

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148718.5 + phase: 0
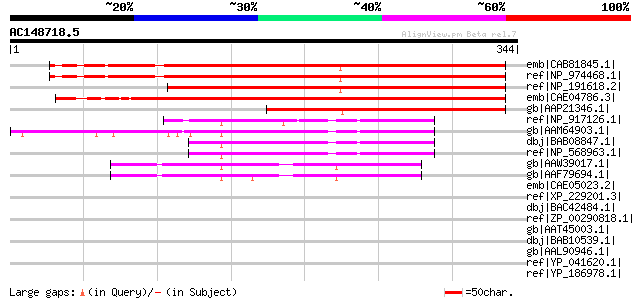
(344 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB81845.1| putative protein [Arabidopsis thaliana] gi|42570... 372 e-102
ref|NP_974468.1| expressed protein [Arabidopsis thaliana] 372 e-102
ref|NP_191618.2| expressed protein [Arabidopsis thaliana] 343 6e-93
emb|CAE04786.3| OSJNBb0020O11.15 [Oryza sativa (japonica cultiva... 326 5e-88
gb|AAP21346.1| At3g60590 [Arabidopsis thaliana] gi|27311683|gb|A... 238 3e-61
ref|NP_917126.1| P0421H07.28 [Oryza sativa (japonica cultivar-gr... 76 1e-12
gb|AAM64903.1| unknown [Arabidopsis thaliana] 69 2e-10
dbj|BAB08847.1| unnamed protein product [Arabidopsis thaliana] g... 65 3e-09
ref|NP_568963.1| expressed protein [Arabidopsis thaliana] 65 3e-09
gb|AAW39017.1| At1g48460 [Arabidopsis thaliana] gi|56381889|gb|A... 59 2e-07
gb|AAF79694.1| T1N15.7 [Arabidopsis thaliana] gi|25405928|pir||F... 55 3e-06
emb|CAE05023.2| OSJNBa0044M19.10 [Oryza sativa (japonica cultiva... 39 0.23
ref|XP_229201.3| PREDICTED: similar to G protein-coupled recepto... 39 0.30
dbj|BAC42484.1| unknown protein [Arabidopsis thaliana] 38 0.50
ref|ZP_00290818.1| hypothetical protein Mmc102000477 [Magnetococ... 37 0.66
gb|AAT45003.1| unknown [Xerophyta humilis] 37 0.66
dbj|BAB10539.1| unnamed protein product [Arabidopsis thaliana] g... 37 0.66
gb|AAL90946.1| AT5g52420/K24M7_17 [Arabidopsis thaliana] gi|1514... 37 0.66
ref|YP_041620.1| FecCD transport family protein [Staphylococcus ... 37 1.1
ref|YP_186978.1| iron compound ABC transporter, permease protein... 37 1.1
>emb|CAB81845.1| putative protein [Arabidopsis thaliana]
gi|42570499|ref|NP_850729.2| expressed protein
[Arabidopsis thaliana] gi|11281196|pir||T47870
hypothetical protein T8B10.250 - Arabidopsis thaliana
Length = 329
Score = 372 bits (956), Expect = e-102
Identities = 182/312 (58%), Positives = 232/312 (74%), Gaps = 17/312 (5%)
Query: 28 LKSGKPFQSRRFNFPSIRLNQSFICCTKLTPWEPSPGVAYAPTDNQSDNFLQSTASVFET 87
LK+G S F F ++R+ +C KL+ WEPSP + +A + +D L TA+VFE+
Sbjct: 31 LKTG----SCNFRFRNLRV----LCTPKLSQWEPSPFI-HASAEEAADIVLDKTANVFES 81
Query: 88 LESSKVDESPTANVEGLVEEKDRPGPELQLFKWPMWLLGPSILLATGMVPTLWLPISSIF 147
+ S +E + + R ++Q+ KWP+WLLGPS+LL +GM PTLWLP+SS+F
Sbjct: 82 IVSESAEEEKVD-----MSAQQRTNSQVQVLKWPIWLLGPSVLLTSGMAPTLWLPLSSVF 136
Query: 148 LGPNIASLLSLIGLDCIFNLGATLFLLMADSCSRPKNPTQEIKSKAPFSYQFWNIVATLT 207
