

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146559.13 - phase: 0 /pseudo
(661 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
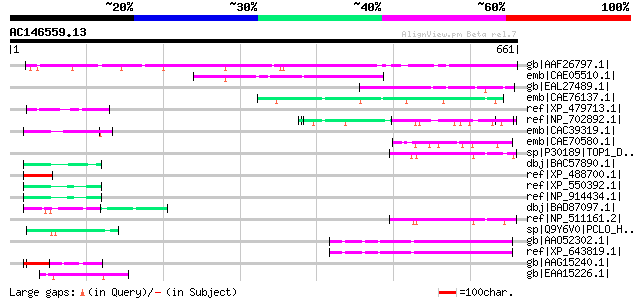
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF26797.1| hypothetical protein [Arabidopsis thaliana] gi|15... 319 2e-85
emb|CAE05510.1| OSJNBa0038P21.3 [Oryza sativa (japonica cultivar... 164 1e-38
gb|EAL27489.1| GA19837-PA [Drosophila pseudoobscura] 60 2e-07
emb|CAE76137.1| hypothetical protein [Neurospora crassa] gi|3241... 58 9e-07
ref|XP_479713.1| putative pherophorin-dz1 protein [Oryza sativa ... 58 1e-06
ref|NP_702892.1| Plasmodium falciparum trophozoite antigen r45-l... 57 1e-06
emb|CAC39319.1| VMP4 protein [Volvox carteri f. nagariensis] 57 1e-06
emb|CAE70580.1| Hypothetical protein CBG17237 [Caenorhabditis br... 57 1e-06
sp|P30189|TOP1_DROME DNA topoisomerase I gi|1772834|gb|AAC24158.... 57 2e-06
dbj|BAC57890.1| pherophorin [Volvox carteri f. nagariensis] 57 2e-06
ref|XP_488700.1| PREDICTED: hypothetical protein XP_488700 [Mus ... 57 2e-06
ref|XP_550392.1| putative leucine-rich repeat/extensin 1 [Oryza ... 57 2e-06
ref|NP_914434.1| putative extensin-like protein [Oryza sativa (j... 57 2e-06
dbj|BAD87097.1| putative receptor protein kinase PERK1 [Oryza sa... 56 4e-06
ref|NP_511161.2| CG6146-PA, isoform A [Drosophila melanogaster] ... 56 4e-06
sp|Q9Y6V0|PCLO_HUMAN Piccolo protein (Aczonin) 56 4e-06
gb|AAO52302.1| similar to Homo sapiens (Human). Dentin sialophos... 55 6e-06
ref|XP_643819.1| hypothetical protein DDB0203095 [Dictyostelium ... 55 6e-06
gb|AAG15240.1| ENOD2 [Styphnolobium japonicum] 55 6e-06
gb|EAA15226.1| developmental protein DG1037 [Plasmodium yoelii y... 55 6e-06
>gb|AAF26797.1| hypothetical protein [Arabidopsis thaliana]
gi|15229239|ref|NP_187066.1| expressed protein
[Arabidopsis thaliana]
Length = 712
Score = 319 bits (817), Expect = 2e-85
Identities = 241/734 (32%), Positives = 358/734 (47%), Gaps = 127/734 (17%)
Query: 21 NPPPA---QLPPPSQLP-----PPLSPPPQQHPP--PLLPPPPPLN-LNTTLSSLTNLLT 69
NP P PPP+ P PP PPP Q+P ++P PPP+ L+ TLSSL +LL+
Sbjct: 12 NPNPNLFYHYPPPNSNPNFFFRPP--PPPLQNPNNYSIVPSPPPIRELSGTLSSLKSLLS 69
Query: 70 FSTQILSTTPQ----------PQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
+ L + Q + + C F+ NH +PP +LFLH LRCP++
Sbjct: 70 ECQRTLDSLSQNLALDHSSLLQKDENGCFVRCPFDSNHFMPPEALFLHSLRCPNTL---- 125
Query: 120 HIDHLLTSLS-YPKTLQNPTSIHLSS-----------YLDF-TNFFYNNCPGVVTFSDAN 166
+ HLL S S Y TL+ P + L++ DF +NFFY +CPG V FS+ +
Sbjct: 126 DLIHLLESFSSYRNTLELPCELQLNNGDGDLCISLDDLADFGSNFFYRDCPGAVKFSELD 185
Query: 167 SVAQTATFHLPDFLSRECSTVCSPTPNHKPSP------IVPSEYYYITREIESWNNFPIT 220
+T T LP LS ECS K ++PS+ + EI+ W +FP +
Sbjct: 186 GKKRTLT--LPHVLSVECSDFVGSDEKVKKIVLDKCLGVLPSDLCAMKNEIDQWRDFPSS 243
Query: 221 YSNSVFRAIFGIGIAKESEMVNWLLSNSSRYGIVIDTSMQKHIFLLFCLGFKAILRDAF- 279
YS+SV +I G + + S + W+L NS+RYG++IDT M+ HIFLLF L K+ +++A
Sbjct: 244 YSSSVLSSIVGSKVVEISALRKWILVNSTRYGVIIDTFMRDHIFLLFRLCLKSAVKEACG 303
Query: 280 --------------------------LLNNVLMWLESQVSILYGV-SGKLFVLNFVKKCI 312
+ VL WL SQ+++LYG +GK F L+ K+CI
Sbjct: 304 FRMESDATDVGEQKIMSCKSSTFECPVFIQVLSWLASQLAVLYGEGNGKFFALDMFKQCI 363
Query: 313 LVGASALLLFPLGNEGGESVGSKEGKSDTNCIRERKILM------------------PQV 354
+ AS ++LF L + G E D +R + ++M PQV
Sbjct: 364 VESASQVMLFRLEGTRSKCSGVVEDLDDAR-LRNKDVIMEKPFENSSGGECGKTLDSPQV 422
Query: 355 V------AAVAALHERALLERKIKEFWFSHPSNNYQLKAEHCYLSDKAKEERNKRADYRP 408
+ AAVAAL+ER+LLE KI+ ++ P YQ AE +++ KA EERN+R YRP
Sbjct: 423 ISVSRVSAAVAALYERSLLEEKIRAVRYAQPLTRYQRAAELGFMTAKADEERNRRCSYRP 482
Query: 409 IIEYDWVHQQHSHHQETQKEKTREELLAEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEY 468
II++D +Q S +Q+ K KTREELLAEERDYKRRRMSYRGKK ++ QV+ D+I EY
Sbjct: 483 IIDHDGRPRQRSLNQDMDKMKTREELLAEERDYKRRRMSYRGKKVKRTPRQVLHDMIEEY 542
Query: 469 MEEIKLAAGVKSPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSHDSPAVTISNPDHH 528
EEIKLA G+ + GMP L S I + S S P
Sbjct: 543 TEEIKLAGGIG-----CFEKGMP-----LQSRSPIGNDQKES--------DFGYSIPSTD 584
Query: 529 EQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQENHRSHASTSPERHR 588
+Q N +D + + + D +R + G +R Q++HRS+ + +
Sbjct: 585 KQWKGENRADIEYPIDNRQNSDKVKR----HDEYDSGSSQR---QQSHRSYKHSDRRDDK 637
Query: 589 SRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEH 648
R R + +++ K+++ + +++ K+ + R DP + R
Sbjct: 638 LRDRRKDKHNDRRDDEFTRTKRHSIEGESYQNYRSSREKSSSDYKTKRDDPYDRRSQQPR 697
Query: 649 DIS-SDDKYIKPDK 661
+ + +D+YI +K
Sbjct: 698 NQNLFEDRYIPTEK 711
>emb|CAE05510.1| OSJNBa0038P21.3 [Oryza sativa (japonica cultivar-group)]
Length = 395
Score = 164 bits (414), Expect = 1e-38
Identities = 107/276 (38%), Positives = 153/276 (54%), Gaps = 34/276 (12%)
