

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147472.6 + phase: 0 /pseudo
(180 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
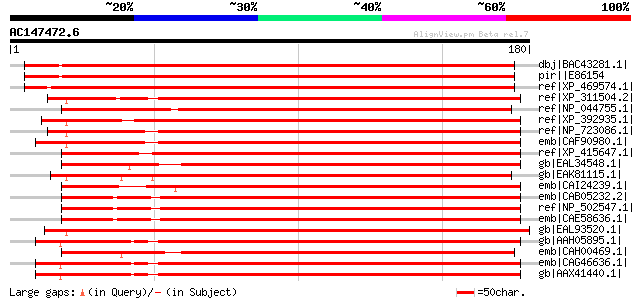
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC43281.1| unknown protein [Arabidopsis thaliana] gi|289508... 282 3e-75
pir||E86154 hypothetical protein T6A9.10 - Arabidopsis thaliana ... 282 3e-75
ref|XP_469574.1| putative cytochrome c oxidase assembly protein ... 270 2e-71
ref|XP_311504.2| ENSANGP00000022656 [Anopheles gambiae str. PEST... 186 2e-46
ref|NP_044755.1| component involved in Haem biosynthesis [Reclin... 186 2e-46
ref|XP_392935.1| PREDICTED: similar to ENSANGP00000022656 [Apis ... 182 4e-45
ref|NP_723086.1| CG31648-PA [Drosophila melanogaster] gi|2294567... 178 7e-44
emb|CAF90980.1| unnamed protein product [Tetraodon nigroviridis] 177 1e-43
ref|XP_415647.1| PREDICTED: similar to RIKEN cDNA 2010004I09 [Ga... 174 1e-42
gb|EAL34548.1| GA16364-PA [Drosophila pseudoobscura] 174 1e-42
gb|EAK81115.1| hypothetical protein UM00726.1 [Ustilago maydis 5... 174 1e-42
emb|CAI24239.1| novel protein (2010004I09Rik) [Mus musculus] gi|... 170 1e-41
emb|CAB05232.2| Hypothetical protein JC8.5 [Caenorhabditis elegans] 169 3e-41
ref|NP_502547.1| cytochrome-c oxidase assembly protein (4O11) [C... 169 3e-41
emb|CAE58636.1| Hypothetical protein CBG01804 [Caenorhabditis br... 169 4e-41
gb|EAL93520.1| cytochrome c oxidase assembly protein cox11 [Aspe... 168 6e-41
gb|AAH05895.1| COX11 homolog [Homo sapiens] 168 6e-41
emb|CAH00469.1| unnamed protein product [Kluyveromyces lactis NR... 167 9e-41
emb|CAG46636.1| COX11 [Homo sapiens] gi|60814946|gb|AAX36326.1| ... 167 2e-40
gb|AAX41440.1| COX11-like cytochrome c oxidase assembly protein ... 167 2e-40
>dbj|BAC43281.1| unknown protein [Arabidopsis thaliana] gi|28950841|gb|AAO63344.1|
At1g02410 [Arabidopsis thaliana]
gi|15217755|ref|NP_171743.1| cytochrome c oxidase
assembly protein CtaG / Cox11 family [Arabidopsis
thaliana]
Length = 287
Score = 282 bits (722), Expect = 3e-75
Identities = 137/170 (80%), Positives = 149/170 (87%), Gaps = 1/170 (0%)
Query: 6 LTGLAFGMGVLTIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQ 65
LT + FGM LT +A+ PLYRTFCQAT Y GTV R+E VEEKIARH + TVT REIVVQ
Sbjct: 97 LTAVVFGMVGLT-YAAVPLYRTFCQATGYGGTVQRKETVEEKIARHSESGTVTEREIVVQ 155
Query: 66 FNADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFN 125
FNAD++DGM WKF PTQREVRVKPGESALAFYT EN+SS PITGVSTYNVTPMK GVYFN
Sbjct: 156 FNADVADGMQWKFTPTQREVRVKPGESALAFYTAENKSSAPITGVSTYNVTPMKAGVYFN 215
Query: 126 KIQCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFK 175
KIQCFCFEEQRLLPGE+IDMPVFFYIDPE E DP+M+GINN+ILSYTFFK
Sbjct: 216 KIQCFCFEEQRLLPGEQIDMPVFFYIDPEFETDPRMDGINNLILSYTFFK 265
>pir||E86154 hypothetical protein T6A9.10 - Arabidopsis thaliana