LG N+ SLLSLIGLDCIFNLGATLFLLMADSC+RPK+P+Q SK PFSY+FWN+ + +
Sbjct: 137 LGSNVVSLLSLIGLDCIFNLGATLFLLMADSCARPKDPSQSCNSKPPFSYKFWNMFSLII 196
Query: 208 GFIVPSLLMFGSQKGF---LQPQLPFISSAVLLGPYLLLLSVQILTELLTWYWQSPVWLV 264
GF+VP LL+FGSQ G LQPQ+PF+SSAV+L PY +LL+VQ LTE+LTW+WQSPVWLV
Sbjct: 197 GFLVPMLLLFGSQSGLLASLQPQIPFLSSAVILFPYFILLAVQTLTEILTWHWQSPVWLV 256
Query: 265 TPIIYEAYRILQLMRGLKLGAELTAPAWMMHTIRGLVCWWVLILGLQLMRVAWFAGLSAR 324
TP++YEAYRILQLMRGL L AE+ AP W++H +RGLV WWVLILG+QLMRVAWFAG ++R
Sbjct: 257 TPVVYEAYRILQLMRGLTLSAEVNAPVWVVHMLRGLVSWWVLILGMQLMRVAWFAGFASR 316
Query: 325 ARKDQSSSSEAS 336
Q S AS
Sbjct: 317 TTTGQQPQSVAS 328
>ref|NP_974468.1| expressed protein [Arabidopsis thaliana]
Length = 404
Score = 372 bits (956), Expect = e-102
Identities = 182/312 (58%), Positives = 232/312 (74%), Gaps = 17/312 (5%)
Query: 28 LKSGKPFQSRRFNFPSIRLNQSFICCTKLTPWEPSPGVAYAPTDNQSDNFLQSTASVFET 87
LK+G S F F ++R+ +C KL+ WEPSP + +A + +D L TA+VFE+
Sbjct: 106 LKTG----SCNFRFRNLRV----LCTPKLSQWEPSPFI-HASAEEAADIVLDKTANVFES 156
Query: 88 LESSKVDESPTANVEGLVEEKDRPGPELQLFKWPMWLLGPSILLATGMVPTLWLPISSIF 147
+ S +E + + R ++Q+ KWP+WLLGPS+LL +GM PTLWLP+SS+F
Sbjct: 157 IVSESAEEEKVD-----MSAQQRTNSQVQVLKWPIWLLGPSVLLTSGMAPTLWLPLSSVF 211
Query: 148 LGPNIASLLSLIGLDCIFNLGATLFLLMADSCSRPKNPTQEIKSKAPFSYQFWNIVATLT 207
LG N+ SLLSLIGLDCIFNLGATLFLLMADSC+RPK+P+Q SK PFSY+FWN+ + +
Sbjct: 212 LGSNVVSLLSLIGLDCIFNLGATLFLLMADSCARPKDPSQSCNSKPPFSYKFWNMFSLII 271
Query: 208 GFIVPSLLMFGSQKGF---LQPQLPFISSAVLLGPYLLLLSVQILTELLTWYWQSPVWLV 264
GF+VP LL+FGSQ G LQPQ+PF+SSAV+L PY +LL+VQ LTE+LTW+WQSPVWLV
Sbjct: 272 GFLVPMLLLFGSQSGLLASLQPQIPFLSSAVILFPYFILLAVQTLTEILTWHWQSPVWLV 331
Query: 265 TPIIYEAYRILQLMRGLKLGAELTAPAWMMHTIRGLVCWWVLILGLQLMRVAWFAGLSAR 324
TP++YEAYRILQLMRGL L AE+ AP W++H +RGLV WWVLILG+QLMRVAWFAG ++R
Sbjct: 332 TPVVYEAYRILQLMRGLTLSAEVNAPVWVVHMLRGLVSWWVLILGMQLMRVAWFAGFASR 391
Query: 325 ARKDQSSSSEAS 336
Q S AS
Sbjct: 392 TTTGQQPQSVAS 403
>ref|NP_191618.2| expressed protein [Arabidopsis thaliana]
Length = 236
Score = 343 bits (879), Expect = 6e-93
Identities = 157/232 (67%), Positives = 192/232 (82%), Gaps = 3/232 (1%)
Query: 108 KDRPGPELQLFKWPMWLLGPSILLATGMVPTLWLPISSIFLGPNIASLLSLIGLDCIFNL 167
+ R ++Q+ KWP+WLLGPS+LL +GM PTLWLP+SS+FLG N+ SLLSLIGLDCIFNL
Sbjct: 4 QQRTNSQVQVLKWPIWLLGPSVLLTSGMAPTLWLPLSSVFLGSNVVSLLSLIGLDCIFNL 63
Query: 168 GATLFLLMADSCSRPKNPTQEIKSKAPFSYQFWNIVATLTGFIVPSLLMFGSQKGF---L 224
GATLFLLMADSC+RPK+P+Q SK PFSY+FWN+ + + GF+VP LL+FGSQ G L