Query: 240 MVNWLLSNSSRYGIVIDTSMQKHIFLLFCLGFKAILRDAFL------------------- 280
M W+L+ S RYG+VID + + H+++L L KA +A
Sbjct: 1 MRRWVLAASQRYGVVIDAATRDHVWVLLRLCLKAAASEARCSLEDARGAEGGGSGFVDPR 60
Query: 281 --------LNNVLMWLESQVSILYGVS-GKLFVLNFVKKCILVGASALLLFPLGNEGGES 331
L + + WL +Q+ ILYG S G+ F + V++ IL S L + G +GG
Sbjct: 61 AIRFECPRLVDAVSWLGTQLRILYGESSGRSFAIAAVREAILRAGSCLAV---GVDGGGG 117
Query: 332 VGSKEGKSDTNCIRERKILMPQVVAAVAALHERALLERKIKEFWFSHPSNNYQLKAEHCY 391
G EG SD + + + QV AA+AALHER LE KIK P+ +QL E+
Sbjct: 118 SGV-EG-SDFGNVGTPSVSVAQVAAAIAALHERFSLEEKIKALRAPRPAK-FQLLLEYSK 174
Query: 392 LSDKAKEERNKRADYRPIIEYDWVHQQHSHHQETQKEKTREELLAEERDYKRRRMSYRGK 451
++ +EER+KR +YR ++EYD + + QE + KTREELLAEERDYKRRR SYRGK
Sbjct: 175 ALERGREERSKRPNYRAVLEYDGIISRRVDSQEFGRVKTREELLAEERDYKRRRTSYRGK 234
Query: 452 KRNQSTIQVMRDLIGEYMEEIKLAAGVKSPVNVSED 487
K ++ +++RD+I E+MEEIK A G+ ++V D
Sbjct: 235 KAKRNPKEILRDIIDEHMEEIKQAGGIGCHLDVPGD 270
>gb|EAL27489.1| GA19837-PA [Drosophila pseudoobscura]
Length = 695
Score = 60.1 bits (144), Expect = 2e-07
Identities = 51/209 (24%), Positives = 89/209 (42%), Gaps = 13/209 (6%)
Query: 457 TIQVMRDLIGEYMEEIK-LAAGVKSPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSH 515
T+ +R GE + L K+ V E+ P P +SH + +N R S
Sbjct: 53 TVNALRKFNGEVGVAARTLVTRWKAMVAAEEEPAEPTPSAIGTTSHEEDSGKSNGRRSSE 112
Query: 516 DSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQEN 575
D P + + + S K K HD +S + + + S HH + +D+E+
Sbjct: 113 DEPELEYKGSNSSSGGEDGHSSSKRKSGHDDSSS--KNKSERSSSSRHHSKSSK-SDKES 169
Query: 576 HRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKD------RWQNDTHKNH 629
+ H S+ +R R + + E E +++ KK N +TKD + +H++
Sbjct: 170 DKKHKSSRHDRSRDKDKDKERESRKESKEHRD-KKSNGEHKTKDSSSPNKSSSSSSHRSS 228
Query: 630 TSDSFSRSDPSESRGVGEHDISSDDKYIK 658
TS S+++ +S EH S+ +K IK
Sbjct: 229 TSQHTSKAESHKSD--KEHRKSNHEKSIK 255
>emb|CAE76137.1| hypothetical protein [Neurospora crassa]
gi|32414851|ref|XP_327905.1| predicted protein
[Neurospora crassa] gi|28917819|gb|EAA27507.1| predicted
protein [Neurospora crassa]
Length = 1015
Score = 58.2 bits (139), Expect = 9e-07
Identities = 83/381 (21%), Positives = 142/381 (36%), Gaps = 67/381 (17%)
Query: 324 LGNEGGESVGSKEGKSDTNCIRE---RKILMPQVVAAVAALHERAL-LERKIKEFWFSHP 379
+G+E E + K+D + E R+ +V + + E+ LER+IKE
Sbjct: 152 IGDEAAEEERIRGSKTDEDYAAETAARRAERERVREELRLVEEKKRRLEREIKEKEEQKR 211
Query: 380 SNNYQLKA-EHCYLSDKAKEERNKRADYRPIIEYDWVHQQHSHHQETQKEKTREELLAEE 438
+ K E + +K +EER R + R E + + H+E + ++R+ +
Sbjct: 212 REERRKKQLEEDAIREKEREERRARREQR---ERERELSRERRHRERDRNRSRDRDRHRD 268
Query: 439 RDYKRRRMSYRGKKRNQS-------------------------------TIQVMRDLIGE 467
RD R R RG+ R++S ++ R+ + +
Sbjct: 269 RDRDRDRDRGRGRSRDRSRGRDRRLSRDAKARNDHGRPEDVKTQLSKEDNERLEREALAD 328
Query: 468 YMEEIKLAAGVKSPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSH--------DSPA 519
+ E K A + + V D+ + PPP + + I S +VS P
Sbjct: 329 LLRESKRVAAKQPELEV--DTTLAPPPRRTKPASAIDPIRRASPKVSDVKKTADTAGKPG 386
Query: 520 VTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHHH----------GGDER 569
VT D E + T+ D + A+ D + K G+ H G +R
Sbjct: 387 VTEETKDVTEPKEVTDTKDIKEPKETASRVDSRRSKSPSPGTRGHRDKEKEKDREGEKDR 446
Query: 570 GTDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNH 629
G D + R ++ R R RS HR + R++ + S DR ++D ++H
Sbjct: 447 GRDYDRRRKDNRSASPRARRIERSRSHRREDSRSRLRDRRERSRSRPRLDR-RDDRREDH 505
Query: 630 TSD-------SFSRSDPSESR 643
D + R D S SR
Sbjct: 506 VRDRSRSRRRATDRRDRSRSR 526
>ref|XP_479713.1| putative pherophorin-dz1 protein [Oryza sativa (japonica
cultivar-group)] gi|42408241|dbj|BAD09398.1| putative
pherophorin-dz1 protein [Oryza sativa (japonica
cultivar-group)]
Length = 342
Score = 57.8 bits (138), Expect = 1e-06
Identities = 38/110 (34%), Positives = 46/110 (41%), Gaps = 16/110 (14%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTPQP 81
PPP + PPP +PPP PP + PPP PPPPP S P P
Sbjct: 97 PPPRRAPPPPSMPPP-PPPRRAPPPPATPPPPPRRAPPPPSPPIR-----------PPPP 144
Query: 82 QTPTTNLIPCLFNPNH-LIPPPSLFLHHLRCPSSPRPLPHIDHLLTSLSY 130
TP P P+H L PPP P P P PHI ++ +S+
Sbjct: 145 PTPRPYAPP---PPSHPLAPPPPHISPPAPVPPPPSPPPHIVIIVVFVSF 191
Score = 43.5 bits (101), Expect = 0.022
Identities = 18/37 (48%), Positives = 21/37 (56%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P M P P PPP + PPP +PP + PP PPPP
Sbjct: 35 PVMPPRPQAPPPPQRFPPPPAPPIRPPSPPGRAPPPP 71
Score = 42.4 bits (98), Expect = 0.049
Identities = 37/125 (29%), Positives = 43/125 (33%), Gaps = 37/125 (29%)
Query: 18 PTMNPPPAQL------------------PPPSQLPPPLS----PPPQQHPPPLLPPPPPL 55
P PPP + PPP + PPP S PP + PPP LPPPPP
Sbjct: 41 PQAPPPPQRFPPPPAPPIRPPSPPGRAPPPPGRAPPPPSQAPPPPRRAPPPPALPPPPPR 100
