

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147472.2 - phase: 0
(363 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
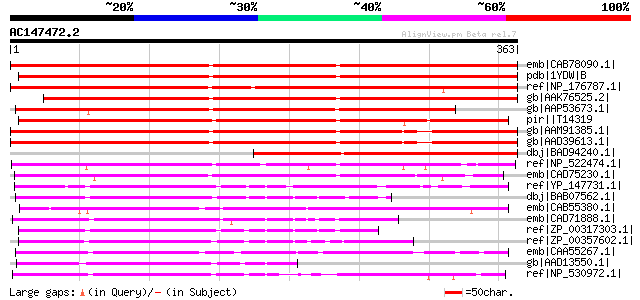
emb|CAB78090.1| AX110P-like protein [Arabidopsis thaliana] gi|45... 369 e-100
pdb|1YDW|B Chain B, X-Ray Structure Of Gene Product From Arabido... 356 7e-97
ref|NP_176787.1| oxidoreductase N-terminal domain-containing pro... 353 6e-96
gb|AAK76525.2| putative AX110P protein [Arabidopsis thaliana] 342 1e-92
gb|AAP53673.1| putative oxidoreductase [Oryza sativa (japonica c... 333 5e-90
pir||T14319 protein AX110P - carrot gi|285739|dbj|BAA03455.1| AX... 332 1e-89
gb|AAM91385.1| At1g34200/F23M19.12 [Arabidopsis thaliana] gi|136... 309 1e-82
gb|AAD39613.1| Similar to gb|D14605 AX110P embryogenesis-associa... 309 1e-82
dbj|BAD94240.1| AX110P like protein [Arabidopsis thaliana] 180 7e-44
ref|NP_522474.1| hypothetical protein RS01685 [Ralstonia solanac... 176 1e-42
emb|CAD75230.1| putative oxidoreductase [Rhodopirellula baltica ... 166 8e-40
ref|YP_147731.1| oxidoreductase [Geobacillus kaustophilus HTA426... 136 1e-30
dbj|BAB07562.1| oxidoreductase [Bacillus halodurans C-125] gi|15... 131 4e-29
emb|CAB55380.1| putative oxidoreductase [Leishmania major] 129 1e-28
emb|CAD71888.1| oxidoreductase [Rhodopirellula baltica SH 1] gi|... 121 4e-26
ref|ZP_00317303.1| COG0673: Predicted dehydrogenases and related... 117 7e-25
ref|ZP_00357602.1| COG0673: Predicted dehydrogenases and related... 115 2e-24
emb|CAA55267.1| unnamed protein product [Sinorhizobium meliloti]... 112 2e-23
gb|AAD13550.1| oxidoreductase homolog [Streptomyces cyanogenus] 111 3e-23
ref|NP_530972.1| oxidoreductase [Agrobacterium tumefaciens str. ... 109 1e-22
>emb|CAB78090.1| AX110P-like protein [Arabidopsis thaliana]
gi|4538897|emb|CAB39634.1| AX110P-like protein
[Arabidopsis thaliana] gi|21554042|gb|AAM63123.1|
AX110P-like protein [Arabidopsis thaliana]
gi|26983872|gb|AAN86188.1| putative AX110P protein
[Arabidopsis thaliana] gi|15233990|ref|NP_192705.1|
oxidoreductase family protein [Arabidopsis thaliana]
gi|7485600|pir||T04014 hypothetical protein F17A8.20 -
Arabidopsis thaliana
Length = 362
Score = 369 bits (946), Expect = e-100
Identities = 186/363 (51%), Positives = 253/363 (69%), Gaps = 4/363 (1%)
Query: 1 MTEKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIY 60
M + +R G++GCA IARK++RAI L PNATIS + SRS+ KA FA N+ P S +I+
Sbjct: 1 MATETQIRIGVMGCADIARKVSRAIHLAPNATISGVASRSLEKAKAFATANNYPESTKIH 60
Query: 61 GSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEA 120
GSY+ ++ED +DA+Y+PLPTSLHV WA+ AA K KH+L+EKP A++V E D I++ACEA
Sbjct: 61 GSYESLLEDPEIDALYVPLPTSLHVEWAIKAAEKGKHILLEKPVAMNVTEFDKIVDACEA 120
Query: 121 NGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPE 180
NGVQ MDG MW+H+PRT+ +K LS+S+ G ++ + S + ++FL+N+IRVKP
Sbjct: 121 NGVQIMDGTMWVHNPRTALLKEF--LSDSERFGQLKTVQSCFSFAGDEDFLKNDIRVKPG 178
Query: 181 LDALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTV 240
LD LGALGD WY I A+L A ++LP TVTA P N AGVILS SL W+ +
Sbjct: 179 LDGLGALGDAGWYAIRATLLANNFELPKTVTAFPGAVLNEAGVILSCGASLSWE--DGRT 236
Query: 241 ANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRPE 300
A I+CSF ++ +M+++ G+ G++ + DFIIPYKET ASF + A F DL W P
Sbjct: 237 ATIYCSFLANLTMEITAIGTKGTLRVHDFIIPYKETEASFTTSTKAWFNDLVTAWVSPPS 296
Query: 301 EVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKSLELGCKP 360
E V +LPQEA MV+EFA LV I+++G++P WP ISRKTQLVVDAVK+S++ +
Sbjct: 297 EHTVKTELPQEACMVREFARLVGEIKNNGAKPDGYWPSISRKTQLVVDAVKESVDKNYQQ 356
Query: 361 VSL 363
+SL
Sbjct: 357 ISL 359
>pdb|1YDW|B Chain B, X-Ray Structure Of Gene Product From Arabidopsis Thaliana
At4g09670 gi|60594285|pdb|1YDW|A Chain A, X-Ray
Structure Of Gene Product From Arabidopsis Thaliana
At4g09670
Length = 362
Score = 356 bits (913), Expect = 7e-97
Identities = 181/357 (50%), Positives = 245/357 (67%), Gaps = 4/357 (1%)