gi|9857538|gb|AAG00893.1| Similar to cytochrome-c
oxidase assembly protein [Arabidopsis thaliana]
Length = 216
Score = 282 bits (722), Expect = 3e-75
Identities = 137/170 (80%), Positives = 149/170 (87%), Gaps = 1/170 (0%)
Query: 6 LTGLAFGMGVLTIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQ 65
LT + FGM LT +A+ PLYRTFCQAT Y GTV R+E VEEKIARH + TVT REIVVQ
Sbjct: 26 LTAVVFGMVGLT-YAAVPLYRTFCQATGYGGTVQRKETVEEKIARHSESGTVTEREIVVQ 84
Query: 66 FNADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFN 125
FNAD++DGM WKF PTQREVRVKPGESALAFYT EN+SS PITGVSTYNVTPMK GVYFN
Sbjct: 85 FNADVADGMQWKFTPTQREVRVKPGESALAFYTAENKSSAPITGVSTYNVTPMKAGVYFN 144
Query: 126 KIQCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFK 175
KIQCFCFEEQRLLPGE+IDMPVFFYIDPE E DP+M+GINN+ILSYTFFK
Sbjct: 145 KIQCFCFEEQRLLPGEQIDMPVFFYIDPEFETDPRMDGINNLILSYTFFK 194
>ref|XP_469574.1| putative cytochrome c oxidase assembly protein [Oryza sativa
(japonica cultivar-group)] gi|28301938|gb|AAO38831.1|
putative cytochrome c oxidase assembly protein [Oryza
sativa (japonica cultivar-group)]
Length = 244
Score = 270 bits (689), Expect = 2e-71
Identities = 129/170 (75%), Positives = 144/170 (83%), Gaps = 1/170 (0%)
Query: 6 LTGLAFGMGVLTIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQ 65
L G+A M V +A+ PLYR FCQAT Y GTV RRE VEEKI+RH + T TSREI+VQ
Sbjct: 72 LLGVAAAM-VGASYAAVPLYRRFCQATGYGGTVQRRESVEEKISRHARDGTTTSREIIVQ 130
Query: 66 FNADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFN 125
FNAD++DGMPWKF+PTQREV+VKPGESALAFYT EN+SS PITGVSTYNV PMK +YFN
Sbjct: 131 FNADVADGMPWKFIPTQREVKVKPGESALAFYTAENRSSAPITGVSTYNVAPMKAAIYFN 190
Query: 126 KIQCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFK 175
KIQCFCFEEQ LLPGE+IDMPVFFYIDPE E DPKM G+NNI+LSYTFFK
Sbjct: 191 KIQCFCFEEQTLLPGEQIDMPVFFYIDPEFETDPKMEGVNNIVLSYTFFK 240
>ref|XP_311504.2| ENSANGP00000022656 [Anopheles gambiae str. PEST]
gi|55242715|gb|EAA07179.2| ENSANGP00000022656 [Anopheles
gambiae str. PEST]
Length = 195
Score = 186 bits (473), Expect = 2e-46
Identities = 93/167 (55%), Positives = 119/167 (70%), Gaps = 7/167 (4%)
Query: 14 GVLTI---FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADI 70
GVLT+ +A+ PLYR FCQA SY GT T + EK+ +S Q V R I ++FNADI
Sbjct: 19 GVLTVGLSYAAVPLYRMFCQAYSYGGT-TSQGHDGEKV---ESMQKVKDRVIKIKFNADI 74
Query: 71 SDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCF 130
M W F P Q E+RV PGE+ALAFY+ +N + P+ G+STYNV P + G YFNKIQCF
Sbjct: 75 GASMRWNFKPQQTEIRVVPGETALAFYSAKNPTDHPVVGISTYNVIPFEAGAYFNKIQCF 134
Query: 131 CFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFEEQ+L P E++D+PVFFYIDP+ +DPKM +++I LSYTFF+ K
Sbjct: 135 CFEEQQLNPHEEVDLPVFFYIDPDFIEDPKMELVDSITLSYTFFESK 181
>ref|NP_044755.1| component involved in Haem biosynthesis [Reclinomonas americana]
gi|7446861|pir||S78137 cytochrome-c oxidase assembly
protein cox11 - Reclinomonas americana (ATCC 50394)