Query: 56 NLNTTLSSLTNLLTFSTQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSP 115
S + P P TP P PPPS +R P P
Sbjct: 101 RAPPPPS-----MPPPPPPRRAPPPPATPPP-------PPRRAPPPPS---PPIRPPPPP 145
Query: 116 RPLPH 120
P P+
Sbjct: 146 TPRPY 150
Score = 42.4 bits (98), Expect = 0.049
Identities = 18/35 (51%), Positives = 20/35 (56%), Gaps = 2/35 (5%)
Query: 19 TMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPP 53
T PPP + PPP PP+ PPP P P PPPP
Sbjct: 123 TPPPPPRRAPPPPS--PPIRPPPPPTPRPYAPPPP 155
Score = 36.2 bits (82), Expect = 3.5
Identities = 15/33 (45%), Positives = 17/33 (51%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P P +PP Q PPP P PP+ PP PP
Sbjct: 32 PKPPVMPPRPQAPPPPQRFPPPPAPPIRPPSPP 64
Score = 36.2 bits (82), Expect = 3.5
Identities = 18/35 (51%), Positives = 20/35 (56%), Gaps = 1/35 (2%)
Query: 21 NPPP-AQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
NP P A L P + PP + P PQ PPP PPPP
Sbjct: 20 NPNPYAPLNPNAPKPPVMPPRPQAPPPPQRFPPPP 54
Score = 35.4 bits (80), Expect = 5.9
Identities = 21/59 (35%), Positives = 25/59 (41%), Gaps = 8/59 (13%)
Query: 18 PTMNPPPAQLPPPSQLPPP----LSPPPQQHPPPLLP----PPPPLNLNTTLSSLTNLL 68
P + PPP P P PPP PPP PP +P PPP + + S LL
Sbjct: 137 PPIRPPPPPTPRPYAPPPPSHPLAPPPPHISPPAPVPPPPSPPPHIVIIVVFVSFGGLL 195
>ref|NP_702892.1| Plasmodium falciparum trophozoite antigen r45-like protein
[Plasmodium falciparum 3D7] gi|23498309|emb|CAD49281.1|
Plasmodium falciparum trophozoite antigen r45-like
protein [Plasmodium falciparum 3D7]
Length = 1222
Score = 57.4 bits (137), Expect = 1e-06
Identities = 63/287 (21%), Positives = 116/287 (39%), Gaps = 28/287 (9%)
Query: 383 YQLKAEHCYLSDKAKEERNKRADYRPIIEYDWVHQQHSHHQETQKEKTREELLA---EER 439
Y+ + +C DK E N + D + + D H+ S+H+ K+ + +
Sbjct: 373 YEPECANCNEEDKNMSENNHKKDSKH--KGDSNHKSDSNHKSDSNHKSDSNHKSGSNHKS 430
Query: 440 DYKRRRMS-YRGKKRNQSTIQVMRDLIGEYMEEIKLAAGVKSPVNVSEDSGMPPPPPKLL 498
D + S ++ +QS M D + K + KS + DS K
Sbjct: 431 DCNHKSGSNHKSDSNHQSDCNHMSDHNHKSDNNHKSDSSHKSDSSHKSDSSH-----KSG 485
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSH---TNYSDKSKVVHDATSRDYEQRK 555
S+H ++ N+ + SH S + S+ +H +H +N+ ++S +++ ++ K
Sbjct: 486 SNH--KSDNNHKSDSSHKSGSNHKSDHNHKSDSNHKSDSNHKNESNHKNESNHKNESNHK 543
Query: 556 Q--GHQGSHHHGGDERGTDQENHRS-HASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYN 612
H+ +H D NH S H S H+S H H H + + K N
Sbjct: 544 NESNHKNESNHKNDSNHKSDSNHMSDHNHKSDNNHKS---DHNHMSDHNHKSDNNHKSDN 600
Query: 613 NSSRTKDRWQNDTHK---NHTSDSFSRSDPSESRGVGEHDISSDDKY 656
N + ++ HK NH SD+ +SD + + +H+ SD+ +
Sbjct: 601 NHMSDHNHKSDNNHKSDNNHKSDNNHKSDHNH---MSDHNHKSDNNH 644
Score = 56.6 bits (135), Expect = 2e-06
Identities = 57/292 (19%), Positives = 112/292 (37%), Gaps = 30/292 (10%)
Query: 377 SHPSNNYQLKAEHCYLSD-------KAKEERNKRADYRPIIEYDWV--HQQHSHHQETQK 427
+H S+N K++H ++SD K + N ++D + +++ + H S H
Sbjct: 619 NHKSDNNH-KSDHNHMSDHNHKSDNNHKSDHNHKSDSNHMSDHNHMSDHNHKSDHNHKSD 677
Query: 428 EKTREELLAEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIKLAAGVKSPVNVSED 487
+ + + + +++ ++S M D K KS N D
Sbjct: 678 HNHKSDNNHKSDSNHKSDSNHKSDHNHKSDSNHMSDHNHMSDHNHKSDHNHKSDNNHKSD 737
Query: 488 SGMPPPPPKLLSSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKS-KVVHDA 546
S +++N+ + +H S + +S DH+ H + SD + K H+
Sbjct: 738 SNH-------------KSDSNHKSDHNHKSDSNHMS--DHNHMSDHNHMSDHNHKSDHNH 782
Query: 547 TSRDYEQRKQGHQGSHHHGGDERGTDQENHRS-HASTSPERHRSRSRSHEHRVHHEKQDY 605
S + + H+ +H D NH+S H S H+S + H ++
Sbjct: 783 KSDNNHKSDSNHKSDSNHKSDSNHKSDHNHKSDHKHMSDNNHKSDNNHKSDHNHKSDNNH 842
Query: 606 SGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHDISSDDKYI 657
+ + S K + + NH SDS +SD + +H+ +SD ++
Sbjct: 843 KSDHNHKSDSNHKSDSNHKSDSNHKSDSNHKSDNNHK---SDHNHNSDSNHM 891
Score = 53.9 bits (128), Expect = 2e-05
Identities = 63/292 (21%), Positives = 111/292 (37%), Gaps = 27/292 (9%)
Query: 381 NNYQLKAEHCYLSDKAKEERNKRADYRPIIEYDWVHQQHSHHQETQKEKTREELLAEERD 440
NN++ + H S+ K + N ++D + D H+ S+H+ K E
Sbjct: 492 NNHKSDSSHKSGSNH-KSDHNHKSDSNH--KSDSNHKNESNHKNESNHKNESNHKNESNH 548
Query: 441 YKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIKLAAGVKSPVNVSEDSGMPPPPPKLLSS 500
+ +++ ++S M D + K S N D+ + S
Sbjct: 549 --KNESNHKNDSNHKSDSNHMSDHNHKSDNNHKSDHNHMSDHNHKSDNNHKSDNNHM-SD 605
Query: 501 HTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKS-KVVHDATSRDYEQRKQGHQ 559
H ++ N+ + +H S DH+ H + SD + K H+ S H
Sbjct: 606 HNHKSDNNHKSDNNHKSD--NNHKSDHNHMSDHNHKSDNNHKSDHNHKSDSNHMSDHNHM 663
Query: 560 GSHHHGGDERGTDQENHRS---HASTSPERHRSRSRS-HEHRV-------HHEKQDYSGR 608
H+H D NH+S H S S + S +S H H+ H+ D++ +
Sbjct: 664 SDHNHKSDHNHKSDHNHKSDNNHKSDSNHKSDSNHKSDHNHKSDSNHMSDHNHMSDHNHK 723
Query: 609 KKYNNSSRTKDRWQNDTHK---NHTSDSFSRSDPSESRGVGEHDISSDDKYI 657