Sbjct: 64 GATLFLLMADSCARPKDPSQSCNSKPPFSYKFWNMFSLIIGFLVPMLLLFGSQSGLLASL 123
Query: 225 QPQLPFISSAVLLGPYLLLLSVQILTELLTWYWQSPVWLVTPIIYEAYRILQLMRGLKLG 284
QPQ+PF+SSAV+L PY +LL+VQ LTE+LTW+WQSPVWLVTP++YEAYRILQLMRGL L
Sbjct: 124 QPQIPFLSSAVILFPYFILLAVQTLTEILTWHWQSPVWLVTPVVYEAYRILQLMRGLTLS 183
Query: 285 AELTAPAWMMHTIRGLVCWWVLILGLQLMRVAWFAGLSARARKDQSSSSEAS 336
AE+ AP W++H +RGLV WWVLILG+QLMRVAWFAG ++R Q S AS
Sbjct: 184 AEVNAPVWVVHMLRGLVSWWVLILGMQLMRVAWFAGFASRTTTGQQPQSVAS 235
>emb|CAE04786.3| OSJNBb0020O11.15 [Oryza sativa (japonica cultivar-group)]
gi|50926722|ref|XP_473308.1| OSJNBb0020O11.15 [Oryza
sativa (japonica cultivar-group)]
Length = 511
Score = 326 bits (836), Expect = 5e-88
Identities = 164/306 (53%), Positives = 212/306 (68%), Gaps = 12/306 (3%)
Query: 32 KPFQSRRFNFPSIRLNQSFICCTKLTPWEPSPGVAYAPTDNQSDNFLQSTASVFETLESS 91
KP S F +R CT+ WE S + YA ++ + N ++ T V E +++
Sbjct: 37 KPLHSGHFENIVLR-------CTQNLSWEAS--LPYASAEDGA-NIIKGT-EVVEPIDTE 85
Query: 92 KVDESPTANVE-GLVEEKDRPGPELQLFKWPMWLLGPSILLATGMVPTLWLPISSIFLGP 150
+ E P + VE P +L FK P+WLLGPSILL T +VPTLWLP+SS+FLGP
Sbjct: 86 EAPEIPILQSDQDFVEVIKEPSMQLTTFKLPIWLLGPSILLVTSIVPTLWLPLSSVFLGP 145
Query: 151 NIASLLSLIGLDCIFNLGATLFLLMADSCSRPKNPTQEIKSKAPFSYQFWNIVATLTGFI 210
NIA LLSL+GLD IFN+GA LF LMAD+C RP+ + E+ + P SY+FWN+ A++ GF+
Sbjct: 146 NIAGLLSLVGLDFIFNMGAMLFFLMADACGRPEANSSELIKQIPTSYRFWNLAASIVGFL 205
Query: 211 VPSLLMFGSQKGFLQPQLPFISSAVLLGPYLLLLSVQILTELLTWYWQSPVWLVTPIIYE 270
VP L F S KG LQP +PFI AVLLGPYLLLLSVQ+LTE+LTW+W+SPVWLV P++YE
Sbjct: 206 VPLALFFASHKGTLQPHIPFIPFAVLLGPYLLLLSVQVLTEMLTWHWKSPVWLVAPVVYE 265
Query: 271 AYRILQLMRGLKLGAELTAPAWMMHTIRGLVCWWVLILGLQLMRVAWFAGLSARARKDQS 330
YR+LQLMRGL+L E+TAP WM+ ++RGLV WWVL+LG+QLMRVAWFAGL +
Sbjct: 266 GYRVLQLMRGLQLADEITAPGWMVQSLRGLVSWWVLVLGIQLMRVAWFAGLKFASTSSTK 325
Query: 331 SSSEAS 336
S+A+
Sbjct: 326 CYSDAT 331
>gb|AAP21346.1| At3g60590 [Arabidopsis thaliana] gi|27311683|gb|AAO00807.1|
putative protein [Arabidopsis thaliana]
Length = 166
Score = 238 bits (606), Expect = 3e-61
Identities = 109/165 (66%), Positives = 134/165 (81%), Gaps = 3/165 (1%)
Query: 175 MADSCSRPKNPTQEIKSKAPFSYQFWNIVATLTGFIVPSLLMFGSQKGFL---QPQLPFI 231
MADSC+RPK+P+Q SK PFSY+FWN+ + + GF+VP LL+FGSQ G L QPQ+PF+
Sbjct: 1 MADSCARPKDPSQSCNSKPPFSYKFWNMFSLIIGFLVPMLLLFGSQSGLLASLQPQIPFL 60
Query: 232 SSAVLLGPYLLLLSVQILTELLTWYWQSPVWLVTPIIYEAYRILQLMRGLKLGAELTAPA 291
SSAV+L PY +LL+VQ LTE+LTW+WQSPVWLVTP++YEAYRILQLMRGL L AE+ AP
