

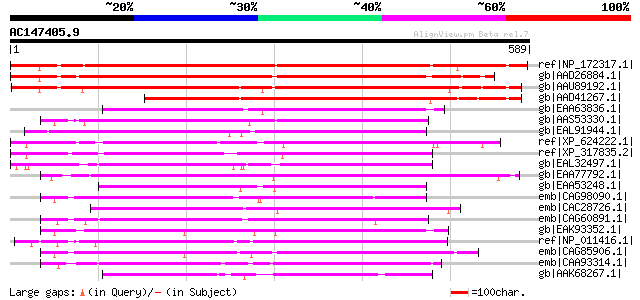
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147405.9 + phase: 0
(589 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_172317.1| GTP-binding family protein [Arabidopsis thalian... 733 0.0
gb|AAD26884.1| putative nucleotide-binding protein [Arabidopsis ... 692 0.0
gb|AAU89192.1| expressed protein [Oryza sativa (japonica cultiva... 668 0.0
gb|AAD41267.1| unknown [Zea mays] 561 e-158
gb|EAA63836.1| hypothetical protein AN2263.2 [Aspergillus nidula... 335 3e-90
gb|AAS53330.1| AFL042Cp [Ashbya gossypii ATCC 10895] gi|45198477... 333 7e-90
gb|EAL91944.1| GTP binding protein, putative [Aspergillus fumiga... 330 1e-88
ref|XP_624222.1| PREDICTED: similar to ENSANGP00000014391 [Apis ... 329 2e-88
ref|XP_317835.2| ENSANGP00000014391 [Anopheles gambiae str. PEST... 328 2e-88
gb|EAL32497.1| GA13246-PA [Drosophila pseudoobscura] 326 1e-87
gb|EAA77792.1| hypothetical protein FG07194.1 [Gibberella zeae P... 323 7e-87
gb|EAA53248.1| hypothetical protein MG07525.4 [Magnaporthe grise... 323 7e-87
emb|CAG98090.1| unnamed protein product [Kluyveromyces lactis NR... 322 3e-86
emb|CAC28726.1| conserved hypothetical protein [Neurospora crass... 321 4e-86
emb|CAG60891.1| unnamed protein product [Candida glabrata CBS138... 315 2e-84
gb|EAK93352.1| hypothetical protein CaO19.10967 [Candida albican... 313 1e-83
ref|NP_011416.1| Putative GTPase involved in 60S ribosomal subun... 311 5e-83
emb|CAG85906.1| unnamed protein product [Debaryomyces hansenii C... 310 8e-83
emb|CAA93314.1| SPAC3F10.16c [Schizosaccharomyces pombe] gi|1911... 304 5e-81
gb|AAK68267.1| Hypothetical protein C53H9.2a [Caenorhabditis ele... 296 2e-78
>ref|NP_172317.1| GTP-binding family protein [Arabidopsis thaliana]
gi|66792666|gb|AAY56435.1| At1g08410 [Arabidopsis
thaliana] gi|6664306|gb|AAF22888.1| T27G7.9 [Arabidopsis
thaliana]
Length = 589
Score = 733 bits (1891), Expect = 0.0
Identities = 390/597 (65%), Positives = 453/597 (75%), Gaps = 21/597 (3%)
Query: 1 MGKNQKTELGRALVKQHNQMIQQTKEKGKIYK---KKFLESFTEVSDIDAIIEQADEDVD 57
MGK++KT LGR+LVK HN MIQ++K+KGK YK KK LES TEVSDIDAIIEQA+E
Sbjct: 1 MGKSEKTSLGRSLVKHHNHMIQESKDKGKYYKNLQKKVLESVTEVSDIDAIIEQAEE--- 57
Query: 58 EQQLLDIPLPPPTAL-INLDHASGSDFNGLTVEEMKKEQKIEEALHASSLRVPRRPFWSA 116
++L I T L INLD S S + EE +++QKIEEALHASSL+VPRRP W+
Sbjct: 58 AERLYTINHSSSTPLSINLDTNSSSSV--IAAEEWREQQKIEEALHASSLQVPRRPPWTP 115
Query: 117 EMSADELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRVVERSDLLVMVV 176
EMS +EL ANE Q FL WRR L LEEN+KLVLTPFEKNLDIWRQLWRV+ERSDL+VMVV
Sbjct: 116 EMSVEELDANEKQAFLNWRRMLVSLEENEKLVLTPFEKNLDIWRQLWRVLERSDLIVMVV 175
Query: 177 DSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHDILFIFWSA 236
D+RDPLFYRCPDLEAYA+E+D HK +LLVNKADLLP VREKWAEYFR ++ILF+FWSA
Sbjct: 176 DARDPLFYRCPDLEAYAQEIDEHKKIMLLVNKADLLPTDVREKWAEYFRLNNILFVFWSA 235
Query: 237 KAATAVLEGKKLGSS--QADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSK 294
AATA LEGK L Q DN+ D+PD IYGRDELL+RLQ EA+ IV +R S +S
Sbjct: 236 IAATATLEGKVLKEQWRQPDNLQKTDDPDIMIYGRDELLSRLQFEAQEIVKVRNSRAASV 295
Query: 295 SSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEK 354
SS VVGFVGYPNVGKSSTINALVGQK+TGVTSTPGKTKHFQTLIIS++
Sbjct: 296 SSQSWTG-EYQRDQAVVGFVGYPNVGKSSTINALVGQKRTGVTSTPGKTKHFQTLIISDE 354
Query: 355 LILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLP 414
L+LCDCPGLVFPSFSSSRYEMI GVLPIDRMT+HRE +QVVA++VPR VIE +YNISLP
Sbjct: 355 LMLCDCPGLVFPSFSSSRYEMIASGVLPIDRMTEHREAIQVVADKVPRRVIESVYNISLP 414
Query: 415 KPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQILKDYIDGKLPHYEMPPG 474
KPK+YE QSRPP A+ELL++YCASRG SSGLPDET+A+R ILKDYI GKLPHY MPPG
Sbjct: 415 KPKTYERQSRPPHAAELLKSYCASRGYVASSGLPDETKAARLILKDYIGGKLPHYAMPPG 474
Query: 475 LSTQELASEDSNEHDQVNPHVSDASDIEDSSV---VETELAPKLEHVLDDLSSFDMANGL 531
+ A E E Q + + S+ +DS+V E E P ++ VLDDLSSFD+ANGL
Sbjct: 475 MPQ---ADEPDIEDTQELEDILEGSESDDSAVGDETENEQVPGIDDVLDDLSSFDLANGL 531
Query: 532 -ASNKVAPKKTKESQKHHRKPPRTKNRSWRAGNAGKDDTDGMPIARFHQKPVNSGPL 587
+S KV KK S K H+KP R K+R+WR N +D DGMP + QKP N+GPL
Sbjct: 532 KSSKKVTAKKQTASHKQHKKPQRKKDRTWRVQNT--EDGDGMPSVKVFQKPANTGPL 586
>gb|AAD26884.1| putative nucleotide-binding protein [Arabidopsis thaliana]
gi|25407868|pir||A84670 probable nucleotide-binding
protein [imported] - Arabidopsis thaliana
gi|15225856|ref|NP_180288.1| GTP-binding family protein
[Arabidopsis thaliana] gi|63003826|gb|AAY25442.1|
At2g27200 [Arabidopsis thaliana]
Length = 537
Score = 692 bits (1787), Expect = 0.0
Identities = 370/558 (66%), Positives = 427/558 (76%), Gaps = 30/558 (5%)
Query: 1 MGKNQKTELGRALVKQHNQMIQQTKEKGKIYK---KKFLESFTEVSDIDAIIEQADEDVD 57
MGKN+KT LGRALVK HN MIQ+TKEKGK YK KK LES TEVSDIDAIIEQA+E
Sbjct: 1 MGKNEKTSLGRALVKHHNHMIQETKEKGKSYKDQHKKVLESVTEVSDIDAIIEQAEE--- 57
Query: 58 EQQLLDIPLPPPTAL-INLDHASGSDFNGLTVEEMKKEQKIEEALHASSLRVPRRPFWSA 116
++L I T + IN+D +GS +G+T +E K+++ EEALHASSL+VPRRP W+
Sbjct: 58 AERLFAIHHDSATPVPINMD--TGSSSSGITAKEWKEQRMREEALHASSLQVPRRPHWTP 115
Query: 117 EMSADELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRVVERSDLLVMVV 176
+M+ ++L ANE Q FLTWRR LA LEEN+KLVLTPFEKNLDIWRQLWRV+ERSDL+VMVV
Sbjct: 116 KMNVEKLDANEKQAFLTWRRKLASLEENEKLVLTPFEKNLDIWRQLWRVLERSDLIVMVV 175
Query: 177 DSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHDILFIFWSA 236
D+RDPLFYRCPDLEAYA+E+D HK T+LLVNKADLLP+ VREKWAEYF ++ILF+FWSA