+N+ S + + HK NH SD +SD S + +H+ SD ++
Sbjct: 724 SDHNHKSDNNHK-SDSNHKSDSNHKSDHNHKSD---SNHMSDHNHMSDHNHM 771
Score = 50.8 bits (120), Expect = 1e-04
Identities = 42/181 (23%), Positives = 75/181 (41%), Gaps = 20/181 (11%)
Query: 499 SSHTISTEANNSREVSH--DSPAVTISN--PDHHEQQSHTNYSDKSKVVHDATSRDYEQR 554
S H ++ N+ + +H DS + SN DH+ + H + SD + + D+ +
Sbjct: 778 SDHNHKSDNNHKSDSNHKSDSNHKSDSNHKSDHNHKSDHKHMSDNNHKSDNNHKSDHNHK 837
Query: 555 KQG-HQGSHHHGGDERGTDQENHRS---HASTSPER--------HRSRSRSHEHRVHHEK 602
H+ H+H D NH+S H S S + H S S+ H+ K
Sbjct: 838 SDNNHKSDHNHKSDSNHKSDSNHKSDSNHKSDSNHKSDNNHKSDHNHNSDSNHMSDHNHK 897
Query: 603 QDYSGRKKYNNSSRTKDRWQNDTHK---NHTSDSFSRSDPSESRGVGEHDISSDDKYIKP 659
D++ + +N+ S + N+ HK NH SD ++ ++ ++D S +
Sbjct: 898 SDHNHKSDHNHKSDNNHKSDNN-HKSDHNHKSDHKKNNNNNKDNKNDDNDDSDASDAVHE 956
Query: 660 D 660
D
Sbjct: 957 D 957
Score = 49.7 bits (117), Expect = 3e-04
Identities = 34/164 (20%), Positives = 71/164 (42%), Gaps = 17/164 (10%)
Query: 498 LSSHTISTEANNSREVSHDSPAVTISNPDHHEQQSH-TNYSDKSKVVHDATSRDYEQRKQ 556
+S H ++ N+ + SH S + S+ H +H ++ + KS H + S
Sbjct: 453 MSDHNHKSDNNHKSDSSHKSDSSHKSDSSHKSGSNHKSDNNHKSDSSHKSGSN------- 505
Query: 557 GHQGSHHHGGDERGTDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSR 616
H+ H+H D NH++ ++ E + +H++ +H+ + + N S
Sbjct: 506 -HKSDHNHKSDSNHKSDSNHKNESNHKNESNHKNESNHKNESNHKNES-----NHKNDSN 559
Query: 617 TKDRWQNDTHKNHTSDSFSRSDP---SESRGVGEHDISSDDKYI 657
K + + NH SD+ +SD S+ +++ SD+ ++
Sbjct: 560 HKSDSNHMSDHNHKSDNNHKSDHNHMSDHNHKSDNNHKSDNNHM 603
Score = 49.3 bits (116), Expect = 4e-04
Identities = 53/264 (20%), Positives = 102/264 (38%), Gaps = 20/264 (7%)
Query: 377 SHPSNNYQLKAEHCYLSD-KAKEERNKRADYRPIIEYDWVHQQHSHHQETQKEKTREELL 435
+H S++ + ++H ++SD K + N ++D + D H+ S+H+ K+ +
Sbjct: 703 NHKSDSNHM-SDHNHMSDHNHKSDHNHKSDNNH--KSDSNHKSDSNHKSDHNHKSDSNHM 759
Query: 436 AEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIKLAAGVKSPVNVSEDSGMPPPPP 495
++ ++ ++S D + K + KS N D
Sbjct: 760 SDHNHMSDH--NHMSDHNHKSDHNHKSDNNHKSDSNHKSDSNHKSDSNHKSDHNHK---- 813
Query: 496 KLLSSHTISTEANNSREVSHDSPAVTISNPDH---HEQQSHTNYSDKSKVVHDATSRDYE 552
S H ++ N+ + +H S S+ +H H +S +N+ S D+ +
Sbjct: 814 ---SDHKHMSDNNHKSDNNHKSDHNHKSDNNHKSDHNHKSDSNHKSDSNHKSDSNHKSDS 870
Query: 553 QRKQ--GHQGSHHHGGDERGTDQENHRS-HASTSPERHRSRSRSHEHRVHHEKQDYSGRK 609
K H+ H+H D NH+S H S H+S + H ++
Sbjct: 871 NHKSDNNHKSDHNHNSDSNHMSDHNHKSDHNHKSDHNHKSDNNHKSDNNHKSDHNHKSDH 930
Query: 610 KYNNSSRTKDRWQNDTHKNHTSDS 633
K NN++ KD +D + SD+
Sbjct: 931 KKNNNN-NKDNKNDDNDDSDASDA 953
>emb|CAC39319.1| VMP4 protein [Volvox carteri f. nagariensis]
Length = 1143
Score = 57.4 bits (137), Expect = 1e-06
Identities = 40/120 (33%), Positives = 49/120 (40%), Gaps = 19/120 (15%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
P+ PPP+ PPPS PPP PPP PPP PPPPP S
Sbjct: 523 PSPPPPPSPPPPPSPPPPPSPPPPPSPPPPPSPPPPP---------------SPPPPPSP 567
Query: 78 TPQPQTPTTNLIPCLFNPNH-LIPPPSLFLHHLRCPSSPR---PLPHIDHLLTSLSYPKT 133
P P P P +P H PPP + PS P P P D+++ ++ P T
Sbjct: 568 PPPPSPPPPPSPPPPPSPRHPPSPPPRPRPPRPQPPSPPSPSPPSPRTDNVVAWITQPDT 627
Score = 52.4 bits (124), Expect = 5e-05
Identities = 34/100 (34%), Positives = 36/100 (36%), Gaps = 19/100 (19%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
P PP+ PPPS PPP PPP PPP PPPPP
Sbjct: 517 PPRPSPPSPPPPPSPPPPPSPPPPPSPPPPPSPPPPPSP-------------------PP 557
Query: 78 TPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRP 117
P P P + P P PPP H P PRP
Sbjct: 558 PPSPPPPPSPPPPPSPPPPPSPPPPPSPRHPPSPPPRPRP 597
Score = 42.7 bits (99), Expect = 0.037
Identities = 22/45 (48%), Positives = 24/45 (52%), Gaps = 8/45 (17%)
Query: 18 PTMNPPPAQL----PPPSQLPP----PLSPPPQQHPPPLLPPPPP 54
P + PPP+ L PPS PP P PPP PPP PPPPP
Sbjct: 497 PFVPPPPSPLLTSPRPPSPRPPRPSPPSPPPPPSPPPPPSPPPPP 541
>emb|CAE70580.1| Hypothetical protein CBG17237 [Caenorhabditis briggsae]
Length = 631
Score = 57.4 bits (137), Expect = 1e-06
Identities = 53/194 (27%), Positives = 84/194 (42%), Gaps = 45/194 (23%)
Query: 500 SHTISTEANNSREVSHDSPAVTISNPDH----HEQQSHTNYSDKSKVVHDA----TSRDY 551
SH ++ SR+ S +SP P+H HE++ H + + + D+ +SRD
Sbjct: 46 SHKRKHRSSGSRD-SRESP-----RPNHKKRRHEKRRHEDDDRRDEHYGDSRPKSSSRDK 99
Query: 552 EQRKQG----------HQGSHHHGGDERGTDQENHRSHASTSPERHRS------------ 589
++R Q H G H G+ RG +N RS + P +HRS
Sbjct: 100 DRRDQRRHRDDYLEDYHCGKRRHNGESRGKSPKNIRSDSRDEP-KHRSSGSRDSRESPRP 158
Query: 590 --RSRSHEHRVHHE---KQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSF---SRSDPSE 641
+ R HE R H + + ++ G + +SSR KDR H++ + + R D E
Sbjct: 159 NHKKRRHEKRRHEDDDRRDEHYGDSRPKSSSRDKDRRDQRRHRDDYLEDYHCRKRRDNGE 218
Query: 642 SRGVGEHDISSDDK 655
SRG +I SD +
Sbjct: 219 SRGKSPKNIRSDSR 232
Score = 41.6 bits (96), Expect = 0.083
