

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
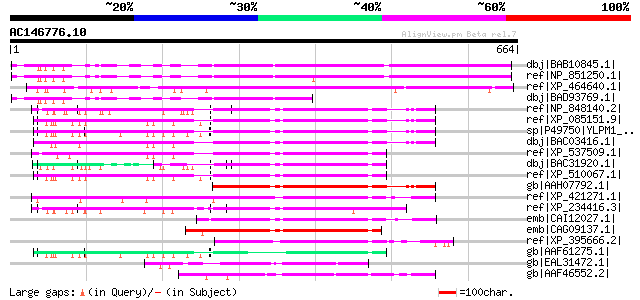
Query= AC146776.10 - phase: 0
(664 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB10845.1| unnamed protein product [Arabidopsis thaliana] 549 e-155
ref|NP_851250.1| nuclear protein ZAP-related [Arabidopsis thaliana] 538 e-151
ref|XP_464640.1| nuclear protein ZAP-like [Oryza sativa (japonic... 530 e-149
dbj|BAD93769.1| ZAP - like protein [Arabidopsis thaliana] gi|306... 271 5e-71
ref|NP_848140.2| YLP motif containing 1 [Mus musculus] gi|601684... 234 7e-60
ref|XP_085151.9| PREDICTED: YLP motif containing 1 [Homo sapiens] 225 3e-57
sp|P49750|YLPM1_HUMAN YLP motif containing protein 1 (Nuclear pr... 225 3e-57
dbj|BAC03416.1| FLJ00353 protein [Homo sapiens] 225 3e-57
ref|XP_537509.1| PREDICTED: similar to Nuclear protein ZAP3 (ZAP... 221 6e-56
dbj|BAC31920.1| unnamed protein product [Mus musculus] 219 2e-55
ref|XP_510067.1| PREDICTED: YLP motif containing 1 [Pan troglody... 219 3e-55
gb|AAH07792.1| YLPM1 protein [Homo sapiens] 214 7e-54
ref|XP_421271.1| PREDICTED: similar to FLJ00353 protein [Gallus ... 213 1e-53
ref|XP_234416.3| PREDICTED: similar to YLP motif containing prot... 213 2e-53
emb|CAI12027.1| novel protein similar to vertebrate YLP motif co... 210 1e-52
emb|CAG09137.1| unnamed protein product [Tetraodon nigroviridis] 206 3e-51
ref|XP_395666.2| PREDICTED: similar to YLPM1 protein [Apis melli... 168 5e-40
gb|AAF61275.1| unknown [Homo sapiens] 157 8e-37
gb|EAL31472.1| GA17072-PA [Drosophila pseudoobscura] 142 3e-32
gb|AAF46552.2| CG32685-PC [Drosophila melanogaster] gi|24640949|... 128 5e-28
>dbj|BAB10845.1| unnamed protein product [Arabidopsis thaliana]
Length = 644
Score = 549 bits (1415), Expect = e-155
Identities = 336/722 (46%), Positives = 407/722 (55%), Gaps = 167/722 (23%)
Query: 3 RQWRPNPNLNPRPIQYQTNLCPNCLTPHFPFCPN---PS-----PNWPPPP--------- 45
+QWRP P Q N+CP C PHFPFCP PS PN+PPPP
Sbjct: 18 QQWRPAPT--------QPNICPICTVPHFPFCPPYPPPSSFAYNPNFPPPPHLNSPRPGF 69
Query: 46 -----PPPNPTINNY------------------DSDFDRTFKKPRIE------------- 69
PP P N+Y D + DR++K+ RI+
Sbjct: 70 DSFTGPPVRPPQNHYPPWQPHHGNQWRPVAVDVDREADRSYKRARIDTIAGGSPGYGVSE 129
Query: 70 --------DDERRLKLIRDHGLNPPQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQH 121
++ERRLK++RDHG Y P N + H Q+
Sbjct: 130 SPSPRISWENERRLKMVRDHG----------------YGLAAPS------NIEMNH--QY 165
Query: 122 HSEFNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQM 181
SEF + P P HHP PY Y +GSN
Sbjct: 166 GSEFRNGGQFNGVAPLPPPP--------------PHHPPPYGG-YFSGSNG--------- 201
Query: 182 EASRFYRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPE 241
+PPLP SPPPPLP P SLFPVT+++
Sbjct: 202 ------QPPLPVSPPPPLP---------SSHPSSLFPVTTNSS----------------- 229
Query: 242 FSTAFPTEEPSKQY--LGDAQPFSINPLAAEKPKFVDASQLFRNPLRTSRPDHFVIILRG 299
PT PS Y + +A P S LA + K +D S L + P R++RPDHFVIILRG
Sbjct: 230 -----PTIPPSSSYPQMPNASPSSAQ-LAPTRSKVIDVSHLLKPPHRSTRPDHFVIILRG 283
Query: 300 FPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTEVEKVEDSDGPKSSSSGRNKRPVT 359
PGSGK+YLAK+LRD+EVENGG APRIHSMDDYFMTEVEKVE+SD S SSGR+KRP+
Sbjct: 284 LPGSGKSYLAKLLRDVEVENGGSAPRIHSMDDYFMTEVEKVEESDS-TSLSSGRSKRPIV 342
Query: 360 KKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFIIVDDRNLRVADFAQFWATAKRSGY 419
K VMEYCYEPEMEEAYRSSMLKAFK+T+E+G F+F+IVDDRNLRVADF QFWATAKRSGY
Sbjct: 343 KTVMEYCYEPEMEEAYRSSMLKAFKRTLEDGAFSFVIVDDRNLRVADFTQFWATAKRSGY 402
Query: 420 EVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQWEEAPSLYLQLDMKSLFHGDDLKESR 479
E YILEATYKDP GCAARNVHG T ++++MAEQWEEAPSLY+QLD+KS DDLKE+
Sbjct: 403 EAYILEATYKDPTGCAARNVHGITVDQVQQMAEQWEEAPSLYMQLDIKSFTRWDDLKENE 462
Query: 480 IQEVDMDMDDDLDDALVAAQGREADKAVVRPVKDG-EGSIKDGKRWDAEEEHPT-EVREL 537
IQEVDMDM+DD R++D + K EGS K +WDAE T EV+EL
Sbjct: 463 IQEVDMDMEDDF-----GLPERKSDNSTQSEEKGATEGSYKSESKWDAESGSRTEEVKEL 517
Query: 538 GKSKWSEDFEDDIDQTEGMKGNINALSGLIHQYGKERKSVHWGDQVGKTGFSIGTARKVT 597
+SKWS ED+ + ++ M+ N +L + ++ KSV WGD+ G GFSIG AR +
Sbjct: 518 SRSKWSNVEEDETENSQSMRRNSKSLPKSSQERQRKGKSVWWGDKGGDAGFSIGAARNMN 577
Query: 598 TLSLVIGPGAGYNLKSNPLPKEESPTRNSV--ETKKRSTFQERIRAERESFKAVFDGRRP 655
SL+IGPG+GYN+KSNPL EES + K R FQ+++RAERESFKAVFD R
Sbjct: 578 MPSLIIGPGSGYNVKSNPLSAEESRALADAIGKAKVRGIFQDQLRAERESFKAVFDKRHV 637
Query: 656 RI 657
RI
Sbjct: 638 RI 639
>ref|NP_851250.1| nuclear protein ZAP-related [Arabidopsis thaliana]
Length = 661
Score = 538 bits (1387), Expect = e-151
Identities = 336/739 (45%), Positives = 407/739 (54%), Gaps = 184/739 (24%)
Query: 3 RQWRPNPNLNPRPIQYQTNLCPNCLTPHFPFCPN---PS-----PNWPPPP--------- 45
+QWRP P Q N+CP C PHFPFCP PS PN+PPPP
Sbjct: 18 QQWRPAPT--------QPNICPICTVPHFPFCPPYPPPSSFAYNPNFPPPPHLNSPRPGF 69
Query: 46 -----PPPNPTINNY------------------DSDFDRTFKKPRIE------------- 69
PP P N+Y D + DR++K+ RI+
Sbjct: 70 DSFTGPPVRPPQNHYPPWQPHHGNQWRPVAVDVDREADRSYKRARIDTIAGGSPGYGVSE 129
Query: 70 --------DDERRLKLIRDHGLNPPQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQH 121
++ERRLK++RDHG Y P N + H Q+
Sbjct: 130 SPSPRISWENERRLKMVRDHG----------------YGLAAPS------NIEMNH--QY 165
Query: 122 HSEFNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQM 181
SEF + P P HHP PY Y +GSN
Sbjct: 166 GSEFRNGGQFNGVAPLPPPP--------------PHHPPPYGG-YFSGSNG--------- 201
Query: 182 EASRFYRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPE 241
+PPLP SPPPPLP P SLFPVT+++
Sbjct: 202 ------QPPLPVSPPPPLP---------SSHPSSLFPVTTNSS----------------- 229
Query: 242 FSTAFPTEEPSKQY--LGDAQPFSINPLAAEKPKFVDASQLFRNPLRTSRPDHFVIILRG 299
PT PS Y + +A P S LA + K +D S L + P R++RPDHFVIILRG
Sbjct: 230 -----PTIPPSSSYPQMPNASPSSAQ-LAPTRSKVIDVSHLLKPPHRSTRPDHFVIILRG 283
Query: 300 FPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTEVEKVEDSDGPKSSSSGRNKRPVT 359
PGSGK+YLAK+LRD+EVENGG APRIHSMDDYFMTEVEKVE+SD S SSGR+KRP+
Sbjct: 284 LPGSGKSYLAKLLRDVEVENGGSAPRIHSMDDYFMTEVEKVEESDS-TSLSSGRSKRPIV 342
Query: 360 KKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFII-----------------VDDRNL 402
K VMEYCYEPEMEEAYRSSMLKAFK+T+E+G F+F+I VDDRNL
Sbjct: 343 KTVMEYCYEPEMEEAYRSSMLKAFKRTLEDGAFSFVIVCFLELTVSCWYMSLILVDDRNL 402
Query: 403 RVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQWEEAPSLYL 462
RVADF QFWATAKRSGYE YILEATYKDP GCAARNVHG T ++++MAEQWEEAPSLY+
Sbjct: 403 RVADFTQFWATAKRSGYEAYILEATYKDPTGCAARNVHGITVDQVQQMAEQWEEAPSLYM 462
Query: 463 QLDMKSLFHGDDLKESRIQEVDMDMDDDLDDALVAAQGREADKAVVRPVKDG-EGSIKDG 521
QLD+KS DDLKE+ IQEVDMDM+DD R++D + K EGS K
Sbjct: 463 QLDIKSFTRWDDLKENEIQEVDMDMEDDF-----GLPERKSDNSTQSEEKGATEGSYKSE 517
Query: 522 KRWDAEEEHPT-EVRELGKSKWSEDFEDDIDQTEGMKGNINALSGLIHQYGKERKSVHWG 580
+WDAE T EV+EL +SKWS ED+ + ++ M+ N +L + ++ KSV WG
Sbjct: 518 SKWDAESGSRTEEVKELSRSKWSNVEEDETENSQSMRRNSKSLPKSSQERQRKGKSVWWG 577
Query: 581 DQVGKTGFSIGTARKVTTLSLVIGPGAGYNLKSNPLPKEESPTRNSV--ETKKRSTFQER 638
D+ G GFSIG AR + SL+IGPG+GYN+KSNPL EES + K R FQ++
Sbjct: 578 DKGGDAGFSIGAARNMNMPSLIIGPGSGYNVKSNPLSAEESRALADAIGKAKVRGIFQDQ 637
Query: 639 IRAERESFKAVFDGRRPRI 657
+RAERESFKAVFD R RI
Sbjct: 638 LRAERESFKAVFDKRHVRI 656
>ref|XP_464640.1| nuclear protein ZAP-like [Oryza sativa (japonica cultivar-group)]
gi|49387952|dbj|BAD25050.1| nuclear protein ZAP-like
[Oryza sativa (japonica cultivar-group)]
Length = 753
Score = 530 bits (1366), Expect = e-149
Identities = 334/752 (44%), Positives = 423/752 (55%), Gaps = 132/752 (17%)
Query: 22 LCPNCLTPHFPFCP-------------------------NPSPNWPPPP-------PPP- 48
LCP C PHFPFCP +P P PPPP PPP
Sbjct: 13 LCPVCAAPHFPFCPPPPHPFPYDLHPPPPPPPPEYHAPFHPPPPPPPPPEYHVPFHPPPP 72
Query: 49 ---NPTINNYD-SDFDRTFKKPRIED----------------------DERRLKLIRDHG 82
P + Y+ D + K+ R+ D +R L LIRDHG
Sbjct: 73 MWAPPGPHPYEVLDMEAPHKRMRVGDPYGDGMPPPLPPPPPPGMVPVEGDRLLGLIRDHG 132
Query: 83 LNPPQFQHPPPPPPPQYHPPPPP-----------PPPHIHNTPIYH----PQQHHSEFNS 127
P PPPP P PPP PPP ++ P H P H+
Sbjct: 133 RPP----FPPPPGMLHGEPYPPPDRFGYGGGRGYPPPPNYDNPYAHEGSFPDYEHAGRFP 188
Query: 128 PFHD------PNFHHFQPQP-VDNHIHNHN-FNARES--HHPYPYPNPYPNGSNNGFSDN 177
P H+ +F PQ NH H +N F ES P P P YP + + + +
Sbjct: 189 PAHERLAALGSSFVPGGPQEGYFNHDHRYNRFQRSESPVAPPLPPPARYPE-ARSRYDSH 247
Query: 178 NAQMEASRFYRPPLPTSPPPPLPMDPPMYFSAP--KKPP------SLFPVTSSA------ 223
EA PPPP P++PP+ S+ KPP SLFP+ S +
Sbjct: 248 GWHPEAD---------VPPPPPPLEPPVPSSSDYHAKPPLQAVKSSLFPIHSGSPAATAR 298
Query: 224 PHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYL--------GDAQPFSINPLAAEKPKFV 275
P H Q + ++ E P Y G Q + + + K +
Sbjct: 299 PPSSHTLHQAHLMPNANRYNGPIHNEVPGLAYQPHLEQHLGGGRQTQAQHSINNAKISVI 358
Query: 276 DASQLFRNPLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMT 335
A LF+ PLR SRPDH VIILRG PGSGK+YLAK LRDLEVENGG+APRIHSMDDYFM
Sbjct: 359 SACDLFKQPLRGSRPDHIVIILRGLPGSGKSYLAKALRDLEVENGGNAPRIHSMDDYFMI 418
Query: 336 EVE-KVEDSDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTF 394
EVE KVED++G KSSS+ + ++ +TKKV+EYCYEPEMEE YRSSML AFKKT++EG FTF
Sbjct: 419 EVEKKVEDNEGSKSSSTSKGRKQLTKKVIEYCYEPEMEETYRSSMLNAFKKTLDEGNFTF 478
Query: 395 IIVDDRNLRVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQW 454
+IVDDRNLRVADFAQFWA+AK+SGYEVY+LEA YKDP GCAARNVHGFT ++ KMA W
Sbjct: 479 VIVDDRNLRVADFAQFWASAKKSGYEVYLLEAPYKDPTGCAARNVHGFTVDDVNKMAADW 538
Query: 455 EEAPSLYLQLDMKSLFHGDDLKESRIQEVDMDMDDDLDDALVAAQGREAD---KAVVRPV 511