Query: 7 VRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEV 66
+R G+ GCA IARK++RAI L PNATIS + SRS+ KA FA N+ P S +I+GSY+ +
Sbjct: 7 IRIGVXGCADIARKVSRAIHLAPNATISGVASRSLEKAKAFATANNYPESTKIHGSYESL 66
Query: 67 VEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFM 126
+ED +DA+Y+PLPTSLHV WA+ AA K KH+L+EKP A +V E D I++ACEANGVQ
Sbjct: 67 LEDPEIDALYVPLPTSLHVEWAIKAAEKGKHILLEKPVAXNVTEFDKIVDACEANGVQIX 126
Query: 127 DGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELDALGA 186
DG W+H+PRT+ +K LS+S+ G ++ + S + ++FL+N+IRVKP LD LGA
Sbjct: 127 DGTXWVHNPRTALLKEF--LSDSERFGQLKTVQSCFSFAGDEDFLKNDIRVKPGLDGLGA 184
Query: 187 LGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVANIHCS 246
LGD WY I A+L A ++LP TVTA P N AGVILS SL W+ + A I+CS
Sbjct: 185 LGDAGWYAIRATLLANNFELPKTVTAFPGAVLNEAGVILSCGASLSWE--DGRTATIYCS 242
Query: 247 FHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRPEEVHVVN 306
F ++ + +++ G+ G++ + DFIIPYKET ASF + A F DL W P E V
Sbjct: 243 FLANLTXEITAIGTKGTLRVHDFIIPYKETEASFTTSTKAWFNDLVTAWVSPPSEHTVKT 302
Query: 307 KLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKSLELGCKPVSL 363
+LPQEA V+EFA LV I+++G++P WP ISRKTQLVVDAVK+S++ + +SL
Sbjct: 303 ELPQEACXVREFARLVGEIKNNGAKPDGYWPSISRKTQLVVDAVKESVDKNYQQISL 359
>ref|NP_176787.1| oxidoreductase N-terminal domain-containing protein [Arabidopsis
thaliana] gi|12322603|gb|AAG51297.1| oxidoreductase,
putative [Arabidopsis thaliana] gi|25404587|pir||A96686
probable oxidoreductase F15E12.2 [imported] -
Arabidopsis thaliana
Length = 364
Score = 353 bits (905), Expect = 6e-96
Identities = 186/368 (50%), Positives = 246/368 (66%), Gaps = 9/368 (2%)
Query: 1 MTEKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSL-PASVRI 59
M++ TVRFG+LGC A K R +T + NA I AI S+ A FA N+L P +V I
Sbjct: 1 MSDTKTVRFGLLGCIRFASKFVRTVTESSNAVIIAISDPSLETAKTFATGNNLSPETVTI 60
Query: 60 YGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACE 119
YGSY+E++ D+ VDAVY+ +P + RWAV AA KKKHVLVEKP A D EL+ I+EACE
Sbjct: 61 YGSYEELLNDANVDAVYLTMPVTQRARWAVTAAEKKKHVLVEKPPAQDATELEKIVEACE 120
Query: 120 ANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKP 179
NGVQFMDG +WLHH RT ++ + +S +G V+ ++ST T P ++ LE K
Sbjct: 121 YNGVQFMDGTIWLHHQRTVKIRD--TMFDSGLLGDVRHMYSTMTTPVPEQVLER--LTKE 176
Query: 180 ELDALGALGDLAWYCIGASLWAKGYKLPTTVTALP-DVTRNMAGVILSITTSLQWDQPNQ 238
+ GA+G+L WY IGA+LWA Y++P +V ALP V+ N G ILS T SLQ+
Sbjct: 177 AMGLAGAIGELGWYPIGAALWAMSYQMPISVRALPSSVSTNSVGTILSCTASLQFGSTAT 236
Query: 239 TVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVR 298
T A +HCSF S S DL+ISGS GS+ + D++IPYKE A F++T GAKFVD+HIGWNV
Sbjct: 237 TTAIVHCSFLSQLSTDLAISGSKGSIQMNDYVIPYKEDKAWFEYTSGAKFVDMHIGWNVT 296
Query: 299 PEEVHV-VNKLP--QEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKSLE 355
PE V V + P QEA+M++EF LV I+ + +WP+IS+KTQL+VDAVKKS++
Sbjct: 297 PERVTVDCSGTPETQEAMMLREFTRLVEGIKRGDLEADRRWPDISKKTQLIVDAVKKSVD 356
Query: 356 LGCKPVSL 363
+GC+ V L
Sbjct: 357 IGCEVVHL 364
>gb|AAK76525.2| putative AX110P protein [Arabidopsis thaliana]
Length = 338
Score = 342 bits (877), Expect = 1e-92
Identities = 174/339 (51%), Positives = 235/339 (68%), Gaps = 4/339 (1%)
Query: 25 ITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEVVEDSGVDAVYIPLPTSLH 84
I L PNATIS + SRS+ KA FA N+ P S +I+GSY+ ++ED +DA+Y+PLPTSLH
Sbjct: 1 IHLAPNATISGVASRSLEKAKAFATANNYPESTKIHGSYESLLEDPEIDALYVPLPTSLH 60
Query: 85 VRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFMDGCMWLHHPRTSNMKHLL 144
V WA+ AA K KH+L+EKP A++V E D I++ACEANGVQ MDG MW+H+PRT+ +K
Sbjct: 61 VEWAIKAAEKGKHILLEKPVAMNVTEFDKIVDACEANGVQIMDGTMWVHNPRTALLKEF- 119
Query: 145 DLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELDALGALGDLAWYCIGASLWAKGY 204
LS+S+ G ++ + S + ++FL+N+IRVKP LD LGALGD WY I A+L A +
Sbjct: 120 -LSDSERFGQLKTVQSCFSFAGDEDFLKNDIRVKPGLDGLGALGDAGWYAIRATLLANNF 178
Query: 205 KLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVANIHCSFHSHTSMDLSISGSNGSM 264