mitochondrion gi|8134373|sp|O21243|COXZ_RECAM CYTOCHROME
C OXIDASE ASSEMBLY PROTEIN CTAG
gi|2258336|gb|AAD11870.1| component involved in Haem
biosynthesis [Reclinomonas americana]
Length = 182
Score = 186 bits (473), Expect = 2e-46
Identities = 89/156 (57%), Positives = 111/156 (71%), Gaps = 2/156 (1%)
Query: 19 FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADISDGMPWKF 78
+ S PLYR FCQ T + GT + + + D Q +R I V+FN D+SD MPWKF
Sbjct: 24 YGSVPLYRIFCQVTGFGGTTQVADLESDILTLKDEQQE--NRIITVRFNGDVSDTMPWKF 81
Query: 79 LPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQRLL 138
P Q+E++V GE+ALAFY+ EN + + I G+STYNV P + G+YFNKIQCFCFEEQRL
Sbjct: 82 HPIQQEIKVMVGETALAFYSAENPTDSSIIGISTYNVNPQQAGIYFNKIQCFCFEEQRLK 141
Query: 139 PGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFF 174
P E IDMPVFF+IDP I DDPKM+ I++I LSYTFF
Sbjct: 142 PHETIDMPVFFFIDPAILDDPKMSDIDSITLSYTFF 177
>ref|XP_392935.1| PREDICTED: similar to ENSANGP00000022656 [Apis mellifera]
gi|66557694|ref|XP_623232.1| PREDICTED: similar to
ENSANGP00000022656 [Apis mellifera]
Length = 224
Score = 182 bits (462), Expect = 4e-45
Identities = 90/169 (53%), Positives = 113/169 (66%), Gaps = 7/169 (4%)
Query: 12 GMGVLTI---FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNA 68
G G+L I +AS PLYR FCQ+ +Y GT++ V + S + + R I ++FNA
Sbjct: 47 GFGILVIGFTYASVPLYRIFCQSYNYGGTLS----VNHDNTKVQSMKPIKDRVIKIKFNA 102
Query: 69 DISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQ 128
DI M W F P Q ++V PGE+ALAFY +N PITG+STYNV P + YFNKIQ
Sbjct: 103 DIGAMMQWNFKPQQNFIKVMPGETALAFYRAQNPLDIPITGISTYNVVPYEAAQYFNKIQ 162
Query: 129 CFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFCFEEQRL P E++DMPVFFYIDPE +DPKM + I+LSYTFF+ K
Sbjct: 163 CFCFEEQRLNPHEEVDMPVFFYIDPEFVNDPKMEDVEEIVLSYTFFEAK 211
>ref|NP_723086.1| CG31648-PA [Drosophila melanogaster] gi|22945671|gb|AAF52256.2|
CG31648-PA [Drosophila melanogaster]
gi|40882501|gb|AAR96162.1| RE57459p [Drosophila
melanogaster]
Length = 241
Score = 178 bits (451), Expect = 7e-44
Identities = 87/167 (52%), Positives = 111/167 (66%), Gaps = 7/167 (4%)
Query: 14 GVLTI---FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADI 70
GVL + +A+ PLY FCQA SY GT T+ E+ + + + R + ++FNADI
Sbjct: 61 GVLIVGLSYAAVPLYSIFCQAYSYGGTTTQGHDAEKV----EHMKKIEDRVLKIRFNADI 116
Query: 71 SDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCF 130
M W F P Q E++V PGE+ALAFYT N + P+ G+STYNV P + G YFNKIQCF
Sbjct: 117 GSSMRWNFKPQQYEIKVAPGETALAFYTARNPTDKPVIGISTYNVIPFEAGAYFNKIQCF 176
Query: 131 CFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFEEQ+L P E++DMPVFFYIDPEI DP + + I LSYTFF+ K
Sbjct: 177 CFEEQQLNPHEEVDMPVFFYIDPEITADPALETCDTITLSYTFFEAK 223
>emb|CAF90980.1| unnamed protein product [Tetraodon nigroviridis]
Length = 263
Score = 177 bits (449), Expect = 1e-43
Identities = 88/169 (52%), Positives = 114/169 (67%), Gaps = 5/169 (2%)