EEAP LYL+LD+ SLF+ D+L+E IQEVDM+ +D D A A EA+ KAV +
Sbjct: 539 EEAPPLYLRLDIHSLFNDDNLREHSIQEVDME-TEDTDGASNTATSTEAENTQKAVSESL 597
Query: 512 KDGEGSIKDGKRWDAEEEHPTE-VRELGKSKWSEDFEDDIDQTEGMKGNINALSGLIHQY 570
+G GK+WD+ EE + +ELG+SKWS+DF++D ++++ +G+ +ALSGL Y
Sbjct: 598 DNGHDQ-GAGKKWDSSEEDDLDGYKELGQSKWSKDFDEDTEKSDHAEGSTHALSGLAQTY 656
Query: 571 GKERKSVHWGDQVGKTGFSIGTARKVTTLSLVIGPGAGYNLKSNPLPKEESPTRNSV--- 627
RK+V WGD++ K GFSIG A++ T SL+IGPG+GYNL SNPL ++ S V
Sbjct: 657 STHRKTVTWGDRLEKGGFSIGAAKRRLTSSLIIGPGSGYNLVSNPLAEDNSKQAKGVINT 716
Query: 628 ETKKRSTFQERIRAERESFKAVFDGRRPRIGL 659
+TKKR F E++R E ESF+AVFD RR R+G+
Sbjct: 717 DTKKR--FSEQLRDEGESFRAVFDKRRQRVGV 746
>dbj|BAD93769.1| ZAP - like protein [Arabidopsis thaliana]
gi|30697735|ref|NP_568961.3| nuclear protein ZAP-related
[Arabidopsis thaliana]
Length = 383
Score = 271 bits (693), Expect = 5e-71
Identities = 186/457 (40%), Positives = 225/457 (48%), Gaps = 158/457 (34%)
Query: 3 RQWRPNPNLNPRPIQYQTNLCPNCLTPHFPFCPN---PS-----PNWPPPP--------- 45
+QWRP P Q N+CP C PHFPFCP PS PN+PPPP
Sbjct: 18 QQWRPAPT--------QPNICPICTVPHFPFCPPYPPPSSFAYNPNFPPPPHLNSPRPGF 69
Query: 46 -----PPPNPTINNY------------------DSDFDRTFKKPRIE------------- 69
PP P N+Y D + DR++K+ RI+
Sbjct: 70 DSFTGPPVRPPQNHYPPWQPHHGNQWRPVAVDVDREADRSYKRARIDTIAGGSPGYGVSE 129
Query: 70 --------DDERRLKLIRDHGLNPPQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQH 121
++ERRLK++RDHG Y P N + H Q+
Sbjct: 130 SPSPRISWENERRLKMVRDHG----------------YGLAAPS------NIEMNH--QY 165
Query: 122 HSEFNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQM 181
SEF + P P HHP PY Y +GSN
Sbjct: 166 GSEFRNGGQFNGVAPLPPPP--------------PHHPPPYGG-YFSGSNG--------- 201
Query: 182 EASRFYRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPE 241
+PPLP SPPPPLP P SLFPVT+++
Sbjct: 202 ------QPPLPVSPPPPLP---------SSHPSSLFPVTTNSS----------------- 229
Query: 242 FSTAFPTEEPSKQY--LGDAQPFSINPLAAEKPKFVDASQLFRNPLRTSRPDHFVIILRG 299
PT PS Y + +A P S LA + K +D S L + P R++RPDHFVIILRG
Sbjct: 230 -----PTIPPSSSYPQMPNASPSSAQ-LAPTRSKVIDVSHLLKPPHRSTRPDHFVIILRG 283
Query: 300 FPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTEVEKVEDSDGPKSSSSGRNKRPVT 359
PGSGK+YLAK+LRD+EVENGG APRIHSMDDYFMTEVEKVE+SD S SSGR+KRP+
Sbjct: 284 LPGSGKSYLAKLLRDVEVENGGSAPRIHSMDDYFMTEVEKVEESDS-TSLSSGRSKRPIV 342
Query: 360 KKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFII 396
K VMEYCYEPEMEEAYRSSMLKAFK+T+E+G F+F+I
Sbjct: 343 KTVMEYCYEPEMEEAYRSSMLKAFKRTLEDGAFSFVI 379
>ref|NP_848140.2| YLP motif containing 1 [Mus musculus] gi|6016842|dbj|BAA85182.1|
nuclear protein ZAP [Mus musculus]
gi|18203404|sp|Q9R0I7|YLPM_MOUSE YLP motif containing
protein 1 (Nuclear protein ZAP3)
Length = 1386
Score = 234 bits (597), Expect = 7e-60
Identities = 189/584 (32%), Positives = 255/584 (43%), Gaps = 120/584 (20%)
Query: 29 PHFPFCPNPSPNWPPPPPPPNPTINNYDS-------DFDRTFKKPRIEDDE-------RR 74
P P P P P PPPPPPP P I + S D D F DD+ R
Sbjct: 783 PPVPIPPPPPP--PPPPPPPPPVIKSKTSSVKQERWDEDSFFGLWDTNDDQGLNSEFKRD 840
Query: 75 LKLIRDHGLNPPQFQH---PPPPPPPQYHPPPPPPPPHIHNTPIYH-------------- 117
I + PP H PPP P P PP PPP H H
Sbjct: 841 TATIPSAPVLPPPPVHSSIPPPGPMPMGMPPMSKPPPVQHTVDYGHGRDMPTNKVEQIPY 900
Query: 118 ---------PQQHHSEFNSPF--------HDPNFHHFQPQPVDNHIHNHNFNARESHHPY 160
P S F++ D + + +P + H RE+H
Sbjct: 901 GERITLRPDPLPERSTFDADHAGQRDRYDRDRDREPYFDRPSNITDHRDFKRDRETHRDR 960
Query: 161 PYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVT 220
+ + + + ++ YR S D P Y +PP P
Sbjct: 961 DRDRVLDYERDRFDRERRPRDDRNQSYRDKKDHSSSRRGGFDRPSYDRKSDRPPYEGPPM 1020
Query: 221 SSA-----PHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYLGDAQPFSINPLAAEKPKFV 275
P E P P P P P +KP+
Sbjct: 1021 FGGERRTYPEERMPLPAPALGHQPPPV-----------------------PRVEKKPESK 1057
Query: 276 DASQLFRNPLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMT 335
+ + + P R SRP+ V+I+RG PGSGKT++AK++RD EVE GG APR+ S+DDYF+
Sbjct: 1058 NVDDILKPPGRESRPERIVVIMRGLPGSGKTHVAKLIRDKEVEFGGPAPRVLSLDDYFIA 1117
Query: 336 EVEKVEDSDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFI 395
EVEK E K SG+ V KKVMEY YE +MEE YR+SM K FKKT+++G F FI
Sbjct: 1118 EVEKEE-----KDPDSGKK---VKKKVMEYEYEADMEETYRTSMFKTFKKTLDDGFFPFI 1169
Query: 396 IVDDRNLRVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQWE 455
I+D N RV F QFW+ AK G+EVY+ E + D C RN+HG +EI KMAE WE
Sbjct: 1170 ILDAINDRVRHFDQFWSAAKTKGFEVYLAEMS-ADNQTCGKRNIHGRKLKEINKMAEHWE 1228
Query: 456 EAPSLYLQLDMKSLFHGDDLKESRIQEVDM-DMDDDLDDALVAAQGREADKAVVRPVKDG 514
AP ++LD++SL L+++ I+EV+M D D +++D
Sbjct: 1229 VAPRHMMRLDIRSL-----LQDAAIEEVEMEDFDANIEDQ-------------------- 1263
Query: 515 EGSIKDGKRWDAEEEHPTEVRELGKSKWSED-FEDDIDQTEGMK 557
K+ K+ DAEEE +E+ KSKW D E +D+ +G++
Sbjct: 1264 ----KEEKK-DAEEEE-SELGYTPKSKWEMDTSEAKLDKLDGLR 1301
Score = 52.8 bits (125), Expect = 4e-05
Identities = 82/324 (25%), Positives = 108/324 (33%), Gaps = 88/324 (27%)
Query: 37 PSPNWPPPPPP-------PNPTINNY------DSDFDRTFKKPRIED--------DERRL 75
P P PPPPPP P P ++ S +F++ + ++
Sbjct: 15 PPPPVPPPPPPVALPEASPGPGYSSSTAPAAPSSSGFMSFREQHLAQLQQLQQMHQKQMQ 74
Query: 76 KLIRDHGLNPPQFQHPPPPPPP-----QYHPPPPPPPPHIHNTPIYHPQQ-------HHS 123
+++ H L PP PPPP P + PPPPP PP Y QQ HH
Sbjct: 75 SVLQPHHLPPPPL--PPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKHQMIHHQ 132
Query: 124 EFNSPFHDPNFHHFQPQ--PVDNHIHNHNFNARESHHPYPYPNP--YPNGSNNGFSDNNA 179
P P P+ PV + +S+ P P P P YP S
Sbjct: 133 RDGPPGLVPMELESPPESPPVPPGSY---MPPSQSYMPPPQPPPSYYPPSS--------- 180
Query: 180 QMEASRFYRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHE--------PHPFP 231
++ Y PP SP P PP S K P+ S A ++ P P P
Sbjct: 181 ----AQPYLPPAQPSPSKPQLPPPPSIPSGNKTAIQQEPLESGAKNKTAEQKQAAPEPDP 236
Query: 232 --------QPYFHTT----------------KPEFSTAFPTEEPSKQYL-GDAQPFSINP 266
Q Y++ K TA EP K+ G A P
Sbjct: 237 STMTPQEQQQYWYRQHLLSLQQRTKVHLPGHKKGLVTAKDVPEPIKEEAPGPAASQVAEP 296
Query: 267 LAAEKPKFVDASQLFRNPLRTSRP 290
LAAEKP ++ PL P
Sbjct: 297 LAAEKPPLPPPNEEAPPPLSPEEP 320
Score = 48.1 bits (113), Expect = 9e-04
Identities = 52/190 (27%), Positives = 65/190 (33%), Gaps = 35/190 (18%)
Query: 90 HPPPPPPPQYHPPPPPPPPHIHNTPIYH----PQQHHSEFNSPFHDPNFHHFQPQPVDNH 145
H PPPP P PPPP P P Y P S F + H Q Q +
Sbjct: 13 HYPPPPVP--PPPPPVALPEASPGPGYSSSTAPAAPSSSGFMSFREQ--HLAQLQQLQQM 68
Query: 146 IHNHNFNARESHH--PYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLP--- 200
+ + HH P P P P P G+ D ++PP P PPPP P
Sbjct: 69 HQKQMQSVLQPHHLPPPPLPPP-PVMPGGGYGD----------WQPPPPPMPPPPGPALS 117
Query: 201 ------MDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQ 254
M PP L P+ +P E P P + P + P +P
Sbjct: 118 YQKQQQYKHQMIHHQRDGPPGLVPMELESPPESPPVPPGSY---MPPSQSYMPPPQPPPS 174
Query: 255 YL--GDAQPF 262
Y AQP+
Sbjct: 175 YYPPSSAQPY 184
Score = 43.9 bits (102), Expect = 0.017
Identities = 49/187 (26%), Positives = 65/187 (34%), Gaps = 52/187 (27%)
Query: 29 PHFPFCPNPSPNWPPPPPPPNPTINNYDSDFDRTFKKPRIEDDERRLKLIRDHGLNPPQF 88
P P P ++ PPPP P + +P + + R P
Sbjct: 741 PSRPGMYPPPGSYRPPPPMGKPP---------GSIVRPSAPPARSSIPMTR------PPV 785
Query: 89 QHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEFNSPFHDPNFHHFQPQPVDNHIHN 148
PPPPPPP PPPPPPPP I + Q+ E D+
Sbjct: 786 PIPPPPPPP---PPPPPPPPVIKSKTSSVKQERWDE------------------DSFFGL 824
Query: 149 HNFNARESHHPYPYPNPYPNGSNNGFSDNNAQM-EASRFYRPPLPTSPPPPLPMDPPMYF 207
+ N + G N+ F + A + A PP+ +S PPP PM PM
Sbjct: 825 WDTNDDQ-------------GLNSEFKRDTATIPSAPVLPPPPVHSSIPPPGPM--PMGM 869
Query: 208 SAPKKPP 214
KPP
Sbjct: 870 PPMSKPP 876
Score = 42.0 bits (97), Expect = 0.064
Identities = 50/223 (22%), Positives = 74/223 (32%), Gaps = 46/223 (20%)
Query: 70 DDERRLKLIRDHGLNPPQFQHPPP--------------PPPPQYHPPPP---PP------ 106
+ + +L+ ++D G P H PP PPP Y PPPP PP
Sbjct: 710 EQKEQLQKLKDFGSEPQMADHLPPPDSRLQNPSRPGMYPPPGSYRPPPPMGKPPGSIVRP 769
Query: 107 --PPHIHNTPIYHPQQHHSEFNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPN 164
PP + P+ P P P P P I + + ++
Sbjct: 770 SAPPARSSIPMTRPP-------VPIPPPPPPPPPPPPPPPVIKSKTSSVKQER------- 815
Query: 165 PYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAP 224
+ S G D N + ++ T P P+ PP++ S P PP P+
Sbjct: 816 -WDEDSFFGLWDTNDDQGLNSEFKRDTATIPSAPVLPPPPVHSSIP--PPGPMPMGM--- 869
Query: 225 HEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYLGDAQPFSINPL 267
P P P HT PT + + G+ +PL
Sbjct: 870 -PPMSKPPPVQHTVDYGHGRDMPTNKVEQIPYGERITLRPDPL 911
Score = 40.8 bits (94), Expect = 0.14
Identities = 37/133 (27%), Positives = 47/133 (34%), Gaps = 40/133 (30%)
Query: 17 QYQTNLCPNCLTPHFPFCPNP----------SPNWPPPPPPPNPTIN------------N 54
Q Q+ L P+ L P P P P P PP PPPP P ++ +
Sbjct: 72 QMQSVLQPHHLPPP-PLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKHQMIH 130
Query: 55 YDSDFDRTFKKPRIEDDERRLKLIRDHGLNPPQFQHPPPPPPPQYHPP-------PP--- 104
+ D +E + + P Q PPP PPP Y+PP PP
Sbjct: 131 HQRDGPPGLVPMELESPPESPPVPPGSYMPPSQSYMPPPQPPPSYYPPSSAQPYLPPAQP 190
Query: 105 -------PPPPHI 110
PPPP I
Sbjct: 191 SPSKPQLPPPPSI 203
Score = 37.4 bits (85), Expect = 1.6
Identities = 22/52 (42%), Positives = 25/52 (47%), Gaps = 5/52 (9%)
Query: 189 PPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKP 240
PPLPT PPP LP P P PP+L P + P P P P T+ P
Sbjct: 492 PPLPTMPPPVLPPSLP----PPVMPPAL-PTSIPPPGMPPPVMPPSLPTSVP 538
>ref|XP_085151.9| PREDICTED: YLP motif containing 1 [Homo sapiens]
Length = 2140
Score = 225 bits (574), Expect = 3e-57