Sbjct: 176 DARDPLFYRCPDLEAYAQEIDEHKKTMLLVNKADLLPSYVREKWAEYFSRNNILFVFWSA 235
Query: 237 KAATAVLEGKKLGSS--QADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSK 294
KAATA LEGK L D DNP K+YGRD+LL RL+ EA IV MR+S G S
Sbjct: 236 KAATATLEGKPLKEQWRAPDTTQKTDNPAVKVYGRDDLLDRLKLEALEIVKMRKSRGVSA 295
Query: 295 SSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEK 354
+S + S VVVGFVGYPNVGKSSTINALVGQK+TGVTSTPGKTKHFQTLIISE
Sbjct: 296 TSTE-----SHCEQVVVGFVGYPNVGKSSTINALVGQKRTGVTSTPGKTKHFQTLIISED 350
Query: 355 LILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLP 414
L+LCDCPGLVFPSFSSSRYEM+ GVLPIDRMT+H E ++VVA VPRH IE++YNISLP
Sbjct: 351 LMLCDCPGLVFPSFSSSRYEMVASGVLPIDRMTEHLEAIKVVAELVPRHAIEDVYNISLP 410
Query: 415 KPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQILKDYIDGKLPHYEMPPG 474
KPKSYE QSRPPLASELLRTYC SRG SSGLPDETRA+RQILKDYI+GKLPH+ MPP
Sbjct: 411 KPKSYEPQSRPPLASELLRTYCLSRGYVASSGLPDETRAARQILKDYIEGKLPHFAMPP- 469
Query: 475 LSTQELASEDSNE--HDQVNPHVSDASDIEDSSVVETELAPKLEHVLDDLSSFDMANGLA 532
E+ +D NE D + + S E L L+ VLDDLSSFD+ANGL
Sbjct: 470 ----EITRDDENETADDTLGAETREGSQTEKKGEEAPSLG--LDQVLDDLSSFDLANGLV 523
Query: 533 SNKVAPKKTKESQKHHRK 550
S+ KTK+ +K HRK
Sbjct: 524 SS-----KTKQHKKSHRK 536
>gb|AAU89192.1| expressed protein [Oryza sativa (japonica cultivar-group)]
Length = 605
Score = 668 bits (1723), Expect = 0.0
Identities = 357/598 (59%), Positives = 438/598 (72%), Gaps = 31/598 (5%)
Query: 3 KNQKTELGRALVKQHNQMIQQTKEKGKIYK-----KKFLESFTEVSDIDAIIEQADEDVD 57
K++ LGRAL +Q N+ KE+G ++ LES EVSDIDA++++A E
Sbjct: 11 KDRGEGLGRALTRQRNKAAAAAKERGHALALARRARQPLESVIEVSDIDAVLQRAAE--- 67
Query: 58 EQQLLDIPLPPPTALINLDHASGS---DFNGL--TVEEMKKEQKIEEALHASSLRVPRRP 112
E LL + L + GS D +G T EE + ++ +EALHA SL+VPRRP
Sbjct: 68 EYLLLGGGGGDGAGDVALSASLGSGLIDLDGTVETEEERRWLREEQEALHAGSLKVPRRP 127
Query: 113 FWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRVVERSDLL 172
W+ +M+ +EL ANE + FL WRR+LARLEEN+KLVLTPFEKN+DIWRQLWRV+ERSDLL
Sbjct: 128 PWTPQMTVEELDANEKRAFLEWRRNLARLEENEKLVLTPFEKNIDIWRQLWRVLERSDLL 187
Query: 173 VMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHDILFI 232
VMVVD+RDPLFYRCPDLE YA+E+D HK TLLLVNKADLLP +VR++WAEYF+ HDIL++
Sbjct: 188 VMVVDARDPLFYRCPDLEVYAQEIDEHKRTLLLVNKADLLPLNVRQRWAEYFKQHDILYL 247
Query: 233 FWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVD----MRR 288
FWSAKAATA LEGKKL S +N +AD DTKIYGRDELL RLQ EAE IV+ +R
Sbjct: 248 FWSAKAATADLEGKKLSSYSMENWNTADL-DTKIYGRDELLVRLQGEAEYIVNQKGALRA 306
Query: 289 SSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQT 348
G S D SVS+ HVVVGFVGYPNVGKSSTINALVGQK+TGVTSTPGKTKHFQT
Sbjct: 307 EEGHESSRSD--SVSTRPKHVVVGFVGYPNVGKSSTINALVGQKRTGVTSTPGKTKHFQT 364
Query: 349 LIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRVPRHVIEEI 408
L+ISE+LILCDCPGLVFPSFSSSR+EM+ CGVLPIDRMT+HR +QVVANRVPR+V+E+I
Sbjct: 365 LVISEELILCDCPGLVFPSFSSSRHEMVACGVLPIDRMTKHRGAIQVVANRVPRNVLEQI 424
Query: 409 YNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQILKDYIDGKLPH 468
Y I+LPKPK+YE SRPP A+ELLR YC SRG + +GLPDETRA+RQILKDY+DGK+PH
Sbjct: 425 YKITLPKPKAYEQLSRPPTAAELLRAYCTSRGHVSHAGLPDETRAARQILKDYLDGKIPH 484
Query: 469 YEMPPGLSTQELASEDSNEHDQVNPHVSD-----ASDIEDSSVVETELAPKLEHVLDDLS 523
+E+PPG + E E++ + + + V ASD +D + + + P + HVL DL
Sbjct: 485 FELPPGDTDSETDPEETTDLEGSDTAVGATADHCASDEQDEEISQAD--PNISHVLSDLE 542
Query: 524 SFDMANGLASNKVAPKKTKESQKHHRKPPRTKNRSWRAGNAGKDDTDGMPIARFHQKP 581
SFD+A+ ++ N KK + S KHH+KP R K+RSWR GN D DG + R QKP
Sbjct: 543 SFDLASEVSKNS-TKKKKEASYKHHKKPQRKKDRSWRVGN---DGADGSAVVRVFQKP 596
>gb|AAD41267.1| unknown [Zea mays]
Length = 444
Score = 561 bits (1446), Expect = e-158
Identities = 282/435 (64%), Positives = 338/435 (76%), Gaps = 14/435 (3%)
Query: 154 KNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLP 213
+N+DIWRQLWRV+ERSDLLVMVVD+RDPLFYRCPDLEAYAKE+D HK T+LLVNKADLLP
Sbjct: 4 ENIDIWRQLWRVLERSDLLVMVVDARDPLFYRCPDLEAYAKEIDEHKRTILLVNKADLLP 63
Query: 214 ASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELL 273
++R++WA+YF+AHDIL++FWSAKAATA LEGKKL AS D DTKIY RDELL
Sbjct: 64 LNIRKRWADYFKAHDILYVFWSAKAATATLEGKKLSGYSEGESASLDL-DTKIYERDELL 122
Query: 274 ARLQSEAEAIVDMRRSSGS--SKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQ 331
+LQ+EAE+IV RR S + + + SVSS + HVVVGFVGYPNVGKSSTINALVG+
Sbjct: 123 MKLQAEAESIVAQRRISPTVDDHEASSSDSVSSVTKHVVVGFVGYPNVGKSSTINALVGE 182
Query: 332 KKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRE 391
K+TGVT TPGKTKHFQTLIISE+L LCDCPGLVFPSFSSSR+EM+ CGVLPIDRMT+HRE
Sbjct: 183 KRTGVTHTPGKTKHFQTLIISEELTLCDCPGLVFPSFSSSRHEMVACGVLPIDRMTKHRE 242
Query: 392 CVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDET 451
+QVVA+RVPR ++E+IY I+LPKPK YE QSRPP A+ELLR YCASRG + +GLPDET
Sbjct: 243 AIQVVADRVPRDILEQIYKIALPKPKPYEPQSRPPTAAELLRAYCASRGHVSHAGLPDET 302
Query: 452 RASRQILKDYIDGKLPHYEMPPGLS-----TQELASEDSNEHDQVNPHVSDASDIEDSSV 506
RA+RQILKDYIDGK+PH+E+PPG++ +++A +S N V+D D ED
Sbjct: 303 RAARQILKDYIDGKIPHFELPPGVTGPEVDFEQIAGSESPTTSAANESVTDDLDEEDDDA 362
Query: 507 VETELAPKLEHVLDDLSSFDMANGLASNKVAPKKTKESQKHHRKPPRTKNRSWRAGNAGK 566
V+ + VLDDL SFD+ NG +K KK + S KHH+KP R K+RSWR GN
Sbjct: 363 VD-PAESNMRDVLDDLESFDLGNG--GSKTTAKKKEASHKHHKKPQRKKDRSWRVGN--- 416
Query: 567 DDTDGMPIARFHQKP 581
D DG + R +QKP
Sbjct: 417 DGGDGTAVVRVYQKP 431
>gb|EAA63836.1| hypothetical protein AN2263.2 [Aspergillus nidulans FGSC A4]