Query: 10 AFGMGVLTI-FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNA 68
A G+G++ + +A+ PLYR +CQA+ GT ++ ++ + V R I V FNA
Sbjct: 88 AAGVGMIGLSYAAVPLYRLYCQASGLGGTAVAGHNADQV----ETMKPVPERVIKVTFNA 143
Query: 69 DISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQ 128
D M W F P Q EV V PGE+ALAFY +N + PITG+STYNV P + G YFNKIQ
Sbjct: 144 DRHASMQWNFRPQQTEVFVVPGETALAFYKAKNPTDKPITGISTYNVVPFEAGQYFNKIQ 203
Query: 129 CFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFCFEEQRL P E++DMPVFFYIDPE ++DP+M ++ I LSYTFF+ K
Sbjct: 204 CFCFEEQRLNPHEEVDMPVFFYIDPEFDEDPRMARVDTITLSYTFFEAK 252
>ref|XP_415647.1| PREDICTED: similar to RIKEN cDNA 2010004I09 [Gallus gallus]
Length = 429
Score = 174 bits (441), Expect = 1e-42
Identities = 85/159 (53%), Positives = 106/159 (66%), Gaps = 4/159 (2%)
Query: 19 FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADISDGMPWKF 78
+A+ PLYR +CQAT GT E R + + V R + V FNAD+ G+ W F
Sbjct: 264 YAAVPLYRLYCQATGLGGTTGAGRGAE----RLEEMRPVRERVLKVTFNADVHAGLQWNF 319
Query: 79 LPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQRLL 138
P Q E+ V PGE+ALAFY +N + PI G+STYNV P + G YFNKIQCFCFEEQ L
Sbjct: 320 RPQQSEIYVVPGETALAFYKAKNPTDKPIIGISTYNVVPFEAGQYFNKIQCFCFEEQWLN 379
Query: 139 PGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
P E++DMPVFFYIDPE +DPKM +++I LSYTFF+ K
Sbjct: 380 PQEEVDMPVFFYIDPEFAEDPKMAKVDSITLSYTFFEAK 418
>gb|EAL34548.1| GA16364-PA [Drosophila pseudoobscura]
Length = 245
Score = 174 bits (441), Expect = 1e-42
Identities = 85/162 (52%), Positives = 108/162 (66%), Gaps = 10/162 (6%)
Query: 19 FASPPLYRTFCQATSYDGTVTR---RERVEEKIARHDSNQTVTSREIVVQFNADISDGMP 75
+A+ PLY FCQA SY GT ++ E+VE + +D R + ++FNADI M
Sbjct: 73 YAAVPLYSIFCQAYSYGGTTSQGHDAEKVEHMVKVND-------RVLKIRFNADIGSSMR 125
Query: 76 WKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQ 135
W F P Q E++V PGE+ALAFYT N + + G+STYNV P + G YFNKIQCFCFEEQ
Sbjct: 126 WNFKPQQYEIKVAPGETALAFYTARNPTDKAVIGISTYNVIPFEAGAYFNKIQCFCFEEQ 185
Query: 136 RLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
+L P E++DMPVFFYIDPEI DP + + I LSYTFF+ K
Sbjct: 186 QLNPHEEVDMPVFFYIDPEITQDPALEHCDTITLSYTFFEAK 227
>gb|EAK81115.1| hypothetical protein UM00726.1 [Ustilago maydis 521]
gi|49068104|ref|XP_398341.1| hypothetical protein
UM00726.1 [Ustilago maydis 521]
Length = 349
Score = 174 bits (440), Expect = 1e-42
Identities = 83/167 (49%), Positives = 116/167 (68%), Gaps = 7/167 (4%)
Query: 15 VLTI---FASPPLYRTFCQATSYDGT-VTRRERVEEKI---ARHDSNQTVTSREIVVQFN 67
VLT+ +A+ PLYR FC AT + GT +T ER A D + ++ I V FN
Sbjct: 157 VLTLGFSYAAVPLYRAFCSATGFSGTPITDPERFAPSRLVPAYLDPSTNAPTKRIRVTFN 216
Query: 68 ADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKI 127
AD SD +PW F P Q+E+ V PGE+ALAFYT +N+S I G++TYNV+P ++ YF K+
Sbjct: 217 ADASDAIPWSFTPVQKEIYVLPGETALAFYTAKNKSDKDIIGIATYNVSPDRIAPYFAKV 276
