

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148718.4 + phase: 0
(324 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
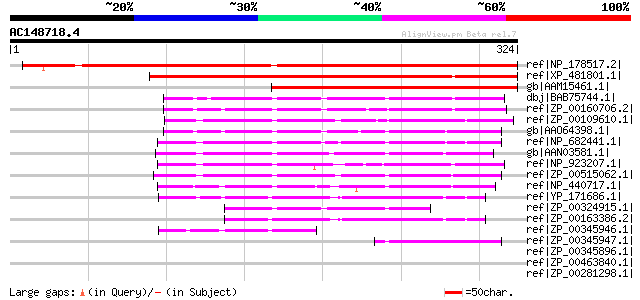
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_178517.2| expressed protein [Arabidopsis thaliana] 358 1e-97
ref|XP_481801.1| unknown protein [Oryza sativa (japonica cultiva... 344 2e-93
gb|AAM15461.1| hypothetical protein [Arabidopsis thaliana] gi|45... 227 3e-58
dbj|BAB75744.1| alr4045 [Nostoc sp. PCC 7120] gi|17231537|ref|NP... 130 7e-29
ref|ZP_00160706.2| hypothetical protein Avar03003027 [Anabaena v... 127 4e-28
ref|ZP_00109610.1| hypothetical protein Npun02003258 [Nostoc pun... 125 1e-27
gb|AAO64398.1| hypothetical protein [Nodularia spumigena] 124 5e-27
ref|NP_682441.1| hypothetical protein tll1651 [Thermosynechococc... 117 4e-25
gb|AAN03581.1| hypothetical protein [Synechococcus sp. PCC 7002] 113 9e-24
ref|NP_923207.1| hypothetical protein gll0261 [Gloeobacter viola... 99 2e-19
ref|ZP_00515062.1| conserved hypothetical protein [Crocosphaera ... 94 5e-18
ref|NP_440717.1| hypothetical protein slr1215 [Synechocystis sp.... 94 5e-18
ref|YP_171686.1| hypothetical protein syc0976_c [Synechococcus e... 86 1e-15
ref|ZP_00324915.1| hypothetical protein Tery02005319 [Trichodesm... 80 6e-14
ref|ZP_00163386.2| hypothetical protein Selo03002036 [Synechococ... 77 5e-13
ref|ZP_00345946.1| hypothetical protein Npun02000227 [Nostoc pun... 55 4e-06
ref|ZP_00345947.1| hypothetical protein Npun02000228 [Nostoc pun... 50 1e-04
ref|ZP_00345896.1| hypothetical protein Npun02000293 [Nostoc pun... 40 0.071
ref|ZP_00463840.1| Binding-protein-dependent transport systems i... 37 0.79
ref|ZP_00281298.1| COG0395: ABC-type sugar transport system, per... 37 0.79
>ref|NP_178517.2| expressed protein [Arabidopsis thaliana]
Length = 320
Score = 358 bits (919), Expect = 1e-97
Identities = 176/319 (55%), Positives = 222/319 (69%), Gaps = 10/319 (3%)
Query: 9 NLISCNFPSLKT--KKSIQPHITVVASGCCCRLSSFKPLVAMKTPF-VAATTSFRRKSSS 65
+LISCNF L K S +P +S L S A +P +A + +K +
Sbjct: 6 SLISCNFSRLPRLLKPSTRPQTLTQSSSLLLLLQSR----ASSSPHRIALFPKYEKKVTF 61
Query: 66 SSIVICNDSNKQNSSVEEKEEVKRDWTTSILLFLLWAALIYYVSFLSPNQTPSRDMYFLK 125
I + +N + + ++ RDW++SILLF LW AL+YY L+P+QTP++D+YFLK
Sbjct: 62 FHICKSSSNNPEEPEKTQIQDEGRDWSSSILLFALWGALLYYCFNLAPDQTPTQDLYFLK 121
Query: 126 KLLNLKGDDGFRMNEVLVSEWYIMGFWPLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGI 185
KLLNLKGDDGFRMN++LV WYIMG WPLVY+MLLLPTG S P WPF+ SFFGG+
Sbjct: 122 KLLNLKGDDGFRMNQILVGLWYIMGLWPLVYAMLLLPTGTSK---TPAWPFVVLSFFGGV 178
Query: 186 YALLPYFVLWKPPPPPVEEAELKTWPLNFLESKITAMILLASGIGIIIYAGLAGEDVWKE 245