Sbjct: 61 SSAVILFPYFILLAVQTLTEILTWHWQSPVWLVTPVVYEAYRILQLMRGLTLSAEVNAPV 120
Query: 292 WMMHTIRGLVCWWVLILGLQLMRVAWFAGLSARARKDQSSSSEAS 336
W++H +RGLV WWVLILG+QLMRVAWFAG ++R Q S AS
Sbjct: 121 WVVHMLRGLVSWWVLILGMQLMRVAWFAGFASRTTTGQQPQSVAS 165
>ref|NP_917126.1| P0421H07.28 [Oryza sativa (japonica cultivar-group)]
Length = 351
Score = 76.3 bits (186), Expect = 1e-12
Identities = 60/189 (31%), Positives = 94/189 (48%), Gaps = 15/189 (7%)
Query: 105 VEEKDRPGPELQLFKWPMWLLGPSILLATGMVPTLWLP--ISSIFLGPNIASLLSLIGLD 162
VE PG E +W +GP+IL+A ++P+L+L +S++F + L L +
Sbjct: 120 VESVPHPGKEASRL---VWFVGPTILVAFLVLPSLYLRKVLSAVFEDSLLTDFLILFFTE 176
Query: 163 CIFNLGATLFLLMADSCSRPKN---PTQEIKSKAPFSYQFWNIVATLTGFIVPSLLMFGS 219
+F G +F+L+ D RP P I SK+ F ++ ++ + ++P L M
Sbjct: 177 ALFYGGVAIFVLLIDKVWRPLQQVAPKSYIWSKSRF-FRISSVTTMVLSLMIPLLTM--- 232
Query: 220 QKGFLQPQLPFISSAVLLGPYLLLLSVQILTELLTWYWQSPVWLVTPIIYEAYRILQLMR 279
G + P +SA L PYL+ L VQ E + +SP W V PII++ YR+ QL R
Sbjct: 233 --GMVWPWTGPAASATL-APYLVGLVVQFAFEQYARHRKSPSWPVIPIIFKIYRLHQLNR 289
Query: 280 GLKLGAELT 288
+L LT
Sbjct: 290 AAQLVTALT 298
>gb|AAM64903.1| unknown [Arabidopsis thaliana]
Length = 367
Score = 69.3 bits (168), Expect = 2e-10
Identities = 80/322 (24%), Positives = 137/322 (41%), Gaps = 41/322 (12%)
Query: 1 MASSLIH--THYKLHTSTFERAYPRSHGLLKSGKPFQSRRFNFPSIRLNQSFICCTKLT- 57
MA+ L+ T L S F PRS +S + + +PS+ L I ++
Sbjct: 1 MAAKLVSSSTSSSLVPSIFSPFQPRSSFQFRSPRVQPAHLHCYPSLHLQTDKIGSLHISS 60
Query: 58 ----PWEPSPGVAYAP--TDNQSDNFLQSTASVFETLESSKVDESPTANVEGLVE---EK 108
P E S Y P T + ++ + T + +T+ E+ + + ++
Sbjct: 61 RGQKPSEVSRKRTYLPLATSEEKFHYTEDTTNDPDTVSPQIGTEATPRDDDSTIQYNRSD 120
Query: 109 DRPG-----------------PELQLFKWP--MWLLGPSILLATGMVPTLWLP--ISSIF 147
+PG PE Q W +WL+GP++L+++ ++P ++L +S++F
Sbjct: 121 GKPGFISFYNPRNKTEDIIIPPETQS-PWGRLLWLIGPAVLVSSFILPPVYLRRIVSAVF 179
Query: 148 LGPNIASLLSLIGLDCIFNLGATLFLLMADSCSRPKNPTQEIKSKAPFSYQFWNIVATLT 207
+ L L + +F G FLL+ D + + + P Q + VATL
Sbjct: 180 EDSLLTDFLILFFTEALFYCGVAAFLLIIDRSRKTSGKVPQNRINPPQLGQRISSVATLV 239
Query: 208 -GFIVPSLLMFGSQKGFLQPQLPFISSAVLLGPYLLLLSVQILTELLTWYWQSPVWLVTP 266
++P + M GF+ P +SA L PYL+ + VQ E Y SP + P
Sbjct: 240 LSLMIPMVTM-----GFVWPWTGPAASATL-APYLVGIVVQFAFEQYARYRNSPSSPIIP 293
Query: 267 IIYEAYRILQLMRGLKLGAELT 288
II++ YR+ QL R +L L+
Sbjct: 294 IIFQVYRLHQLNRAAQLVTALS 315
>dbj|BAB08847.1| unnamed protein product [Arabidopsis thaliana]
gi|42573776|ref|NP_974984.1| expressed protein
[Arabidopsis thaliana]