Identities = 184/583 (31%), Positives = 258/583 (43%), Gaps = 120/583 (20%)
Query: 32 PFCPNPSPNWPPPPPPPNPTIN---------NYDSD-----FDRTFKKPRIEDDERRLKL 77
P P P P PPP PPP P I +D D +D ++ + +
Sbjct: 1542 PPVPIPPPPPPPPLPPPPPVIKPQTSAVEQERWDEDSFYGLWDTNDEQGLNSEFKSETAA 1601
Query: 78 IRDHGLNPPQFQH---PPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEF--NSPFHDP 132
I + PP H PPP P P PP PPP H + + P+ +
Sbjct: 1602 IPSAPVLPPPPVHSSIPPPGPVPMGMPPMSKPPPVQQTVDYGHGRDISTNKVEQIPYGER 1661
Query: 133 NFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNN-------------- 178
P P + + R+ + PY + +N +D+
Sbjct: 1662 ITLRPDPLPERSTFETEHAGQRDRYDRERDREPYFDRQSNVIADHRDFKRDRETHRDRDR 1721
Query: 179 ----AQMEASRF-------------YRPPLPTSPPPPLPMDPPMYFSAPKKP----PSLF 217
+ RF YR S D P Y +P PS+F
Sbjct: 1722 DRGVIDYDRDRFDRERRPRDDRAQSYRDKKDHSSSRRGGFDRPSYDRKSDRPVYEGPSMF 1781
Query: 218 P-VTSSAPHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYLGDAQPFSINPLAAEKPKFVD 276
+ P E P P P P P +KP+ +
Sbjct: 1782 GGERRTYPEERMPLPAPSLSHQPPPA-----------------------PRVEKKPESKN 1818
Query: 277 ASQLFRNPLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTE 336
+ + P R SRP+ V+I+RG PGSGKT++AK++RD EVE GG APR+ S+DDYF+TE
Sbjct: 1819 VDDILKPPGRESRPERIVVIMRGLPGSGKTHVAKLIRDKEVEFGGPAPRVLSLDDYFITE 1878
Query: 337 VEKVEDSDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFII 396
VEK E K SG+ V KKVMEY YE EMEE YR+SM K FKKT+++G F FII
Sbjct: 1879 VEKEE-----KDPDSGKK---VKKKVMEYEYEAEMEETYRTSMFKTFKKTLDDGFFPFII 1930
Query: 397 VDDRNLRVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQWEE 456
+D N RV F QFW+ AK G+EVY+ E + D C RN+HG +EI KMA+ WE
Sbjct: 1931 LDAINDRVRHFDQFWSAAKTKGFEVYLAEMS-ADNQTCGKRNIHGRKLKEINKMADHWET 1989
Query: 457 APSLYLQLDMKSLFHGDDLKESRIQEVDM-DMDDDLDDALVAAQGREADKAVVRPVKDGE 515
AP ++LD++SL L+++ I+EV+M D D ++++
Sbjct: 1990 APRHMMRLDIRSL-----LQDAAIEEVEMEDFDANIEEQ--------------------- 2023
Query: 516 GSIKDGKRWDAEEEHPTEVRELGKSKWSED-FEDDIDQTEGMK 557
K+ K+ DAEEE +E+ + KSKW D E +D+ +G++
Sbjct: 2024 ---KEEKK-DAEEEE-SELGYIPKSKWEMDTSEAKLDKLDGLR 2061
Score = 53.9 bits (128), Expect = 2e-05
Identities = 65/255 (25%), Positives = 86/255 (33%), Gaps = 74/255 (29%)
Query: 37 PSPNWPPPPP------PPNPTINNY------DSDFDRTFKKPRIEDDERRLKLIRDH--- 81
P P PPPPP P P ++ S +F++ + ++ ++ +
Sbjct: 15 PPPPVPPPPPVALPEASPGPGYSSSTTPAAPSSSGFMSFREQHLAQLQQLQQMHQKQMQC 74
Query: 82 GLNPPQFQHPPPPPPP--------QYHPPPPPPPPHIHNTPIYHPQQHHSEFNSPFHDPN 133
L P PP PPPP + PPPPP PP Y QQ +
Sbjct: 75 VLQPHHLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKH-------QM 127
Query: 134 FHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPT 193
HH + P E P P P P GS Y PP +
Sbjct: 128 LHHQRDGP-------PGLVPMELESP-PESPPVPPGS----------------YMPPSQS 163
Query: 194 SPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPEFSTAFPT----- 248
PPP P PPS +P TSS P+ P P P + P S PT
Sbjct: 164 YMPPPQP------------PPSYYPPTSSQPYLPPAQPSP--SQSPPSQSYLAPTPSYSS 209
Query: 249 -EEPSKQYLGDAQPF 262
S+ YL +Q +
Sbjct: 210 SSSSSQSYLSHSQSY 224
Score = 46.6 bits (109), Expect = 0.003
Identities = 45/192 (23%), Positives = 66/192 (33%), Gaps = 42/192 (21%)
Query: 30 HFPFCPNPSP---------NWPPPPPP----PNPTIN------------NYDSDFDRTFK 64
H P P P P +W PPPPP P P ++ ++ D
Sbjct: 80 HLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKHQMLHHQRDGPPGLV 139
Query: 65 KPRIEDDERRLKLIRDHGLNPPQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSE 124
+E + + P Q PPP PPP Y+PP P Y P S
Sbjct: 140 PMELESPPESPPVPPGSYMPPSQSYMPPPQPPPSYYPPTSSQP--------YLPPAQPSP 191
Query: 125 FNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEAS 184
SP P+ + P P +++ ++ S + Y S S + S
Sbjct: 192 SQSP---PSQSYLAPTP------SYSSSSSSSQSYLSHSQSYLPSSQASPSRPSQGHSKS 242
Query: 185 RFYRPPLPTSPP 196
+ PP P++PP
Sbjct: 243 QLLAPPPPSAPP 254
Score = 43.1 bits (100), Expect = 0.029
Identities = 59/260 (22%), Positives = 84/260 (31%), Gaps = 44/260 (16%)
Query: 13 PRPIQYQ-----TNLCPNCLTPHFPFCPNPSPNWPPPPPPPNPTINNYDSDFDRTFKKPR 67
P P++ + T+ P + P P P+ PPP PP P + + D R
Sbjct: 327 PEPVKEEVTVPATSQVPESPSSEEPPLPPPNEEVPPPLPPEEPQSEDPEED-------AR 379
Query: 68 IEDDERRLKLIRDHGLNPPQFQ-------HPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQ 120
++ + + H + FQ H Q + PPPHI + +
Sbjct: 380 LKQLQAAAAHWQQHQQHRVGFQYQGIMQKHTQLQQILQQYQQIIQPPPHIQTMSVDMQLR 439
Query: 121 HHSEFNSPF---HDPNFHHFQPQPVDNHIHNHNFNARESHHPY-------PYPNPYPNGS 170
H+ F + FQ H + H +E + Y
Sbjct: 440 HYEMQQQQFQHLYQEWEREFQLWEEQLHSYPHKDQLQEYEKQWKTWQGHMKATQSYLQEK 499
Query: 171 NNGFSDNNAQMEASRF----------YRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVT 220
N F + Q + PPLPT PPP LP P P PP+L P T
Sbjct: 500 VNSFQNMKNQYMGNMSMPPPFVPYSQMPPPLPTMPPPVLPPSLP----PPVMPPAL-PAT 554
Query: 221 SSAPHEPHPFPQPYFHTTKP 240
P P P P T+ P
Sbjct: 555 VPPPGMPPPVMPPSLPTSVP 574
Score = 41.2 bits (95), Expect = 0.11
Identities = 58/235 (24%), Positives = 77/235 (32%), Gaps = 41/235 (17%)
Query: 85 PPQFQHPPPPPPPQYHPPPPPP------PPHIHNTPIYHPQQHHSEFNSPFHDPNFHHFQ 138
PP PPP PP PP PP PP P+ P S P P+
Sbjct: 528 PPLPTMPPPVLPPSLPPPVMPPALPATVPPPGMPPPVMPPSLPTS-VPPPGMPPSLSSAG 586
Query: 139 PQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPP 198
P PV P P P ++ + PP P PPP
Sbjct: 587 PPPV--------LPPPSLSSAGPPPVLPPPSLSSTAPPPVMPLPPLSSATPP-PGIPPPG 637
Query: 199 LPMDPPMYFSAPKKPPSLFPVTSSAPHEPHP--FPQPYFH------TTKPEFSTAFPTEE 250
+P P +A PP+ +S P +P P P P T P +A P+E+
Sbjct: 638 VPQGIPPQLTAAPVPPASSSQSSQVPEKPRPALLPTPVSFGSAPPTTYHPPLQSAGPSEQ 697
Query: 251 -----------------PSKQYLGDAQPFSINPLAAEKPKFVDASQLFRNPLRTS 288
S Q LG++ P+ A K V + L +P R+S
Sbjct: 698 VNSKAPLSKSALPYSSFSSDQGLGESSAAPSQPITAVKDMPVRSGGLLPDPPRSS 752
Score = 40.0 bits (92), Expect = 0.24
Identities = 43/191 (22%), Positives = 64/191 (32%), Gaps = 44/191 (23%)
Query: 68 IEDDERRLKLIRDHGLNPPQFQHPPP---------------PPPPQYHPPPP---PP--- 106
+ + + +L+ ++D G P H PP PPP Y PPPP PP
Sbjct: 1466 LREQKEQLQKMKDFGSEPQMADHLPPQESRLQNTSSRPGMYPPPGSYRPPPPMGKPPGSI 1525
Query: 107 -----PPHIHNTPIYHPQQHHSEFNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYP 161
PP + P+ P P P P P + +A E
Sbjct: 1526 VRPSAPPARSSVPVTRP---------PVPIPPPPPPPPLPPPPPVIKPQTSAVEQER--- 1573
Query: 162 YPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTS 221
+ S G D N + + ++ P P+ PP++ S P PP P+
Sbjct: 1574 ----WDEDSFYGLWDTNDEQGLNSEFKSETAAIPSAPVLPPPPVHSSIP--PPGPVPMGM 1627
Query: 222 SAPHEPHPFPQ 232
+P P Q
Sbjct: 1628 PPMSKPPPVQQ 1638
>sp|P49750|YLPM1_HUMAN YLP motif containing protein 1 (Nuclear protein ZAP3) (ZAP113)
Length = 1951
Score = 225 bits (574), Expect = 3e-57
Identities = 184/583 (31%), Positives = 258/583 (43%), Gaps = 120/583 (20%)
Query: 32 PFCPNPSPNWPPPPPPPNPTIN---------NYDSD-----FDRTFKKPRIEDDERRLKL 77
P P P P PPP PPP P I +D D +D ++ + +
Sbjct: 1347 PPVPIPPPPPPPPLPPPPPVIKPQTSAVEQERWDEDSFYGLWDTNDEQGLNSEFKSETAA 1406
Query: 78 IRDHGLNPPQFQH---PPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEF--NSPFHDP 132
I + PP H PPP P P PP PPP H + + P+ +
Sbjct: 1407 IPSAPVLPPPPVHSSIPPPGPVPMGMPPMSKPPPVQQTVDYGHGRDISTNKVEQIPYGER 1466
Query: 133 NFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNN-------------- 178
P P + + R+ + PY + +N +D+
Sbjct: 1467 ITLRPDPLPERSTFETEHAGQRDRYDRERDREPYFDRQSNVIADHRDFKRDRETHRDRDR 1526
Query: 179 ----AQMEASRF-------------YRPPLPTSPPPPLPMDPPMYFSAPKKP----PSLF 217
+ RF YR S D P Y +P PS+F
Sbjct: 1527 DRGVIDYDRDRFDRERRPRDDRAQSYRDKKDHSSSRRGGFDRPSYDRKSDRPVYEGPSMF 1586
Query: 218 P-VTSSAPHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYLGDAQPFSINPLAAEKPKFVD 276
+ P E P P P P P +KP+ +
Sbjct: 1587 GGERRTYPEERMPLPAPSLSHQPPPA-----------------------PRVEKKPESKN 1623
Query: 277 ASQLFRNPLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTE 336
+ + P R SRP+ V+I+RG PGSGKT++AK++RD EVE GG APR+ S+DDYF+TE
Sbjct: 1624 VDDILKPPGRESRPERIVVIMRGLPGSGKTHVAKLIRDKEVEFGGPAPRVLSLDDYFITE 1683
Query: 337 VEKVEDSDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFII 396
VEK E K SG+ V KKVMEY YE EMEE YR+SM K FKKT+++G F FII
Sbjct: 1684 VEKEE-----KDPDSGKK---VKKKVMEYEYEAEMEETYRTSMFKTFKKTLDDGFFPFII 1735
Query: 397 VDDRNLRVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQWEE 456
+D N RV F QFW+ AK G+EVY+ E + D C RN+HG +EI KMA+ WE
Sbjct: 1736 LDAINDRVRHFDQFWSAAKTKGFEVYLAEMS-ADNQTCGKRNIHGRKLKEINKMADHWET 1794
Query: 457 APSLYLQLDMKSLFHGDDLKESRIQEVDM-DMDDDLDDALVAAQGREADKAVVRPVKDGE 515
AP ++LD++SL L+++ I+EV+M D D ++++
Sbjct: 1795 APRHMMRLDIRSL-----LQDAAIEEVEMEDFDANIEEQ--------------------- 1828
Query: 516 GSIKDGKRWDAEEEHPTEVRELGKSKWSED-FEDDIDQTEGMK 557
K+ K+ DAEEE +E+ + KSKW D E +D+ +G++
Sbjct: 1829 ---KEEKK-DAEEEE-SELGYIPKSKWEMDTSEAKLDKLDGLR 1866
Score = 53.9 bits (128), Expect = 2e-05
Identities = 65/255 (25%), Positives = 86/255 (33%), Gaps = 74/255 (29%)
Query: 37 PSPNWPPPPP------PPNPTINNY------DSDFDRTFKKPRIEDDERRLKLIRDH--- 81
P P PPPPP P P ++ S +F++ + ++ ++ +
Sbjct: 15 PPPPVPPPPPVALPEASPGPGYSSSTTPAAPSSSGFMSFREQHLAQLQQLQQMHQKQMQC 74
Query: 82 GLNPPQFQHPPPPPPP--------QYHPPPPPPPPHIHNTPIYHPQQHHSEFNSPFHDPN 133
L P PP PPPP + PPPPP PP Y QQ +
Sbjct: 75 VLQPHHLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKH-------QM 127
Query: 134 FHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPT 193
HH + P E P P P P GS Y PP +
Sbjct: 128 LHHQRDGP-------PGLVPMELESP-PESPPVPPGS----------------YMPPSQS 163