gi|67523615|ref|XP_659867.1| hypothetical protein
AN2263_2 [Aspergillus nidulans FGSC A4]
gi|49089362|ref|XP_406400.1| hypothetical protein
AN2263.2 [Aspergillus nidulans FGSC A4]
Length = 650
Score = 335 bits (858), Expect = 3e-90
Identities = 175/414 (42%), Positives = 247/414 (59%), Gaps = 39/414 (9%)
Query: 106 LRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRV 165
L VPRRP W A + +EL A E + F+ WRR LA L+EN L++TPFE+NL++WRQLWRV
Sbjct: 124 LTVPRRPAWDASTTRNELDAMERESFMDWRRGLAELQENNDLLMTPFERNLEVWRQLWRV 183
Query: 166 VERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFR 225
+ERSD++V +VD+R+PL +R DLE Y KE+D K LLLVNKAD+L RE WA+YF
Sbjct: 184 IERSDIVVQIVDARNPLLFRSEDLETYVKEIDPKKRNLLLVNKADMLTDKQREMWADYFE 243
Query: 226 AHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVD 285
+ I F F+SA+ A E ++ D + + + L + EA+ VD
Sbjct: 244 RNQIEFRFFSAQMAKEANEARENEGEDEDTKSLTEGTENL-----NLQESKEKEADGGVD 298
Query: 286 -------------------------MRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVG 320
+ + ++K +D S V+G VGYPNVG
Sbjct: 299 LPSGTKAPSPKRTNILDVDELEELFLSNAPDATKDDEDEQDGDSKPRKTVIGLVGYPNVG 358
Query: 321 KSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGV 380
KSSTINAL+G KK V+STPGKTKHFQTL +S +++LCDCPGLVFP+F+S++ E++ GV
Sbjct: 359 KSSTINALLGAKKVSVSSTPGKTKHFQTLYLSPEIMLCDCPGLVFPNFASTKAELVVNGV 418
Query: 381 LPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRG 440
LPID+ + +VA R+P+H +E++Y +++ E + P A +LLR Y +RG
Sbjct: 419 LPIDQQREFTGPAGLVAKRIPKHFLEDVYGVTIHTRPIEEGGTGEPTAHDLLRAYARARG 478
Query: 441 -QTTSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNP 493
TT G PDE+RA+R ILKDY++GKL PP + ++ +Q++P
Sbjct: 479 FATTGQGQPDESRAARYILKDYVNGKLLFCHPPP--------ANEAEGEEQIDP 524
>gb|AAS53330.1| AFL042Cp [Ashbya gossypii ATCC 10895] gi|45198477|ref|NP_985506.1|
AFL042Cp [Eremothecium gossypii]
Length = 641
Score = 333 bits (855), Expect = 7e-90
Identities = 195/453 (43%), Positives = 272/453 (59%), Gaps = 23/453 (5%)
Query: 36 LESFTEVSDIDAII---EQADEDVDEQQLLDIPLPPPTALINLDHASGSDF---NGLTVE 89
L S T+ S +DA + E A++D + ++ + I +D +G+D +G+ +
Sbjct: 66 LRSVTQESSLDAFLSTAELAEKDFTADRHSNVKI------IRMD--AGADVATSHGIMLS 117
Query: 90 EMKKEQKIE-EALHASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARL-EENKKL 147
++ E + H++ L VPRRP+W EM+ EL E + FL WRR LA L EEN L
Sbjct: 118 NQQRAALNEKQRAHSAELIVPRRPYWDEEMTRFELERQEKEAFLNWRRKLATLQEENDDL 177
Query: 148 VLTPFEKNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVN 207
+LTPFE+N+++WRQLWRVVERSDL+V +VD+RDPL +R DLE Y KE+D K LLLVN
Sbjct: 178 LLTPFERNIEVWRQLWRVVERSDLVVQIVDARDPLLFRSTDLEEYVKELDERKQNLLLVN 237
Query: 208 KADLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIY 267
KADLL R WA+YF A I F F+SA+ A +LE +K D P +
Sbjct: 238 KADLLTTKQRIIWAKYFIAKGIAFTFFSARRANELLELQKELGEDYVQREEQDEPVAMVD 297
Query: 268 GRD---ELLARLQS-EAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSS 323
G E+L R++ + E + D+ S S+ + + +G VGYPNVGKSS
Sbjct: 298 GETVDAEVLERIRILKIEELEDLFLSKAPSEPLQEPRP--GHEPLIQIGLVGYPNVGKSS 355
Query: 324 TINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPI 383
TINALVG KK V+STPGKTKHFQT+ +S++++LCDCPGLVFP+F+ ++ E++ GVLPI
Sbjct: 356 TINALVGAKKVSVSSTPGKTKHFQTIKLSDRVMLCDCPGLVFPNFAYNKGELVCNGVLPI 415
Query: 384 DRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTT 443
D++ + +VA R+P++ +E +Y I + E P A ELL Y +RG T
Sbjct: 416 DQLRDYIGPSTLVAERIPKYFLEAVYGIHIETRSEEEGGGEHPSAQELLVAYARARGYMT 475
Query: 444 SS-GLPDETRASRQILKDYIDGKLPHYEMPPGL 475
G DE RA+R ILKDY++GKL + PP L
Sbjct: 476 QGFGSADEPRAARYILKDYVNGKLLYINPPPHL 508
>gb|EAL91944.1| GTP binding protein, putative [Aspergillus fumigatus Af293]
Length = 676
Score = 330 bits (845), Expect = 1e-88
Identities = 192/497 (38%), Positives = 278/497 (55%), Gaps = 47/497 (9%)
Query: 17 HNQMIQQTKEKGKIYKKKFLESFTEVSDIDAIIEQA--DEDVDEQQLLDIPLPPPT-ALI 73
HN I +T + G Y + + V + +I EQA DE + +L + I
Sbjct: 34 HNAAITRTAQDGSTYITNAAKEASWVK-MRSITEQAALDEFLSTAELAGTDFTAEKMSNI 92
Query: 74 NLDHASGSDFNGLTVEEMKKEQKIEEALHASSLRVPRRPFWSAEMSADELHANETQHFLT 133
+ H+ + L+ E K +K + + L VPRRP W A + D+L A E + FL
Sbjct: 93 KIIHSDQKNPYLLSASEEKSARKKHQQ-NKGKLTVPRRPKWDATTTRDQLEAMEKESFLE 151
Query: 134 WRRSLARLEENKKLVLTPFEKNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYA 193
WRR LA L+EN L++TPFE+NL++WRQLWRV+ERSD++V +VD+R+PL +R DLE Y
Sbjct: 152 WRRGLAELQENNDLLMTPFERNLEVWRQLWRVIERSDVVVQIVDARNPLMFRSEDLENYV 211
Query: 194 KEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKL----- 248
KE+D K LLLVNKAD+L RE WA+YF ++I F F+SA A E + L
Sbjct: 212 KEIDPKKQNLLLVNKADMLTEKQREMWADYFERNNINFRFFSAHLAKERNEARLLEEESD 271
Query: 249 ---GSSQADNMASADN----------------------------PDTKIYGRDELLARLQ 277
+A+++A+A + P+ + R ++L
Sbjct: 272 SESEEKEAEDLANATSSMNINDPQDAKNASIEDGQPEHDGGLKLPEYRKSRRTDIL---- 327
Query: 278 SEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVT 337
+ E + ++ SS D +G VGYPNVGKSSTINAL+G KK V+
Sbjct: 328 -DVEELEELFLSSTPDTLPDSGNGAGQRKQKTTIGLVGYPNVGKSSTINALLGAKKVSVS 386
Query: 338 STPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVA 397
+TPGKTKHFQTL +S +++LCDCPGLVFP+F++++ E++ GVLPID+ + +VA
Sbjct: 387 ATPGKTKHFQTLYLSPEIMLCDCPGLVFPNFATTKAELVVNGVLPIDQQREFTGPAGLVA 446
Query: 398 NRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRG-QTTSSGLPDETRASRQ 456
R+P+H +E++Y + + E + P A +LLR Y +RG TT G PDE+RA+R
Sbjct: 447 QRIPKHFLEDVYGVKIHTRPLEEGGTGIPSAHDLLRAYARARGFATTGQGQPDESRAARY 506