Length = 366
Score = 65.1 bits (157), Expect = 3e-09
Identities = 49/170 (28%), Positives = 84/170 (48%), Gaps = 9/170 (5%)
Query: 122 MWLLGPSILLATGMVPTLWLP--ISSIFLGPNIASLLSLIGLDCIFNLGATLFLLMADSC 179
+WL+GP++L+++ ++P ++L +S++F + L L + +F G FLL+ D
Sbjct: 151 LWLIGPAVLVSSFILPPVYLRRIVSAVFEDSLLTDFLILFFTEALFYCGVAAFLLIIDRS 210
Query: 180 SRPKNPTQEIKSKAPFSYQFWNIVATLT-GFIVPSLLMFGSQKGFLQPQLPFISSAVLLG 238
+ + + Q + VATL ++P + M GF+ P +SA L
Sbjct: 211 RKGSGKVPQNRINPSQLGQRISSVATLVLSLMIPMVTM-----GFVWPWTGPAASATL-A 264
Query: 239 PYLLLLSVQILTELLTWYWQSPVWLVTPIIYEAYRILQLMRGLKLGAELT 288
PYL+ + VQ E Y SP + PII++ YR+ QL R +L L+
Sbjct: 265 PYLVGIVVQFAFEQYARYRNSPSSPIIPIIFQVYRLHQLNRAAQLVTALS 314
>ref|NP_568963.1| expressed protein [Arabidopsis thaliana]
Length = 366
Score = 65.1 bits (157), Expect = 3e-09
Identities = 49/170 (28%), Positives = 84/170 (48%), Gaps = 9/170 (5%)
Query: 122 MWLLGPSILLATGMVPTLWLP--ISSIFLGPNIASLLSLIGLDCIFNLGATLFLLMADSC 179
+WL+GP++L+++ ++P ++L +S++F + L L + +F G FLL+ D
Sbjct: 151 LWLIGPAVLVSSFILPPVYLRRIVSAVFEDSLLTDFLILFFTEALFYCGVAAFLLIIDRS 210
Query: 180 SRPKNPTQEIKSKAPFSYQFWNIVATLT-GFIVPSLLMFGSQKGFLQPQLPFISSAVLLG 238
+ + + Q + VATL ++P + M GF+ P +SA L
Sbjct: 211 RKGSGKVPQNRINPSQLGQRISSVATLVLSLMIPMVTM-----GFVWPWTGPAASATL-A 264
Query: 239 PYLLLLSVQILTELLTWYWQSPVWLVTPIIYEAYRILQLMRGLKLGAELT 288
PYL+ + VQ E Y SP + PII++ YR+ QL R +L L+
Sbjct: 265 PYLVGIVVQFAFEQYARYRNSPSSPIIPIIFQVYRLHQLNRAAQLVTALS 314
>gb|AAW39017.1| At1g48460 [Arabidopsis thaliana] gi|56381889|gb|AAV85663.1|
At1g48460 [Arabidopsis thaliana]
gi|15221197|ref|NP_175279.1| expressed protein
[Arabidopsis thaliana]
Length = 340
Score = 59.3 bits (142), Expect = 2e-07
Identities = 52/220 (23%), Positives = 93/220 (41%), Gaps = 21/220 (9%)
Query: 69 PTDNQSDNFLQSTASVFETLESSKVDESPTANVEGLVEEKDRPGPELQLFKWP-MWLLGP 127
P QSD + A +F S T LVEE Q K +W+L P
Sbjct: 70 PEKKQSDKSNYARAELFRGKSGSVSFNGLTHQ---LVEESKLVSAPFQEEKGSFLWVLAP 126
Query: 128 SILLATGMVPTLWLP--ISSIFLGPNIASLLSLIGLDCIFNLGATLFLLMADSCSRPKNP 185
+L+++ ++P +L I + F +A +++ + +F G +FL + D RP
Sbjct: 127 VVLISSLILPQFFLSGIIEATFKNDTVAEIVTSFCFETVFYAGLAIFLSVTDRVQRPY-- 184
Query: 186 TQEIKSKAPFSYQFWNIVATLTGFIVPSLLMFGSQ------KGFLQPQLPFISSAVLLGP 239
FS + W ++ L G++ + L G + ++ I + + + P
Sbjct: 185 -------LDFSSKRWGLITGLRGYLTSAFLTMGLKVVVPVFAVYMTWPALGIDALIAVLP 237
Query: 240 YLLLLSVQILTELLTWYWQSPVWLVTPIIYEAYRILQLMR 279
+L+ +VQ + E S W + PI++E YR+ Q+ R
Sbjct: 238 FLVGCAVQRVFEARLERRGSSCWPIVPIVFEVYRLYQVTR 277