YALLPYF LW PP PPV E EL+ WPLN LESK+TA + L +G+GII+Y+ + W E
Sbjct: 179 YALLPYFALWNPPSPPVSETELRQWPLNVLESKVTAGVTLVAGLGIILYSVVGNAGDWTE 238
Query: 246 FFQYCRESKFIHITSIDFTLLSTLAPFWVYNDMTARKWFDKGSWLLPVSLIPLLGPALYI 305
F+QY RESKFIH+TS+DF LLS APFWVYNDMT RKWFDKGSWLLPVS+IP LGP+LY+
Sbjct: 239 FYQYFRESKFIHVTSLDFCLLSAFAPFWVYNDMTTRKWFDKGSWLLPVSVIPFLGPSLYL 298
Query: 306 LLRPSLSTSAVAQSPAESE 324
LLRP++S + + A S+
Sbjct: 299 LLRPAVSETIAPKDTASSD 317
>ref|XP_481801.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|29647433|dbj|BAC75435.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 357
Score = 344 bits (883), Expect = 2e-93
Identities = 156/235 (66%), Positives = 190/235 (80%), Gaps = 1/235 (0%)
Query: 90 DWTTSILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIM 149
DWTTS+L+F +WA L+YY+ L+PNQTP RD YFL+KL NLKGDDGFRMN+VLV WYIM
Sbjct: 122 DWTTSVLIFGIWAGLMYYIFQLAPNQTPYRDTYFLQKLCNLKGDDGFRMNDVLVPLWYIM 181
Query: 150 GFWPLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKT 209
G WPLVYSMLLLPTGRSSKS +PVWPFL S GG YAL+PYFVLWKPPPPP++E E+
Sbjct: 182 GLWPLVYSMLLLPTGRSSKSKIPVWPFLILSCIGGAYALIPYFVLWKPPPPPIDEEEIGQ 241
Query: 210 WPLNFLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTL 269
WPL FLESK+TA + A G+G+I+YA AG + W+EF +Y RESK IHIT +DF LLS
Sbjct: 242 WPLKFLESKLTAGVTFAVGLGLIVYAAKAGGEDWQEFIRYFRESKLIHITCLDFCLLSAF 301
Query: 270 APFWVYNDMTARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTSAVAQSPAESE 324
+PFWVYND+TAR+W GSWLLP++LIP +GP+LY+LLRPSLS+ A P++ +
Sbjct: 302 SPFWVYNDLTARRW-KNGSWLLPLALIPFVGPSLYLLLRPSLSSLLAATGPSDDK 355
>gb|AAM15461.1| hypothetical protein [Arabidopsis thaliana]
gi|4587614|gb|AAD25842.1| hypothetical protein
[Arabidopsis thaliana] gi|25411076|pir||F84456
hypothetical protein At2g04360 [imported] - Arabidopsis
thaliana
Length = 295
Score = 227 bits (579), Expect = 3e-58
Identities = 100/157 (63%), Positives = 122/157 (77%)
Query: 168 KSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNFLESKITAMILLAS 227
+S P WPF+ SFFGG+YALLPYF LW PP PPV E EL+ WPLN LESK+TA + L +
Sbjct: 136 ESKTPAWPFVVLSFFGGVYALLPYFALWNPPSPPVSETELRQWPLNVLESKVTAGVTLVA 195
Query: 228 GIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWVYNDMTARKWFDKG 287
G+GII+Y+ + W EF+QY RESKFIH+TS+DF LLS APFWVYNDMT RKWFDKG
Sbjct: 196 GLGIILYSVVGNAGDWTEFYQYFRESKFIHVTSLDFCLLSAFAPFWVYNDMTTRKWFDKG 255
Query: 288 SWLLPVSLIPLLGPALYILLRPSLSTSAVAQSPAESE 324
SWLLPVS+IP LGP+LY+LLRP++S + + A S+
Sbjct: 256 SWLLPVSVIPFLGPSLYLLLRPAVSETIAPKDTASSD 292
>dbj|BAB75744.1| alr4045 [Nostoc sp. PCC 7120] gi|17231537|ref|NP_488085.1|
hypothetical protein alr4045 [Nostoc sp. PCC 7120]
gi|25341264|pir||AF2311 hypothetical protein alr4045
[imported] - Nostoc sp. (strain PCC 7120)