Query: 457 ILKDYIDGKLPHYEMPP 473
+LKDY++GKL PP
Sbjct: 507 VLKDYVNGKLLFCHPPP 523
>ref|XP_624222.1| PREDICTED: similar to ENSANGP00000014391 [Apis mellifera]
Length = 608
Score = 329 bits (843), Expect = 2e-88
Identities = 213/615 (34%), Positives = 317/615 (50%), Gaps = 79/615 (12%)
Query: 2 GKNQKTELGRALVKQH-----------NQMIQQTK-EKGKIYKKKFLESFTEVSDIDAII 49
GK + LG++L++ + M+ T+ G + + L+S TE + +
Sbjct: 5 GKIKSGNLGKSLIRDRFSSSRNKRNVDSSMLHTTELNDGYDWGRLNLQSVTEENSFQEFL 64
Query: 50 EQADEDVDE--QQLLDIPLPPPTALINLDHASGSDFNGLTVEEMKKEQKIEEALHASSLR 107
A+ E + L+I P + I L D +E KK + + L+
Sbjct: 65 STAELAGTEFHAEKLNIKFVNPKSGIGL---LSKDEKEKVLESQKKNKTL--------LK 113
Query: 108 VPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRVVE 167
+PRRP W + +A EL + E + FL WRRSL+ L+E + L+LTP+EKNL+ WRQLWRVVE
Sbjct: 114 IPRRPKWDSSTTAHELQSKEREEFLEWRRSLSMLQETEGLMLTPYEKNLEFWRQLWRVVE 173
Query: 168 RSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAH 227
RSD++V +VD+R+PL +RC DLE+Y KEVD K ++L+NKAD L R+ WAEYF
Sbjct: 174 RSDIVVQIVDARNPLLFRCEDLESYVKEVDSKKMNIILINKADFLTEEQRQIWAEYFTNI 233
Query: 228 DILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVDMR 287
+ F+SA AV + K + D + N D + D+ L + +E
Sbjct: 234 KLKVAFFSA--ILAVEKEKIKDIIEEDYSENEQNSDIENDDSDKSLYTSEFASE------ 285
Query: 288 RSSGSSKSSDDNASVSSSSSHV-----------------------VVGFVGYPNVGKSST 324
S S +DN + +SS + +G VGYPNVGKSST
Sbjct: 286 -SEYESADDEDNTEIKNSSELLSRDQLVLFFKTIYKGETYTKGITTIGLVGYPNVGKSST 344
Query: 325 INALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPID 384
INAL+ KK V++TPGKTKHFQTL + + L+LCDCPGLV PSF ++ EMI G+LPID
Sbjct: 345 INALLMDKKVSVSATPGKTKHFQTLFLDKDLLLCDCPGLVMPSFVCTKAEMIIHGILPID 404
Query: 385 RMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTS 444
+M H + ++ +PRH+IE++Y I +P P E R P A E+L Y SRG T
Sbjct: 405 QMKDHVPSITLLGTLIPRHIIEDLYGIMIPLPLEGEDSDRAPTAEEILNAYGYSRGFMTQ 464
Query: 445 SGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQE---------LASED---------SN 486
+G PD R++R +LKD+++G+L + PP + ++ + SE+ +N
Sbjct: 465 NGQPDNPRSARYLLKDFVNGRLLYCVAPPTIEQKKFHTFPPRKRIISENKHLPPRTVRAN 524
Query: 487 EHDQVNPHVSDASDIEDS-SVVETELAPKLEHVLDDLSSFDMANGLASNK---VAPKKTK 542
+ ++ P D +++ S + + H L SS D+ + S + + K K
Sbjct: 525 KGSKITPEDVDKVFFQNNISNIHVKGVIGKMHSLCRSSSNDIGSITGSTQSLLLEEKPWK 584
Query: 543 ESQKHHRKPPRTKNR 557
+ KH K R K R
Sbjct: 585 KINKHSNKKKREKTR 599
>ref|XP_317835.2| ENSANGP00000014391 [Anopheles gambiae str. PEST]
gi|55237002|gb|EAA13064.2| ENSANGP00000014391 [Anopheles
gambiae str. PEST]
Length = 581
Score = 328 bits (842), Expect = 2e-88
Identities = 188/499 (37%), Positives = 278/499 (55%), Gaps = 43/499 (8%)
Query: 1 MGKNQKTELGRALVKQH-----------NQMIQQTK-EKGKIYKKKFLESFTEVSDIDAI 48
MGK + +LGR+L+K M+ T+ + G + + L+S TE S
Sbjct: 1 MGKKKANQLGRSLIKDRFGQGNRKTVADGSMLHTTEVQDGYDWGRLNLQSVTEESSFQEF 60
Query: 49 IEQADEDVDEQQLLDIPLPPPTALINLDHASGSDFNGLTVEEMKKEQKIEEALHASSLRV 108
+ A+ E Q + + +N G VE +KK+ +++ L ++
Sbjct: 61 LRTAELAGTEFQAEKLNI----TYVNPKSKVGLLTTNERVEIIKKQVDMKDLL-----KI 111
Query: 109 PRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRVVER 168
PRRP W+ + +A+EL E FL WRR L L+E +++TP+EKNLD WRQLWRVVER
Sbjct: 112 PRRPKWTKDTTAEELLLAENASFLEWRRGLVALQEQDGMLMTPYEKNLDFWRQLWRVVER 171
Query: 169 SDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHD 228
SD++V +VD+R+PL +R DLE Y +EVD +K ++L+NK+D L A R WA+YF
Sbjct: 172 SDIVVQIVDARNPLLFRTEDLERYVQEVDPNKMNMILLNKSDFLTAEQRVHWAKYFDGQG 231
Query: 229 ILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRR 288
+ F+SA+ L S D + + +E + L E +A R
Sbjct: 232 VRVAFYSARLGNLKL--------------SVDRAEKTLEKIEEKIEDLAREEDAEPGAIR 277
Query: 289 SSGSSKSSDDNASVSSSSSH--------VVVGFVGYPNVGKSSTINALVGQKKTGVTSTP 340
+S ++ + ++ S V VG VGYPNVGKSSTINA+ +KK V++TP
Sbjct: 278 NSSKLLTNAELIALFKSLHRAERVTKDVVTVGLVGYPNVGKSSTINAVFLEKKVSVSATP 337
Query: 341 GKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRV 400
GKTKHFQTL + +L+ CDCPGLV PSF ++ +MI G+LPID+M H V ++ +
Sbjct: 338 GKTKHFQTLYVDSELMFCDCPGLVMPSFCITKADMILNGILPIDQMRDHVPPVNLLCTLI 397
Query: 401 PRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQILKD 460
PRH++E+ Y I + KP E +RPP + ELL + +RG T++G PD++R SR +LKD
Sbjct: 398 PRHILEDTYGIMISKPLEGEDPNRPPYSEELLLAFAYNRGFMTANGQPDQSRGSRYVLKD 457
Query: 461 YIDGKLPHYEMPPGLSTQE 479
Y++GKL + PPG+ E
Sbjct: 458 YVNGKLLYCYAPPGVVQDE 476
>gb|EAL32497.1| GA13246-PA [Drosophila pseudoobscura]
Length = 602
Score = 326 bits (836), Expect = 1e-87
Identities = 204/525 (38%), Positives = 286/525 (53%), Gaps = 67/525 (12%)
Query: 1 MGKNQK---TELGRALVKQ---HNQ--------MIQQTK-EKGKIYKKKFLESFTEVSDI 45
MGK K LGR L+K H Q M+ T+ + G + + L S TE S
Sbjct: 1 MGKKNKGTTPNLGRTLIKDRFGHTQRRKVDNDTMLHTTELQDGYDWGRLNLSSVTEESSF 60
Query: 46 DAIIEQADEDVDEQQL--LDIPLPPPTALINLDHASGSDFNGLTVEEMKKEQKIEEALHA 103
A + A+ E Q L+I P + L + +E + QK E H
Sbjct: 61 QAFLRTAELAGTEFQAEKLNITFVNPKQRVGLLSKT---------QEQRMHQKHHE--HR 109
Query: 104 SSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLW 163
L+VPRRP W SA++L E + FL WRR LA L+E++++++TP+EKNL+ WRQLW
Sbjct: 110 EHLKVPRRPKWDKNTSAEDLERAENEAFLNWRRDLALLQEDEEILMTPYEKNLEFWRQLW 169
Query: 164 RVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEY 223
RVVERSD++V +VD+R+PL +R DLE+Y KEV K ++LVNK+DLL R+ WAEY
Sbjct: 170 RVVERSDVVVQIVDARNPLLFRSTDLESYVKEVKSTKMNMILVNKSDLLTEEQRKHWAEY 229
