = 565
Score = 176 bits (446), Expect = 9e-43
Identities = 114/354 (32%), Positives = 175/354 (49%), Gaps = 56/354 (15%)
Query: 1 MHFGNGITKCRDVLASEFE---FC-SDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHD 56
+H+ G KC +A++ + C +D + ++ + E PD + TGD I+G ++ D
Sbjct: 238 LHYSTGFGKCLQHVAADTDPEGACQADPLSLQHIEAFLDRENPDMVVLTGDQIYGSAAPD 297
Query: 57 AAESMFKAFGPAMESGLPWAAILGNHDQEST-LNREELMSLISLMDYSVSQINPSADSLT 115
A ++ K P + +PWAA+ GNHD E T +NR + M+L+ + YS+SQ P
Sbjct: 298 AETALLKVLAPLIRRKVPWAAVFGNHDHEETNMNRAQQMALMESLPYSLSQAGP------ 351
Query: 116 NSAKGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGI-RTYDWIKD 174
+DG GNY L+V AP S ++ + L+FLD+ + Q + YDW+++
Sbjct: 352 --------EDVDGVGNYWLQVL-APKS---DNPAVTLYFLDTHAKHPNQKLFPGYDWVRE 399
Query: 175 SQLHWL--RHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVGQF 232
SQL WL H +P + +H ++AFFHIP E R K+++GQ+
Sbjct: 400 SQLEWLEKEHKQLQPLQNKYTHIHL-----------SMAFFHIPTTEYRNARGKKMIGQW 448
Query: 233 QEGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFC---------GNLDGIWFCYGGGF 283
+EG A + NS V + +G V + +GHDH NDFC ++ +W CYGGG
Sbjct: 449 KEGAAAPKHNSGVRKLLEEIG-VSVISVGHDHVNDFCMWDDVTAHKDDIPPMWLCYGGGL 507
Query: 284 GYHGY-GKAGWPRRARIILAELQKGKESWTSVQKIMTWKRLDDEKMSKIDEQIL 336
G GY G G+ RR R+ E T I +WKR + D Q+L
Sbjct: 508 GEGGYGGYGGYVRRMRVF--------EIDTEANSITSWKRKVSDYDETFDRQVL 553
>gb|EAL89522.1| phosphoesterase, putative [Aspergillus fumigatus Af293]
Length = 551
Score = 167 bits (423), Expect = 4e-40
Identities = 112/353 (31%), Positives = 173/353 (48%), Gaps = 53/353 (15%)
Query: 1 MHFGNGITKCRDVLASE----FEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHD 56
+H G+ CRD + E + +D T F++R++ +E PDF+ +GD + G +S D
Sbjct: 226 LHLSTGLGVCRDPIPVEPVPGRKCEADPRTLEFVERLLDEEKPDFVVLSGDQVNGETSRD 285
Query: 57 AAESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTN 116
A ++FK+ ++ +P+AAI GNHD E L+RE+LM+++ + YS+S P
Sbjct: 286 AQSALFKSVKLLVDRKIPYAAIFGNHDDEGNLSREQLMTILEDLPYSLSTAGP------- 338
Query: 117 SAKGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGD-RVVYQGIRTYDWIKDS 175
+DG GNY + V G + S L L+ LDS + R YDWIK S
Sbjct: 339 -------EDVDGVGNYIVEVLGRGTTA---HSALTLYLLDSHSYSPDERQFRGYDWIKPS 388
Query: 176 QLHWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQL--FYKQIVGQFQ 233
Q+ W ++ +Q + + + H H+ +AF HIP+PE R +Y+ G +
Sbjct: 389 QIRWFKNTAQSLKTKHHEYSHM--HMN-------MAFIHIPLPEYRDSSNYYR---GNWS 436
Query: 234 EGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFC------GNLDGIWFCYGGGFGYHG 287
E NS G + V GHDH ND+C +W CYGGG G+ G
Sbjct: 437 EAPTAPGFNSGFKDALEEEG-ILFVSCGHDHVNDYCMLNKDRDQKPSLWMCYGGGAGFGG 495
Query: 288 Y-GKAGWPRRARIILAELQKGKESWTSVQKIMTWKRLD-DEKMSKIDEQILWD 338
Y G G+ RR R ++ G +++T+KRL+ E +KIDE ++ D
Sbjct: 496 YGGYGGYVRRVRFYDFDMNPG--------RVVTYKRLEYGEVEAKIDEMMIID 540
>emb|CAH02509.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50304333|ref|XP_452116.1| unnamed protein product
[Kluyveromyces lactis]
Length = 565
Score = 166 bits (420), Expect = 9e-40
Identities = 112/348 (32%), Positives = 162/348 (46%), Gaps = 44/348 (12%)
Query: 1 MHFGNGITKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPS-SHDAAE 59
+HF G CRD +D T F+ +V+ E P + FTGD I G D+