Identities = 24/97 (24%), Positives = 38/97 (38%), Gaps = 3/97 (3%)
Query: 562 HHHGGDERGTDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRW 621
HHH + + S S P + R H + ++ G + +SSR KDR
Sbjct: 43 HHHSHKRKHRSSGSRDSRESPRPNHKKRRHEKRRHEDDDRRDEHYGDSRPKSSSRDKDRR 102
Query: 622 QNDTHKNHTSDSF---SRSDPSESRGVGEHDISSDDK 655
H++ + + R ESRG +I SD +
Sbjct: 103 DQRRHRDDYLEDYHCGKRRHNGESRGKSPKNIRSDSR 139
>sp|P30189|TOP1_DROME DNA topoisomerase I gi|1772834|gb|AAC24158.1| DNA topoisomerase I
[Drosophila melanogaster] gi|158643|gb|AAA28951.1|
topoisomerase I
Length = 972
Score = 57.0 bits (136), Expect = 2e-06
Identities = 53/198 (26%), Positives = 89/198 (44%), Gaps = 38/198 (19%)
Query: 496 KLLSSHTISTEANNSREVSHDSPAVTI----SNPDHH---EQQSHTNYSDKSKVVHDATS 548
+++ S+ ++T + H S + + S+ D H E++ ++ S S H ++S
Sbjct: 20 EVVQSNGVTTNGHGHHHHHHSSSSSSSKHKSSSKDKHRDREREHKSSNSSSSSKEHKSSS 79
Query: 549 RDYEQRKQGHQGSHHHGGD-ERGTDQENHRSHASTS-PERHRSRSRSHEHRVHHEKQ--- 603
RD ++ K S H D ER +HRS +S+S ++ S S H+ H K+
Sbjct: 80 RDKDRHKSSSSSSKHRDKDKERDGSSNSHRSGSSSSHKDKDGSSSSKHKSSSGHHKRSSK 139
Query: 604 -------------DYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHDI 650
S R K ++SSR K+R + +HK+ +S S S+S S SR H
Sbjct: 140 DKERRDKDKDRGSSSSSRHKSSSSSRDKER-SSSSHKSSSSSSSSKSKHSSSR----HSS 194
Query: 651 SSDDK--------YIKPD 660
SS K ++KP+
Sbjct: 195 SSSSKDHPSYDGVFVKPE 212
Score = 37.7 bits (86), Expect = 1.2
Identities = 24/72 (33%), Positives = 32/72 (44%), Gaps = 2/72 (2%)
Query: 591 SRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEH-D 649
+ H H HH S K+ +SS+ K R + HK+ S S S+ S SR H
Sbjct: 29 TNGHGHH-HHHHSSSSSSSKHKSSSKDKHRDREREHKSSNSSSSSKEHKSSSRDKDRHKS 87
Query: 650 ISSDDKYIKPDK 661
SS K+ DK
Sbjct: 88 SSSSSKHRDKDK 99
>dbj|BAC57890.1| pherophorin [Volvox carteri f. nagariensis]
Length = 606
Score = 57.0 bits (136), Expect = 2e-06
Identities = 36/102 (35%), Positives = 41/102 (39%), Gaps = 30/102 (29%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
P+ PPP PPPS PPP PPP PPP PPPPP S
Sbjct: 215 PSPPPPPPPPPPPSPPPPPPPPPPPSPPPPPPPPPPP---------------------SP 253
Query: 78 TPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
P P P+ + P +P+ PPP P SP P P
Sbjct: 254 PPPPPPPSPSPPPPPPSPSPPPPPP---------PPSPPPAP 286
Score = 44.7 bits (104), Expect = 0.010
Identities = 33/98 (33%), Positives = 34/98 (34%), Gaps = 34/98 (34%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTPQP 81
PPP PPP PPP PPP PP PPPPP P P
Sbjct: 206 PPPPPPPPPPSPPPPPPPPPPPSPP---PPPPP-----------------------PPPP 239
Query: 82 QTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
P P +P PPPS PS P P P
Sbjct: 240 SPPPPPPPPPPPSPPPPPPPPS--------PSPPPPPP 269
Score = 40.8 bits (94), Expect = 0.14
Identities = 20/37 (54%), Positives = 22/37 (59%), Gaps = 5/37 (13%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P+ +PPP PPPS PPP PPP PPP PP P
Sbjct: 260 PSPSPPP---PPPSPSPPP--PPPPPSPPPAPPPFVP 291
Score = 40.4 bits (93), Expect = 0.18
Identities = 19/37 (51%), Positives = 20/37 (53%), Gaps = 2/37 (5%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP+ PPP PP SPPP PP P PPP
Sbjct: 254 PPPPPPPSPSPPPP--PPSPSPPPPPPPPSPPPAPPP 288
Score = 38.9 bits (89), Expect = 0.54
Identities = 16/31 (51%), Positives = 17/31 (54%)
Query: 24 PAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P + Q PPP PPP PPP PPPPP
Sbjct: 196 PQNIVVEPQPPPPPPPPPPPSPPPPPPPPPP 226
Score = 37.4 bits (85), Expect = 1.6
Identities = 16/26 (61%), Positives = 16/26 (61%), Gaps = 3/26 (11%)
Query: 28 PPPSQLPPPLSPPPQQHPPPLLPPPP 53
PPP PPP PPP PPP PPPP
Sbjct: 205 PPP---PPPPPPPPSPPPPPPPPPPP 227
>ref|XP_488700.1| PREDICTED: hypothetical protein XP_488700 [Mus musculus]
Length = 456
Score = 57.0 bits (136), Expect = 2e-06
Identities = 27/40 (67%), Positives = 29/40 (72%), Gaps = 2/40 (5%)
Query: 18 PTMNPPPAQLP-PPSQLPPPLSPPPQQHPPPL-LPPPPPL 55
PT+ PPP P PPS PPPL PPP + PPPL LPPPPPL
Sbjct: 98 PTILPPPPAPPRPPSPAPPPLPPPPPRAPPPLPLPPPPPL 137
Score = 42.4 bits (98), Expect = 0.049
Identities = 28/75 (37%), Positives = 33/75 (43%), Gaps = 18/75 (24%)
Query: 19 TMNPPPAQLPP--PSQLPPPLSPP-PQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQIL 75
T NP P P P+ LPPP +PP P PP LPPPPP + L
Sbjct: 85 TTNPIPKPTIPGSPTILPPPPAPPRPPSPAPPPLPPPPPR---------------APPPL 129
Query: 76 STTPQPQTPTTNLIP 90
P P P T+L+P
Sbjct: 130 PLPPPPPLPPTSLVP 144
Score = 42.0 bits (97), Expect = 0.063
Identities = 24/58 (41%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Query: 18 PTMNPPPAQLPPPSQLPP-PLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQI 74
P+ PPP PPP PP PL PPP P L+P PP L S LL T +
Sbjct: 111 PSPAPPPLPPPPPRAPPPLPLPPPPPLPPTSLVPLPPAEPSKEVLLSTVVLLLMLTPV 168