+LP TVTA P N AGVILS SL W+ + A I+CSF ++ +M+++ G+ G++
Sbjct: 179 ELPKTVTAFPGAVLNEAGVILSCGASLSWE--DGRTATIYCSFLANLTMEITAIGTKGTL 236
Query: 265 HLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRPEEVHVVNKLPQEALMVQEFAMLVAS 324
+ DFIIPYKET ASF + A F DL W P E V +LPQEA MV+EFA LV
Sbjct: 237 RVHDFIIPYKETEASFTTSTKAWFNDLVTAWVSPPSEHTVKTELPQEACMVREFARLVGE 296
Query: 325 IRDSGSQPSIKWPEISRKTQLVVDAVKKSLELGCKPVSL 363
I+++G++P WP ISRKTQLVVDAVK+S++ + +SL
Sbjct: 297 IKNNGAKPDGYWPSISRKTQLVVDAVKESVDKNYQQISL 335
>gb|AAP53673.1| putative oxidoreductase [Oryza sativa (japonica cultivar-group)]
gi|37534168|ref|NP_921386.1| putative oxidoreductase
[Oryza sativa (japonica cultivar-group)]
gi|21671919|gb|AAM74281.1| Putative oxidoreductase
[Oryza sativa (japonica cultivar-group)]
Length = 337
Score = 333 bits (854), Expect = 5e-90
Identities = 170/317 (53%), Positives = 226/317 (70%), Gaps = 6/317 (1%)
Query: 5 DTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPA--SVRIYGS 62
D VRFGI+GCA+IARKLARA+ L P A ++A+GSRS +KA FA E L + R++GS
Sbjct: 22 DPVRFGIMGCASIARKLARAMLLAPGAAVAAVGSRSEAKARAFAEETGLLLRHAPRLHGS 81
Query: 63 YDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANG 122
Y+ ++ D GVDAVY+PLPTSLHVRWA AAA KHVL+EKP AL A+LD IL AC+A G
Sbjct: 82 YEALLADPGVDAVYLPLPTSLHVRWATAAAAAGKHVLLEKPTALCAADLDAILAACDAAG 141
Query: 123 VQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELD 182
VQFMD MW+HHPRT+ M+ L +++ G V+ I+S + +EFL+N+IRVKP+LD
Sbjct: 142 VQFMDATMWMHHPRTAKMREL--VADEATTGDVRVINSLFSFRANEEFLQNDIRVKPDLD 199
Query: 183 ALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVAN 242
ALG LGD WY I A LWA Y+LP TV AL + RN AGV+L+ +L W + +A
Sbjct: 200 ALGTLGDAGWYSIRAILWAVDYELPKTVIALRNPVRNQAGVLLACGATLYW--ADGKIAT 257
Query: 243 IHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRPEEV 302
+CSF ++ +MD++I G+NG++H+ DF+IPY+E A+F+ +KF +LHIGW+ P +
Sbjct: 258 FNCSFLTNLTMDMTIVGTNGTLHVTDFVIPYEEKYAAFNMASKSKFAELHIGWDPLPSKH 317
Query: 303 HVVNKLPQEALMVQEFA 319
V LPQEALMVQEF+
Sbjct: 318 VVSTDLPQEALMVQEFS 334
>pir||T14319 protein AX110P - carrot gi|285739|dbj|BAA03455.1| AX110P [Daucus
carota] gi|740202|prf||2004427A embryogenesis-associated
protein
Length = 390
Score = 332 bits (850), Expect = 1e-89
Identities = 175/355 (49%), Positives = 243/355 (68%), Gaps = 9/355 (2%)
Query: 7 VRFGILGCATIARKLARAITLTP-NATISAIGSRSISKAAKFAVENSLPASVRIYGSYDE 65
V+ GILGCA IARKL+RAI L P + ISAIGSRS SKA +FA N P S ++YGSYD
Sbjct: 9 VKIGILGCAEIARKLSRAIHLVPEDVCISAIGSRSESKAIQFAAANGFPVSAKVYGSYDA 68
Query: 66 VVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQF 125
V+ED VDA+Y+PLPTSLH++WAV AA K+KH+LVEKP A+ V ELD ILEAC+ NGVQ+
Sbjct: 69 VLEDPDVDAIYMPLPTSLHLKWAVLAAQKQKHLLVEKPVAMHVDELDAILEACDDNGVQY 128
Query: 126 MDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELDALG 185
MDG M HHPR++ M+ LD +++ G ++ I S T + ++L NNIR KP+LD LG
Sbjct: 129 MDGTMLQHHPRSAKMREYLD--DAEHFGQLRSIISCFTFAASPDYLANNIRTKPDLDGLG 186
Query: 186 ALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVANIHC 245
ALGD+ W+CI + LWA ++LP ALP N AGVI+S SL W+ + A ++C
Sbjct: 187 ALGDIGWHCIRSILWAADFELPKFAIALPGPVLNKAGVIISCGASLHWE--DGKTATLNC 244
Query: 246 SFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFD--FTFGAKFVDLHIGWNVRPEEVH 303
SF + +MD+++ G+ G++H+ DF IP++E A+F G + + G + P +
Sbjct: 245 SFLENLTMDVTLVGTKGTLHMHDFNIPFEEGRATFSTGTKVGVQMICQPGGRSSTP-SIR 303
Query: 304 VVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKS-LELG 357
+ LPQE LM++EF+ LVA+I+ +G++P KW SRKTQL +D +K+S L++G
Sbjct: 304 SLLILPQECLMLKEFSRLVANIKSNGAKPEKKWATYSRKTQLTMDPIKESVLQMG 358
>gb|AAM91385.1| At1g34200/F23M19.12 [Arabidopsis thaliana]
gi|13605869|gb|AAK32920.1| F23M19.12/F23M19.12
[Arabidopsis thaliana] gi|18399170|ref|NP_564441.1|
oxidoreductase family protein [Arabidopsis thaliana]
Length = 352
Score = 309 bits (791), Expect = 1e-82
Identities = 165/364 (45%), Positives = 230/364 (62%), Gaps = 16/364 (4%)