Sbjct: 241 LHFSVGKGVCRDEFPQHETCEADPKTLQFIDQVLDIEKPQMVVFTGDQIMGDECKQDSET 300
Query: 60 SMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSAK 119
++ K P + +PWA + GNHD E +LNR +L S + YS+ +I P DS N
Sbjct: 301 ALLKVLAPVISRKIPWAMVWGNHDDEGSLNRWQLSEFASKLPYSLFEIGP-RDSKDNQF- 358
Query: 120 GHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIRTYDWIKDSQLHW 179
G GNY V G G ++ + L+FLDS + YDW+K+ Q +
Sbjct: 359 --------GLGNYVREVKGGDG-----TTNIALYFLDSHKYSKSKAFPGYDWVKEEQWEY 405
Query: 180 LRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYK----QIVGQFQEG 235
+ + + +Q HS D + ++AFFHIP+PE R + ++VG ++EG
Sbjct: 406 MEEYLESHDSIKQ-AKHSGDLI-------SMAFFHIPLPEYRNFPQESGSNRVVGTYKEG 457
Query: 236 VACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFC------GNLDGIWFCYGGGFGYHGY- 288
+ R NS ++T +G V +GHDH ND+C D IW CYGG G GY
Sbjct: 458 ITAPRYNSEGVKTLHKLG-VSVTSVGHDHCNDYCLLDDFNDGEDKIWLCYGGAAGEGGYA 516
Query: 289 GKAGWPRRARIILAELQKGKESWTSVQKIMTWKRLDDEKMSKIDEQIL 336
G G RR R+ + K + I +WKRL+ + D Q L
Sbjct: 517 GYGGTERRIRVYEIDALK--------KDIYSWKRLNGSPENTFDHQKL 556
>emb|CAG57991.1| unnamed protein product [Candida glabrata CBS138]
gi|50285325|ref|XP_445091.1| unnamed protein product
[Candida glabrata]
Length = 578
Score = 162 bits (411), Expect = 1e-38
Identities = 120/357 (33%), Positives = 170/357 (47%), Gaps = 53/357 (14%)
Query: 1 MHFGNGITKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAES 60
+H G G +C D + +D T F++ V+ E P F+ F+GD I G S +ES
Sbjct: 256 LHLGVGKNRCLDEYPHHDKCEADSKTLKFVEEVLDIEKPGFVVFSGDQIMGDRSLQDSES 315
Query: 61 -MFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSAK 119
++KA P + +PWA + GNHD E +L+R EL L + YS QI+P D+ N+
Sbjct: 316 VLYKAVDPVIRRRIPWAMVWGNHDDEGSLSRWELSKLAMKLPYSRFQISPH-DTKDNTF- 373
Query: 120 GHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLD----SGDRVVYQGIRTYDWIKDS 175
G GNY +++ +A L+L+F+D S +Y G YDW+K+
Sbjct: 374 --------GVGNYAHQIFYENDPEVA---ALSLYFMDSHKYSKTGKIYLG---YDWLKEE 419
Query: 176 QLHWLRHVSQEPQAQEQDPLHSTDHVTSPI-TPPALAFFHIPIPEVRQLFYK-------Q 227
QL +++ + + H+ I A+ F HIP+PE L K +
Sbjct: 420 QLEYIQSLYERGM---------KSHIKENIHRHAAMTFIHIPLPEYLNLDSKKRPGESNE 470
Query: 228 IVGQFQEGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLD-----GIWFCYGGG 282
++G F+EGV R NS L +G V V GHDH ND+C + D IW CYGGG
Sbjct: 471 LIGTFKEGVTAPRYNSGGLVALDKIG-VDVVGCGHDHCNDYCLHDDSTSNKNIWLCYGGG 529
Query: 283 FGYHGY-GKAGWPRRARIILAELQKGKESWTSVQKIMTWKRLDDEKMSKIDEQILWD 338
G GY G G RR R + Q G KI TWKRL+ + D QI+ D
Sbjct: 530 AGEGGYAGYGGTERRIRTYKFDPQTG--------KITTWKRLNSKPSEIFDFQIVKD 578
>gb|EAA67353.1| hypothetical protein FG01846.1 [Gibberella zeae PH-1]
gi|46109928|ref|XP_382022.1| hypothetical protein
FG01846.1 [Gibberella zeae PH-1]
Length = 549
Score = 159 bits (403), Expect = 9e-38
Identities = 110/356 (30%), Positives = 172/356 (47%), Gaps = 57/356 (16%)
Query: 1 MHFGNGITKCRDVLASEFEF--C-SDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDA 57
+H G+ CR+ + + C +D T F+ RV+ DE PD + +GD + G ++ DA
Sbjct: 222 LHLSTGVGACREAVPDSYNGGKCEADPRTLDFVNRVLDDEKPDLVVLSGDQVNGDTAPDA 281
Query: 58 AESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNS 117