Query: 128 QCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFF 174
+CFCFEEQ+LL GE++D+PVFF+ID ++ DDP G+++++LSYTFF
Sbjct: 277 ECFCFEEQKLLAGEEVDLPVFFFIDSDVLDDPSTKGVDDVVLSYTFF 323
>emb|CAI24239.1| novel protein (2010004I09Rik) [Mus musculus]
gi|56206634|emb|CAI23913.1| novel protein
(2010004I09Rik) [Mus musculus]
gi|38511926|gb|AAH61233.1| COX11 homolog, cytochrome c
oxidase assembly protein [Mus musculus]
gi|39841021|ref|NP_950173.1| COX11 homolog, cytochrome c
oxidase assembly protein [Mus musculus]
Length = 275
Score = 170 bits (431), Expect = 1e-41
Identities = 84/164 (51%), Positives = 106/164 (64%), Gaps = 14/164 (8%)
Query: 19 FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQT-----VTSREIVVQFNADISDG 73
+A+ PLYR +CQ T G+ +A H S+Q V R I V FNAD+
Sbjct: 110 YAAVPLYRLYCQTTGLGGSA---------VAGHSSDQIENMVPVKDRVIKVTFNADVHAS 160
Query: 74 MPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFE 133
+ W F P Q E+ V PGE+ALAFY +N + P+ G+STYNV P + G YFNKIQCFCFE
Sbjct: 161 LQWNFRPQQTEIYVVPGETALAFYKAKNPTDKPVIGISTYNVVPFEAGQYFNKIQCFCFE 220
Query: 134 EQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
EQRL P E++DMPVFFYIDPE +DP+M ++ I LSYTFF+ K
Sbjct: 221 EQRLNPQEEVDMPVFFYIDPEFAEDPRMVNVDLITLSYTFFEAK 264
>emb|CAB05232.2| Hypothetical protein JC8.5 [Caenorhabditis elegans]
Length = 260
Score = 169 bits (428), Expect = 3e-41
Identities = 82/159 (51%), Positives = 112/159 (69%), Gaps = 4/159 (2%)
Query: 19 FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADISDGMPWKF 78
FA+ P YR FC+ TS+ G +T+ + +KIA + + R I VQFN+D+ M W+F
Sbjct: 77 FAAIPAYRIFCEQTSFGG-LTQVAKDFDKIA---NMKKCEDRLIRVQFNSDVPSSMRWEF 132
Query: 79 LPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQRLL 138
P Q E+ V PGE+ALAFYT N + PI G+S+YN+TP + YFNKIQCFCFEEQ L
Sbjct: 133 KPQQHEIYVHPGETALAFYTARNPTDKPIIGISSYNLTPFQAAYYFNKIQCFCFEEQILN 192
Query: 139 PGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
PGE++D+PVFFYIDP+ +DP + +++I+LSYTFF+ K
Sbjct: 193 PGEQVDLPVFFYIDPDYVNDPALEYLDSILLSYTFFEAK 231
>ref|NP_502547.1| cytochrome-c oxidase assembly protein (4O11) [Caenorhabditis
elegans]
Length = 195
Score = 169 bits (428), Expect = 3e-41
Identities = 82/159 (51%), Positives = 112/159 (69%), Gaps = 4/159 (2%)
Query: 19 FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADISDGMPWKF 78
FA+ P YR FC+ TS+ G +T+ + +KIA + + R I VQFN+D+ M W+F
Sbjct: 12 FAAIPAYRIFCEQTSFGG-LTQVAKDFDKIA---NMKKCEDRLIRVQFNSDVPSSMRWEF 67
Query: 79 LPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQRLL 138
P Q E+ V PGE+ALAFYT N + PI G+S+YN+TP + YFNKIQCFCFEEQ L
Sbjct: 68 KPQQHEIYVHPGETALAFYTARNPTDKPIIGISSYNLTPFQAAYYFNKIQCFCFEEQILN 127
Query: 139 PGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
PGE++D+PVFFYIDP+ +DP + +++I+LSYTFF+ K
Sbjct: 128 PGEQVDLPVFFYIDPDYVNDPALEYLDSILLSYTFFEAK 166
>emb|CAE58636.1| Hypothetical protein CBG01804 [Caenorhabditis briggsae]