Query: 194 SPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPEFSTAFPT----- 248
PPP P PPS +P TSS P+ P P P + P S PT
Sbjct: 164 YMPPPQP------------PPSYYPPTSSQPYLPPAQPSP--SQSPPSQSYLAPTPSYSS 209
Query: 249 -EEPSKQYLGDAQPF 262
S+ YL +Q +
Sbjct: 210 SSSSSQSYLSHSQSY 224
Score = 48.1 bits (113), Expect = 9e-04
Identities = 53/190 (27%), Positives = 68/190 (34%), Gaps = 40/190 (21%)
Query: 99 YHPPPPPPPPHIHNTPIYHPQQHHSEFNSPF--HDPNFHHFQPQPVD-----NHIHNHNF 151
++PPPP PPP P P +S +P F F+ Q + +H
Sbjct: 13 HYPPPPVPPPPPVALPEASPGPGYSSSTTPAAPSSSGFMSFREQHLAQLQQLQQMHQKQM 72
Query: 152 N-ARESHH--PYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLP-------- 200
+ HH P P P P P G+ D ++PP P PPPP P
Sbjct: 73 QCVLQPHHLPPPPLPPP-PVMPGGGYGD----------WQPPPPPMPPPPGPALSYQKQQ 121
Query: 201 -MDPPMYFSAPKKPPSLFPVTSSAPHEPHPFP--------QPYFHTTKPEFSTAFPTEEP 251
M PP L P+ +P E P P Q Y +P S PT
Sbjct: 122 QYKHQMLHHQRDGPPGLVPMELESPPESPPVPPGSYMPPSQSYMPPPQPPPSYYPPTS-- 179
Query: 252 SKQYLGDAQP 261
S+ YL AQP
Sbjct: 180 SQPYLPPAQP 189
Score = 46.6 bits (109), Expect = 0.003
Identities = 45/192 (23%), Positives = 66/192 (33%), Gaps = 42/192 (21%)
Query: 30 HFPFCPNPSP---------NWPPPPPP----PNPTIN------------NYDSDFDRTFK 64
H P P P P +W PPPPP P P ++ ++ D
Sbjct: 80 HLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKHQMLHHQRDGPPGLV 139
Query: 65 KPRIEDDERRLKLIRDHGLNPPQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSE 124
+E + + P Q PPP PPP Y+PP P Y P S
Sbjct: 140 PMELESPPESPPVPPGSYMPPSQSYMPPPQPPPSYYPPTSSQP--------YLPPAQPSP 191
Query: 125 FNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEAS 184
SP P+ + P P +++ ++ S + Y S S + S
Sbjct: 192 SQSP---PSQSYLAPTP------SYSSSSSSSQSYLSHSQSYLPSSQASPSRPSQGHSKS 242
Query: 185 RFYRPPLPTSPP 196
+ PP P++PP
Sbjct: 243 QLLAPPPPSAPP 254
Score = 43.9 bits (102), Expect = 0.017
Identities = 52/231 (22%), Positives = 77/231 (32%), Gaps = 47/231 (20%)
Query: 86 PQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEFNSPFHDPNFHH---FQPQPV 142
P + PP PPP + PPP PP P + + + + H FQ Q +
Sbjct: 346 PSSEEPPLPPPNEEVPPPLPPEEPQSEDPEEDARLKQLQAAAAHWQQHQQHRVGFQYQGI 405
Query: 143 DNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLPMD 202
H + + P P+ + P P PPP +P
Sbjct: 406 MQK-HTQLQQILQQYQQIIQPPPHIQATT------------------PPPGIPPPGVPQG 446
Query: 203 PPMYFSAPKKPPSLFPVTSSAPHEPHP--FPQPYFH------TTKPEFSTAFPTEE---- 250
P +A PP+ +S P +P P P P T P +A P+E+
Sbjct: 447 IPPQLTAAPVPPASSSQSSQVPEKPRPALLPTPVSFGSAPPTTYHPPLQSAGPSEQVNSK 506
Query: 251 -------------PSKQYLGDAQPFSINPLAAEKPKFVDASQLFRNPLRTS 288
S Q LG++ P+ A K V + L +P R+S
Sbjct: 507 APLSKSALPYSSFSSDQGLGESSAAPSQPITAVKDMPVRSGGLLPDPPRSS 557
Score = 40.0 bits (92), Expect = 0.24
Identities = 43/191 (22%), Positives = 64/191 (32%), Gaps = 44/191 (23%)
Query: 68 IEDDERRLKLIRDHGLNPPQFQHPPP---------------PPPPQYHPPPP---PP--- 106
+ + + +L+ ++D G P H PP PPP Y PPPP PP
Sbjct: 1271 LREQKEQLQKMKDFGSEPQMADHLPPQESRLQNTSSRPGMYPPPGSYRPPPPMGKPPGSI 1330
Query: 107 -----PPHIHNTPIYHPQQHHSEFNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYP 161
PP + P+ P P P P P + +A E
Sbjct: 1331 VRPSAPPARSSVPVTRP---------PVPIPPPPPPPPLPPPPPVIKPQTSAVEQER--- 1378
Query: 162 YPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTS 221
+ S G D N + + ++ P P+ PP++ S P PP P+
Sbjct: 1379 ----WDEDSFYGLWDTNDEQGLNSEFKSETAAIPSAPVLPPPPVHSSIP--PPGPVPMGM 1432
Query: 222 SAPHEPHPFPQ 232
+P P Q
Sbjct: 1433 PPMSKPPPVQQ 1443
>dbj|BAC03416.1| FLJ00353 protein [Homo sapiens]
Length = 1766
Score = 225 bits (574), Expect = 3e-57
Identities = 184/583 (31%), Positives = 258/583 (43%), Gaps = 120/583 (20%)
Query: 32 PFCPNPSPNWPPPPPPPNPTIN---------NYDSD-----FDRTFKKPRIEDDERRLKL 77
P P P P PPP PPP P I +D D +D ++ + +
Sbjct: 1168 PPVPIPPPPPPPPLPPPPPVIKPQTSAVEQERWDEDSFYGLWDTNDEQGLNSEFKSETAA 1227
Query: 78 IRDHGLNPPQFQH---PPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEF--NSPFHDP 132
I + PP H PPP P P PP PPP H + + P+ +
Sbjct: 1228 IPSAPVLPPPPVHSSIPPPGPVPMGMPPMSKPPPVQQTVDYGHGRDISTNKVEQIPYGER 1287
Query: 133 NFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNN-------------- 178
P P + + R+ + PY + +N +D+
Sbjct: 1288 ITLRPDPLPERSTFETEHAGQRDRYDRERDREPYFDRQSNVIADHRDFKRDRETHRDRDR 1347
Query: 179 ----AQMEASRF-------------YRPPLPTSPPPPLPMDPPMYFSAPKKP----PSLF 217
+ RF YR S D P Y +P PS+F
Sbjct: 1348 DRGVIDYDRDRFDRERRPRDDRAQSYRDKKDHSSSRRGGFDRPSYDRKSDRPVYEGPSMF 1407
Query: 218 P-VTSSAPHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYLGDAQPFSINPLAAEKPKFVD 276
+ P E P P P P P +KP+ +
Sbjct: 1408 GGERRTYPEERMPLPAPSLSHQPPPA-----------------------PRVEKKPESKN 1444
Query: 277 ASQLFRNPLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTE 336
+ + P R SRP+ V+I+RG PGSGKT++AK++RD EVE GG APR+ S+DDYF+TE
Sbjct: 1445 VDDILKPPGRESRPERIVVIMRGLPGSGKTHVAKLIRDKEVEFGGPAPRVLSLDDYFITE 1504
Query: 337 VEKVEDSDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFII 396
VEK E K SG+ V KKVMEY YE EMEE YR+SM K FKKT+++G F FII
Sbjct: 1505 VEKEE-----KDPDSGKK---VKKKVMEYEYEAEMEETYRTSMFKTFKKTLDDGFFPFII 1556
Query: 397 VDDRNLRVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQWEE 456
+D N RV F QFW+ AK G+EVY+ E + D C RN+HG +EI KMA+ WE
Sbjct: 1557 LDAINDRVRHFDQFWSAAKTKGFEVYLAEMS-ADNQTCGKRNIHGRKLKEINKMADHWET 1615
Query: 457 APSLYLQLDMKSLFHGDDLKESRIQEVDM-DMDDDLDDALVAAQGREADKAVVRPVKDGE 515
AP ++LD++SL L+++ I+EV+M D D ++++
Sbjct: 1616 APRHMMRLDIRSL-----LQDAAIEEVEMEDFDANIEEQ--------------------- 1649
Query: 516 GSIKDGKRWDAEEEHPTEVRELGKSKWSED-FEDDIDQTEGMK 557
K+ K+ DAEEE +E+ + KSKW D E +D+ +G++
Sbjct: 1650 ---KEEKK-DAEEEE-SELGYIPKSKWEMDTSEAKLDKLDGLR 1687
Score = 41.2 bits (95), Expect = 0.11
Identities = 58/235 (24%), Positives = 77/235 (32%), Gaps = 41/235 (17%)
Query: 85 PPQFQHPPPPPPPQYHPPPPPP------PPHIHNTPIYHPQQHHSEFNSPFHDPNFHHFQ 138
PP PPP PP PP PP PP P+ P S P P+
Sbjct: 154 PPLPTMPPPVLPPSLPPPVMPPALPATVPPPGMPPPVMPPSLPTS-VPPPGMPPSLSSAG 212
Query: 139 PQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPP 198
P PV P P P ++ + PP P PPP
Sbjct: 213 PPPV--------LPPPSLSSAGPPPVLPPPSLSSTAPPPVMPLPPLSSATPP-PGIPPPG 263
Query: 199 LPMDPPMYFSAPKKPPSLFPVTSSAPHEPHP--FPQPYFH------TTKPEFSTAFPTEE 250
+P P +A PP+ +S P +P P P P T P +A P+E+
Sbjct: 264 VPQGIPPQLTAAPVPPASSSQSSQVPEKPRPALLPTPVSFGSAPPTTYHPPLQSAGPSEQ 323
Query: 251 -----------------PSKQYLGDAQPFSINPLAAEKPKFVDASQLFRNPLRTS 288
S Q LG++ P+ A K V + L +P R+S
Sbjct: 324 VNSKAPLSKSALPYSSFSSDQGLGESSAAPSQPITAVKDMPVRSGGLLPDPPRSS 378
Score = 40.0 bits (92), Expect = 0.24
Identities = 43/191 (22%), Positives = 64/191 (32%), Gaps = 44/191 (23%)
Query: 68 IEDDERRLKLIRDHGLNPPQFQHPPP---------------PPPPQYHPPPP---PP--- 106
+ + + +L+ ++D G P H PP PPP Y PPPP PP
Sbjct: 1092 LREQKEQLQKMKDFGSEPQMADHLPPQESRLQNTSSRPGMYPPPGSYRPPPPMGKPPGSI 1151
Query: 107 -----PPHIHNTPIYHPQQHHSEFNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYP 161
PP + P+ P P P P P + +A E
Sbjct: 1152 VRPSAPPARSSVPVTRP---------PVPIPPPPPPPPLPPPPPVIKPQTSAVEQER--- 1199
Query: 162 YPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTS 221
+ S G D N + + ++ P P+ PP++ S P PP P+
Sbjct: 1200 ----WDEDSFYGLWDTNDEQGLNSEFKSETAAIPSAPVLPPPPVHSSIP--PPGPVPMGM 1253
Query: 222 SAPHEPHPFPQ 232
+P P Q
Sbjct: 1254 PPMSKPPPVQQ 1264
Score = 38.9 bits (89), Expect = 0.54
Identities = 23/52 (44%), Positives = 25/52 (47%), Gaps = 5/52 (9%)
Query: 189 PPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKP 240
PPLPT PPP LP P P PP+L P T P P P P T+ P
Sbjct: 154 PPLPTMPPPVLPPSLP----PPVMPPAL-PATVPPPGMPPPVMPPSLPTSVP 200
>ref|XP_537509.1| PREDICTED: similar to Nuclear protein ZAP3 (ZAP113) [Canis
familiaris]
Length = 1956
Score = 221 bits (563), Expect = 6e-56
Identities = 171/520 (32%), Positives = 234/520 (44%), Gaps = 94/520 (18%)
Query: 29 PHFPFCPNPSPNWPPPPPPPNPTINNYDSDFDR------TFKKPRIEDDERRLK------ 76
P P P P P PPPPPPP P I S ++ +F +DE+ L
Sbjct: 1375 PPIPIPPPPPP--PPPPPPPPPVIKPQTSAVEQERWDEDSFYGLWDTNDEQGLNSEFKPE 1432
Query: 77 --LIRDHGLNPPQFQH--PPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEF--NSPFH 130
I + PP PPP P P PP PPP H + + P+
Sbjct: 1433 TGTIPSAPVLPPPLHPSIPPPGPLPMGMPPMSKPPPVQQTVDYGHGRDMSTNKVEQIPYG 1492
Query: 131 DPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNN------------ 178
+ P P + + R+ + PY + +N +D+
Sbjct: 1493 ERITLRPDPLPERSTFETEHAGQRDRYDRDRDREPYFDRQSNVIADHRDFKRDRETHRDR 1552
Query: 179 ------AQMEASRF-------------YRPPLPTSPPPPLPMDPPMYFSAPKKP----PS 215
+ RF YR S D P Y +P PS
Sbjct: 1553 DRDRGVIDYDRDRFDRERRPRDDRTQSYRDKKDHSSSRRGGFDRPSYDRKSDRPAYEGPS 1612
Query: 216 LFP-VTSSAPHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYLGDAQPFSINPLAAEKPKF 274
+F + P E P P P P P +KP+
Sbjct: 1613 MFGGERRTYPEERMPLPAPSLSHQPPPA-----------------------PRVEKKPES 1649
Query: 275 VDASQLFRNPLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFM 334
+ + + P R SRP+ V+I+RG PGSGKT++AK++RD EVE GG APR+ S+DDYF+
Sbjct: 1650 KNVDDILKPPGRESRPERIVVIMRGLPGSGKTHVAKLIRDKEVEFGGPAPRVLSLDDYFI 1709
Query: 335 TEVEKVEDSDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTF 394
TEVEK E K SG+ V KKVMEY YE EMEE YR+SM K FKKT+++G F F
Sbjct: 1710 TEVEKEE-----KDPDSGKK---VKKKVMEYEYEAEMEETYRTSMFKTFKKTLDDGFFPF 1761
Query: 395 IIVDDRNLRVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQW 454
II+D N RV F QFW+ AK G+EVY+ E + D C RN+HG +EI KMA+ W