>ref|XP_550392.1| putative leucine-rich repeat/extensin 1 [Oryza sativa (japonica
cultivar-group)] gi|55296299|dbj|BAD68079.1| putative
leucine-rich repeat/extensin 1 [Oryza sativa (japonica
cultivar-group)] gi|55296121|dbj|BAD67840.1| putative
leucine-rich repeat/extensin 1 [Oryza sativa (japonica
cultivar-group)]
Length = 520
Score = 56.6 bits (135), Expect = 2e-06
Identities = 35/102 (34%), Positives = 37/102 (35%), Gaps = 35/102 (34%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
P +PPP PPPS PPP SPPP PPP PPP P +
Sbjct: 390 PPPSPPPPSPPPPSTSPPPPSPPPPSPPPPSPPPPAP--------------------IFH 429
Query: 78 TPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
PQP P PPP H CP P P P
Sbjct: 430 PPQPPPP---------------PPPPAPQPHPPCPELPPPPP 456
Score = 43.5 bits (101), Expect = 0.022
Identities = 36/113 (31%), Positives = 41/113 (35%), Gaps = 31/113 (27%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
P +PP PPP P P P P+ PPP PPPPP +T LS
Sbjct: 426 PIFHPPQPPPPPPPPAPQPHPPCPELPPPP--PPPPPCG-------------GATPALSP 470
Query: 78 TPQPQTPTTNLIPCLFNPNHLI----PPPSLFLHHLRCP-------SSPRPLP 119
P P P + P H + PPP HH P SP P P
Sbjct: 471 PPPPP-----YYPGPWPPVHGVPYGSPPPPPLQHHSSWPPIHSVPYGSPPPPP 518
>ref|NP_914434.1| putative extensin-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 579
Score = 56.6 bits (135), Expect = 2e-06
Identities = 35/102 (34%), Positives = 37/102 (35%), Gaps = 35/102 (34%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILST 77
P +PPP PPPS PPP SPPP PPP PPP P +
Sbjct: 390 PPPSPPPPSPPPPSTSPPPPSPPPPSPPPPSPPPPAP--------------------IFH 429
Query: 78 TPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLP 119
PQP P PPP H CP P P P
Sbjct: 430 PPQPPPP---------------PPPPAPQPHPPCPELPPPPP 456
Score = 40.8 bits (94), Expect = 0.14
Identities = 18/37 (48%), Positives = 20/37 (53%), Gaps = 2/37 (5%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P +PP PPP P P P P+ PPP PPPPP
Sbjct: 426 PIFHPPQPPPPPPPPAPQPHPPCPELPPPP--PPPPP 460
Score = 39.3 bits (90), Expect = 0.41
Identities = 22/47 (46%), Positives = 26/47 (54%), Gaps = 11/47 (23%)
Query: 18 PTMNPPPAQLPPPSQLPPP-------LSPPPQQHPPPLLPPP-PPLN 56
P +PP +LPPP PPP LSPPP PPP P P PP++
Sbjct: 442 PQPHPPCPELPPPPPPPPPCGGATPALSPPP---PPPYYPGPWPPVH 485
Score = 39.3 bits (90), Expect = 0.41
Identities = 21/45 (46%), Positives = 21/45 (46%), Gaps = 8/45 (17%)
Query: 18 PTMNPPPAQLPPPSQLPP-PLSPPPQQHPPP-------LLPPPPP 54
P PPP P P PP P PPP PPP L PPPPP
Sbjct: 430 PPQPPPPPPPPAPQPHPPCPELPPPPPPPPPCGGATPALSPPPPP 474
Score = 37.7 bits (86), Expect = 1.2
Identities = 24/58 (41%), Positives = 25/58 (42%), Gaps = 21/58 (36%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPP-PQQHPP------------------PLL--PPPPP 54
P PPPA + P Q PPP PP PQ HPP P L PPPPP
Sbjct: 418 PPSPPPPAPIFHPPQPPPPPPPPAPQPHPPCPELPPPPPPPPPCGGATPALSPPPPPP 475
>dbj|BAD87097.1| putative receptor protein kinase PERK1 [Oryza sativa (japonica
cultivar-group)]
Length = 682
Score = 55.8 bits (133), Expect = 4e-06
Identities = 62/197 (31%), Positives = 77/197 (38%), Gaps = 27/197 (13%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHP--------PPLLPPPPPLNLNTTLSSLTNLLT 69
P ++PPP PP S PPP++PPP P PP PPP P T S +
Sbjct: 51 PPVSPPPVTPPPVS--PPPVTPPPVTPPTPVAPPPVPPSPPPPTPTPTPVTPSPPPPVTP 108
Query: 70 FSTQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPLPHIDHLLTSLS 129
+++ P P PT P NP P PS P+SP P P I SLS
Sbjct: 109 SPPPPVASPPPPDVPTAP--PPSNNPPSPPPSPS------NVPASPPP-PRI-----SLS 154
Query: 130 YPKTLQNPTSIHLSSYLDFTNFFYNNCPGVVTFSDANSVAQTATFHLPDFLSRECSTVCS 189
P PT SS +N GV A V A + F+S+
Sbjct: 155 PPPPPSTPTQSGASSGSKSSNNGTVVAVGVAV--AAVVVLGLAAGLIYFFVSKRRRRRQH 212
Query: 190 PTPNHKPS-PIVPSEYY 205
P P H P P P+E+Y
Sbjct: 213 PPPPHHPGYPPFPAEFY 229
Score = 48.9 bits (115), Expect = 5e-04
Identities = 38/106 (35%), Positives = 45/106 (41%), Gaps = 16/106 (15%)
Query: 18 PTMNPP--PAQLPPPSQLPPPLSPPPQQHPPPLLPP---PPPLNLNTTLSSLTNLLTFST 72
P PP P PPP PPP+SPPP PPP+ PP PPP+ S +
Sbjct: 12 PAAPPPQTPPVTPPPVTAPPPVSPPPVT-PPPVTPPPVSPPPV-------SPPPVTPPPV 63
Query: 73 QILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSPRPL 118
TP P TP T + P P+ PPP PS P P+
Sbjct: 64 SPPPVTPPPVTPPTPVAPPPVPPS---PPPPTPTPTPVTPSPPPPV 106
>ref|NP_511161.2| CG6146-PA, isoform A [Drosophila melanogaster]
gi|62473712|ref|NP_001014742.1| CG6146-PC, isoform C
[Drosophila melanogaster] gi|61677900|gb|AAX52496.1|
CG6146-PC, isoform C [Drosophila melanogaster]
gi|7293054|gb|AAF48440.1| CG6146-PA, isoform A
[Drosophila melanogaster]
Length = 974
Score = 55.8 bits (133), Expect = 4e-06
Identities = 51/196 (26%), Positives = 88/196 (44%), Gaps = 32/196 (16%)