Query: 224 FRAHDILFIFWSAKAATAVLEGKKLGSSQ------------ADNMASADN---------- 261
F I F+SA L+ + + Q AD + + N
Sbjct: 230 FDCEGIRTAFYSATLVEEELKREAEAARQESFPALKELRDAADEIQQSLNKVEGALDAIN 289
Query: 262 ----PD---TKIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFV 314
PD ++ G D+ R+ S E I +RR + +D HV +G V
Sbjct: 290 RKVKPDDVAEQLLG-DKNSPRVLSRTEMIEFLRRIYTGPRHTD---------QHVTIGMV 339
Query: 315 GYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYE 374
GYPNVGKSSTIN+L+ KK V++TPGKTK FQTL + ++LCDCPGLV PSF ++ +
Sbjct: 340 GYPNVGKSSTINSLMTVKKVSVSATPGKTKRFQTLYLDNDIMLCDCPGLVMPSFVLTKAD 399
Query: 375 MITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRT 434
M+ G+LPID+M H V ++ R+PRHV+E+ Y I + KP E R P + ELL
Sbjct: 400 MLLNGILPIDQMRDHVPAVNLLCERIPRHVLEDKYGIVIAKPVEGEDMERAPHSEELLLA 459
Query: 435 YCASRGQTTSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQE 479
Y +RG TS+G PD+ R++R +LKDY++GKL + PP ++ E
Sbjct: 460 YGYNRGFMTSNGQPDQARSARYVLKDYVNGKLLYAMSPPSVTQAE 504
>gb|EAA77792.1| hypothetical protein FG07194.1 [Gibberella zeae PH-1]
gi|46125633|ref|XP_387370.1| hypothetical protein
FG07194.1 [Gibberella zeae PH-1]
Length = 666
Score = 323 bits (829), Expect = 7e-87
Identities = 203/570 (35%), Positives = 306/570 (53%), Gaps = 41/570 (7%)
Query: 36 LESFTEVSDID---AIIEQADEDVDEQQLLDIPLPPPTALINLDHASGSDFNGLTVEEMK 92
+ S TE D+D A E A D ++ T + + H + L+ +E +
Sbjct: 59 MRSVTEQGDLDEFLATAELAGTDFTAEK---------TNNVKIIHTDQKNPYLLSAQE-E 108
Query: 93 KEQKIEEALHASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLTPF 152
K+ + H L VPRRP W + + DEL E + FL WRR LA L+EN L++TPF
Sbjct: 109 KDVLGKHKQHKGRLSVPRRPKWDSTTTPDELDRLERESFLNWRRGLAELQENNDLLMTPF 168
Query: 153 EKNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLL 212
E+NL++WRQLWRV+ERSDL+V +VD+R+PL +R DL+ Y KE+D K LLL+NKAD++
Sbjct: 169 ERNLEVWRQLWRVIERSDLIVQIVDARNPLLFRSEDLDDYVKEIDPKKQNLLLINKADMM 228
Query: 213 PASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDEL 272
R WA++ I + F+SA+ A A E + L S + + S + G+ E
Sbjct: 229 TPKQRLAWAKHLTEAGIAYRFFSAELAKAENEARNLEDSDDEAVDSPVEEQGESSGQQEQ 288
Query: 273 LARLQSEAEAIVDMRRSSGSSKSSDD-----------------NASVSSSSSH-VVVGFV 314
+E + + + ++ +D+ +A + S H + VG V
Sbjct: 289 EGASLTEEPEESQIDQEAKVAEETDEIDTQILTVEELEGIFLKHAPADAGSGHKLQVGLV 348
Query: 315 GYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYE 374
GYPNVGKSSTINAL+G KK V+STPGKTKHFQT+ +S ++LCDCPGLVFP+F++++ +
Sbjct: 349 GYPNVGKSSTINALIGSKKVSVSSTPGKTKHFQTIHLSPDVVLCDCPGLVFPNFATTKAD 408
Query: 375 MITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRT 434
++ G+LPID++ + V +V RVP+ +E IY I + E + P ASELLR
Sbjct: 409 LVCNGILPIDQLREFLGPVGLVTQRVPQPFLEAIYGIHVRTRAIEEGGTGIPTASELLRA 468
Query: 435 YCASRG-QTTSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNP 493
Y +RG QT G PDE+RA+R ILKDY+ GKL PPG+ + + + +
Sbjct: 469 YARARGFQTQGLGQPDESRAARYILKDYVAGKLLFVSPPPGIDDAADFNRGLYDEEHLPE 528
Query: 494 HVSDASDIEDSSVVETELAPKLEHVLDDLSSFDMANGLASNKVAPKKTKESQKHHRKPPR 553
A S + + A + DD + D+A+ ++ A K+++ K P
Sbjct: 529 KRRAALSAAMSHLSVVDPASSEPFLADDDDAVDLASTISVPLPAGPKSQKLDKGFFGPGS 588
Query: 554 -----TKNRSWRAGNAGKDDTDGMPIARFH 578
+K +++ + GK+D P+A H
Sbjct: 589 NQGHVSKPFNYQYSDQGKED----PLAGKH 614
>gb|EAA53248.1| hypothetical protein MG07525.4 [Magnaporthe grisea 70-15]
gi|39972447|ref|XP_367614.1| hypothetical protein
MG07525.4 [Magnaporthe grisea 70-15]
Length = 650
Score = 323 bits (829), Expect = 7e-87
Identities = 174/398 (43%), Positives = 237/398 (58%), Gaps = 32/398 (8%)
Query: 102 HASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQ 161
H L VPRRP W + ++L A E + FL WRR LA L E+ L++TPFE+NL++WRQ
Sbjct: 118 HKQRLTVPRRPKWDETTTPEQLDAMERESFLNWRRGLAELAESNDLLMTPFERNLEVWRQ 177
Query: 162 LWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWA 221
LWRV+ERSDL+V +VD+R+PL +R DL+ Y K VD K LLL+NKAD++ R++WA
Sbjct: 178 LWRVIERSDLVVQIVDARNPLLFRSEDLDHYVKSVDAKKENLLLINKADMMTLKQRQEWA 237
Query: 222 EYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASAD---------NPDTKIYGRDEL 272
Y + I F F+SA A + E + L S D+ A NPD + E
Sbjct: 238 RYLKGAKIAFKFFSASYAKELNEARDLESESEDDEAEESSVADGGAKLNPDASVPQSAEK 297
Query: 273 LARLQSEAEAIVDMRRSSGSSKSSDDN----------------ASVSSSSSHVVVGFVGY 316
EAE SS + SD S + + VG VGY
Sbjct: 298 EESGDQEAE------ESSLEEEDSDTRIITVEELESIFLKHAPESSADPKQKMQVGLVGY 351
Query: 317 PNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMI 376
PNVGKSSTINAL+G KK V+STPGKTKHFQT+ +SEK++LCDCPGLVFP+F+S++ +++
Sbjct: 352 PNVGKSSTINALIGAKKVSVSSTPGKTKHFQTIHLSEKVVLCDCPGLVFPNFASTKADLV 411
Query: 377 TCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYC 436
GVLPID + + +VA R+P+ +E +Y I++ + E + P A ELL Y
Sbjct: 412 CNGVLPIDELREFTGPAALVARRIPKAYLEAVYGINIKTRPAEEGGTGVPTAEELLVAYA 471
Query: 437 ASRGQT-TSSGLPDETRASRQILKDYIDGKLPHYEMPP 473
+RG T T + PDE++A+R ILKDY++GKL + E PP
Sbjct: 472 RARGFTKTGNASPDESKAARHILKDYVNGKLLYCEPPP 509
>emb|CAG98090.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50310721|ref|XP_455382.1| unnamed protein product
[Kluyveromyces lactis]
Length = 648
Score = 322 bits (824), Expect = 3e-86
Identities = 197/460 (42%), Positives = 271/460 (58%), Gaps = 34/460 (7%)
Query: 36 LESFTEVSDIDAII---EQADEDVDEQQLLDIPLPPPTALINLDH-ASGSDFNGLTV-EE 90
L S T+ S +D + E AD+D + ++ + I +D+ A + G ++ E
Sbjct: 65 LRSVTQESALDEFLSTAELADKDFTADRHSNVKI------IRMDNGADAATSQGFSLTNE 118
Query: 91 MKKEQKIEEALHASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEE-NKKLVL 149