Length = 226
Score = 130 bits (326), Expect = 7e-29
Identities = 80/218 (36%), Positives = 116/218 (52%), Gaps = 12/218 (5%)
Query: 99 LLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPLVYSM 158
+LW +Y L+P P D + L K NL +N ++++ + +MG WP++YS
Sbjct: 9 VLWLGFTFYAFVLAPPDQP--DTFELIK--NLSTGQWQGVNPLIIALFNLMGIWPMIYSA 64
Query: 159 LLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNFLESK 218
L+ GR K +P W F +ASF GI+ALLPY L +P P + K + L+S+
Sbjct: 65 LMFIDGRGQK--IPAWLFATASFGVGIFALLPYLTLREPNP---QFPGTKNTLIKLLDSR 119
Query: 219 ITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWVYNDM 278
+T MIL ++ Y G+ W F Q + +FIH+ S+DF LLS L P + +DM
Sbjct: 120 VTGMILTIVAAILVAYGLQQGD--WSNFLQQWQTKRFIHVMSLDFCLLSLLFPALLGDDM 177
Query: 279 TARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTSAV 316
T R W K +LIPL GP LY+ LRP L ++V
Sbjct: 178 TRRGW-QKPQLFWVFALIPLFGPLLYLCLRPPLPEASV 214
>ref|ZP_00160706.2| hypothetical protein Avar03003027 [Anabaena variabilis ATCC 29413]
Length = 226
Score = 127 bits (319), Expect = 4e-28
Identities = 81/219 (36%), Positives = 119/219 (53%), Gaps = 12/219 (5%)
Query: 99 LLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPLVYSM 158
+LW +Y L+P P D + L K L++ G +N ++++ + +MG WP++YS
Sbjct: 9 VLWLGFTFYAFVLAPPDQP--DTFELIKNLSIGQWQG--INPLIIALFSLMGIWPMIYSA 64
Query: 159 LLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNFLESK 218
++ GR K +P W F +ASF GI+ALLPY L +P P K + L+S+
Sbjct: 65 IMFIDGRGQK--IPAWLFTTASFGVGIFALLPYLTLREPNP---HFPGTKNTLIKLLDSR 119
Query: 219 ITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWVYNDM 278
+T +IL GI++ GL D W F + ++FIHI S+DF LLS L P + +DM
Sbjct: 120 VTGVILTIVA-GILVAYGLQQGD-WSNFVHEWKTNRFIHIMSLDFCLLSLLFPALLRDDM 177
Query: 279 TARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTSAVA 317
R W K +LIPL GP LY+ LRP L ++VA
Sbjct: 178 ARRGW-QKPQLFWLFALIPLFGPLLYLCLRPPLPEASVA 215
>ref|ZP_00109610.1| hypothetical protein Npun02003258 [Nostoc punctiforme PCC 73102]
Length = 225
Score = 125 bits (315), Expect = 1e-27
Identities = 76/223 (34%), Positives = 123/223 (55%), Gaps = 12/223 (5%)
Query: 100 LWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPLVYSML 159
LW Y FL+P + P + + NL +N ++++ + +MG WPLVYS +
Sbjct: 10 LWLGFTIYAFFLAPPEQPGT----FELIKNLSTGQWQGVNPLVIALFNLMGIWPLVYSAV 65
Query: 160 LLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNFLESKI 219
+ G+ K +P W F++ASF GI+ALLPY L +P V + + L L+S++
Sbjct: 66 VFIDGKGQK--IPAWLFVTASFGVGIFALLPYLALREPNNQFVGKKNIL---LKLLDSRV 120
Query: 220 TAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWVYNDMT 279
T ++L + + ++ Y GL G D W F Q + ++FI++ S+DF+LLS L P + +DM
Sbjct: 121 TGIVLTVATVLLVAY-GLKGGD-WGNFVQQWQTNRFINVMSLDFSLLSLLFPALLGDDMA 178