Length = 426
Score = 169 bits (427), Expect = 4e-41
Identities = 82/159 (51%), Positives = 112/159 (69%), Gaps = 4/159 (2%)
Query: 19 FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADISDGMPWKF 78
FA+ P YR FC+ TS+ G +T+ + +KIA + + R I VQFN+D+ M W+F
Sbjct: 243 FAAIPAYRIFCEQTSFGG-LTQVAKDFDKIA---NMKKCEDRLIRVQFNSDVPSSMRWEF 298
Query: 79 LPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQRLL 138
P Q E+ V PGE+ALAFYT N + PI G+S+YN+TP + YFNKIQCFCFEEQ L
Sbjct: 299 KPQQHEIFVHPGETALAFYTARNPTDKPIIGISSYNLTPFQAAYYFNKIQCFCFEEQILN 358
Query: 139 PGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
PGE++D+PVFFYIDP+ +DP + +++I+LSYTFF+ K
Sbjct: 359 PGEQVDLPVFFYIDPDYVNDPALEYLDSILLSYTFFEAK 397
>gb|EAL93520.1| cytochrome c oxidase assembly protein cox11 [Aspergillus fumigatus
Af293]
Length = 226
Score = 168 bits (426), Expect = 6e-41
Identities = 82/169 (48%), Positives = 107/169 (62%), Gaps = 1/169 (0%)
Query: 13 MGVLTI-FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADIS 71
+G L + + S PLY+ CQ ++G R + +R + + FN +S
Sbjct: 48 VGTLALAYGSVPLYKMVCQQIGWNGQPVLTHRTGDGDTSSRVTPVTDARRLRITFNGSVS 107
Query: 72 DGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFC 131
D +PWKF P QREV V PGE+ALAFYT N+ T I GV+TY+VTP ++ YF+KIQCFC
Sbjct: 108 DVLPWKFTPQQREVCVLPGETALAFYTATNKGPTDIIGVATYSVTPGQVAPYFSKIQCFC 167
Query: 132 FEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDKVSE 180
FEEQ+L GE +DMPVFF+IDP+ DP M GI+ I LSYTFFK K +
Sbjct: 168 FEEQKLNAGESVDMPVFFFIDPDFVKDPAMKGIDTITLSYTFFKAKYDD 216
>gb|AAH05895.1| COX11 homolog [Homo sapiens]
Length = 276
Score = 168 bits (426), Expect = 6e-41
Identities = 84/169 (49%), Positives = 111/169 (64%), Gaps = 5/169 (2%)
Query: 10 AFGMGVL-TIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNA 68
A +G+L +A+ PLYR +CQ T G+ +KI ++ V R I + FNA
Sbjct: 101 AVAVGMLGASYAAVPLYRLYCQTTGLGGSAVAGH-ASDKI---ENMVPVKDRIIKISFNA 156
Query: 69 DISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQ 128
D+ + W F P Q E+ V PGE+ALAFY V+N + P+ G+STYN+ P + G YFNKIQ
Sbjct: 157 DVHASLQWNFRPQQTEIYVVPGETALAFYRVKNPTDKPVIGISTYNIVPFEAGQYFNKIQ 216
Query: 129 CFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFCFEEQRL P E++DMPVFFYIDPE +DP+M ++ I LSYTFF+ K
Sbjct: 217 CFCFEEQRLNPQEEVDMPVFFYIDPEFAEDPRMIKVDLITLSYTFFEAK 265
>emb|CAH00469.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50306795|ref|XP_453373.1| unnamed protein product
[Kluyveromyces lactis]
Length = 261
Score = 167 bits (424), Expect = 9e-41
Identities = 80/159 (50%), Positives = 108/159 (67%), Gaps = 7/159 (4%)
Query: 19 FASPPLYRTFCQATSYDGT--VTRRERVEEKIARHDSNQTVTSREIVVQFNADISDGMPW 76
+A+ PLYR C T Y G +R+ ++K+ DSN + I + F +++S +PW
Sbjct: 86 YAAVPLYRAICARTGYGGIPITDKRKFTDDKLIPVDSN-----KRIRISFTSEVSQILPW 140
Query: 77 KFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQR 136