Query: 496 KLLSSHTISTEANNSREVSHDSPAVTISNP------DHH---EQQSHTNYSDKSKVVHDA 546
+++ S+ ++T + H S + + S+ D H E++ ++ S S H +
Sbjct: 20 EVVQSNGVTTNGHGHHHHHHHSSSSSSSSKHKSSSKDKHRDREREHKSSNSSSSSKEHKS 79
Query: 547 TSRDYEQRKQGHQGSHHHGGD-ERGTDQENHRSHASTS-PERHRSRSRSHEHRVHHEKQ- 603
+SRD ++ K S H D ER +HRS +S+S ++ S S H+ H K+
Sbjct: 80 SSRDKDRHKSSSSSSKHRDKDKERDGSSNSHRSGSSSSHKDKDGSSSSKHKSSSGHHKRS 139
Query: 604 ---------------DYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESR----G 644
S R K ++SSR K+R + +HK+ +S S S+S S SR
Sbjct: 140 SKDKERRDKDKDRGSSSSSRHKSSSSSRDKER-SSSSHKSSSSSSSSKSKHSSSRHSSSS 198
Query: 645 VGEHDISSDDKYIKPD 660
+ S D ++KP+
Sbjct: 199 SSKDQPSYDGVFVKPE 214
Score = 38.9 bits (89), Expect = 0.54
Identities = 24/74 (32%), Positives = 32/74 (42%), Gaps = 2/74 (2%)
Query: 589 SRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEH 648
+ H H HH S K+ +SS+ K R + HK+ S S S+ S SR H
Sbjct: 29 TNGHGHHHH-HHHSSSSSSSSKHKSSSKDKHRDREREHKSSNSSSSSKEHKSSSRDKDRH 87
Query: 649 -DISSDDKYIKPDK 661
SS K+ DK
Sbjct: 88 KSSSSSSKHRDKDK 101
>sp|Q9Y6V0|PCLO_HUMAN Piccolo protein (Aczonin)
Length = 5183
Score = 55.8 bits (133), Expect = 4e-06
Identities = 46/132 (34%), Positives = 52/132 (38%), Gaps = 18/132 (13%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPP---PPLNLN-----TTLSSLTNLLTF--S 71
PPP PPP PPP PPP PPP P P P L TT + L + +T +
Sbjct: 2336 PPPPPPPPPPPPPPPPPPPPPPLPPPTSPKPTILPKKKLTVASPVTTATPLFDAVTTLET 2395
Query: 72 TQILSTTPQPQTPTTNLIPCLFNPNHLIPPPSLFLHHLRCPSSP--RPLPHIDHLLTSLS 129
T +L + P T P P P L H PS P P P I L
Sbjct: 2396 TAVLRSNGLPVTRICTTAPPPVPPKPSSIPSGLVFTHRPEPSKPPIAPKPVIPQL----- 2450
Query: 130 YPKTLQNPTSIH 141
P T Q PT IH
Sbjct: 2451 -PTTTQKPTDIH 2461
Score = 35.4 bits (80), Expect = 5.9
Identities = 18/35 (51%), Positives = 18/35 (51%), Gaps = 1/35 (2%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPP 52
P PAQ PPSQ P PPPQQ P P P P
Sbjct: 434 PGPTKTPAQTKPPSQQPGSTKPPPQQ-PGPAKPSP 467
>gb|AAO52302.1| similar to Homo sapiens (Human). Dentin sialophosphoprotein
precursor [Contains: Dentin phosphoprotein (Dentin
phosphophoryn) (DPP); Dentin sialoprotein (DSP)]
[Dictyostelium discoideum]
Length = 1206
Score = 55.5 bits (132), Expect = 6e-06
Identities = 53/241 (21%), Positives = 101/241 (40%), Gaps = 12/241 (4%)
Query: 418 QHSHHQETQKEKTREELLAEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIKLAAG 477
Q S++ + E + E S+ N+S+ Q L EY E +
Sbjct: 768 QSSNNLSSNNESSNNE---SSNQGSNNESSHSESSNNESSNQ---SLNSEYNNEPSQGSN 821
Query: 478 VKSPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYS 537
+S N S + + SSH ++E++N+ SH S + +N Q +NYS
Sbjct: 822 SESS-NESSNQSLNSESNNDSSSHGSNSESSNNESSSHGSNNESSNN--ESSSQDSSNYS 878
Query: 538 DKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQ--ENHRSHASTSPERHRSRSRSHE 595
+ ++ +S + +QGS++ + + Q N SH S + + S ++E
Sbjct: 879 SNNGSSNNESSSQGSNNESSNQGSNNESSNNESSSQGSNNESSHNEPSNQSSNNESSNNE 938
Query: 596 HRVHHEKQDYSGRKKYNN-SSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHDISSDD 654
+ + S K NN SS + Q +++ ++S + S +ES G ++ SS++
Sbjct: 939 SSNNESSNNESSSKGSNNESSNNESSNQGSNNESSNNESNNESSNNESSSQGSNNESSNN 998
Query: 655 K 655
+
Sbjct: 999 E 999
Score = 45.4 bits (106), Expect = 0.006
Identities = 26/159 (16%), Positives = 72/159 (44%), Gaps = 2/159 (1%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
S+H S+ ++ E SH+ + SN + +S S+ ++++++ +
Sbjct: 620 SAHNESSNQGSNNESSHNESSNQGSNNESSNNESSNQGSNNESSHNESSNQGSNNESSNN 679
Query: 559 QGSHHHGGDERGTDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTK 618
+ S+ +E ++ +++ + S S S+ H+E + + +++ +
Sbjct: 680 ESSNQGSNNESSHNETSNQGSNNESSHNESSSQGSNNESAHNESSNQGSNNESSHNESSS 739
Query: 619 DRWQNDTHKNHTSDSFSRSDPS--ESRGVGEHDISSDDK 655
N++ N +++ S ++PS ES +++SS+++
Sbjct: 740 QGSNNESSNNDSNNQNSNNEPSNNESSNQSSNNLSSNNE 778
Score = 45.1 bits (105), Expect = 0.007
Identities = 33/159 (20%), Positives = 72/159 (44%), Gaps = 5/159 (3%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSH-TNYSDKSKVVHDATSRDYEQRKQG 557
S++ S + +N+ SH S + S ++E SH +N S+ ++ +S + +
Sbjct: 1015 SNNESSNQGSNNESSSHGSNNESSSKGSNNESSSHGSNNESSSQGSNNESSNNESSNQNS 1074
Query: 558 HQGSHHHGGDERGTDQENHRSHASTSPERHR-SRSRSHEHRVHHE---KQDYSGRKKYNN 613
+ S ++ +G++ E+ + +S+ + S + S+ ++E K++ ++
Sbjct: 1075 NNESSNNESSSKGSNNESSNNESSSQGSNNESSNNESNNQNSNNESSHKEESESNSHQSS 1134
Query: 614 SSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHDISS 652
+ +KD+ N N +S+S S SE E SS
Sbjct: 1135 ETESKDQVSNSGSMNQSSESSESSSLSEQSSAFESSSSS 1173
Score = 42.7 bits (99), Expect = 0.037
Identities = 37/167 (22%), Positives = 67/167 (39%), Gaps = 13/167 (7%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
S++ S +++N+ +++S SN + + SH S+ S S + E +