Query: 280 ARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTSAVAQSPAE 322
R W + + L ++LIPL GP +Y+ +RP L V P +
Sbjct: 179 RRGWKNPQIFWL-IALIPLFGPLIYLCVRPPLPEVGVEAIPGQ 220
>gb|AAO64398.1| hypothetical protein [Nodularia spumigena]
Length = 219
Score = 124 bits (310), Expect = 5e-27
Identities = 80/217 (36%), Positives = 124/217 (56%), Gaps = 14/217 (6%)
Query: 99 LLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPLVYSM 158
LLW + Y +P P D + L K L++ +G +N ++++ + +MG WP++YS
Sbjct: 9 LLWLGFLTYAFLFAPPDQP--DTFELIKNLSVGKWEG--INPLVIALFNLMGIWPVIYSA 64
Query: 159 LLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNFLESK 218
+L GR K +P WPF +ASF G +ALLPY VL +P E + K+ L L+S+
Sbjct: 65 VLFMDGRGQK--IPAWPFATASFAVGAFALLPYLVLREPNQ---EFSGKKSLWLKLLDSR 119
Query: 219 ITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWVYNDM 278
+T +I L G I++ GL G+ W F Q + S+FIH+ S+DF +LS L P + +DM
Sbjct: 120 VTGLI-LTIGAAILVRYGLQGD--WGNFVQQWQTSRFIHVMSLDFCVLSLLFPALLGDDM 176
Query: 279 TARKWFDKGSWLLPVSLIPLLGPALYILLR-PSLSTS 314
AR+ + ++LIPL GP +Y+ +R P L T+
Sbjct: 177 -ARRGMKNQVFFWLIALIPLFGPLIYLSVRSPLLETN 212
>ref|NP_682441.1| hypothetical protein tll1651 [Thermosynechococcus elongatus BP-1]
gi|22295376|dbj|BAC09203.1| tll1651 [Thermosynechococcus
elongatus BP-1]
Length = 232
Score = 117 bits (294), Expect = 4e-25
Identities = 77/220 (35%), Positives = 118/220 (53%), Gaps = 13/220 (5%)
Query: 95 ILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPL 154
+ L +LW LI Y +P P L + L + +N ++VS + +MG WPL
Sbjct: 21 LFLAVLWVGLIVYSFGFAPPDQPQT----LTLIQRLATGNWQGINPLIVSLFNLMGVWPL 76
Query: 155 VYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNF 214
+Y+ LL+ G+ + P WPF + SF G +ALLPY +L +P P + AEL +
Sbjct: 77 IYTALLVVDGQGQR--FPAWPFAAFSFVVGAFALLPYLILRQPAP--LFTAELNRG-VRL 131
Query: 215 LESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWV 274
ES+IT + + I ++ Y AG+ W++F+Q S+FIH+ ++DF LLS L P +
Sbjct: 132 WESRITGIGIFLLAIVLLSYGLFAGD--WQDFWQQWHSSRFIHVMTLDFCLLSLLLPVLL 189
Query: 275 YNDMTARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTS 314
+D R+ +K W SL+P +G Y+LLRP L S
Sbjct: 190 VDDW-QRRGLEKSPWRW-WSLVPFVGALGYLLLRPPLILS 227
>gb|AAN03581.1| hypothetical protein [Synechococcus sp. PCC 7002]
Length = 231
Score = 113 bits (282), Expect = 9e-24
Identities = 73/217 (33%), Positives = 115/217 (52%), Gaps = 13/217 (5%)
Query: 94 SILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWP 153
SI L LW A + Y +P P L+ + L G +N ++++ + IMG +P
Sbjct: 4 SIFLGSLWLAFVLYAFLGAPPNQPET----LELITALSGGQWDGINPLIIALFNIMGIFP 59
Query: 154 LVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLN 213
++Y LL G K +P W F + SFF G +ALLPY L + P + K W L