KF+P QREV V PGE+ALAFY +N S I G++TY++TP + YFNKIQCFCFEEQ+
Sbjct: 141 KFVPQQREVYVLPGETALAFYKAKNNSDKDIIGMATYSITPGESSPYFNKIQCFCFEEQK 200
Query: 137 LLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFK 175
L GE++DMPVFF+IDP+ DP M I++IIL YTFFK
Sbjct: 201 LAAGEEVDMPVFFFIDPDFASDPSMRNIDDIILHYTFFK 239
>emb|CAG46636.1| COX11 [Homo sapiens] gi|60814946|gb|AAX36326.1| COX11-like
cytochrome c oxidase assembly protein [synthetic
construct] gi|4758034|ref|NP_004366.1| COX11 homolog
[Homo sapiens] gi|60416378|sp|Q9Y6N1|COX11_HUMAN
Cytochrome c oxidase assembly protein COX11,
mitochondrial precursor gi|3170264|gb|AAD08645.1|
cytochrome c oxidase assembly protein COX11 [Homo
sapiens]
Length = 276
Score = 167 bits (422), Expect = 2e-40
Identities = 83/169 (49%), Positives = 110/169 (64%), Gaps = 5/169 (2%)
Query: 10 AFGMGVL-TIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNA 68
A +G+L +A+ PLYR +CQ T G+ +KI ++ V R I + FNA
Sbjct: 101 AVAVGMLGASYAAVPLYRLYCQTTGLGGSAVAGH-ASDKI---ENMVPVKDRIIKISFNA 156
Query: 69 DISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQ 128
D+ + W F P Q E+ V PGE+ALAFY +N + P+ G+STYN+ P + G YFNKIQ
Sbjct: 157 DVHASLQWNFRPQQTEIYVVPGETALAFYRAKNPTDKPVIGISTYNIVPFEAGQYFNKIQ 216
Query: 129 CFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFCFEEQRL P E++DMPVFFYIDPE +DP+M ++ I LSYTFF+ K
Sbjct: 217 CFCFEEQRLNPQEEVDMPVFFYIDPEFAEDPRMIKVDLITLSYTFFEAK 265
>gb|AAX41440.1| COX11-like cytochrome c oxidase assembly protein [synthetic
construct]
Length = 276
Score = 167 bits (422), Expect = 2e-40
Identities = 83/169 (49%), Positives = 110/169 (64%), Gaps = 5/169 (2%)
Query: 10 AFGMGVL-TIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNA 68
A +G+L +A+ PLYR +CQ T G+ +KI ++ V R I + FNA
Sbjct: 101 AVAVGMLGASYAAVPLYRLYCQTTGLGGSAVAGH-ASDKI---ENMVPVKDRIIKISFNA 156
Query: 69 DISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQ 128
D+ + W F P Q E+ V PGE+ALAFY +N + P+ G+STYN+ P + G YFNKIQ
Sbjct: 157 DVHASLQWNFRPQQTEIYVVPGETALAFYRAKNPTDKPVIGISTYNIVPFEAGQYFNKIQ 216
Query: 129 CFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFCFEEQRL P E++DMPVFFYIDPE +DP+M ++ I LSYTFF+ K
Sbjct: 217 CFCFEEQRLNPQEEVDMPVFFYIDPEFAEDPRMIKVDLITLSYTFFEAK 265
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 302,414,904
Number of Sequences: 2540612
Number of extensions: 11673341
Number of successful extensions: 29451
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 133
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 29222
Number of HSP's gapped (non-prelim): 144
length of query: 180
length of database: 863,360,394
effective HSP length: 120
effective length of query: 60
effective length of database: 558,486,954
effective search space: 33509217240
effective search space used: 33509217240
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC147472.6