Query: 1 MTEKDTVRFGILGCATIARKLARAITLTPNATISAIGSRS-ISKAAKFAVENSLPASVRI 59
M+ + +R G+L C RKL+RAI L+PNATISAI + S I +A FA N+ P + ++
Sbjct: 1 MSGDNQIRIGLLSCTNKVRKLSRAINLSPNATISAIATTSSIEEAKSFAKSNNFPPNTKL 60
Query: 60 YGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACE 119
+ SY+ ++ED VDAVY P+PT LHV WA AA K KH+L++KP AL+VAE D I+EACE
Sbjct: 61 HSSYESLLEDPDVDAVYFPIPTRLHVEWATLAAIKGKHILLDKPVALNVAEFDQIVEACE 120
Query: 120 ANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKP 179
NGVQFMDG W+H PRT +K +++ + G ++ ++S + + +FL+++IRVKP
Sbjct: 121 VNGVQFMDGTQWMHSPRTDKIKEF--VNDLESFGQIKSVYSCFSFAASDDFLKHDIRVKP 178
Query: 180 ELDALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQT 239
LD LGALGD WY I A L +KLP TVTAL N AG++LS W+ +
Sbjct: 179 GLDGLGALGDAGWYTIQAILLVNNFKLPKTVTALSGSVLNDAGIVLSCGALFDWE--DGV 236
Query: 240 VANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRP 299
A I+CSF ++ +M+++ G+ GS+ + DF+IPY ET A F T G V H
Sbjct: 237 NATIYCSFLANLTMEITAIGTKGSLRVHDFVIPYMETEAVFT-TSGGGCVSEH------- 288
Query: 300 EEVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKSLELGCK 359
V+ +L QEA MV EFA LV I++ G++P W SR TQL+VDAVK+S++ +
Sbjct: 289 ---KVMTELSQEACMVMEFARLVGEIKNRGAKPDGFWMRFSRNTQLLVDAVKESIDNNLE 345
Query: 360 PVSL 363
PV L
Sbjct: 346 PVFL 349
>gb|AAD39613.1| Similar to gb|D14605 AX110P embryogenesis-associated protein from
Daucus carota and is a member of the PF|01408
Oxidoreductase family. ESTs gb|Z35057, gb|T20683 and
gb|Z48399 come from this gene. [Arabidopsis thaliana]
gi|25518452|pir||C86466 hypothetical protein F23M19.12
[imported] - Arabidopsis thaliana
Length = 450
Score = 309 bits (791), Expect = 1e-82
Identities = 165/364 (45%), Positives = 230/364 (62%), Gaps = 16/364 (4%)
Query: 1 MTEKDTVRFGILGCATIARKLARAITLTPNATISAIGSRS-ISKAAKFAVENSLPASVRI 59
M+ + +R G+L C RKL+RAI L+PNATISAI + S I +A FA N+ P + ++
Sbjct: 1 MSGDNQIRIGLLSCTNKVRKLSRAINLSPNATISAIATTSSIEEAKSFAKSNNFPPNTKL 60
Query: 60 YGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACE 119
+ SY+ ++ED VDAVY P+PT LHV WA AA K KH+L++KP AL+VAE D I+EACE
Sbjct: 61 HSSYESLLEDPDVDAVYFPIPTRLHVEWATLAAIKGKHILLDKPVALNVAEFDQIVEACE 120
Query: 120 ANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKP 179
NGVQFMDG W+H PRT +K +++ + G ++ ++S + + +FL+++IRVKP
Sbjct: 121 VNGVQFMDGTQWMHSPRTDKIKEF--VNDLESFGQIKSVYSCFSFAASDDFLKHDIRVKP 178
Query: 180 ELDALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQT 239
LD LGALGD WY I A L +KLP TVTAL N AG++LS W+ +
Sbjct: 179 GLDGLGALGDAGWYTIQAILLVNNFKLPKTVTALSGSVLNDAGIVLSCGALFDWE--DGV 236
Query: 240 VANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRP 299
A I+CSF ++ +M+++ G+ GS+ + DF+IPY ET A F T G V H
Sbjct: 237 NATIYCSFLANLTMEITAIGTKGSLRVHDFVIPYMETEAVFT-TSGGGCVSEH------- 288
Query: 300 EEVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKSLELGCK 359
V+ +L QEA MV EFA LV I++ G++P W SR TQL+VDAVK+S++ +
Sbjct: 289 ---KVMTELSQEACMVMEFARLVGEIKNRGAKPDGFWMRFSRNTQLLVDAVKESIDNNLE 345
Query: 360 PVSL 363
PV L
Sbjct: 346 PVFL 349
>dbj|BAD94240.1| AX110P like protein [Arabidopsis thaliana]
Length = 190
Score = 180 bits (456), Expect = 7e-44
Identities = 96/189 (50%), Positives = 125/189 (65%), Gaps = 2/189 (1%)
Query: 175 IRVKPELDALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWD 234
+RVKP LD LGALGD WY I A+L A ++LP TVTA P N AGVILS SL W+
Sbjct: 1 MRVKPGLDGLGALGDAGWYAIRATLLANNFELPKTVTAFPGAVLNEAGVILSCGASLSWE 60
Query: 235 QPNQTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIG 294
A I+CSF ++ +M+++ G+ G++ + DFIIPYKET ASF + A F DL
Sbjct: 61 DGR--TATIYCSFLANLTMEITAIGTKGTLRVHDFIIPYKETEASFTTSTKAWFNDLVTA 118
Query: 295 WNVRPEEVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKSL 354
W P E V +LPQEA MV+EFA LV I+++G++P WP ISRKTQLVVDAVK+S+
Sbjct: 119 WVSPPSEHTVKTELPQEACMVREFARLVGEIKNNGAKPDGYWPSISRKTQLVVDAVKESV 178
Query: 355 ELGCKPVSL 363
+ + +SL
Sbjct: 179 DKNYQQISL 187