Sbjct: 60 VMYGALLFLDGDRQK--LPAWVFAAGSFFLGAFALLPYLALRRDNPTFTGQ---KNWWLR 114
Query: 214 FLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFW 273
+ +S++ + L + +G++IY AG+ W +F +FIH+ S+DF LL+ L P
Sbjct: 115 WQDSRLLGLFCLVALLGLLIYGFSAGD--WGDFGTRFWGDRFIHVMSLDFCLLALLFPTL 172
Query: 274 VYNDMTARKWFDKGSWLL-PVSLIPLLGPALYILLRP 309
+ +D +AR+ + +WLL ++ P LG ALY+ LRP
Sbjct: 173 IKDD-SARRGLGEQNWLLWAIAFTPPLGAALYLALRP 208
>ref|NP_923207.1| hypothetical protein gll0261 [Gloeobacter violaceus PCC 7421]
gi|35210821|dbj|BAC88202.1| gll0261 [Gloeobacter
violaceus PCC 7421]
Length = 219
Score = 98.6 bits (244), Expect = 2e-19
Identities = 73/229 (31%), Positives = 112/229 (48%), Gaps = 24/229 (10%)
Query: 95 ILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPL 154
I L LW + Y P +T S L++ +L R + V+ + +MG PL
Sbjct: 8 IALAALWLGFVAYAFLGGPPETRSP----LQQAASLADLALGRGEPLSVALFNLMGLIPL 63
Query: 155 VYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFV------LWKPPPPPVEEAELK 208
Y LL+P K +P WPF ASF G +ALLPY + +W PP A L
Sbjct: 64 AYLFLLVPDAHGEK--LPAWPFAVASFAVGAFALLPYLICRRPHGVWPHIPPAGRAARL- 120
Query: 209 TWPLNFLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLST 268
+S+ A+ ++A G+ + + GL + W F + R S+F+H+ +DF + +
Sbjct: 121 ------FDSRWFALAVMA-GVAGLFWFGLT-QGNWAAFAEQWRTSRFVHVMGLDFLVSTA 172
Query: 269 LAPFWVYNDMTARKWFDKGSWLLPVS-LIPLLGPALYILLRPSLSTSAV 316
L P + +D+ R K +W L ++ +PL GP LY++LRPSL AV
Sbjct: 173 LLPLLIGDDLRRRS--VKRTWPLWIACALPLFGPLLYLVLRPSLPARAV 219
>ref|ZP_00515062.1| conserved hypothetical protein [Crocosphaera watsonii WH 8501]
gi|67856656|gb|EAM51897.1| conserved hypothetical
protein [Crocosphaera watsonii WH 8501]
Length = 215
Score = 94.0 bits (232), Expect = 5e-18
Identities = 61/222 (27%), Positives = 107/222 (47%), Gaps = 12/222 (5%)
Query: 93 TSILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFW 152
+ I + LLW + Y L+P P ++ + NL + +N +++S + +G
Sbjct: 3 SKIFMSLLWVGFLIYGFLLAPPGQPDT----IELIQNLSTGNWDNINPLIISVFNSIGLV 58
Query: 153 PLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPL 212
++Y+ LL GR K + WPF G +A+LPYF+L +P P E L +
Sbjct: 59 TVLYANLLFIDGREQK--IVAWPFAITGLALGTFAILPYFILREPNPKFSGEKNLF---I 113
Query: 213 NFLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPF 272
+S+ ++L + + + + ++G W +F + +FI++ S+D L + P
Sbjct: 114 KLSDSRFLHLLLTITLLVFVFWGAISGS--WVDFVSEWQSRRFINVMSLDLLCLCLIFPL 171
Query: 273 WVYNDMTARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTS 314
+ +DM R D + +SLIPLLG +Y+ RPSL S
Sbjct: 172 IIRDDMLRRN-IDSNTLFWVISLIPLLGTLVYLCFRPSLPQS 212
>ref|NP_440717.1| hypothetical protein slr1215 [Synechocystis sp. PCC 6803]
gi|1652475|dbj|BAA17397.1| slr1215 [Synechocystis sp.