K + ++ LHA L VPRRP W+ M+ EL E + FL WRR LA L+E N+ L+L
Sbjct: 119 QKSKINEKQRLHARELIVPRRPKWNESMTRFELERLEKEAFLEWRRKLAYLQEDNEDLLL 178
Query: 150 TPFEKNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKA 209
TPFE+N+++WRQLWRVVER DL+V +VD+RDPL +R DLE Y KEVD K LLL+NKA
Sbjct: 179 TPFERNIEVWRQLWRVVERCDLVVQIVDARDPLLFRSTDLEKYVKEVDDRKQNLLLINKA 238
Query: 210 DLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKK-LGSSQADNMASADNPDTKIYG 268
DLL R WA+Y + I F F+SA A +LE ++ LG + + D +I
Sbjct: 239 DLLTRKQRIIWAKYLLSRGISFTFFSAAKANEILERQEELGDEYVE-----EEDDEEILA 293
Query: 269 RDELLARLQSEA---EAI-------VDMRRSSGSSKSSDDNA--SVSSSSSHVVVGFVGY 316
+E + L E E + +D K+ +D + + +G VGY
Sbjct: 294 TEEEIEELDGEEVNKEILDKINILKIDQLEELFLDKAPNDPLLPPLPGQEPIIQIGLVGY 353
Query: 317 PNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMI 376
PNVGKSSTINALVG KK V+STPGKTKHFQT+ +S+K+ LCDCPGLVFP+F+ ++ E++
Sbjct: 354 PNVGKSSTINALVGAKKVSVSSTPGKTKHFQTIKLSDKVTLCDCPGLVFPNFAYNKGELV 413
Query: 377 TCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYE--SQSRPPLASELLRT 434
GVLPID++ + +VA RVP++ +E IY I + + KS E P A ELL
Sbjct: 414 CNGVLPIDQLRDYIGPSTLVAERVPKYFLEAIYGIHI-QTKSVEEGGNGEVPTAQELLVA 472
Query: 435 YCASRGQTTSS-GLPDETRASRQILKDYIDGKLPHYEMPP 473
Y +RG T G DE+RASR ILKDY++GKL + PP
Sbjct: 473 YARARGYMTQGFGAADESRASRYILKDYVNGKLLYINPPP 512
>emb|CAC28726.1| conserved hypothetical protein [Neurospora crassa]
gi|32405786|ref|XP_323506.1| hypothetical protein (
(AL513445) conserved hypothetical protein [Neurospora
crassa] ) gi|28922243|gb|EAA31488.1| hypothetical
protein ( (AL513445) conserved hypothetical protein
[Neurospora crassa] )
Length = 653
Score = 321 bits (823), Expect = 4e-86
Identities = 176/446 (39%), Positives = 265/446 (58%), Gaps = 28/446 (6%)
Query: 92 KKEQKI--EEALHASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVL 149
K+EQ + ++ + S L VPRRP W + + DEL E + FL WRR LA L+E + L++
Sbjct: 106 KEEQAVLGKQRANKSRLTVPRRPQWDSTTTRDELDQRERESFLEWRRGLAELQETQDLLM 165
Query: 150 TPFEKNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKA 209
TPFE+NL++WRQLWRV+ERSD++V +VD+R+PL +R DLE Y K+VD K+ LLL+NKA
Sbjct: 166 TPFERNLEVWRQLWRVIERSDVIVQIVDARNPLMFRSEDLEVYVKDVDPKKHNLLLINKA 225
Query: 210 DLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGR 269
DL+ R+ WA Y + I + F+SA+ A ++E S D+ +D + +
Sbjct: 226 DLMTYKQRKMWANYLKGEGIDYRFFSAQLAKEMIEAGGYADSDEDSEDESDAGEGP--SK 283
Query: 270 DELLARLQSEAEAIVDMRRSSGSSKSSDDNASV---------------SSSSSHVVVGFV 314
E + + +A + + ++ +D + + + VG V
Sbjct: 284 QEETPKAEEKASEENEEAKEEEITEHNDPDTHILRVDELEDILLQYQPEGQDRKLQVGLV 343
Query: 315 GYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYE 374
GYPNVGKSSTINAL+G KK V+STPGKTKHFQT+ +S+ ++LCDCPGLVFP+F++++ E
Sbjct: 344 GYPNVGKSSTINALIGAKKVSVSSTPGKTKHFQTIHLSDNVLLCDCPGLVFPNFANTKAE 403
Query: 375 MITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRT 434
++ GVLPID++ ++ +VA R+P+ +E +Y I + E + P A ELL
Sbjct: 404 LVCNGVLPIDQLREYTGPAALVARRIPQPFLEAVYGIKIKTRPLEEGGTGIPTADELLDA 463
Query: 435 YCASRGQTTSS-GLPDETRASRQILKDYIDGKLPHYEMPP--GLSTQELASE-DSNEHDQ 490
Y RG T G PD++RA R ILKDY++GKL + E PP G+ E +E H
Sbjct: 464 YARHRGYMTQGLGQPDQSRAVRYILKDYVNGKLLYCEPPPGSGVDGPEFNAELYDMAHLP 523
Query: 491 VNPHVS-----DASDIEDSSVVETEL 511
N V+ + +DI+D++ + +E+
Sbjct: 524 ENRRVAMLSALEGADIDDTATLTSEM 549
>emb|CAG60891.1| unnamed protein product [Candida glabrata CBS138]
gi|50291015|ref|XP_447940.1| unnamed protein product
[Candida glabrata]
Length = 639
Score = 315 bits (808), Expect = 2e-84
Identities = 188/456 (41%), Positives = 271/456 (59%), Gaps = 28/456 (6%)
Query: 36 LESFTEVSDIDAIIEQA---DEDVDEQQLLDIPLPPPTALINLDHASGSDFN---GLTVE 89
L S T+ S +D + A D+D + ++ + I +D+ GS + LT E
Sbjct: 65 LRSVTQESALDEFLNTAALADKDFTADRHSNVKI------IRIDNGDGSATSQGFSLTNE 118
Query: 90 EMKKEQKIEEALHASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKK-LV 148
+ + AL A+ L VPRRP+W M+ +L E FL WRR LA L+EN + L+
Sbjct: 119 QKSTIDAKQRAL-ANDLIVPRRPYWDESMTKYQLERQENDAFLEWRRKLAHLQENNEDLL 177
Query: 149 LTPFEKNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNK 208
LTPFE+N+++W+QLWRVVERSDL+V +VD+RDPL +R DLE Y EVD K LLL+NK
Sbjct: 178 LTPFERNIEVWKQLWRVVERSDLVVQIVDARDPLLFRSVDLERYVTEVDDRKQNLLLINK 237
Query: 209 ADLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKK-LGSSQADNMASADNPDTKIY 267
ADLL + R WA+YF + + F+SA A +LE +K LG ++ ++ D+
Sbjct: 238 ADLLTRNQRITWAKYFAKKNNSYAFYSALRANQLLEKQKELGDDYSEEHIEEESDDS--- 294
Query: 268 GRDELLARLQSEAEAI--VDMRRSSGSSKSSDDNAS--VSSSSSHVVVGFVGYPNVGKSS 323
+E L E I +D S++ + + + S + +G VGYPNVGKSS
Sbjct: 295 --EETLEESVREGIKILTIDQLEELFLSRAPSEPLTEPLPGQESLLQIGLVGYPNVGKSS 352
Query: 324 TINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPI 383
TIN+LVG KK V+STPGKTKHFQT+ +S+ ++LCDCPGLVF +F+ ++ E++ GVLPI
Sbjct: 353 TINSLVGAKKVSVSSTPGKTKHFQTIKLSDHVMLCDCPGLVFANFAYNKGELVCNGVLPI 412
Query: 384 DRMTQHRECVQVVANRVPRHVIEEIYNISL---PKPKSYESQSRPPLASELLRTYCASRG 440
D++ + +VA R+P++ +E +Y I + + +S + + P A ELL Y +RG
Sbjct: 413 DQLRDYVGPCALVAERIPKYYLEAVYGIHIQTRSREESGDIEPENPTAQELLVAYARARG 472
Query: 441 QTTSS-GLPDETRASRQILKDYIDGKLPHYEMPPGL 475
T G DE+RASR ILKDY++GKL + PP L
Sbjct: 473 YMTQGFGSADESRASRYILKDYVNGKLLYVNPPPHL 508
>gb|EAK93352.1| hypothetical protein CaO19.10967 [Candida albicans SC5314]
gi|46433895|gb|EAK93321.1| hypothetical protein
CaO19.3463 [Candida albicans SC5314]