Sbjct: 1762 IILDAINDRVRHFDQFWSAAKTKGFEVYLAEMS-ADNQTCGKRNIHGRKLKEINKMADHW 1820
Query: 455 EEAPSLYLQLDMKSLFHGDDLKESRIQEVDM-DMDDDLDD 493
E AP ++LD++SL L+++ I+EV+M D D ++++
Sbjct: 1821 ETAPRHMMRLDIRSL-----LQDAAIEEVEMEDFDANIEE 1855
Score = 45.1 bits (105), Expect = 0.008
Identities = 46/182 (25%), Positives = 61/182 (33%), Gaps = 56/182 (30%)
Query: 35 PNPSPNWPPPP--PPPNPTINNYDSDFDRTFKKPRIEDDERRLKLIRDHGLNPPQFQHPP 92
P P PPPP PP + +P + + R P PP
Sbjct: 1340 PPPGSYRPPPPIGKPPGSIV------------RPSAPPARSSVPVTR------PPIPIPP 1381
Query: 93 PPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEFNSPFHDPNFHHFQPQPVDNHIHNHNFN 152
PPPPP PPPPPPP P+ PQ E + + +F+
Sbjct: 1382 PPPPP---PPPPPPP------PVIKPQTSAVE-QERWDEDSFYGL--------------- 1416
Query: 153 ARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLPMDPPMYFSAPKK 212
+ G N+ F + ++ PPL S PPP P+ PM K
Sbjct: 1417 ---------WDTNDEQGLNSEFKPETGTIPSAPVLPPPLHPSIPPPGPL--PMGMPPMSK 1465
Query: 213 PP 214
PP
Sbjct: 1466 PP 1467
Score = 37.4 bits (85), Expect = 1.6
Identities = 22/52 (42%), Positives = 25/52 (47%), Gaps = 5/52 (9%)
Query: 189 PPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKP 240
PPLPT PPP LP P P PP+L P + P P P P T+ P
Sbjct: 359 PPLPTMPPPVLPPSLP----PPVMPPAL-PASVPPPGMPPPVMPPSLPTSVP 405
Score = 36.2 bits (82), Expect = 3.5
Identities = 33/110 (30%), Positives = 41/110 (37%), Gaps = 13/110 (11%)
Query: 120 QHHSEFNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNA 179
Q +F S P+ H PQ D + N + P Y P P G G +
Sbjct: 1307 QKMKDFGSEPQMPD--HLPPQ--DTRLQNASSRPGMYPPPGSYRPPPPIGKPPGSIVRPS 1362
Query: 180 QMEASRFY---RPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHE 226
A RPP+P PPPP P P P PP + P TS+ E
Sbjct: 1363 APPARSSVPVTRPPIPIPPPPPPPPPP------PPPPPVIKPQTSAVEQE 1406
>dbj|BAC31920.1| unnamed protein product [Mus musculus]
Length = 818
Score = 219 bits (559), Expect = 2e-55
Identities = 130/307 (42%), Positives = 175/307 (56%), Gaps = 60/307 (19%)
Query: 189 PPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSA-PHEPHPFPQPYFHTTKPEFSTAFP 247
PPLPT PPP LP P PP + P+ + A H+P P P+
Sbjct: 539 PPLPTMPPPVLP---------PSLPPPVMPLPAPALGHQPPPVPR--------------- 574
Query: 248 TEEPSKQYLGDAQPFSINPLAAEKPKFVDASQLFRNPLRTSRPDHFVIILRGFPGSGKTY 307
+KP+ + + + P R SRP+ V+I+RG PGSGKT+
Sbjct: 575 --------------------VEKKPESKNVDDILKPPGRESRPERIVVIMRGLPGSGKTH 614
Query: 308 LAKMLRDLEVENGGDAPRIHSMDDYFMTEVEKVEDSDGPKSSSSGRNKRPVTKKVMEYCY 367
+AK++RD EVE GG APR+ S+DDYF+ EVEK E K SG+ V KKVMEY Y
Sbjct: 615 VAKLIRDKEVEFGGPAPRVLSLDDYFIAEVEKEE-----KDPDSGKK---VKKKVMEYEY 666
Query: 368 EPEMEEAYRSSMLKAFKKTVEEGVFTFIIVDDRNLRVADFAQFWATAKRSGYEVYILEAT 427
E +MEE YR+SM K FKKT+++G F FII+D N RV F QFW+ AK G+EVY+ E +
Sbjct: 667 EADMEETYRTSMFKTFKKTLDDGFFPFIILDAINDRVRHFDQFWSAAKTKGFEVYLAEMS 726
Query: 428 YKDPVGCAARNVHGFTQQEIEKMAEQWEEAPSLYLQLDMKSLFHGDDLKESRIQEVDM-D 486
D C RN+HG +EI KMAE WE AP ++LD++SL L+++ I+EV+M D
Sbjct: 727 -ADNQTCGKRNIHGRKLKEINKMAEHWEVAPRHMMRLDIRSL-----LQDAAIEEVEMED 780
Query: 487 MDDDLDD 493
D +++D
Sbjct: 781 FDANIED 787
Score = 57.8 bits (138), Expect = 1e-06
Identities = 63/250 (25%), Positives = 86/250 (34%), Gaps = 64/250 (25%)
Query: 37 PSPNWPPPPPP-------PNPTINNY------DSDFDRTFKKPRIEDDERRLKLIRDH-- 81
P P PPPPPP P P ++ S +F++ + ++ ++ +
Sbjct: 15 PPPPVPPPPPPVALPEASPGPGYSSSTAPAAPSSSGFMSFREQHLAQLQQLQQMHQKQMQ 74
Query: 82 -GLNPPQFQHPPPPPPP--------QYHPPPPPPPPHIHNTPIYHPQQHHSEFNSPFHDP 132
L P PP PPPP + PPPPP PP Y QQ +
Sbjct: 75 CVLQPHHLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKH-------Q 127
Query: 133 NFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLP 192
HH + P E P P P P GS Y PP
Sbjct: 128 MIHHQRDGP-------PGLVPMELESP-PESPPVPPGS----------------YMPPSQ 163
Query: 193 TSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPEFSTAFPTEEPS 252
+ PPP P PP Y+ P L P S P P + P +S++ S
Sbjct: 164 SYMPPPQP--PPSYYPPSSAQPYLPPAQPSPSQSP---PSQSYLAPTPSYSSS----SSS 214
Query: 253 KQYLGDAQPF 262
+ YL +QP+
Sbjct: 215 QSYLSHSQPY 224
Score = 53.1 bits (126), Expect = 3e-05
Identities = 85/322 (26%), Positives = 108/322 (33%), Gaps = 96/322 (29%)
Query: 30 HFPFCPNPSP---------NWPPPPPP-PNPTINNYDSDFDRTFKKPRIEDDERRLKLIR 79
H P P P P +W PPPPP P P + +K I R
Sbjct: 81 HLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKHQMIHHQ-------R 133
Query: 80 DH--GLNPPQFQHPP---PPPPPQYHPP-----PPPPPPHIHNTPIYHPQQHHSEFNSPF 129
D GL P + + PP P PP Y PP PPP PP P Y+P + P
Sbjct: 134 DGPPGLVPMELESPPESPPVPPGSYMPPSQSYMPPPQPP-----PSYYPPSSAQPYLPPA 188
Query: 130 HDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRP 189
QP P +S Y P P+ S++ S + + + S+ Y P
Sbjct: 189 --------QPSP------------SQSPPSQSYLAPTPSYSSS--SSSQSYLSHSQPYLP 226
Query: 190 PLPTSPP--------PPLPMDPPMYFSAPKKPPSLFPVTSSAPHE--------PHPFP-- 231
P SP P LP PP S K P+ S A ++ P P P
Sbjct: 227 PSQASPSRSSQGPSKPQLP-PPPSIPSGNKTAIQQEPLESGAKNKTAEQKQAAPEPDPST 285
Query: 232 ------QPYFHTT----------------KPEFSTAFPTEEPSKQYL-GDAQPFSINPLA 268
Q Y++ K TA EP K+ G A PLA
Sbjct: 286 MTPQEQQQYWYRQHLLSLQQRTKVHLPGHKKGLVTAKDVPEPIKEEAPGPAASQVAEPLA 345
Query: 269 AEKPKFVDASQLFRNPLRTSRP 290
AEKP ++ PL P
Sbjct: 346 AEKPPLPPPNEEAPPPLSPEEP 367
Score = 48.5 bits (114), Expect = 7e-04
Identities = 56/210 (26%), Positives = 69/210 (32%), Gaps = 33/210 (15%)
Query: 90 HPPPPPPPQYHPPPPPPPPHIHNTPIYH----PQQHHSEFNSPFHDPNFHHFQPQPVDNH 145
H PPPP P PPPP P P Y P S F + + Q Q H
Sbjct: 13 HYPPPPVP--PPPPPVALPEASPGPGYSSSTAPAAPSSSGFMSFREQHLAQLQ-QLQQMH 69
Query: 146 IHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLP----- 200
+ H P P P P G+ D ++PP P PPPP P
Sbjct: 70 QKQMQCVLQPHHLPPPPLPPPPVMPGGGYGD----------WQPPPPPMPPPPGPALSYQ 119
Query: 201 ----MDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYL 256
M PP L P+ +P E P P + P + P +P Y
Sbjct: 120 KQQQYKHQMIHHQRDGPPGLVPMELESPPESPPVPPGSY---MPPSQSYMPPPQPPPSYY 176
Query: 257 --GDAQPFSINPLAAEKPKFVDASQLFRNP 284
AQP+ P A P SQ + P
Sbjct: 177 PPSSAQPYL--PPAQPSPSQSPPSQSYLAP 204
>ref|XP_510067.1| PREDICTED: YLP motif containing 1 [Pan troglodytes]
Length = 1666
Score = 219 bits (557), Expect = 3e-55
Identities = 168/518 (32%), Positives = 232/518 (44%), Gaps = 93/518 (17%)
Query: 32 PFCPNPSPNWPPPPPPPNPTIN---------NYDSD-----FDRTFKKPRIEDDERRLKL 77
P P P P PPP PPP P I +D D +D ++ + +
Sbjct: 1151 PPVPIPPPPPPPPLPPPPPVIKPQTSAVEQERWDEDSFYGLWDTNDEQGLNSEFKSETAA 1210
Query: 78 IRDHGLNPPQFQH---PPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEF--NSPFHDP 132
I + PP H PPP P P PP PPP H + + P+ +
Sbjct: 1211 IPSAPVLPPPPVHSSIPPPGPVPMGMPPMSKPPPVQQTVDYGHGRDISTNKVEQIPYGER 1270
Query: 133 NFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNN-------------- 178
P P + + R+ + PY + +N +D+
Sbjct: 1271 ITLRPDPLPERSTFETEHAGQRDRYDRERDREPYFDRQSNVIADHRDFKRDRETHRDRDR 1330
Query: 179 ----AQMEASRF-------------YRPPLPTSPPPPLPMDPPMYFSAPKKP----PSLF 217
+ RF YR S D P Y +P PS+F
Sbjct: 1331 DRGVIDYDRDRFDRERRPRDDRAQSYRDKKDHSSSRRGGFDRPSYDRKSDRPVYEGPSMF 1390
Query: 218 P-VTSSAPHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYLGDAQPFSINPLAAEKPKFVD 276
+ P E P P P P P +KP+ +
Sbjct: 1391 GGERRTYPEERMPLPAPSLSHQPPPA-----------------------PRVEKKPESKN 1427
Query: 277 ASQLFRNPLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTE 336
+ + P R SRP+ V+I+RG PGSGKT++AK++RD EVE GG APR+ S+DDYF+TE
Sbjct: 1428 VDDILKPPGRESRPERIVVIMRGLPGSGKTHVAKLIRDKEVEFGGPAPRVLSLDDYFITE 1487
Query: 337 VEKVEDSDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFII 396
VEK E K SG+ V KKVMEY YE EMEE YR+SM K FKKT+++G F FII
Sbjct: 1488 VEKEE-----KDPDSGKK---VKKKVMEYEYEAEMEETYRTSMFKTFKKTLDDGFFPFII 1539
Query: 397 VDDRNLRVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQWEE 456
+D N RV F QFW+ AK G+EVY+ E + D C RN+HG +EI KMA+ WE
Sbjct: 1540 LDAINDRVRHFDQFWSAAKTKGFEVYLAEMS-ADNQTCGKRNIHGRKLKEINKMADHWET 1598
Query: 457 APSLYLQLDMKSLFHGDDLKESRIQEVDM-DMDDDLDD 493
AP ++LD++SL L+++ I+EV+M D D ++++
Sbjct: 1599 APRHMMRLDIRSL-----LQDAAIEEVEMEDFDANIEE 1631
Score = 53.9 bits (128), Expect = 2e-05
Identities = 65/255 (25%), Positives = 86/255 (33%), Gaps = 74/255 (29%)
Query: 37 PSPNWPPPPP------PPNPTINNY------DSDFDRTFKKPRIEDDERRLKLIRDH--- 81
P P PPPPP P P ++ S +F++ + ++ ++ +
Sbjct: 75 PPPPVPPPPPVALPEASPGPGYSSSTTPAAPSSSGFMSFREQHLAQLQQLQQMHQKQMQC 134
Query: 82 GLNPPQFQHPPPPPPP--------QYHPPPPPPPPHIHNTPIYHPQQHHSEFNSPFHDPN 133
L P PP PPPP + PPPPP PP Y QQ +
Sbjct: 135 VLQPHHLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKH-------QM 187
Query: 134 FHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPT 193
HH + P E P P P P GS Y PP +
Sbjct: 188 LHHQRDGP-------PGLVPMELESP-PESPPVPPGS----------------YMPPSQS 223
Query: 194 SPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPEFSTAFPT----- 248
PPP P PPS +P TSS P+ P P P + P S PT
Sbjct: 224 YMPPPQP------------PPSYYPPTSSQPYLPPTQPSP--SQSPPSQSYLAPTSSYSS 269
Query: 249 -EEPSKQYLGDAQPF 262
S+ YL +Q +
Sbjct: 270 SSSSSQSYLSHSQSY 284
Score = 46.6 bits (109), Expect = 0.003
Identities = 52/190 (27%), Positives = 67/190 (34%), Gaps = 40/190 (21%)
Query: 99 YHPPPPPPPPHIHNTPIYHPQQHHSEFNSPF--HDPNFHHFQPQPVD-----NHIHNHNF 151
++PPPP PPP P P +S +P F F+ Q + +H
Sbjct: 73 HYPPPPVPPPPPVALPEASPGPGYSSSTTPAAPSSSGFMSFREQHLAQLQQLQQMHQKQM 132
Query: 152 N-ARESHH--PYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLP-------- 200
+ HH P P P P P G+ D ++PP P PPPP P