PCC 6803] gi|7470388|pir||S77550 hypothetical protein
slr1215 - Synechocystis sp. (strain PCC 6803)
Length = 222
Score = 94.0 bits (232), Expect = 5e-18
Identities = 66/224 (29%), Positives = 108/224 (47%), Gaps = 23/224 (10%)
Query: 95 ILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPL 154
+ L LW AL+ + +P P + + +K L + D +N V+++ + +MG WP+
Sbjct: 6 LALACLWLALVIFAFGFAPPSNP-QTLVLIKSLSLGQWQD---INPVIIALFNLMGIWPM 61
Query: 155 VYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNF 214
Y+ +LL + K P WPF++ SFF G +ALLPY V W PP P + A P N
Sbjct: 62 TYAAMLLGDRQGRK--FPAWPFVAGSFFLGAFALLPYLVFW-PPTAPTDPA-----PSNL 113
Query: 215 LESKIT--------AMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLL 266
SK+T +L I + A G+ W ++ Q + ++FI + S DF L
Sbjct: 114 GSSKLTKFWRSPWLGRVLFLLAIACVSGAVFRGD--WADYGQQLQTNQFIAVMSSDFLCL 171
Query: 267 STLAPFWVYNDMTARKWFDKGSWLLPVSLIPLLGPALYILLRPS 310
+ P + ++ +K L+ S +PL G Y+ +RP+
Sbjct: 172 TLAFPLVLAQELKHQK-ASPSLLLVLGSTVPLFGALAYLSIRPN 214
>ref|YP_171686.1| hypothetical protein syc0976_c [Synechococcus elongatus PCC 6301]
gi|56685944|dbj|BAD79166.1| hypothetical protein
[Synechococcus elongatus PCC 6301]
Length = 230
Score = 85.9 bits (211), Expect = 1e-15
Identities = 67/209 (32%), Positives = 101/209 (48%), Gaps = 13/209 (6%)
Query: 96 LLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPLV 155
L + W +L+ Y + L+P P + L + L+L G +N ++V+ + MG WPL+
Sbjct: 10 LAAIAWLSLVSYTALLAPPDDPQT--WDLIRRLSLGQWQG--INLLIVALFNAMGLWPLL 65
Query: 156 YSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNFL 215
Y LLL G + WPF ASF G +ALLPYFV P + A WP L
Sbjct: 66 YGPLLLLDGH--QRGWRAWPFAIASFGAGAFALLPYFVFRPLPTGSLPVA----WP-PLL 118
Query: 216 ESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWVY 275
+ + L GI I + LA + +F Q + S+FI + ++DF L+ L P +
Sbjct: 119 QKLDRRWLTLPLGIAIALLGLLALQGDLADFVQQFQTSRFIQVMTLDFACLTLLFPVLLL 178
Query: 276 NDMTARKWFDKGSWLLPVSLIPLLGPALY 304
D R+ + W L + +PLLG +Y
Sbjct: 179 ED-AHRRGDRRRIWPL-IGFLPLLGAVIY 205
>ref|ZP_00324915.1| hypothetical protein Tery02005319 [Trichodesmium erythraeum IMS101]
Length = 138
Score = 80.5 bits (197), Expect = 6e-14
Identities = 51/132 (38%), Positives = 75/132 (56%), Gaps = 7/132 (5%)
Query: 138 MNEVLVSEWYIMGFWPLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKP 197
+N ++VS + I+G W L+YS LLL GR K VP W F + SF G +A+LPY L +P
Sbjct: 9 INPLIVSLFNIIGIWSLIYSCLLLSDGRGQK--VPAWLFSTLSFGVGEFAILPYLGLRQP 66
Query: 198 PPPPVEEAELKTWPLNFLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIH 257