>ref|NP_522474.1| hypothetical protein RS01685 [Ralstonia solanacearum GMI1000]
gi|17431385|emb|CAD18064.1| CONSERVED HYPOTHETICAL
PROTEIN [Ralstonia solanacearum]
Length = 376
Score = 176 bits (446), Expect = 1e-42
Identities = 129/375 (34%), Positives = 191/375 (50%), Gaps = 27/375 (7%)
Query: 2 TEKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSL----PASV 57
T + + R+GILG A IARK AI + N I A+GSRS A +F E P +V
Sbjct: 11 TAEPSCRWGILGTAGIARKNWHAILHSGNGRIVAVGSRSAQTAQRFIAECQASAPAPYAV 70
Query: 58 RIYGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEA 117
YD ++ +DA+YIPLPT+L WA+ AA KH+++EKP + A+L I++A
Sbjct: 71 EAVEGYDALIRRPDIDALYIPLPTALRAEWAIKAARAGKHIVIEKPCGVTSADLLCIIDA 130
Query: 118 CEANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRV 177
A GVQFMDG M++H R M+ +LD + +G ++ I S + ++E N V
Sbjct: 131 ANAAGVQFMDGVMFMHSSRLQAMRSVLD--DGASVGDIRRITSHFSFDADAAWVERNSAV 188
Query: 178 KPELDALGALGDLAWYCIGASLWAKGYKLPTTVTA--LPDVTR-NMAGVI-LSITTSLQW 233
+P G LGD+ WY I +L+A Y +P V L V R + AGV+ L +
Sbjct: 189 EP----AGCLGDVGWYSIRLALFAMQYSMPVEVRGRILRSVKRPDGAGVVPTEFVGELLF 244
Query: 234 DQPNQTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASF---DFTFGAKFVD 290
D N A ++ +F + ISGS G +HLKDF++P+ +F + FG
Sbjct: 245 D--NGVTAALYNAFTTSRQQWADISGSKGYLHLKDFVLPHYGNEVAFTVGNDRFGGDGCF 302
Query: 291 LHIGWN---VRPEEVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVV 347
++ + V E ++ QE + + F LV SG + WPE++ KTQ V+
Sbjct: 303 FNLERHEKTVSLPEYSNNHRTAQETGLYRTFGGLVL----SGKREPF-WPEVALKTQRVM 357
Query: 348 DAVKKSLELGCKPVS 362
DA+ S G PV+
Sbjct: 358 DALLLSANAGGGPVA 372
>emb|CAD75230.1| putative oxidoreductase [Rhodopirellula baltica SH 1]
gi|32474689|ref|NP_867683.1| putative oxidoreductase
[Rhodopirellula baltica SH 1]
Length = 373
Score = 166 bits (421), Expect = 8e-40
Identities = 119/371 (32%), Positives = 178/371 (47%), Gaps = 31/371 (8%)
Query: 4 KDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRI---- 59
K R+GILG A IARK +AI ++ N ++A+ SR + +A F + SL I
Sbjct: 3 KSICRWGILGAANIARKNWKAIRMSGNGVVAAVASRDVQRAEAFIHDCSLECPPVITGPD 62
Query: 60 ----------YGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVA 109
G Y ++ +DAVYI LPT+ +W +AAA KHV+ EKP A+
Sbjct: 63 GSAELTRPEAIGDYQALLNRDDIDAVYIALPTTHRKKWVLAAAAAGKHVIAEKPFAVHAE 122
Query: 110 ELDLILEACEANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQE 169
+ + +AC GV MDG M+ H R +KH L++ G ++ I + +
Sbjct: 123 DAIEMADACREAGVNLMDGVMFDHSSRLPELKH--TLTDPVAFGDLKRIQTHFSFCGDDS 180
Query: 170 FLENNIRVKPELDALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITT 229
F E NIR +L+ GALGDL WYCI +LWA G +LPT V+ V + V
Sbjct: 181 FEEANIRASADLEPHGALGDLGWYCIRFTLWAAGDRLPTAVSGQSTVRLDDGRVPGEFQG 240
Query: 230 SLQWDQPNQTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFV 289
+++D + A CSF ++SG+ G L DF++P+ + +S++ F+
Sbjct: 241 EMRFD--DGLTAGFFCSFFCVNQQTATLSGNRGYATLNDFVLPFSGSESSWETHQHELFI 298
Query: 290 DLHIGWNV-RPEEVHVVNKL------PQEALMVQEFAMLVASIRDSGSQPSIKWPEISRK 342
D + WN H V + QE M + FA V R + EI+ K
Sbjct: 299 D-NCRWNFGEHSRKHGVTEHASGEADSQETRMFRHFADEVLHRR-----VEARSSEIAVK 352
Query: 343 TQLVVDAVKKS 353
TQ V+DA+++S
Sbjct: 353 TQQVLDALRRS 363
>ref|YP_147731.1| oxidoreductase [Geobacillus kaustophilus HTA426]
gi|56380255|dbj|BAD76163.1| oxidoreductase [Geobacillus
kaustophilus HTA426]
Length = 331
Score = 136 bits (342), Expect = 1e-30
Identities = 117/355 (32%), Positives = 169/355 (46%), Gaps = 31/355 (8%)
Query: 4 KDTVRFGILGCATIARK-LARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGS 62
K TVRFGI+ A IA++ + AI T +A + AI S S KA + A E +P + Y S
Sbjct: 2 KPTVRFGIISTAAIAKEVMIPAIRRTSHAEVVAIASES-GKAKQAAKELGIP---KAYDS 57
Query: 63 YDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANG 122
Y ++++D VDAVYIPLP SLH W + AA KKKHVL EKPAAL ++ ++E CE NG
Sbjct: 58 YGQLLDDPDVDAVYIPLPNSLHAEWTIKAAKKKKHVLCEKPAALCAEDVRRMIEECEKNG 117
Query: 123 VQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELD 182