Length = 684
Score = 313 bits (801), Expect = 1e-83
Identities = 187/485 (38%), Positives = 273/485 (55%), Gaps = 38/485 (7%)
Query: 36 LESFTEVSDIDAII---EQADED--VDEQQLLDIPLPPPTALINLDHASGSDFNGLTVEE 90
L S T+ + +D + E AD D D+ Q + I T+++N NGL +
Sbjct: 71 LRSVTQENALDEFLSTAELADTDFTADKNQQVKIIKVGNTSIVNS--------NGLLSTD 122
Query: 91 MKKEQKIEEALHASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLT 150
+ + + + L +P+RP W+ S E+ E FL+WRR LA+L EN L+LT
Sbjct: 123 EMLAMRQKHMMFENKLTIPKRPKWNKNQSKIEIDRQENLAFLSWRRELAQLTENNDLLLT 182
Query: 151 PFEKNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEV-----DVHKNTLLL 205
PFE+NL++W+QLWRVVER DL+V +VD+R+PLF+R DL+ Y + + KN LLL
Sbjct: 183 PFERNLEVWKQLWRVVERCDLIVQIVDARNPLFFRSIDLDKYVTSLSDPDNNKAKNNLLL 242
Query: 206 VNKADLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTK 265
VNKAD+L R WAEYF+ I ++F+SA A +LE ++ N + + ++
Sbjct: 243 VNKADMLTRDQRIAWAEYFKLKKINYVFFSAAKANELLEKEREELENIQNSTGSSSSSSR 302
Query: 266 IYGRDELLARL---QSEAEAIVDMR--RSSGSSKSSDDNA-----SVSSSSSHVVVGFVG 315
+ A + Q E EA +R + K D+A + +G VG
Sbjct: 303 NTNNNAAAAAVDAEQEEGEANDSIRILKIEELEKLFMDSAPKFEVDPEFPDRKLQIGLVG 362
Query: 316 YPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEM 375
YPNVGKSSTINALVG KK V+STPGKTKHFQT+ +S ++LCDCPGLVFP+F+ + E+
Sbjct: 363 YPNVGKSSTINALVGSKKVSVSSTPGKTKHFQTIHLSPDVLLCDCPGLVFPNFAYTNAEL 422
Query: 376 ITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYE-SQSRPPLASELLRT 434
+ GVLPID++ +H V +V R+P+ +E +Y I +P K + P A ELL
Sbjct: 423 VCNGVLPIDQLREHIPPVSLVCQRIPKFFLEAVYGIHIPIQKVEDGGNGEYPTARELLNA 482
Query: 435 YCASRGQTTSS-GLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNP 493
Y +RG T G DE RA+R ILKDY++GKL + PP E +N+++ + P
Sbjct: 483 YARARGYMTQGFGSADEPRAARYILKDYVNGKLLYVNPPP--------KEATNKNEWILP 534
Query: 494 HVSDA 498
++ ++
Sbjct: 535 NLQES 539
>ref|NP_011416.1| Putative GTPase involved in 60S ribosomal subunit biogenesis;
required for the release of Nmd3p from 60S subunits in
the cytoplasm; Lsg1p [Saccharomyces cerevisiae]
gi|1322637|emb|CAA96805.1| unnamed protein product
[Saccharomyces cerevisiae]
gi|1723894|sp|P53145|YGJ9_YEAST Hypothetical GTP-binding
protein in SEH1-PRP20 intergenic region
Length = 640
Score = 311 bits (796), Expect = 5e-83
Identities = 204/515 (39%), Positives = 289/515 (55%), Gaps = 36/515 (6%)
Query: 6 KTELGRAL--VKQHNQMIQQ--------TKEKGKIYKKKFLESFTEVSDIDAIIEQA--- 52
K ELGRA+ +Q I+ T +K + K L S T+ S +D + A
Sbjct: 26 KLELGRAIKYARQKENAIEYLPDGEMRFTTDKHEANWVK-LRSVTQESALDEFLSTAALA 84
Query: 53 DEDVDEQQLLDIPLPPPTALINLDHASGSDFNGLTVEEMKKEQK----IEEALHASSLRV 108
D+D + ++ + I +D SG+D M EQ+ ++ A L V
Sbjct: 85 DKDFTADRHSNVKI------IRMD--SGNDSATSQGFSMTNEQRGNLNAKQRALAKDLIV 136
Query: 109 PRRPFWSAEMSADELHANETQHFLTWRRSLARLEE-NKKLVLTPFEKNLDIWRQLWRVVE 167
PRRP W+ MS +L E + FL WRR LA L+E N+ L+LTPFE+N+++W+QLWRVVE
Sbjct: 137 PRRPEWNEGMSKFQLDRQEKEAFLEWRRKLAHLQESNEDLLLTPFERNIEVWKQLWRVVE 196
Query: 168 RSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAH 227
RSDL+V +VD+R+PL +R DLE Y KE D K LLLVNKADLL R WA+YF +
Sbjct: 197 RSDLVVQIVDARNPLLFRSVDLERYVKESDDRKANLLLVNKADLLTKKQRIAWAKYFISK 256
Query: 228 DILFIFWSAKAATAVLEGKK-LGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVDM 286
+I F F+SA A +LE +K +G + + D + + DE + ++ +D
Sbjct: 257 NISFTFYSALRANQLLEKQKEMGEDYREQ--DFEEADKEGFDADEKV--MEKVKILSIDQ 312
Query: 287 RRSSGSSKSSDDNA--SVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTK 344
SK+ ++ + + +G VGYPNVGKSSTIN+LVG KK V+STPGKTK
Sbjct: 313 LEELFLSKAPNEPLLPPLPGQPPLINIGLVGYPNVGKSSTINSLVGAKKVSVSSTPGKTK 372
Query: 345 HFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRVPRHV 404
HFQT+ +S+ ++LCDCPGLVFP+F+ ++ E++ GVLPID++ + +VA R+P++
Sbjct: 373 HFQTIKLSDSVMLCDCPGLVFPNFAYNKGELVCNGVLPIDQLRDYIGPAGLVAERIPKYY 432
Query: 405 IEEIYNISL-PKPKSYESQSRPPLASELLRTYCASRGQTTSS-GLPDETRASRQILKDYI 462
IE IY I + K + P A ELL Y +RG T G DE RASR ILKDY+
Sbjct: 433 IEAIYGIHIQTKSRDEGGNGDIPTAQELLVAYARARGYMTQGYGSADEPRASRYILKDYV 492
Query: 463 DGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSD 497
+GKL + PP L + + E + +V D
Sbjct: 493 NGKLLYVNPPPHLEDDTPYTREECEEFNKDLYVFD 527
>emb|CAG85906.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50418683|ref|XP_457861.1| unnamed protein product
[Debaryomyces hansenii]
Length = 664
Score = 310 bits (794), Expect = 8e-83
Identities = 196/513 (38%), Positives = 288/513 (55%), Gaps = 32/513 (6%)
Query: 36 LESFTEVSDIDAII---EQADEDVDEQQLLDIPLPPPTALINLDHASGSDFNGLTVEEMK 92
L S T+ + +D + E AD D ++ + + I + + S + NGL +E
Sbjct: 73 LRSVTQENALDEFLNTAELADTDFTAEKNQQVKI------IKIGNTSIVNQNGLLTQEET 126
Query: 93 KEQKIEEALHASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLTPF 152
E + + + + L VPRRP W+ E+ E FL WRR LA L EN L+LTPF
Sbjct: 127 LELRRKHKIFENKLTVPRRPQWNKNQLKLEIERQENLAFLQWRRELASLTENNDLLLTPF 186
Query: 153 EKNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEV-----DVHKNTLLLVN 207
E+N+++WRQLWRVVER DL+V +VD+R+PL +R DLE Y +E+ + K LLLVN
Sbjct: 187 ERNIEVWRQLWRVVERCDLIVQIVDARNPLLFRSIDLEKYVQELSKPDDNATKKNLLLVN 246
Query: 208 KADLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKK--LGSSQADNMASADNPDTK 265
KADLL R +W++YF + +I ++F+SA A +LE ++ L SQ D + P+ K
Sbjct: 247 KADLLTREQRIQWSDYFISQNINYVFFSASKANELLEEEQENLEESQRDPNYT---PERK 303
Query: 266 IYGRDELL---ARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKS 322