Sbjct: 133 QCVLQPHHLPPPPLPPP-PVMPGGGYGD----------WQPPPPPMPPPPGPALSYQKQQ 181
Query: 201 -MDPPMYFSAPKKPPSLFPVTSSAPHEPHPFP--------QPYFHTTKPEFSTAFPTEEP 251
M PP L P+ +P E P P Q Y +P S PT
Sbjct: 182 QYKHQMLHHQRDGPPGLVPMELESPPESPPVPPGSYMPPSQSYMPPPQPPPSYYPPTS-- 239
Query: 252 SKQYLGDAQP 261
S+ YL QP
Sbjct: 240 SQPYLPPTQP 249
Score = 43.5 bits (101), Expect = 0.022
Identities = 44/192 (22%), Positives = 65/192 (32%), Gaps = 42/192 (21%)
Query: 30 HFPFCPNPSP---------NWPPPPPP----PNPTIN------------NYDSDFDRTFK 64
H P P P P +W PPPPP P P ++ ++ D
Sbjct: 140 HLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKHQMLHHQRDGPPGLV 199
Query: 65 KPRIEDDERRLKLIRDHGLNPPQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSE 124
+E + + P Q PPP PPP Y+PP P Y P S
Sbjct: 200 PMELESPPESPPVPPGSYMPPSQSYMPPPQPPPSYYPPTSSQP--------YLPPTQPSP 251
Query: 125 FNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEAS 184
SP P+ + P +++ ++ S + Y S S + S
Sbjct: 252 SQSP---PSQSYLAP------TSSYSSSSSSSQSYLSHSQSYLPSSQASPSRPSQGHSKS 302
Query: 185 RFYRPPLPTSPP 196
+ PP P++PP
Sbjct: 303 QLLAPPPPSAPP 314
Score = 43.1 bits (100), Expect = 0.029
Identities = 59/260 (22%), Positives = 84/260 (31%), Gaps = 44/260 (16%)
Query: 13 PRPIQYQ-----TNLCPNCLTPHFPFCPNPSPNWPPPPPPPNPTINNYDSDFDRTFKKPR 67
P P++ + T+ P + P P P+ PPP PP P + + D R
Sbjct: 387 PEPVKEEVTVPATSQVPESPSSEEPPLPPPNEEVPPPLPPEEPQSEDPEED-------AR 439
Query: 68 IEDDERRLKLIRDHGLNPPQFQ-------HPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQ 120
++ + + H + FQ H Q + PPPHI + +
Sbjct: 440 LKQLQAAAAHWQQHQQHRVGFQYQGIMQKHTQLQQILQQYQQIIQPPPHIQTMSVDMQLR 499
Query: 121 HHSEFNSPF---HDPNFHHFQPQPVDNHIHNHNFNARESHHPY-------PYPNPYPNGS 170
H+ F + FQ H + H +E + Y
Sbjct: 500 HYEMQQQQFQHLYQEWEREFQLWEEQLHSYPHKDQLQEYEKQWKTWQGHMKATQSYLQEK 559
Query: 171 NNGFSDNNAQMEASRF----------YRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVT 220
N F + Q + PPLPT PPP LP P P PP+L P T
Sbjct: 560 VNSFQNMKNQYMGNMSMPPPFVPYSQMPPPLPTMPPPVLPPSLP----PPVMPPAL-PAT 614
Query: 221 SSAPHEPHPFPQPYFHTTKP 240
P P P P T+ P
Sbjct: 615 VPPPGMPPPVMPPSLPTSVP 634
Score = 41.2 bits (95), Expect = 0.11
Identities = 58/235 (24%), Positives = 77/235 (32%), Gaps = 41/235 (17%)
Query: 85 PPQFQHPPPPPPPQYHPPPPPP------PPHIHNTPIYHPQQHHSEFNSPFHDPNFHHFQ 138
PP PPP PP PP PP PP P+ P S P P+
Sbjct: 588 PPLPTMPPPVLPPSLPPPVMPPALPATVPPPGMPPPVMPPSLPTS-VPPPGMPPSLSSAG 646
Query: 139 PQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPP 198
P PV P P P ++ + PP P PPP
Sbjct: 647 PPPV--------LPPPSLSSAGPPPVLPPPSLSSTAPPPVMPLPPLSSATPP-PGIPPPG 697
Query: 199 LPMDPPMYFSAPKKPPSLFPVTSSAPHEPHP--FPQPYFH------TTKPEFSTAFPTEE 250
+P P +A PP+ +S P +P P P P T P +A P+E+
Sbjct: 698 VPQGIPPQLTAAPVPPASSSQSSQVPEKPRPALLPTPVSFGSAPPTTYHPPLQSAGPSEQ 757
Query: 251 -----------------PSKQYLGDAQPFSINPLAAEKPKFVDASQLFRNPLRTS 288
S Q LG++ P+ A K V + L +P R+S
Sbjct: 758 VNSKAPLSKSALPYSSFSSDQGLGESSAAPSQPITAVKDMPVRSGGLLPDPPRSS 812
Score = 37.7 bits (86), Expect = 1.2
Identities = 35/131 (26%), Positives = 46/131 (34%), Gaps = 28/131 (21%)
Query: 85 PPQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEFNSPFHDPNFHHFQPQPVDN 144
PP P PP PPPPPPPP P+ PQ E + + +F+
Sbjct: 1140 PPARSSVPVTRPPVPIPPPPPPPPLPPPPPVIKPQTSAVE-QERWDEDSFYGL------- 1191
Query: 145 HIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQM-EASRFYRPPLPTSPPPPLPMDP 203
+ G N+ F A + A PP+ +S PPP P+
Sbjct: 1192 -----------------WDTNDEQGLNSEFKSETAAIPSAPVLPPPPVHSSIPPPGPV-- 1232
Query: 204 PMYFSAPKKPP 214
PM KPP
Sbjct: 1233 PMGMPPMSKPP 1243
>gb|AAH07792.1| YLPM1 protein [Homo sapiens]
Length = 386
Score = 214 bits (545), Expect = 7e-54
Identities = 128/294 (43%), Positives = 179/294 (60%), Gaps = 42/294 (14%)
Query: 266 PLAAEKPKFVDASQLFRNPLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPR 325
P +KP+ + + + P R SRP+ V+I+RG PGSGKT++AK++RD EVE GG APR
Sbjct: 48 PRVEKKPESKNVDDILKPPGRESRPERIVVIMRGLPGSGKTHVAKLIRDKEVEFGGPAPR 107
Query: 326 IHSMDDYFMTEVEKVEDSDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKK 385
+ S+DDYF+TEVEK E K SG+ V KKVMEY YE EMEE YR+SM K FKK
Sbjct: 108 VLSLDDYFITEVEKEE-----KDPDSGKK---VKKKVMEYEYEAEMEETYRTSMFKTFKK 159
Query: 386 TVEEGVFTFIIVDDRNLRVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQ 445
T+++G F FII+D N RV F QFW+ AK G+EVY+ E + D C RN+HG +
Sbjct: 160 TLDDGFFPFIILDAINDRVRHFDQFWSAAKTKGFEVYLAEMS-ADNQTCGKRNIHGRKLK 218
Query: 446 EIEKMAEQWEEAPSLYLQLDMKSLFHGDDLKESRIQEVDM-DMDDDLDDALVAAQGREAD 504
EI KMA+ WE AP ++LD++SL L+++ I+EV+M D D ++++
Sbjct: 219 EINKMADHWETAPRHMMRLDIRSL-----LQDAAIEEVEMEDFDANIEEQ---------- 263
Query: 505 KAVVRPVKDGEGSIKDGKRWDAEEEHPTEVRELGKSKWSED-FEDDIDQTEGMK 557
K+ K+ DAEEE +E+ + KSKW D E +D+ +G++
Sbjct: 264 --------------KEEKK-DAEEEE-SELGYIPKSKWEMDTSEAKLDKLDGLR 301
>ref|XP_421271.1| PREDICTED: similar to FLJ00353 protein [Gallus gallus]
Length = 2402
Score = 213 bits (543), Expect = 1e-53
Identities = 180/577 (31%), Positives = 250/577 (43%), Gaps = 87/577 (15%)
Query: 29 PHFPFCP---NPSPNWPPPPPPPNPTINNYDSD-FDRTFKKPRIEDDERRLKLIRDHGLN 84
PH P PS P P +D D F + + +L + +
Sbjct: 1375 PHTPVSTPTTTPSHGQSLKPSPSAVEQERWDEDSFHGLWDTNEDKGANMEYELRKQESMM 1434
Query: 85 PPQFQHP----------PPPPPPQYHP--PPPPPPPHIHNTPIY-HPQQHHSEFNSPFHD 131
PP P PP P P PP P P I T Y H + +
Sbjct: 1435 PPPVSSPVKVPAVHTSVPPAVPGTLPPVIPPVPKAPVIQQTVDYGHGRDITTSKVEQIPY 1494
Query: 132 PNFHHFQPQPVDNHI-----HNHNFNARESHHPYPYPNPYPNGSNNGFSDN--------N 178
+P+P+ H +N PY N + F +
Sbjct: 1495 GERVTLRPEPLPERQAFQKEHPGRYNRERDREPYFERQGNSNADHRDFKRERELHRDRGS 1554
Query: 179 AQMEASRFY--RPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFH 236
+ RF R P P P P Y P S P+E Y H
Sbjct: 1555 VDYDRERFEKERHPRDDRVLPTAPSRTPSYRDKKDHPSSRRGGFDRPPYERKTDRPAYDH 1614
Query: 237 TTKP--EFST--AFPTEEPSKQYLGDAQPFSIN---------PLAAEKPKFVDASQLFRN 283
E S+ + E + Y + P S P KP+ + + +
Sbjct: 1615 GPSMFGENSSVQSLEFEGDRRNYPEERMPISAPSMPRQPPPAPRVERKPESKNVDDILKP 1674
Query: 284 PLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTEVEKVE-D 342
P R SRP+ VII+RG PGSGKT++AK++RD EVE GG APR+ S+DDYF+TEVEK E D
Sbjct: 1675 PGRDSRPERIVIIMRGLPGSGKTHVAKLIRDKEVECGGPAPRVLSLDDYFITEVEKEERD 1734
Query: 343 SDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFIIVDDRNL 402
D K V KKVMEY YE +MEE YR+SM K FKKT+++G F FII+D N
Sbjct: 1735 PDTGKK---------VKKKVMEYEYEADMEETYRTSMFKTFKKTLDDGFFPFIILDAIND 1785
Query: 403 RVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQWEEAPSLYL 462
RV F QFW+ AK G+EVY+ E + D C+ RN+HG ++I +M++ WE AP +
Sbjct: 1786 RVRHFEQFWSAAKTKGFEVYLAEMS-ADNQTCSKRNIHGRKLKDISRMSDHWEAAPRHMM 1844
Query: 463 QLDMKSLFHGDDLKESRIQEVDM-DMDDDLDDALVAAQGREADKAVVRPVKDGEGSIKDG 521
+LD++SL L+++ I+EV+M D D +++D Q EA K
Sbjct: 1845 RLDIRSL-----LQDAAIEEVEMEDFDANIED-----QKEEAKK---------------- 1878
Query: 522 KRWDAEEEHPTEVRELGKSKWSED-FEDDIDQTEGMK 557
D EE +E+ + KSKW D E +D+ +G++
Sbjct: 1879 ---DTTEEEESELGYIPKSKWEMDTSEAKLDKLDGLR 1912
>ref|XP_234416.3| PREDICTED: similar to YLP motif containing protein 1 (Nuclear protein
ZAP3) (ZAP113) [Rattus norvegicus]
Length = 2469
Score = 213 bits (541), Expect = 2e-53
Identities = 177/558 (31%), Positives = 244/558 (43%), Gaps = 86/558 (15%)
Query: 29 PHFPFCPNPSPNWPPPPPPPNPTINNYDS-------DFDRTFKKPRIEDDE-------RR 74
P P P P P PPPPPPP P I S D D F DD+ R
Sbjct: 1836 PPVPIPPPPPP--PPPPPPPPPVIKPKTSSVKQERWDEDSFFGLWDTNDDQGLNSEFKRD 1893
Query: 75 LKLIRDHGLNPPQFQHPPPPPP---PQYHPPPPPPPPHIHNTPIYHPQQHHSEF--NSPF 129
I + PP HP PPP P PP PPP H H + + P+
Sbjct: 1894 TAAIPSAPVLPPPPVHPSIPPPGPMPMGMPPMSKPPPVQHTVDYGHGRDMPTNKVEQIPY 1953
Query: 130 HDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFS-------------- 175
+ P P + + R+ + PY + +N
Sbjct: 1954 GERITLRPDPLPERSAFDADHAGQRDRYDRDRDREPYFDRQSNMTDHRDFKRDRETHRDR 2013
Query: 176 DNNAQMEASRFYRPPLPTSPPPPLPMDPPMYFSAPK---KPPSLFPVTSSAPHEPHPFPQ 232
D E RF R P D + S+ + PS + P+E P
Sbjct: 2014 DRVLDYERDRFDRERRPRDDRNQSYRDKKDHSSSRRGGFDRPSYDRKSDRPPYEGPPMFG 2073
Query: 233 PYFHTTKPEFSTAFPTEEPSKQYLGDAQPFSINPLAAEKPKFVDASQLFRNPLRTSRPDH 292
T E P PS + QP + P +KP+ + + + P R SRP+
Sbjct: 2074 GERRTYPEE---RMPLPAPSLGH----QPPPV-PRVEKKPESKNVDDILKPPGRESRPER 2125
Query: 293 FVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTEVEKVEDSDGPKSSSSG 352
V+I+RG PGSGKT++AK++RD EVE GG APR+ S+DDYF+ EVEK E K SG
Sbjct: 2126 IVVIMRGLPGSGKTHVAKLIRDKEVEFGGPAPRVLSLDDYFIAEVEKEE-----KDPDSG 2180
Query: 353 RNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFIIVDDRNLRVADFAQFWA 412
+ V KKVMEY YE +MEE YR+SM K FKKT+++G F FII+D N RV F QFW+
Sbjct: 2181 KK---VKKKVMEYEYEADMEETYRTSMFKTFKKTLDDGFFPFIILDAINDRVRHFDQFWS 2237
Query: 413 TAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEK----------------------- 449
AK G+EVY+ E + D C RN+HG +EI K
Sbjct: 2238 AAKTKGFEVYLAEMS-ADNQTCGKRNIHGRKLKEINKVIVQKLCTGEPSITLSALAALSS 2296
Query: 450 ------MAEQWEEAPSLYLQLDMKSLFHGDDLKESRIQEVDMDMDDDLDDALVAAQGR-E 502
MAE WE AP ++LD++SL ++E +++ D +++D ++ A + E
Sbjct: 2297 VFSPALMAEHWEAAPRHMMRLDIRSLLQDAAIEEVEMEDFDANIEDQKEEKKDAEEEESE 2356
Query: 503 AD-KAVVRPVKDGEGSIK 519
D ++ V++ G +K
Sbjct: 2357 LDFSGIIAFVRERYGKLK 2374
Score = 57.8 bits (138), Expect = 1e-06
Identities = 63/250 (25%), Positives = 86/250 (34%), Gaps = 64/250 (25%)
Query: 37 PSPNWPPPPPP-------PNPTINNY------DSDFDRTFKKPRIEDDERRLKLIRDH-- 81
P P PPPPPP P P ++ S +F++ + ++ ++ +