V FM+ M+ HP+ +K LL IG V+++ S + E NIR+ EL
Sbjct: 118 VLFMEAFMYQFHPQHERVKQLLAAGE---IGEVKYMRSHFSFYLQDR--ETNIRMNDELG 172
Query: 183 ALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVAN 242
G+L D+ YC+ ++ + L T L + + +TTS N +A
Sbjct: 173 G-GSLFDVGCYCVHST----RHILDTEPVELFVQSCFDPHHSVDLTTSGWMRMENGVLAQ 227
Query: 243 IHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRPEEV 302
CSF + + G+ G I+ + D G V G NVR E V
Sbjct: 228 FTCSFDMLFQNEYEVVGTKGK------IVVSRAYRPDVDGGEGRITVATADG-NVREETV 280
Query: 303 HVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKSLELG 357
Q AL ++ F+ + P + PE + +DA + S++ G
Sbjct: 281 ----AGDQYALQIEHFSRSILE-----GTPLLYSPERMIQQARTLDACRTSMKTG 326
>dbj|BAB07562.1| oxidoreductase [Bacillus halodurans C-125]
gi|15616405|ref|NP_244710.1| oxidoreductase [Bacillus
halodurans C-125] gi|25328435|pir||C84130 oxidoreductase
BH3843 [imported] - Bacillus halodurans (strain C-125)
Length = 334
Score = 131 bits (329), Expect = 4e-29
Identities = 100/271 (36%), Positives = 142/271 (51%), Gaps = 20/271 (7%)
Query: 5 DTVRFGILGCATIARK-LARAITLTPNATISAIGSRS-ISKAAKFAVENSLPASVRIYGS 62
+ VR+GILG A IA+K L AI NA + A+ SR+ + KA +A E +P + Y
Sbjct: 3 NNVRWGILGTAGIAKKQLIPAIKRASNAEVVAVSSRNDMEKANCYADECDIP---KRYAR 59
Query: 63 YDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANG 122
Y++++ D +DAVYIPLP LH+ WA AA KHVL EKPAAL AE +++AC A+G
Sbjct: 60 YEDLLNDPDIDAVYIPLPNHLHLTWAKKAAEHGKHVLCEKPAALTAAEAKEMVDACAAHG 119
Query: 123 VQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELD 182
V FM+ M+ HP+ +K L+D S IG V+ I S + E NNIR+ E+
Sbjct: 120 VVFMEAFMYQFHPQHGEVKRLID---SGKIGEVRLIRSNFSFQLPLE--ANNIRLNREMG 174
Query: 183 ALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVAN 242
G+L D+ YCI + + G K P + + GV S T LQ + +V
Sbjct: 175 G-GSLYDVGCYCIHLTRYLTG-KEPVHAYGTGYLHPEL-GVDTSFTGVLQMEGGVTSV-- 229
Query: 243 IHCSFHSHTSMDLSISGSNGSMHLKDFIIPY 273
SF + G+ G++ I+PY
Sbjct: 230 FTSSFEQPADDSYEVVGTEGTI-----IVPY 255
>emb|CAB55380.1| putative oxidoreductase [Leishmania major]
Length = 374
Score = 129 bits (325), Expect = 1e-28
Identities = 102/367 (27%), Positives = 166/367 (44%), Gaps = 23/367 (6%)
Query: 8 RFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFA--VENSLP---ASVRIYGS 62
R G+LG A IA + + +++ +G R + + F V ++L A+V S
Sbjct: 7 RVGLLGAANIAWRAWTGVRAN-GMSVTRVGCRDVDRGRNFVKRVCDALKIDEAAVPAVCS 65
Query: 63 YDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANG 122
Y+E+V + VD VYI +P W KKHV+ EKP A D L +EA +
Sbjct: 66 YEELVSAADVDVVYIAIPVKARDHWVRECIKYKKHVVGEKPPATDAEMLRSWIEALDQQN 125
Query: 123 VQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELD 182
+ +MDG M H R +K + G GPV+ I + T+ + FL NNIR PEL+
Sbjct: 126 LLYMDGTMLSHGKR---VKEICAAVKQMG-GPVKHIFANVTLGGDEAFLANNIRTNPELE 181
Query: 183 ALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQ-TVA 241
GALGDL WYCI +L +++PT VT + +N G I+S + ++ + +
Sbjct: 182 PHGALGDLGWYCIRYALHLMDFQMPTEVTG-RILKQNDKGAIISFIGDMTFEVGGEVALV 240
Query: 242 NIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDF-TFGAKFVDLHIGWNVRPE 300
+ SF + L I+ + G + L D +P + + + + ++ N+ E
Sbjct: 241 SFFVSFEAAFEQTLHITTTEGMLQLDDMCLPMTSRPETVYYEVHNSSYDNVCESHNLHTE 300
Query: 301 EVHVVNKLPQEALMVQEFAMLVASIRDSG----------SQPSIKWPEISRKTQLVVDAV 350
H VN + + + + + G + S W +S KTQ V+D +
Sbjct: 301 VKHTVNGDAPNSQCTELWGDVARILYADGEGDARRLKAKEESSRYWATVSWKTQAVMDKM 360
Query: 351 KKSLELG 357
+S +G
Sbjct: 361 LESALIG 367
>emb|CAD71888.1| oxidoreductase [Rhodopirellula baltica SH 1]
gi|32471218|ref|NP_864211.1| oxidoreductase
[Rhodopirellula baltica SH 1]
Length = 371
Score = 121 bits (303), Expect = 4e-26
Identities = 91/281 (32%), Positives = 128/281 (45%), Gaps = 16/281 (5%)
Query: 3 EKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGS 62
E+ +RFGILG I R+L + TP +++AI SR+ +A +A + +V
Sbjct: 24 ERRPIRFGILGTGRITRRLVADLQSTPGVSVTAIASRTSERARWYADSYGIAHAV---SG 80
Query: 63 YDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANG 122