+MFK +E +P+AAI GNHD E T++RE M+++ + +S+S P
Sbjct: 282 PTAMFKIVSLLIERKIPYAAIFGNHDDEKTMSREAQMAIMESLPFSLSTAGP-------- 333
Query: 118 AKGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLD----SGDRVVYQGIRTYDWIK 173
+ IDG GNY + V A G + S L ++ +D + D + G YDW+K
Sbjct: 334 ------ADIDGVGNYYVEVL-ARGK--TDHSALTIYLMDTHAYTPDERNFPG---YDWVK 381
Query: 174 DSQLHWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVGQFQ 233
+Q+ W + + + + ++ H+ +AF HIP+ E + VG+++
Sbjct: 382 PNQIEWFKKTAAGLKKNHNE--YTGRHMD-------IAFIHIPLTEYADPALPR-VGEWK 431
Query: 234 EGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCG---------NLDGIWFCYGGGFG 284
EGV NS V G + V GHDH ND+C + +W CY GG G
Sbjct: 432 EGVTAPVYNSGFRDALVEQG-IVMVSAGHDHCNDYCSLSLAGEGETKIPALWMCYAGGSG 490
Query: 285 YHGY-GKAGWPRRARIILAELQKGKESWTSVQKIMTWKRLDD-EKMSKIDEQILWD 338
+ GY G G+ RR R+ E T+ +I TWKRL+ + S+ID QI+ D
Sbjct: 491 FGGYAGYGGYHRRVRLF--------EVDTNEARIKTWKRLESGDTASRIDLQIIVD 538
>gb|AAS50762.1| ABL009Wp [Ashbya gossypii ATCC 10895] gi|45185221|ref|NP_982938.1|
ABL009Wp [Eremothecium gossypii]
Length = 569
Score = 157 bits (396), Expect = 6e-37
Identities = 111/352 (31%), Positives = 157/352 (44%), Gaps = 54/352 (15%)
Query: 1 MHFGNGITKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGP-SSHDAAE 59
+H G +CRD + +D+ T F+ V+ E PD +TGD I G +DA
Sbjct: 256 LHLVPGKGECRDEFPTTENCEADVKTMKFVNDVLDIERPDMAVYTGDQITGDLCDNDAET 315
Query: 60 SMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSAK 119
+ KAF PA + +P+A I GNHD +LNR +L + + +S+ +I P
Sbjct: 316 PLLKAFAPANQRWIPFAVIWGNHDDAGSLNRLQLSQYVEALPFSLFKIGPR--------- 366
Query: 120 GHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVY-QGIRTYDWIKDSQLH 178
M + G GNY +V G G + +F+D+ +G R YDW+K+ Q
Sbjct: 367 -DTMDRSFGMGNYVHQVLGENG-----HPEITFYFVDTHSYAPNPRGRRVYDWVKEEQWQ 420
Query: 179 WLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQL-------FYKQIVGQ 231
+ H T +LAF HIP+PE + Y Q +G
Sbjct: 421 YFEDC----------------HAKLEHTELSLAFLHIPLPEYLDVKSKKDPQKYNQFLGT 464
Query: 232 FQEGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLD------GIWFCYGGGFGY 285
F+EGV R NS + +G V AV GHDH ND+C D IW CYGG G
Sbjct: 465 FREGVTAPRHNSGGAERLARLG-VSAVTAGHDHCNDYCLQTDFRDIDPKIWMCYGGAAGE 523
Query: 286 HGYGKAGWPRRARIILAELQKGKESWTSVQKIMTWKRLDDEKMSKIDEQILW 337
GYG G R RI + E+ T ++I TWKRL+ K D +++
Sbjct: 524 GGYGGYGGTER-RIRIFEID------TREKRIETWKRLNSSPNDKFDAHLIY 568
>gb|EAA59812.1| hypothetical protein AN3604.2 [Aspergillus nidulans FGSC A4]
gi|67526293|ref|XP_661208.1| hypothetical protein
AN3604_2 [Aspergillus nidulans FGSC A4]
gi|49092560|ref|XP_407741.1| hypothetical protein
AN3604.2 [Aspergillus nidulans FGSC A4]
Length = 799
Score = 156 bits (394), Expect = 1e-36
Identities = 108/351 (30%), Positives = 168/351 (47%), Gaps = 49/351 (13%)
Query: 1 MHFGNGITKCRDVLASEF---EFC-SDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHD 56
+H G+ CRD + E + C +D T F++R++ +E PD + +GD + G +S D
Sbjct: 477 LHLSTGLGHCRDPVPPELIPGQGCEADPRTLDFIERLLDEEQPDLVILSGDQVNGETSRD 536
Query: 57 AAESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTN 116
A +FK+ ++ +P+AAI GNHD E L+R + M+++ + YS+S P
Sbjct: 537 AQSPLFKSVKLLVDRKIPYAAIFGNHDDEGNLDRHQSMAILEDLPYSLSSAGP------- 589