+ K LN L+S I + L I ++IY+ +GE W +F + + S+FIH
Sbjct: 67 NS---IFSGKKNLFLNILDSLILGIALGLGTIFLLIYSFKSGE--WADFIRQWQSSRFIH 121
Query: 258 ITSIDFTLLSTL 269
+ S+DF +L L
Sbjct: 122 VMSLDFCMLCLL 133
>ref|ZP_00163386.2| hypothetical protein Selo03002036 [Synechococcus elongatus PCC
7942]
Length = 206
Score = 77.4 bits (189), Expect = 5e-13
Identities = 56/167 (33%), Positives = 82/167 (48%), Gaps = 9/167 (5%)
Query: 138 MNEVLVSEWYIMGFWPLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKP 197
+N ++V+ + MG WPL+Y LLL G + WPF ASF G +ALLPYFV
Sbjct: 24 INLLIVALFNAMGLWPLLYGPLLLLDGH--QRGWRAWPFAIASFGAGAFALLPYFVFRPL 81
Query: 198 PPPPVEEAELKTWPLNFLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIH 257
P + A WP L+ + L GI I + LA + +F Q + S+FI
Sbjct: 82 PTGSLPVA----WP-PLLQKLDRRWLTLPLGIAIALLGLLALQGDLADFVQQFQTSRFIQ 136
Query: 258 ITSIDFTLLSTLAPFWVYNDMTARKWFDKGSWLLPVSLIPLLGPALY 304
+ ++DF L+ L P + D R+ + W L + +PLLG +Y
Sbjct: 137 VMTLDFACLTLLFPVLLLED-AHRRGDRRRIWPL-IGFLPLLGAVIY 181
>ref|ZP_00345946.1| hypothetical protein Npun02000227 [Nostoc punctiforme PCC 73102]
Length = 113
Score = 54.7 bits (130), Expect = 4e-06
Identities = 30/102 (29%), Positives = 53/102 (51%), Gaps = 8/102 (7%)
Query: 96 LLFLLWAALIYYVSFLSPNQTPSRDMYFL-KKLLNLKGDDGFRMNEVLVSEWYIMGFWPL 154
+L+ +W + Y L+P D + L +KLL L+ + +N ++ + + +MG WP+
Sbjct: 14 ILWAIWLGFVTYTILLAP--VDQADTWSLGQKLLTLQWQE---VNAIIPTIFCLMGIWPM 68
Query: 155 VYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWK 196
+Y+ L+ G+ S WP+ S F G+ LLPY + K
Sbjct: 69 IYACLMFADGK--MQSFRAWPYFIGSNFTGVICLLPYLIFRK 108
>ref|ZP_00345947.1| hypothetical protein Npun02000228 [Nostoc punctiforme PCC 73102]
Length = 94
Score = 49.7 bits (117), Expect = 1e-04
Identities = 27/82 (32%), Positives = 44/82 (52%), Gaps = 3/82 (3%)
Query: 234 YAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFW-VYNDMTARKWFDKGSWLLP 292
YA +AG W+++ Q F+H+ S+DF L+ + P +++D AR+
Sbjct: 3 YAVIAGN--WEDYIQQFFHKPFVHLISLDFCLMCLIFPISSLFDDDMARRGIKDKRIFWV 60
Query: 293 VSLIPLLGPALYILLRPSLSTS 314
V++ PL GP +Y+ LRPS S
Sbjct: 61 VAIFPLFGPLVYLCLRPSFPGS 82
>ref|ZP_00345896.1| hypothetical protein Npun02000293 [Nostoc punctiforme PCC 73102]
Length = 77
Score = 40.4 bits (93), Expect = 0.071
Identities = 23/67 (34%), Positives = 37/67 (54%), Gaps = 1/67 (1%)
Query: 248 QYCRESKFIHITSIDFTLLSTLAPFWVYNDMTARKWFDKGSWLLPVSLIPLLGPALYILL 307
Q + +FI+ S+ F L L P + +D+ R W + + L V+ +PLLGP Y+ L