>gb|AAF79694.1| T1N15.7 [Arabidopsis thaliana] gi|25405928|pir||F96524 protein
T1N15.7 [imported] - Arabidopsis thaliana
Length = 343
Score = 55.1 bits (131), Expect = 3e-06
Identities = 53/223 (23%), Positives = 92/223 (40%), Gaps = 24/223 (10%)
Query: 69 PTDNQSDNFLQSTASVFETLESSKVDESPTANVEGLVEEKDRPGPELQLFKWP-MWLLGP 127
P QSD + A +F S T LVEE Q K +W+L P
Sbjct: 70 PEKKQSDKSNYARAELFRGKSGSVSFNGLTHQ---LVEESKLVSAPFQEEKGSFLWVLAP 126
Query: 128 SILLATGMVPTLWLP--ISSIFLGPNIASLLSLIGLDC---IFNLGATLFLLMADSCSRP 182
+L+++ ++P +L I + F +A ++ C +F G +FL + D RP
Sbjct: 127 VVLISSLILPQFFLSGIIEATFKNDTVAGRSEIVTSFCFETVFYAGLAIFLSVTDRVQRP 186
Query: 183 KNPTQEIKSKAPFSYQFWNIVATLTGFIVPSLLMFGSQ------KGFLQPQLPFISSAVL 236
FS + W ++ L G++ + L G + ++ I + +
Sbjct: 187 Y---------LDFSSKRWGLITGLRGYLTSAFLTMGLKVVVPVFAVYMTWPALGIDALIA 237
Query: 237 LGPYLLLLSVQILTELLTWYWQSPVWLVTPIIYEAYRILQLMR 279
+ P+L+ +VQ + E S W + PI++E YR+ Q+ R
Sbjct: 238 VLPFLVGCAVQRVFEARLERRGSSCWPIVPIVFEVYRLYQVTR 280
>emb|CAE05023.2| OSJNBa0044M19.10 [Oryza sativa (japonica cultivar-group)]
gi|50923831|ref|XP_472276.1| OSJNBa0044M19.10 [Oryza
sativa (japonica cultivar-group)]
Length = 274
Score = 38.9 bits (89), Expect = 0.23
Identities = 44/162 (27%), Positives = 72/162 (44%), Gaps = 22/162 (13%)
Query: 127 PSILLA--TGMVPTLWLPISSIFLGPNIASLLSLIGLDCIFNLGATLFLLMADSCSRPKN 184
PS +L G + + L + + + + + L+ LI D F + TL+LL+ S P
Sbjct: 27 PSTMLGGVMGSLRVIELQLVAFIMVFSASGLVPLI--DLAFPVATTLYLLLLSRLSFPPL 84
Query: 185 ----PTQEIKSKAPFS----YQFWNIVATLTGFIVPSLLMFGSQKGFLQPQLPFISSAVL 236
P+ S+ F +Q + ++ T G +P + G GF + + SA
Sbjct: 85 HSTLPSSSSSSQEIFRGSTWFQAYVVLGTTVGLFLPLAHVLG---GFARGDDGAVRSAT- 140
Query: 237 LGPYLLLLSVQILTE----LLTWYWQSPVWLVTPIIYEAYRI 274
P+L LLS QILTE L + PV + P++Y R+
Sbjct: 141 --PHLFLLSCQILTENVVGALGAAFSPPVRALVPLLYTVRRV 180
>ref|XP_229201.3| PREDICTED: similar to G protein-coupled receptor 112 [Rattus
norvegicus]
Length = 2794
Score = 38.5 bits (88), Expect = 0.30
Identities = 37/109 (33%), Positives = 50/109 (44%), Gaps = 10/109 (9%)
Query: 1 MASSLIHTHYK---LHTSTFERAYPRSHGLLKSGKPFQSRRFNFPSIRLNQSFICCTKLT 57
MA+ + H+ + TS F Y S L KS KP + F +N+S T L
Sbjct: 307 MATEIFHSTTATDFIDTSVFTENYTVSETLTKS-KPAAGKTTLF---LMNESTSIATTLC 362
Query: 58 PWEPSPGVAYAPTDNQSDNFLQSTASVFETLESSKVDE-SPTANVEGLV 105
P S VA PT FL S+A+ T+ S ++E SP A V G+V
Sbjct: 363 PKHKSTEVAILPTSKSQQEFLVSSAA--RTVSWSTLEETSPIATVVGIV 409
>dbj|BAC42484.1| unknown protein [Arabidopsis thaliana]