Y E++ VDAVY+ LP SLH W +A+A KHVL EKP A + +AC
Sbjct: 81 YAELIARDDVDAVYVALPPSLHAEWMIASAAAGKHVLCEKPLATTSQATQEMSDACAKAR 140
Query: 123 VQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQF-----IHSTSTMPTTQEFLENNIRV 177
V ++D WLHH RT + L + G F H +S + F + R
Sbjct: 141 VHWLDATAWLHHDRTDMFRQWLSGATVPQ-GEADFRLGSLRHVSSAVSFFNPFQSGDHRQ 199
Query: 178 KPELDALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPN 237
L G L DL WY G A LP TV A V R+ GV +T + + PN
Sbjct: 200 DAALGG-GCLLDLGWYAAGILRLAND-GLPITVNA-QSVMRD--GVPYRVTAIMNF--PN 252
Query: 238 QTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSA 278
A + C+F + T I+G S+ DF P+ + A
Sbjct: 253 DVSATMSCAFDTATRKWFEIAGEEASIVCDDFTRPWPDKPA 293
>ref|ZP_00317303.1| COG0673: Predicted dehydrogenases and related proteins
[Microbulbifer degradans 2-40]
Length = 328
Score = 117 bits (292), Expect = 7e-25
Identities = 84/260 (32%), Positives = 135/260 (51%), Gaps = 17/260 (6%)
Query: 7 VRFGILGCATIARK-LARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDE 65
+R+GI+ A IAR+ L A+ + +A + A+ SR ++KA FA + +P + YGSY+
Sbjct: 4 IRWGIVSTAKIAREWLIPALHASQHAEVVAVASRDLAKAEAFAEKAGIP---KAYGSYES 60
Query: 66 VVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEAN-GVQ 124
++ D VDA+Y PLP HV ++ A KHVL EKP +D A + +LE AN +
Sbjct: 61 LLADPNVDAIYNPLPNDQHVPVSMQAIKAGKHVLCEKPLGMDAANIQPLLELAAANPQLV 120
Query: 125 FMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELDAL 184
M+ M+ HP+ ++ +++ + +G + + + T ENN+R KP +
Sbjct: 121 VMEAFMYRFHPQWVKVREIIERGD---LGQINAVEADFTYFNRD---ENNVRNKPGIGG- 173
Query: 185 GALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVANIH 244
G L D+ YCI A+ + G + P VT + D+ GV T L + +A +
Sbjct: 174 GGLLDIGCYCISAARFIFG-REPKRVTGMLDIDPQF-GVDRHATGLLDF---GPGMATFY 228
Query: 245 CSFHSHTSMDLSISGSNGSM 264
CS S +S + ISG GS+
Sbjct: 229 CSTQSDSSQWVKISGEKGSL 248
>ref|ZP_00357602.1| COG0673: Predicted dehydrogenases and related proteins
[Chloroflexus aurantiacus]
Length = 346
Score = 115 bits (289), Expect = 2e-24
Identities = 91/284 (32%), Positives = 138/284 (48%), Gaps = 19/284 (6%)
Query: 7 VRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEV 66
+++GIL IAR+ A A+ ++ + A+ +R +A FA E + R Y Y+ +
Sbjct: 1 MKWGILSAGRIARRFASAVAVSATEQVVAVAARDAERATAFAGEFGI---ARAYSDYEAL 57
Query: 67 VEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFM 126
+ D V+AVY LP SLH RW++AA KHVL EKP A +A+ + A +A+ M
Sbjct: 58 LNDPDVEAVYNALPNSLHARWSIAALQAGKHVLCEKPLATTLADGLAMAAAAQAHRRLLM 117
Query: 127 DGCMWLHHPRTSNMKHLLDLSNSDG-IGPVQFIHSTSTMPTTQEFLENNIRVKPELDALG 185
+ M+ HP+T ++ LL +DG IG V+ + NIR+ PEL G
Sbjct: 118 EAFMYRFHPQTITVQRLL----ADGVIGDVKQVRGNFAFLLDD---TTNIRLNPELGG-G 169
Query: 186 ALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVANIHC 245
AL D+ Y + + A G P +A VT + LS L W ++ VA I C
Sbjct: 170 ALYDIGCYPLSYARMAFG-SAPQKASAF--VTGEPVDLSLS---GLLWFAEDR-VATITC 222
Query: 246 SFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFV 289
SF S + + I GS G +HL+ P + + + G + V
Sbjct: 223 SFQSAWNQRIEIIGSAGWIHLERPFNPTPDLATTIAIARGGRHV 266
>emb|CAA55267.1| unnamed protein product [Sinorhizobium meliloti]
gi|1073149|pir||S51570 hypothetical protein 334 -
Rhizobium meliloti gi|1353088|sp|P49305|YMO1_RHIME
Hypothetical 36.4 kDa protein in mocC-mocA intergenic
region (ORF334)
Length = 334
Score = 112 bits (280), Expect = 2e-23
Identities = 107/355 (30%), Positives = 155/355 (43%), Gaps = 29/355 (8%)
Query: 5 DTVRFGILGCATIA-RKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSY 63
D VRFGI+G A IA K+ ++ + AI SR + +A A + S YGSY
Sbjct: 2 DLVRFGIIGTAAIAVEKVIPSMLSAEGLEVVAIASRDLDRARAAATRFGIGRS---YGSY 58
Query: 64 DEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGV 123
DE++ D ++AVYIPLP LHV WA+ AA KHVL EKP ALDV EL +++ + G
Sbjct: 59 DEILADPEIEAVYIPLPNHLHVHWAIRAAEAGKHVLCEKPLALDVEELSRLIDCRDRTGR 118
Query: 124 QFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELDA 183