Sbjct: 15 PPPPVPPPPPPVALPEASPGPGYSSSTAPAAPSSSGFMSFREQHLAQLQQLQQMHQKQMQ 74
Query: 82 -GLNPPQFQHPPPPPPP--------QYHPPPPPPPPHIHNTPIYHPQQHHSEFNSPFHDP 132
L P PP PPPP + PPPPP PP Y QQ +
Sbjct: 75 CVLQPHHLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKH-------Q 127
Query: 133 NFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLP 192
HH + P E P P P P GS Y PP
Sbjct: 128 MIHHQRDGP-------PGLVPMELESP-PESPPVPPGS----------------YMPPSQ 163
Query: 193 TSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPEFSTAFPTEEPS 252
+ PPP P PP Y+ P L P S P P + P +S++ S
Sbjct: 164 SYMPPPQP--PPSYYPPSSAQPYLPPAQPSPSQSP---PSQSYLAPTPSYSSS----SSS 214
Query: 253 KQYLGDAQPF 262
+ YL +QP+
Sbjct: 215 QSYLSHSQPY 224
Score = 48.5 bits (114), Expect = 7e-04
Identities = 56/210 (26%), Positives = 69/210 (32%), Gaps = 33/210 (15%)
Query: 90 HPPPPPPPQYHPPPPPPPPHIHNTPIYH----PQQHHSEFNSPFHDPNFHHFQPQPVDNH 145
H PPPP P PPPP P P Y P S F + + Q Q H
Sbjct: 13 HYPPPPVP--PPPPPVALPEASPGPGYSSSTAPAAPSSSGFMSFREQHLAQLQ-QLQQMH 69
Query: 146 IHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLP----- 200
+ H P P P P G+ D ++PP P PPPP P
Sbjct: 70 QKQMQCVLQPHHLPPPPLPPPPVMPGGGYGD----------WQPPPPPMPPPPGPALSYQ 119
Query: 201 ----MDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYL 256
M PP L P+ +P E P P + P + P +P Y
Sbjct: 120 KQQQYKHQMIHHQRDGPPGLVPMELESPPESPPVPPGSY---MPPSQSYMPPPQPPPSYY 176
Query: 257 --GDAQPFSINPLAAEKPKFVDASQLFRNP 284
AQP+ P A P SQ + P
Sbjct: 177 PPSSAQPYL--PPAQPSPSQSPPSQSYLAP 204
Score = 45.4 bits (106), Expect = 0.006
Identities = 48/196 (24%), Positives = 65/196 (32%), Gaps = 49/196 (25%)
Query: 30 HFPFCPNPSP---------NWPPPPPP----PNPTIN------------NYDSDFDRTFK 64
H P P P P +W PPPPP P P ++ ++ D
Sbjct: 81 HLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKHQMIHHQRDGPPGLV 140
Query: 65 KPRIEDDERRLKLIRDHGLNPPQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSE 124
+E + + P Q PPP PPP Y+PP P Y P S
Sbjct: 141 PMELESPPESPPVPPGSYMPPSQSYMPPPQPPPSYYPPSSAQP--------YLPPAQPSP 192
Query: 125 FNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEAS 184
SP P+ + P P + + S P P+ S+ G S
Sbjct: 193 SQSP---PSQSYLAPTPSYSSSSSSQSYLSHSQPYLPPSQASPSRSSQGPS--------- 240
Query: 185 RFYRPPLPTSPPPPLP 200
+P LP PPP +P
Sbjct: 241 ---KPQLP-PPPPSIP 252
Score = 42.4 bits (98), Expect = 0.049
Identities = 42/152 (27%), Positives = 60/152 (38%), Gaps = 44/152 (28%)
Query: 67 RIEDDERRLKLIRDHGLNPPQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEFN 126
++ +++ ++ G PP PPPPPPP PPPPPPPP I Q+ E
Sbjct: 1818 KLSPEDKNSSILMSCGRWPP-VPIPPPPPPP---PPPPPPPPVIKPKTSSVKQERWDE-- 1871
Query: 127 SPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRF 186
D+ + N + G N+ F + A + ++
Sbjct: 1872 ----------------DSFFGLWDTNDDQ-------------GLNSEFKRDTAAIPSAPV 1902
Query: 187 YRPP--LPTSPPP-PLPMD-PPMYFSAPKKPP 214
PP P+ PPP P+PM PPM KPP
Sbjct: 1903 LPPPPVHPSIPPPGPMPMGMPPM-----SKPP 1929
Score = 38.1 bits (87), Expect = 0.92
Identities = 23/52 (44%), Positives = 25/52 (47%), Gaps = 5/52 (9%)
Query: 189 PPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKP 240
PPLPT PPP LP P P PP+L P T P P P P T+ P
Sbjct: 813 PPLPTMPPPVLPPSLP----PPVMPPAL-PSTIPPPGMPPPVMPPSLPTSVP 859
Score = 35.4 bits (80), Expect = 6.0
Identities = 20/51 (39%), Positives = 24/51 (46%), Gaps = 6/51 (11%)
Query: 176 DNNAQMEASRFYRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHE 226
D N+ + S PP+P PPPP P P P PP + P TSS E
Sbjct: 1823 DKNSSILMSCGRWPPVPIPPPPPPPPPP------PPPPPVIKPKTSSVKQE 1867
>emb|CAI12027.1| novel protein similar to vertebrate YLP motif containing 1 (YLPM1)
[Danio rerio]
Length = 1424
Score = 210 bits (535), Expect = 1e-52
Identities = 126/316 (39%), Positives = 187/316 (58%), Gaps = 45/316 (14%)
Query: 245 AFPTE-EPSKQYLGDAQPFSINPLAAEKPKFVDASQLFRNPLRTSRPDHFVIILRGFPGS 303
++P E +P L + P + P +KP+ +A + + P R+SRPD V+I+RG PGS
Sbjct: 1070 SYPDERQPPPTSLPPSAP--VAPPVEKKPEIKNAEDILKPPGRSSRPDRIVVIMRGLPGS 1127
Query: 304 GKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTEVEKVEDSDGPKSSSSGRNKRPVTKKVM 363
GK+++AK++RD EVE GG PR+ +DDYFMTEVEK+E K SG+ + KV+
Sbjct: 1128 GKSHVAKLIRDKEVECGGAPPRVLGLDDYFMTEVEKIE-----KDPDSGKR---IKTKVL 1179
Query: 364 EYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFIIVDDRNLRVADFAQFWATAKRSGYEVYI 423
EY YEPEME+ YRSSMLK FKKT+++G F FII+D N +V F QFW+ AK G+EVY+
Sbjct: 1180 EYEYEPEMEDTYRSSMLKTFKKTLDDGFFPFIILDAINDKVKYFDQFWSAAKTKGFEVYL 1239
Query: 424 LEATYKDPVGCAARNVHGFTQQEIEKMAEQWEEAPSLYLQLDMKSLFHGDDLKESRIQEV 483
E T D CA RN+HG T ++I K++ WE AP ++LD++SL L+++ I++V
Sbjct: 1240 AEIT-TDQQTCAKRNIHGRTLKDISKLSGGWESAPPHMVRLDIRSL-----LQDAAIEDV 1293
Query: 484 DMDMDDDLDDALVAAQGREADKAVVRPVKDGEGSIKDGKRWDAEEEHPTEVRELGKSKWS 543
+M+ + D+ KA V+ ++ T++ + KSKW
Sbjct: 1294 EMEDFNPSDE----------PKAEVK-----------------NDDDETDLGYVPKSKWE 1326
Query: 544 ED-FEDDIDQTEGMKG 558
D E +D+ +G+ G
Sbjct: 1327 MDTSEAKLDKLDGLVG 1342
Score = 35.0 bits (79), Expect = 7.8
Identities = 38/184 (20%), Positives = 62/184 (33%), Gaps = 32/184 (17%)
Query: 37 PSPNWPPPPPPPNPTINNYDSDF-----DRTFKKPRIEDDERRLKLIRDHGLNPPQFQHP 91
P W PPP NY+ ++ D +++P E +E +HG +
Sbjct: 648 PRERWHGPPPHEE---ENYEEEYPYGAEDDVYRRPPHEYNEDY-----EHGDEYYGSREE 699
Query: 92 PPPPPPQYHPPPPPPPPHIHNTPI---------YHPQQHHSEFNSPFH--DPNFHHFQPQ 140
P+ PP PP + P Y ++ E PF+ DP + +
Sbjct: 700 WDGEQPERDYPPHRPPERVREDPWLEERERSFPYEEDRYREERRGPFYPDDPPYQDRDRE 759
Query: 141 PVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLP 200
P F++R P P P G + S+ + + + P P + PL
Sbjct: 760 PP--------FHSRSDWERPPPPPPPERGYSRSLSETDYEHKLDPLASLPAPQATDSPLD 811
Query: 201 MDPP 204
P
Sbjct: 812 ESSP 815
>emb|CAG09137.1| unnamed protein product [Tetraodon nigroviridis]
Length = 462
Score = 206 bits (523), Expect = 3e-51
Identities = 115/260 (44%), Positives = 162/260 (62%), Gaps = 18/260 (6%)
Query: 231 PQPYFHTTKPEFST-AFPTEE---PSKQYLGDAQPFSINPLAAEKPKFVDASQLFRNPLR 286
P+ Y H+T P ++P + P P P +KP+ + + + P R
Sbjct: 119 PERYGHSTPPYVDRRSYPEDRGPPPPPPVAPHPPPPQPPPRLEKKPEIKNIEDILKPPGR 178
Query: 287 TSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTEVEKVEDSDGP 346
+SRP+ VII+RG PGSGK+++AK++RD EVE GG PR+ +DDYFMTEVEKVE
Sbjct: 179 SSRPERIVIIMRGLPGSGKSHVAKLIRDKEVECGGTPPRVLVLDDYFMTEVEKVE----- 233
Query: 347 KSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFIIVDDRNLRVAD 406
K +GR V KV+E+ YEPEME+ YR+SMLK FKKT+++G F FII+D N +V
Sbjct: 234 KDPDTGRK---VKTKVLEFEYEPEMEDTYRNSMLKTFKKTLDDGFFPFIILDSVNDKVKH 290
Query: 407 FAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQWEEAPSLYLQLDM 466
F QFW+ AK G+EVY+ E T D CA RNVHG ++I KM+ WE +P ++LD+
Sbjct: 291 FDQFWSAAKTKGFEVYVAEIT-ADTQTCAKRNVHGRLLKDIMKMSTSWEPSPRHMMRLDV 349
Query: 467 KSLFHGDDLKESRIQEVDMD 486
+SL L+++ I+EV+M+
Sbjct: 350 RSL-----LQDAAIEEVEME 364
Score = 45.1 bits (105), Expect = 0.008
Identities = 16/29 (55%), Positives = 19/29 (65%)
Query: 86 PQFQHPPPPPPPQYHPPPPPPPPHIHNTP 114
P+ + PPPPPP HPPPP PPP + P
Sbjct: 136 PEDRGPPPPPPVAPHPPPPQPPPRLEKKP 164
>ref|XP_395666.2| PREDICTED: similar to YLPM1 protein [Apis mellifera]
Length = 1860
Score = 168 bits (426), Expect = 5e-40
Identities = 111/326 (34%), Positives = 172/326 (52%), Gaps = 52/326 (15%)
Query: 269 AEKPKFVDASQLFRNPLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHS 328
A+ P L P R SRP IILRG PGSGK+++AK+++D EVE GG APRI S
Sbjct: 1533 AKTPNCTMVDDLLCPPGRQSRPPKIAIILRGPPGSGKSFVAKLIKDKEVEQGGSAPRILS 1592
Query: 329 MDDYFMTEVEKVEDSDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVE 388
+DDYF+ E E +E +D N + VT K M Y YE ME++Y +S++KAFKK V
Sbjct: 1593 LDDYFLVEKE-IESTDD--------NGKKVTIKDMIYEYEEVMEQSYITSLVKAFKKNVT 1643
Query: 389 EGVFTFIIVDDRNLRVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIE 448
+G F FII+D N +++D+ + W+ AK G++VY+ E D C RN+H T+ EI
Sbjct: 1644 DGFFNFIILDCINEKISDYEEMWSFAKTKGFKVYVCEME-MDLQICLKRNIHNRTEDEIN 1702
Query: 449 KMAEQWEEAPSLYLQLDMKSLFHGDDLKESRIQEVDMDMDDDLDDALVAAQGREADKAVV 508
++ + +E PS + +LD+ S+ L+E I+EV M+ ++ + ++Q E
Sbjct: 1703 RIIDYFEPTPSYHQKLDVNSM-----LQEQAIEEVHMEDSEETQEK--SSQQNEDS---- 1751
Query: 509 RPVKDGEGSIKDGKRWDAEEEHPTEVRELGKSKWSE-DFEDDIDQTEGM-----KGNINA 562
+D + ++D +G SKW + ED +D+ +G+ +G +
Sbjct: 1752 ---QDSQDDMQDA---------------IGVSKWERMEAEDKLDRLDGLAKKKNEGKVQT 1793
Query: 563 LSGLIH---QYGKE----RKSVHWGD 581
+ + Y E +K V W D
Sbjct: 1794 MKDFLQVPDYYNMEDTSGKKRVRWAD 1819
>gb|AAF61275.1| unknown [Homo sapiens]
Length = 1822
Score = 157 bits (398), Expect = 8e-37
Identities = 142/518 (27%), Positives = 202/518 (38%), Gaps = 133/518 (25%)
Query: 32 PFCPNPSPNWPPPPPPPNPTIN---------NYDSD-----FDRTFKKPRIEDDERRLKL 77
P P P P PPP PPP P I +D D +D ++ + +
Sbjct: 1347 PPVPIPPPPPPPPLPPPPPVIKPQTSAVEQERWDEDSFYGLWDTNDEQGLNSEFKSETAA 1406
Query: 78 IRDHGLNPPQFQH---PPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEF--NSPFHDP 132
I + PP H PPP P P PP PPP H + + P+ +
Sbjct: 1407 IPSAPVLPPPPVHSSIPPPGPVPMGMPPMSKPPPVQQTVDYGHGRDISTNKVEQIPYGER 1466
Query: 133 NFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNN-------------- 178
P P + + R+ + PY + +N +D+
Sbjct: 1467 ITLRPDPLPERSTFETEHAGQRDRYDRERDREPYFDRQSNVIADHRDFKRDRETHRDRDR 1526
Query: 179 ----AQMEASRF-------------YRPPLPTSPPPPLPMDPPMYFSAPKKP----PSLF 217
+ RF YR S D P Y +P PS+F
Sbjct: 1527 DRGVIDYDRDRFDRERRPRDDRAQSYRDKKDHSSSRRGGFDRPSYDRKSDRPVYEGPSMF 1586