Length = 125
Score = 37.7 bits (86), Expect = 0.50
Identities = 25/67 (37%), Positives = 35/67 (51%), Gaps = 1/67 (1%)
Query: 222 GFLQPQLPFISSAVLLGPYLLLLSVQILTELLTWYWQSPVWLVTPIIYEAYRILQLMRGL 281
GF+ P +SA L PYL+ + VQ E Y SP + PII++ YR+ QL R
Sbjct: 8 GFVWPWTGPAASATL-APYLVGIVVQFAFEQYARYRNSPSSPIIPIIFQVYRLHQLNRAA 66
Query: 282 KLGAELT 288
+L L+
Sbjct: 67 QLVTALS 73
>ref|ZP_00290818.1| hypothetical protein Mmc102000477 [Magnetococcus sp. MC-1]
Length = 379
Score = 37.4 bits (85), Expect = 0.66
Identities = 35/131 (26%), Positives = 55/131 (41%), Gaps = 6/131 (4%)
Query: 117 LFKWPMWLLGPSILLATGMVPTLWLPISSIFLGPNIASLLSLIGLDCIFNLGATLFLLMA 176
L W LG ++L ++P LWL ++ G + LLS + +N L L M
Sbjct: 130 LLLWGGLTLGVTLLSQRSLLPWLWLTLTG---GAGLVKLLSSVAPFMHWNFEQQLLLSML 186
Query: 177 DSCSRPKNPTQEIKSKAP---FSYQFWNIVATLTGFIVPSLLMFGSQKGFLQPQLPFISS 233
+ ++ + P + FW +++ +TG I LL ++ Q P I
Sbjct: 187 LLPAVTSLFILLLRQRWPGMTAALHFWLLLSWITGVIWVDLLATMGERPTFQAAPPLIYG 246
Query: 234 AVLLGPYLLLL 244
V L P LLLL
Sbjct: 247 GVTLLPALLLL 257
>gb|AAT45003.1| unknown [Xerophyta humilis]
Length = 239
Score = 37.4 bits (85), Expect = 0.66
Identities = 33/120 (27%), Positives = 56/120 (46%), Gaps = 16/120 (13%)
Query: 162 DCIFNLGATLFLLMADSCSRPKNPTQEIKSKAPFS----YQFWNIVATLTGFIVPSLLMF 217
D +F + AT + ++ SR PT +++ F +Q + ++ T G +P +
Sbjct: 28 DLVFPVFATAYFIIL---SRVAFPTYHTQAREVFHGSKPFQAYVVIGTAVGLFLPLAYVL 84
Query: 218 GSQKGFLQPQLPFISSAVLLGPYLLLLSVQILTELL---TWYWQSPVWLVTPIIYEAYRI 274
G GF + + +A P+L LLS QILTE + + PV + P++Y RI
Sbjct: 85 G---GFARGDNMAVRAAT---PHLFLLSCQILTENVISGLSLFSPPVRALVPLLYTVRRI 138
>dbj|BAB10539.1| unnamed protein product [Arabidopsis thaliana]
gi|18423389|ref|NP_568771.1| expressed protein
[Arabidopsis thaliana]
Length = 242
Score = 37.4 bits (85), Expect = 0.66
Identities = 36/137 (26%), Positives = 65/137 (47%), Gaps = 16/137 (11%)
Query: 152 IASLLSLIGL----DCIFNLGATLFLLMADSCSRP--KNPTQE--IKSKAPFSYQFWNIV 203
I +LS GL D IF + ++ P NP ++ + S ++ +
Sbjct: 50 IIIVLSASGLVTIQDFIFTILTLIYFFFLSKLIFPPHNNPNRDAPLTSSTNKIFRIYVTA 109
Query: 204 ATLTGFIVPSLLMFGSQKGFLQPQLPFISSAVLLGPYLLLLSVQILTE-LLTWY-WQSPV 261
A + G I+P +F +G ++ +S+A P++ LL+ QI E L T + + +P
Sbjct: 110 AGIVGLIIPICYIF---EGIVEDDKNGVSAAA---PHVFLLASQIFMEGLATMFGFSAPA 163
Query: 262 WLVTPIIYEAYRILQLM 278
++ PI+Y A R+L L+
Sbjct: 164 RILVPIVYNARRVLTLV 180
>gb|AAL90946.1| AT5g52420/K24M7_17 [Arabidopsis thaliana]
gi|15146256|gb|AAK83611.1| AT5g52420/K24M7_17
[Arabidopsis thaliana]