Query: 117 SAKGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGD-RVVYQGIRTYDWIKDS 175
IDG GNY + V G + + S L L+ LDS + R YDWIK +
Sbjct: 590 -------EDIDGVGNYIVEVLGRGNT---DHSALTLYLLDSHSYSPDERQFRGYDWIKPN 639
Query: 176 QLHWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVGQFQEG 235
Q+ W + +Q +A+ Q ++ H+ +AF HIP+PE Q G + E
Sbjct: 640 QIRWFKTTAQGLKAKHQQ--YAYMHMN-------MAFIHIPLPEFAQR-GNYFRGNWSEP 689
Query: 236 VACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFC------GNLDGIWFCYGGGFGYHGY- 288
NS G + V GHDH ND+C +W CYGGG G+ GY
Sbjct: 690 STAPGFNSGFKDALEEEG-ILFVGCGHDHANDYCALSKNEAQKPSLWMCYGGGAGFGGYG 748
Query: 289 GKAGWPRRARIILAELQKGKESWTSVQKIMTWKRLD-DEKMSKIDEQILWD 338
G G+ RR R ++ G +++T+KRL+ ++IDE ++ D
Sbjct: 749 GYGGFIRRVRFFDFDMNPG--------RVVTYKRLEYGNTDARIDEMMIVD 791
>gb|EAL18496.1| hypothetical protein CNBJ1380 [Cryptococcus neoformans var.
neoformans B-3501A] gi|57229430|gb|AAW45863.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58259940|ref|XP_567380.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 731
Score = 151 bits (382), Expect = 2e-35
Identities = 111/381 (29%), Positives = 174/381 (45%), Gaps = 76/381 (19%)
Query: 1 MHFGNGITKCRDVLASEFEFC-SDLNTTLFLKRVIQDETPDFIAFTGDNIFGP-SSHDAA 58
+H+ G +CRD ++ E C D NT +L + E PD + F+GD + G +S+DA
Sbjct: 368 LHYSVGTGECRD---TDIEGCVGDANTAAWLAEALDAENPDLVVFSGDQLNGQRTSYDAR 424
Query: 59 ESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSA 118
+ K P +E +PW A+ GNHD E +R+ M + M YS+S+ P +
Sbjct: 425 SVLAKFAKPVIEREIPWCAVFGNHDSEIYADRDYQMKTLENMPYSLSRAGPKS------- 477
Query: 119 KGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSG---DRVVYQGIRTYDWIKDS 175
+DG GNY ++++ + S N + L+FLDS R + YD++K S
Sbjct: 478 -------VDGVGNYYIKLHSSDAS---NMHIFTLYFLDSHAYQKRTLPWVKPDYDYLKTS 527
Query: 176 QLHWLRHVSQEPQAQEQ-------DPLHST------------DHVTSPITPPALAFFHIP 216
Q+ W R+VS + E+ D L D + P A+ +FHIP
Sbjct: 528 QIDWYRNVSSSIKPIERPFKPDGADDLSGIWSRRSQASRLPRDGSQTLAKPNAMMWFHIP 587
Query: 217 IPEVRQLFYKQIVGQFQEGV-----ACSRVNSAVLQTFVSM---------------GDVK 256
+PE + ++G+ GV S+ NS + +VK
Sbjct: 588 LPEAYNDPDQSLMGELDVGVQMDVAGSSKHNSGFFYNAIKTTYDREEAEGYFSKKTAEVK 647
Query: 257 AVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPRRARII-LAELQKGKESWTSVQ 315
+ GH H D C +DGIW C+ GG + GYG+ G+ RR R+ ++E +
Sbjct: 648 VLSHGHCHNTDRCRRVDGIWMCFDGGSSFSGYGQLGFDRRVRLYRISEYG---------E 698
Query: 316 KIMTWKRLDDEKMSKIDEQIL 336
K+ T+KRL ++ IDEQ+L
Sbjct: 699 KVETYKRLTSGEI--IDEQVL 717
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.137 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 616,259,513
Number of Sequences: 2540612
Number of extensions: 25865291
Number of successful extensions: 58085
Number of sequences better than 10.0: 90
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 57766
Number of HSP's gapped (non-prelim): 122
length of query: 343
length of database: 863,360,394
effective HSP length: 129
effective length of query: 214
effective length of database: 535,621,446
effective search space: 114622989444
effective search space used: 114622989444
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC148819.7