Sbjct: 2 QQFQSDRFINGMSLAFCLFCLLFPTILDDDIARRHWNNPQVFWL-VAFVPLLGPLFYLCL 60
Query: 308 RPSLSTS 314
RP L ++
Sbjct: 61 RPPLPST 67
>ref|ZP_00463840.1| Binding-protein-dependent transport systems inner membrane
component [Burkholderia cenocepacia HI2424]
gi|67658977|ref|ZP_00456344.1| Binding-protein-dependent
transport systems inner membrane component [Burkholderia
cenocepacia AU 1054] gi|67099885|gb|EAM17050.1|
Binding-protein-dependent transport systems inner
membrane component [Burkholderia cenocepacia HI2424]
gi|67093401|gb|EAM10943.1| Binding-protein-dependent
transport systems inner membrane component [Burkholderia
cenocepacia AU 1054]
Length = 283
Score = 37.0 bits (84), Expect = 0.79
Identities = 30/118 (25%), Positives = 53/118 (44%), Gaps = 18/118 (15%)
Query: 91 WTTSILLF--LLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLK------GDDGFRMNEVL 142
W ++LLF + W A+ +F + Q + ++F+ L + + GF N +L
Sbjct: 28 WLIALLLFFPIFWMAI---TAFKTEQQAYASTLFFMPTLDSFREVFARSNYFGFAWNSIL 84
Query: 143 VSEWY----IMGFWPLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWK 196
+S ++ P Y+M P R+ K V +W LS + L+P ++LWK
Sbjct: 85 ISAGVTVISLLFAVPAAYAMAFFPNHRTQK--VLLW-MLSTKMMPSVGVLVPIYLLWK 139
>ref|ZP_00281298.1| COG0395: ABC-type sugar transport system, permease component
[Burkholderia fungorum LB400]
Length = 291
Score = 37.0 bits (84), Expect = 0.79
Identities = 31/118 (26%), Positives = 52/118 (43%), Gaps = 18/118 (15%)
Query: 91 WTTSILLF--LLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLK------GDDGFRMNEVL 142
W ++LLF + W + +F + Q S ++F+ L + + F N VL
Sbjct: 36 WLVALLLFFPIFWMTI---TAFKTEQQAYSSSLFFIPTLDSFREVFARSNYFSFAWNSVL 92
Query: 143 VSEWY----IMGFWPLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWK 196
+S ++ P Y+M PT R+ K V +W LS + L+P ++LWK
Sbjct: 93 ISAGVTILCLLLAVPAAYAMAFFPTRRTQK--VLLW-MLSTKMMPSVGVLVPIYLLWK 147
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.137 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 565,887,142
Number of Sequences: 2540612
Number of extensions: 23727681
Number of successful extensions: 75180
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 75121
Number of HSP's gapped (non-prelim): 33
length of query: 324
length of database: 863,360,394
effective HSP length: 128
effective length of query: 196
effective length of database: 538,162,058
effective search space: 105479763368
effective search space used: 105479763368
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC148718.4