Length = 242
Score = 37.4 bits (85), Expect = 0.66
Identities = 36/137 (26%), Positives = 65/137 (47%), Gaps = 16/137 (11%)
Query: 152 IASLLSLIGL----DCIFNLGATLFLLMADSCSRP--KNPTQE--IKSKAPFSYQFWNIV 203
I +LS GL D IF + ++ P NP ++ + S ++ +
Sbjct: 50 IIIVLSASGLVTIQDFIFTILTLIYFFFLSKLIFPPHNNPNRDAPLTSSTNKIFRIYVTA 109
Query: 204 ATLTGFIVPSLLMFGSQKGFLQPQLPFISSAVLLGPYLLLLSVQILTE-LLTWY-WQSPV 261
A + G I+P +F +G ++ +S+A P++ LL+ QI E L T + + +P
Sbjct: 110 AGIVGLIIPICYIF---EGIVEDDKNGVSAAA---PHVFLLASQIFMEGLATMFGFSAPA 163
Query: 262 WLVTPIIYEAYRILQLM 278
++ PI+Y A R+L L+
Sbjct: 164 RILVPIVYNARRVLALV 180
>ref|YP_041620.1| FecCD transport family protein [Staphylococcus aureus subsp. aureus
MRSA252] gi|49242525|emb|CAG41245.1| FecCD transport
family protein [Staphylococcus aureus subsp. aureus
MRSA252]
Length = 343
Score = 36.6 bits (83), Expect = 1.1
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 5/75 (6%)
Query: 208 GFIVPSLLMFGSQKGFLQPQLPFISSAVLLGPYLLLLSVQILTELLTWYWQSPVWLVTPI 267
G IVP ++ K +L I ++G LLLLS +L+ L+T+ ++SPV +VT
Sbjct: 272 GLIVPHIVKRYVSKNYLV----MIPLTFIIGADLLLLS-DVLSRLITYPYESPVGIVTSF 326
Query: 268 IYEAYRILQLMRGLK 282
+ Y + ++G+K
Sbjct: 327 VGALYFLFITIKGVK 341
>ref|YP_186978.1| iron compound ABC transporter, permease protein [Staphylococcus
aureus subsp. aureus COL] gi|57286379|gb|AAW38473.1|
iron compound ABC transporter, permease protein
[Staphylococcus aureus subsp. aureus COL]
Length = 343
Score = 36.6 bits (83), Expect = 1.1
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 5/75 (6%)
Query: 208 GFIVPSLLMFGSQKGFLQPQLPFISSAVLLGPYLLLLSVQILTELLTWYWQSPVWLVTPI 267
G IVP ++ K +L I ++G LLLLS +L+ L+T+ ++SPV +VT
Sbjct: 272 GLIVPHIVKRYVSKNYLV----MIPLTFIIGADLLLLS-DVLSRLITYPYESPVGIVTSF 326
Query: 268 IYEAYRILQLMRGLK 282
+ Y + ++G+K
Sbjct: 327 VGALYFLFITIKGVK 341
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.137 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 583,802,379
Number of Sequences: 2540612
Number of extensions: 24336213
Number of successful extensions: 66707
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 66678
Number of HSP's gapped (non-prelim): 36
length of query: 344
length of database: 863,360,394
effective HSP length: 129
effective length of query: 215
effective length of database: 535,621,446
effective search space: 115158610890
effective search space used: 115158610890
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC148718.5