Query: 218 P-VTSSAPHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYLGDAQPFSINPLAAEKPKFVD 276
+ P E P P P P P +KP+ +
Sbjct: 1587 GGERRTYPEERMPLPAPSLSHQPPPA-----------------------PRVEKKPESKN 1623
Query: 277 ASQLFRNPLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTE 336
+ + P R SRP+ V+I+RG PGSGKT++AK++R
Sbjct: 1624 VDDILKPPGRESRPERIVVIMRGLPGSGKTHVAKLIR----------------------- 1660
Query: 337 VEKVEDSDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFII 396
VMEY YE EMEE YR+SM K FKKT+++G F FII
Sbjct: 1661 -------------------------VMEYEYEAEMEETYRTSMFKTFKKTLDDGFFPFII 1695
Query: 397 VDDRNLRVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQWEE 456
+D N RV F QFW+ AK G+EVY+ E + D C RN+HG +EI KMA+ WE
Sbjct: 1696 LDAINDRVRHFDQFWSAAKTKGFEVYLAEMS-ADNQTCGKRNIHGRKLKEINKMADHWET 1754
Query: 457 APSLYLQLDMKSLFHGDDLKESRIQEVDM-DMDDDLDD 493
AP ++LD++SL L+++ I+EV+M D D ++++
Sbjct: 1755 APRHMMRLDIRSL-----LQDAAIEEVEMEDFDANIEE 1787
Score = 53.9 bits (128), Expect = 2e-05
Identities = 65/255 (25%), Positives = 86/255 (33%), Gaps = 74/255 (29%)
Query: 37 PSPNWPPPPP------PPNPTINNY------DSDFDRTFKKPRIEDDERRLKLIRDH--- 81
P P PPPPP P P ++ S +F++ + ++ ++ +
Sbjct: 15 PPPPVPPPPPVALPEASPGPGYSSSTTPAAPSSSGFMSFREQHLAQLQQLQQMHQKQMQC 74
Query: 82 GLNPPQFQHPPPPPPP--------QYHPPPPPPPPHIHNTPIYHPQQHHSEFNSPFHDPN 133
L P PP PPPP + PPPPP PP Y QQ +
Sbjct: 75 VLQPHHLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKH-------QM 127
Query: 134 FHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPT 193
HH + P E P P P P GS Y PP +
Sbjct: 128 LHHQRDGP-------PGLVPMELESP-PESPPVPPGS----------------YMPPSQS 163
Query: 194 SPPPPLPMDPPMYFSAPKKPPSLFPVTSSAPHEPHPFPQPYFHTTKPEFSTAFPT----- 248
PPP P PPS +P TSS P+ P P P + P S PT
Sbjct: 164 YMPPPQP------------PPSYYPPTSSQPYLPPAQPSP--SQSPPSQSYLAPTPSYSS 209
Query: 249 -EEPSKQYLGDAQPF 262
S+ YL +Q +
Sbjct: 210 SSSSSQSYLSHSQSY 224
Score = 48.1 bits (113), Expect = 9e-04
Identities = 53/190 (27%), Positives = 68/190 (34%), Gaps = 40/190 (21%)
Query: 99 YHPPPPPPPPHIHNTPIYHPQQHHSEFNSPF--HDPNFHHFQPQPVD-----NHIHNHNF 151
++PPPP PPP P P +S +P F F+ Q + +H
Sbjct: 13 HYPPPPVPPPPPVALPEASPGPGYSSSTTPAAPSSSGFMSFREQHLAQLQQLQQMHQKQM 72
Query: 152 N-ARESHH--PYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLP-------- 200
+ HH P P P P P G+ D ++PP P PPPP P
Sbjct: 73 QCVLQPHHLPPPPLPPP-PVMPGGGYGD----------WQPPPPPMPPPPGPALSYQKQQ 121
Query: 201 -MDPPMYFSAPKKPPSLFPVTSSAPHEPHPFP--------QPYFHTTKPEFSTAFPTEEP 251
M PP L P+ +P E P P Q Y +P S PT
Sbjct: 122 QYKHQMLHHQRDGPPGLVPMELESPPESPPVPPGSYMPPSQSYMPPPQPPPSYYPPTS-- 179
Query: 252 SKQYLGDAQP 261
S+ YL AQP
Sbjct: 180 SQPYLPPAQP 189
Score = 46.6 bits (109), Expect = 0.003
Identities = 45/192 (23%), Positives = 66/192 (33%), Gaps = 42/192 (21%)
Query: 30 HFPFCPNPSP---------NWPPPPPP----PNPTIN------------NYDSDFDRTFK 64
H P P P P +W PPPPP P P ++ ++ D
Sbjct: 80 HLPPPPLPPPPVMPGGGYGDWQPPPPPMPPPPGPALSYQKQQQYKHQMLHHQRDGPPGLV 139
Query: 65 KPRIEDDERRLKLIRDHGLNPPQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSE 124
+E + + P Q PPP PPP Y+PP P Y P S
Sbjct: 140 PMELESPPESPPVPPGSYMPPSQSYMPPPQPPPSYYPPTSSQP--------YLPPAQPSP 191
Query: 125 FNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEAS 184
SP P+ + P P +++ ++ S + Y S S + S
Sbjct: 192 SQSP---PSQSYLAPTP------SYSSSSSSSQSYLSHSQSYLPSSQASPSRPSQGHSKS 242
Query: 185 RFYRPPLPTSPP 196
+ PP P++PP
Sbjct: 243 QLLAPPPPSAPP 254
Score = 43.9 bits (102), Expect = 0.017
Identities = 52/231 (22%), Positives = 77/231 (32%), Gaps = 47/231 (20%)
Query: 86 PQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQQHHSEFNSPFHDPNFHH---FQPQPV 142
P + PP PPP + PPP PP P + + + + H FQ Q +
Sbjct: 346 PSSEEPPLPPPNEEVPPPLPPEEPQSEDPEEDARLKQLQAAAAHWQQHQQHRVGFQYQGI 405
Query: 143 DNHIHNHNFNARESHHPYPYPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLPMD 202
H + + P P+ + P P PPP +P
Sbjct: 406 MQK-HTQLQQILQQYQQIIQPPPHIQATT------------------PPPGIPPPGVPQG 446
Query: 203 PPMYFSAPKKPPSLFPVTSSAPHEPHP--FPQPYFH------TTKPEFSTAFPTEE---- 250
P +A PP+ +S P +P P P P T P +A P+E+
Sbjct: 447 IPPQLTAAPVPPASSSQSSQVPEKPRPALLPTPVSFGSAPPTTYHPPLQSAGPSEQVNSK 506
Query: 251 -------------PSKQYLGDAQPFSINPLAAEKPKFVDASQLFRNPLRTS 288
S Q LG++ P+ A K V + L +P R+S
Sbjct: 507 APLSKSALPYSSFSSDQGLGESSAAPSQPITAVKDMPVRSGGLLPDPPRSS 557
Score = 40.0 bits (92), Expect = 0.24
Identities = 43/191 (22%), Positives = 64/191 (32%), Gaps = 44/191 (23%)
Query: 68 IEDDERRLKLIRDHGLNPPQFQHPPP---------------PPPPQYHPPPP---PP--- 106
+ + + +L+ ++D G P H PP PPP Y PPPP PP
Sbjct: 1271 LREQKEQLQKMKDFGSEPQMADHLPPQESRLQNTSSRPGMYPPPGSYRPPPPMGKPPGSI 1330
Query: 107 -----PPHIHNTPIYHPQQHHSEFNSPFHDPNFHHFQPQPVDNHIHNHNFNARESHHPYP 161
PP + P+ P P P P P + +A E
Sbjct: 1331 VRPSAPPARSSVPVTRP---------PVPIPPPPPPPPLPPPPPVIKPQTSAVEQER--- 1378
Query: 162 YPNPYPNGSNNGFSDNNAQMEASRFYRPPLPTSPPPPLPMDPPMYFSAPKKPPSLFPVTS 221
+ S G D N + + ++ P P+ PP++ S P PP P+
Sbjct: 1379 ----WDEDSFYGLWDTNDEQGLNSEFKSETAAIPSAPVLPPPPVHSSIP--PPGPVPMGM 1432
Query: 222 SAPHEPHPFPQ 232
+P P Q
Sbjct: 1433 PPMSKPPPVQQ 1443
>gb|EAL31472.1| GA17072-PA [Drosophila pseudoobscura]
Length = 1903
Score = 142 bits (359), Expect = 3e-32
Identities = 94/303 (31%), Positives = 144/303 (47%), Gaps = 44/303 (14%)
Query: 177 NNAQMEASRFYRPPLPTSPP-------PPLPMDPPMYFS---APKKPPSLFPVTSSAPHE 226
NN+ ++ P +P P +P PM S AP P SLFP + A +
Sbjct: 1511 NNSISYGGESFKKAFPANPQRRMLGEAPLIPKTEPMPSSSVDAPSAPQSLFPSEAKANED 1570
Query: 227 PHPFPQPYFHTTKPEFSTAFPTEEPSKQYLGDAQPFSINPLAAEKPKFVDASQLFRNPLR 286
P H P + PT E + ++ +P R
Sbjct: 1571 V-----PAGHVPAPHLEMSVPTNENHNT--------------------ISIDEILLSPGR 1605
Query: 287 TSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHSMDDYFMTEVEKVEDSDGP 346
+RP +ILRG PGSGK+Y+A+++++ E+E GG PRI S+DDYF+ E + E P
Sbjct: 1606 LTRPKRICVILRGPPGSGKSYVARLIKEKELEMGGATPRILSIDDYFIIENDYEEKC--P 1663
Query: 347 KSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVEEGVFTFIIVDDRNLRVAD 406
K+ + + KK + Y Y+ MEE Y ++K+FKKT+ + ++ FIIVD N +
Sbjct: 1664 KTG------KKIPKKELLYEYDDTMEETYMQYLIKSFKKTLSDNLYDFIIVDCNNNSLRT 1717
Query: 407 FAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIEKMAEQWEEAPSLYLQLDM 466
+F+ AK S + YI++ D C RN H T +EI+ + W E P Y++LD+
Sbjct: 1718 LNEFYCHAKDSNFVPYIVDLLC-DVETCLGRNAHKRTTEEIQLALDNWNETPLQYIKLDV 1776
Query: 467 KSL 469
+L
Sbjct: 1777 ATL 1779
Score = 37.7 bits (86), Expect = 1.2
Identities = 46/211 (21%), Positives = 64/211 (29%), Gaps = 53/211 (25%)
Query: 9 PNLNPRPIQYQTNLCPNCLTPHFPFCPNPSPNWPPPPPPPNPTINNYDSDFDRTFKKPRI 68
P + P P+Q N + P PPPPP N Y +P+
Sbjct: 125 PAVAPVPVQVPPNPAVAGMPGKIMAQPQMYSQPPPPPPQKNANPGGY-------MHQPQH 177
Query: 69 EDDERRLKLIRDHGLNPPQFQHPPPPPPPQYHPPPPPPPPHIHNTPIYHPQ-----QHHS 123
+ + H NP + + PP Q H PPP H + PQ Q +
Sbjct: 178 Q---------QQHQANPNMWPNVPPAAQQQQHLQQPPPASHYNQMAQPQPQIRGYSQPPN 228
Query: 124 EFN---SPF------------HDPNFHHFQPQPVDNHIHNHNFNARESHHPY-------- 160
E +PF DP P ++N + + + P
Sbjct: 229 EMQGNMNPFPNLQQPPPAFMGADPKMRQPPPNAMENQWNQQQHQPPQRNAPRWEGPTDGP 288
Query: 161 ---------PYPNPYPNGSNNGFSDNNAQME 182
P P P P N +S NN ME
Sbjct: 289 GPGPGPGLGPGPGPGPGPRMNRWSTNNPDME 319
>gb|AAF46552.2| CG32685-PC [Drosophila melanogaster] gi|24640949|ref|NP_727393.1|
CG32685-PC [Drosophila melanogaster]
Length = 1884
Score = 128 bits (322), Expect = 5e-28
Identities = 100/350 (28%), Positives = 167/350 (47%), Gaps = 36/350 (10%)
Query: 222 SAPHEPHPFPQPYFHTTKPEFSTAFPTEEPSKQYL---GDAQPFSINPLAAEKPKFVDAS 278
+A + +PFP ++S P+ +A + +P+ A + A+
Sbjct: 1473 NAQEQYNPFPVNPLRRQLGDYSDKLEAPSPAMSLFPSAANANDRTSDPMKASNLEMCVAT 1532
Query: 279 QLFRN----------PLRTSRPDHFVIILRGFPGSGKTYLAKMLRDLEVENGGDAPRIHS 328
RN P R +RP IILRG PG GK+++A+++++ E+E GG PRI S
Sbjct: 1533 SENRNTFSLDEVLLYPGRLNRPKRICIILRGPPGCGKSHVARLIKEKELEMGGANPRILS 1592
Query: 329 MDDYFMTEVEKVEDSDGPKSSSSGRNKRPVTKKVMEYCYEPEMEEAYRSSMLKAFKKTVE 388
+DDYF+ +E D PK+ + + KK + Y Y+ MEE Y ++K+FKKT+
Sbjct: 1593 IDDYFL--IENDYDEKCPKTG------KKIPKKEILYEYDDTMEETYMQYLIKSFKKTLS 1644
Query: 389 EGVFTFIIVDDRNLRVADFAQFWATAKRSGYEVYILEATYKDPVGCAARNVHGFTQQEIE 448
+ ++ F+IVD N + +F+ AK S + YI++ + D C RN H + +I
Sbjct: 1645 DNLYDFVIVDCNNNSLRTLNEFYCHAKDSNFVPYIVD-LHCDLETCLGRNSHQRSVNDIR 1703
Query: 449 KMAEQWEEAPSLYLQLDMKSLFHGDDLKESRIQEVDMDMDDDLDDALVAAQGREADKAVV 508
+ + W + P+ Y++LD+ SL +++ E E DM DD+ + A G A
Sbjct: 1704 VVLDNWCQTPTHYIKLDVSSLL--ENVVEMEDVE-DMATDDNACADVGATVGESA----- 1755
Query: 509 RPVKDGEGSIKDGKRWDAEEEHPTEVRELGKSKWSED-FEDDIDQTEGMK 557
++D D D+ + KSKW D ED++ + +G K
Sbjct: 1756 -ALEDAAVEETD----DSNSADASNYCGFLKSKWERDTTEDNLARLDGTK 1800
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,479,018,011
Number of Sequences: 2540612
Number of extensions: 84558912
Number of successful extensions: 975503
Number of sequences better than 10.0: 8573
Number of HSP's better than 10.0 without gapping: 2489
Number of HSP's successfully gapped in prelim test: 6618
Number of HSP's that attempted gapping in prelim test: 599575
Number of HSP's gapped (non-prelim): 96456
length of query: 664
length of database: 863,360,394
effective HSP length: 135
effective length of query: 529
effective length of database: 520,377,774
effective search space: 275279842446
effective search space used: 275279842446
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 79 (35.0 bits)
Medicago: description of AC146776.10