+ + M HP+ + D+ S IG V+ I T+ L+ V
Sbjct: 119 RIQEAVMIRAHPQ---WDEIFDIVASGEIGEVRAIQGV----FTEVNLDPKSIVNDASIG 171
Query: 184 LGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVANI 243
GAL DL Y I A+ + P V A+ D+ + TS P A +
Sbjct: 172 GGALYDLGVYPIAAARMVFAAE-PERVFAVSDLDPVFG---IDRLTSAVLLFPGGRQATL 227
Query: 244 HCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRPEEVH 303
S ++ I G+ S+ LK+ P + G+K E
Sbjct: 228 VVSTQLALRHNVEIFGTRKSISLKNPFNPTPDDHCRIVLDNGSKLA-------AAAAETR 280
Query: 304 VVNKLPQEALMVQEFAMLVASIRDSGSQP-SIKWPEISRKTQLVVDAVKKSLELG 357
V Q L + F+ A+IR P ++W S T V++A+++S E G
Sbjct: 281 RVAPADQYRLQAERFS---AAIRSGSPLPIELEW---SLGTMKVLNAIQRSAERG 329
>gb|AAD13550.1| oxidoreductase homolog [Streptomyces cyanogenus]
Length = 321
Score = 111 bits (278), Expect = 3e-23
Identities = 72/203 (35%), Positives = 109/203 (53%), Gaps = 14/203 (6%)
Query: 6 TVRFGILGCATIARK-LARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYD 64
TVR G+LGCA IAR+ + A+ P TI+A+ SR ++A + A R Y
Sbjct: 3 TVRIGVLGCADIARRRMLPAMAAEPGITIAAVASRDSARAGELAGR----FGARPVTGYA 58
Query: 65 EVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQ 124
E++E +DAVY+PLP +LH RW AA + KHVL EKP +AE ++ + +G+
Sbjct: 59 ELLERDDIDAVYVPLPAALHARWTEAALRRGKHVLAEKPLTTSLAESRRLIALAQESGLA 118
Query: 125 FMDGCMWLHHPRTSNMKHLLDLSNSDG-IGPVQFIHSTSTMPTTQEFLENNIRVKPELDA 183
M+ M++HH + ++ L+ DG IG ++ H+ +P +++IR PEL
Sbjct: 119 LMENVMFIHHSQHDAVRKLV----WDGVIGELRAFHAAFAIP---RLPDDDIRYVPELGG 171
Query: 184 LGALGDLAWYCIGASLWAKGYKL 206
GAL D Y + A+L G L
Sbjct: 172 -GALWDTGVYPVRAALHFLGDNL 193
>ref|NP_530972.1| oxidoreductase [Agrobacterium tumefaciens str. C58]
gi|17738597|gb|AAL41288.1| oxidoreductase [Agrobacterium
tumefaciens str. C58] gi|25328456|pir||AB2609
oxidoreductase Atu0266 [imported] - Agrobacterium
tumefaciens (strain C58, Dupont)
Length = 341
Score = 109 bits (272), Expect = 1e-22
Identities = 101/366 (27%), Positives = 160/366 (43%), Gaps = 56/366 (15%)
Query: 3 EKDTVRFGILGCATIARK-LARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYG 61
E +RFGI+ A IA+ + AI N +SAI SR ++A A S+P + +G
Sbjct: 12 ENGMLRFGIISTAKIAQDHVIPAIQDAENCVVSAIASRDHARARAVAERFSVPHA---FG 68
Query: 62 SYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEAN 121
SY+E++ +DAVYIPLPT+ HV W + AAN KHVL EKP AL E+D ++ A + N
Sbjct: 69 SYEEMLASDVIDAVYIPLPTAQHVEWTIKAANAGKHVLCEKPIALKAEEIDTLIAARDRN 128
Query: 122 GVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPEL 181
GV + M + P + +K LL S IG ++ + + N+R PEL
Sbjct: 129 GVIVSEAFMVTYSPVWAKVKELL---ASGAIGTLKHVQGAFSYFNRD---PGNMRNIPEL 182
Query: 182 DALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVA 241
GAL D+ Y PT VTR G +P + A
Sbjct: 183 GG-GALPDIGVY-------------PTI------VTRFATGA-----------EPRRVQA 211
Query: 242 NIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVR--- 298
+ T + S+ G+ L ++ + F ++++ +N
Sbjct: 212 TVERDREFGTDIYSSVRADFGTFELSFYLATQLASRQLMVFHGTDGYIEVKSPFNANRWG 271
Query: 299 PEEVHVVNKLPQEALMVQ---------EFAMLVASIRDSGSQPSIKWPEISRKTQLVVDA 349
EE+ + N+ ++ + + E ++R G ++ E SRK QL +DA
Sbjct: 272 AEEIELTNQAHNQSQVFRFQDSRQYKLEAEAFARAVRGEGEVVTL---ENSRKNQLFIDA 328
Query: 350 VKKSLE 355
+ ++ E
Sbjct: 329 IYRAAE 334
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 590,851,033
Number of Sequences: 2540612
Number of extensions: 23775050
Number of successful extensions: 58391
Number of sequences better than 10.0: 969
Number of HSP's better than 10.0 without gapping: 770
Number of HSP's successfully gapped in prelim test: 199
Number of HSP's that attempted gapping in prelim test: 57303
Number of HSP's gapped (non-prelim): 1003
length of query: 363
length of database: 863,360,394
effective HSP length: 129
effective length of query: 234
effective length of database: 535,621,446
effective search space: 125335418364
effective search space used: 125335418364
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC147472.2