+ E + R+ + E SS S + S++ + +G VGYPNVGKS
Sbjct: 304 VEDEPEEVDSKIRILTINELENLFLTSSPSFEKSEE-----FPDRKLQIGLVGYPNVGKS 358
Query: 323 STINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLP 382
STINALVG KK V++TPGKTKHFQT+ +S +++LCDCPGLVFP+F+ + E++ GVLP
Sbjct: 359 STINALVGSKKVSVSATPGKTKHFQTIHLSPEVLLCDCPGLVFPNFAYTNGELVCNGVLP 418
Query: 383 IDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRP-PLASELLRTYCASRG- 440
ID++ +H V +V +RVP+ +E Y I +P K + + A ELL Y +RG
Sbjct: 419 IDQLREHIPPVSIVCHRVPKFYLEAFYGIHIPIQKVEDGGNGVYASARELLNAYARARGF 478
Query: 441 QTTSSGLPDETRASRQILKDYIDGKLPHYEMPP-GLSTQELASEDSNEHDQVNPHVSDAS 499
T G DE+RASR ILKDY+ GKL PP ++ E + E ++N +
Sbjct: 479 MTQGFGSADESRASRYILKDYVSGKLLFVSPPPRKVNDDEWDLPSNEEARELNKELYSLR 538
Query: 500 DIEDSSVVETELAPKLEHVLDDLSSFDMANGLA 532
+ +S + ++ L + FD++ GLA
Sbjct: 539 TLPESR--QHQILSALSSKNVNPEDFDLSKGLA 569
>emb|CAA93314.1| SPAC3F10.16c [Schizosaccharomyces pombe]
gi|19114860|ref|NP_593948.1| hypothetical protein
SPAC3F10.16c [Schizosaccharomyces pombe 972h-]
gi|7492623|pir||T38717 probable GTP-binding protein -
fission yeast (Schizosaccharomyces pombe)
gi|1723260|sp|Q10190|YAWG_SCHPO Hypothetical GTP-binding
protein C3F10.16c in chromosome I
Length = 616
Score = 304 bits (779), Expect = 5e-81
Identities = 177/460 (38%), Positives = 265/460 (57%), Gaps = 28/460 (6%)
Query: 36 LESFTEVSDIDAIIEQAD----EDVDEQQLLDIPLPPPTALINLDHASGSDFNGLTVEEM 91
L S T +D+D + A+ E + E+Q + + + + + F E
Sbjct: 46 LRSVTHETDLDEFLNTAELGEVEFIAEKQNVTV----------IQNPEQNPFLLSKEEAA 95
Query: 92 KKEQKIEEALHASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLTP 151
+ +QK E+ + L +PRRP W +A EL E + FL WRR+LA+L++ + ++TP
Sbjct: 96 RSKQKQEK--NKDRLTIPRRPHWDQTTTAVELDRMERESFLNWRRNLAQLQDVEGFIVTP 153
Query: 152 FEKNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADL 211
FE+NL+IWRQLWRV+ERSD++V +VD+R+PLF+R LE Y KEV K LLVNKAD+
Sbjct: 154 FERNLEIWRQLWRVIERSDVVVQIVDARNPLFFRSAHLEQYVKEVGPSKKNFLLVNKADM 213
Query: 212 LPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDE 271
L R W+ YF ++I F+F+SA+ A E + G +++ N + DE
Sbjct: 214 LTEEQRNYWSSYFNENNIPFLFFSARMAA---EANERGEDLETYESTSSNEIPESLQADE 270
Query: 272 LLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQ 331
+ I ++ G + +++ + + G VGYPNVGKSSTINALVG
Sbjct: 271 ----NDVHSSRIATLKVLEGIFEKF--ASTLPDGKTKMTFGLVGYPNVGKSSTINALVGS 324
Query: 332 KKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRE 391
KK V+STPGKTKHFQT+ +SEK+ L DCPGLVFPSF++++ +++ GVLPID++ ++
Sbjct: 325 KKVSVSSTPGKTKHFQTINLSEKVSLLDCPGLVFPSFATTQADLVLDGVLPIDQLREYTG 384
Query: 392 CVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRG-QTTSSGLPDE 450
++A R+P+ V+E +Y I + E + P A E+L + SRG G PD+
Sbjct: 385 PSALMAERIPKEVLETLYTIRIRIKPIEEGGTGVPSAQEVLFPFARSRGFMRAHHGTPDD 444
Query: 451 TRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQ 490
+RA+R +LKDY++GKL + PP SE + EH Q
Sbjct: 445 SRAARILLKDYVNGKLLYVHPPPNYPNS--GSEFNKEHHQ 482
>gb|AAK68267.1| Hypothetical protein C53H9.2a [Caenorhabditis elegans]
gi|25143542|ref|NP_740787.1| GTP-binding protein,
HSR1-related (1C307) [Caenorhabditis elegans]
Length = 506
Score = 296 bits (757), Expect = 2e-78
Identities = 168/378 (44%), Positives = 221/378 (58%), Gaps = 37/378 (9%)
Query: 106 LRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRV 165
LR+PRRP + ++L E + FL WR L+ L+E LVLTPFE+N D+WR+LWRV
Sbjct: 70 LRIPRRPAKELWENMEDLTKLENEAFLQWRSDLSELQEVDGLVLTPFERNPDMWRELWRV 129
Query: 166 VERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFR 225
VE+SD++V +VD+R+PL +R DL+ Y KEVD K LLLVNKADLL + W EYF
Sbjct: 130 VEKSDIIVQIVDARNPLLFRSKDLDDYVKEVDPAKQILLLVNKADLLKPEQQASWREYFE 189
Query: 226 AHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTK----IYGRDELLARLQSEAE 281
+I IFWS A VL+ ++ NP T + +DEL+A+ +
Sbjct: 190 KENIKVIFWS--AIDEVLD------PIDEDAVETSNPSTSTHMFVTNKDELIAKFKELG- 240
Query: 282 AIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPG 341
+ S S+ V+VG VGYPNVGKSSTIN L G KK V++TPG
Sbjct: 241 -----------------HVSDEPSAKPVMVGMVGYPNVGKSSTINKLAGGKKVSVSATPG 283
Query: 342 KTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRVP 401
KT+HFQT+ I +L LCDCPGLV PSFS R EM G+LP+D+M H ++ +RVP
Sbjct: 284 KTRHFQTIHIDSQLCLCDCPGLVMPSFSFGRSEMFLNGILPVDQMRDHFGPTSLLLSRVP 343
Query: 402 RHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQILKDY 461
HVIE Y+I LP+ +S P A LL + RG SSG+PD +RA+R + KD
Sbjct: 344 VHVIEATYSIMLPEMQS-------PSAINLLNSLAFMRGFMASSGIPDCSRAARLMFKDV 396
Query: 462 IDGKLPHYEMPPGLSTQE 479
+ GKL PPG+ +E
Sbjct: 397 VSGKLIWAAAPPGVEQEE 414
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.313 0.130 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 997,841,105
Number of Sequences: 2540612
Number of extensions: 41635781
Number of successful extensions: 113161
Number of sequences better than 10.0: 1380
Number of HSP's better than 10.0 without gapping: 769
Number of HSP's successfully gapped in prelim test: 611
Number of HSP's that attempted gapping in prelim test: 111113
Number of HSP's gapped (non-prelim): 2086
length of query: 589
length of database: 863,360,394
effective HSP length: 134
effective length of query: 455
effective length of database: 522,918,386
effective search space: 237927865630
effective search space used: 237927865630
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 78 (34.7 bits)
Medicago: description of AC147405.9