Sbjct: 761 SNNESSNQSSNNLSSNNESSNNESSNQGSNNESSH---SESSNNESSNQSLNSEYNNEPS 817
Query: 559 QGSHHHGGDERGTDQENHRSHASTSPERHRSRSRSHEHRVH----------HEKQDYSGR 608
QGS+ +E N S+ +S S S ++E H QD S
Sbjct: 818 QGSNSESSNESSNQSLNSESNNDSSSHGSNSESSNNESSSHGSNNESSNNESSSQDSSNY 877
Query: 609 KKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHDISSDDK 655
N SS + Q +++ S + S +ES G ++ SS ++
Sbjct: 878 SSNNGSSNNESSSQGSNNESSNQGSNNESSNNESSSQGSNNESSHNE 924
Score = 42.7 bits (99), Expect = 0.037
Identities = 27/158 (17%), Positives = 70/158 (44%), Gaps = 2/158 (1%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
SSH ++ ++ E SH+ + SN + +S S+ ++++++ +
Sbjct: 592 SSHNETSNQGSNNESSHNESSSQGSNNESAHNESSNQGSNNESSHNESSNQGSNNESSNN 651
Query: 559 QGSHHHGGDERGTDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTK 618
+ S+ +E ++ +++ + S S S+ H+E + + +++ +
Sbjct: 652 ESSNQGSNNESSHNESSNQGSNNESSNNESSNQGSNNESSHNETSNQGSNNESSHNESSS 711
Query: 619 DRWQNDTHKNHTSDSFSRSDPS--ESRGVGEHDISSDD 654
N++ N +S+ S ++ S ES G ++ SS++
Sbjct: 712 QGSNNESAHNESSNQGSNNESSHNESSSQGSNNESSNN 749
Score = 42.7 bits (99), Expect = 0.037
Identities = 28/155 (18%), Positives = 70/155 (45%), Gaps = 5/155 (3%)
Query: 504 STEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHH 563
S++ +N+ +++S +N H + S+ +++S H+ +S + H S +
Sbjct: 570 SSQGSNNESSNNESSNQGSNNESSHNETSNQGSNNESS--HNESSSQGSNNESAHNESSN 627
Query: 564 HGGDERGTDQENHRSHASTSPERHRSRSR-SHEHRVHHEKQDYSGRKKYNNSSRTKDRWQ 622
G + + E+ ++ + S ++ S+ H+E + + +N+ +
Sbjct: 628 QGSNNESSHNESSNQGSNNESSNNESSNQGSNNESSHNESSNQGSNNESSNNESSNQGSN 687
Query: 623 NDTHKNHTSDSFSRSDPS--ESRGVGEHDISSDDK 655
N++ N TS+ S ++ S ES G ++ S+ ++
Sbjct: 688 NESSHNETSNQGSNNESSHNESSSQGSNNESAHNE 722
Score = 41.2 bits (95), Expect = 0.11
Identities = 37/175 (21%), Positives = 76/175 (43%), Gaps = 18/175 (10%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
SSH S+ ++ E +H+ SN + + SH N S +++++ D + +
Sbjct: 704 SSHNESSSQGSNNESAHNES----SNQGSNNESSH-NESSSQGSNNESSNNDSNNQNSNN 758
Query: 559 QGSHHHGGDERGTD-QENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNN---- 613
+ S++ ++ + N+ S + S + + SH ++E + S +YNN
Sbjct: 759 EPSNNESSNQSSNNLSSNNESSNNESSNQGSNNESSHSESSNNESSNQSLNSEYNNEPSQ 818
Query: 614 ------SSRTKDRWQNDTHKNHTSDSFSRSDPS--ESRGVGEHDISSDDKYIKPD 660
S+ + ++ N N +S S S+ S ES G ++ SS+++ D
Sbjct: 819 GSNSESSNESSNQSLNSESNNDSSSHGSNSESSNNESSSHGSNNESSNNESSSQD 873
Score = 40.4 bits (93), Expect = 0.18
Identities = 42/229 (18%), Positives = 86/229 (37%), Gaps = 7/229 (3%)
Query: 416 HQQHSHHQETQKEKTREELLAEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIKLA 475
+ + SH++ + + E E + + K N + G E
Sbjct: 917 NNESSHNEPSNQSSNNESSNNESSNNESSNNESSSKGSNNESSNNESSNQGSNNESSNNE 976
Query: 476 AGVKSPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSHDSPAVTISNPDHHEQQSH-T 534
+ +S N S G SS+ S+ +++ S++ + SN +E SH +
Sbjct: 977 SNNESSNNESSSQGSNNESSNNESSNNESSSQSSNNGSSNNESSNQGSN---NESSSHGS 1033
Query: 535 NYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQENHRSHASTSPERHRSRSRSH 594
N SK ++ +S + QGS++ + + N S+ +S S+ ++
Sbjct: 1034 NNESSSKGSNNESSSHGSNNESSSQGSNNESSNN---ESSNQNSNNESSNNESSSKGSNN 1090
Query: 595 EHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESR 643
E + S + NN S ++ +HK + + +S +ES+
Sbjct: 1091 ESSNNESSSQGSNNESSNNESNNQNSNNESSHKEESESNSHQSSETESK 1139
Score = 40.0 bits (92), Expect = 0.24
Identities = 24/154 (15%), Positives = 61/154 (39%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
SSH ++ ++ E SH+ + SN + +S S+ ++++S+ +
Sbjct: 690 SSHNETSNQGSNNESSHNESSSQGSNNESAHNESSNQGSNNESSHNESSSQGSNNESSNN 749
Query: 559 QGSHHHGGDERGTDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTK 618
++ + +E ++ +++S + S S + S ++E +++
Sbjct: 750 DSNNQNSNNEPSNNESSNQSSNNLSSNNESSNNESSNQGSNNESSHSESSNNESSNQSLN 809
Query: 619 DRWQNDTHKNHTSDSFSRSDPSESRGVGEHDISS 652
+ N+ + S+S + S +D SS
Sbjct: 810 SEYNNEPSQGSNSESSNESSNQSLNSESNNDSSS 843
Score = 39.7 bits (91), Expect = 0.31
Identities = 41/247 (16%), Positives = 96/247 (38%), Gaps = 14/247 (5%)
Query: 416 HQQHSHHQETQKEKTREELLAEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIKLA 475
++ H T E + + E + + + + T + + E
Sbjct: 556 NESSDDHSSTNNESSSQGSNNESSNNESSNQGSNNESSHNETSNQGSNNESSHNESSSQG 615
Query: 476 AGVKSPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSHDSPAVTISNPD--HHE--QQ 531
+ +S N S + G SSH S+ ++ E S++ + SN + H+E Q
Sbjct: 616 SNNESAHNESSNQGSNNE-----SSHNESSNQGSNNESSNNESSNQGSNNESSHNESSNQ 670
Query: 532 SHTNYSDKSKVVHDATSRDYEQRKQGHQGSH----HHGGDERGTDQEN-HRSHASTSPER 586
N S ++ + ++ + + +QGS+ H+ +G++ E+ H ++
Sbjct: 671 GSNNESSNNESSNQGSNNESSHNETSNQGSNNESSHNESSSQGSNNESAHNESSNQGSNN 730
Query: 587 HRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVG 646
S + S ++E + + +N+ + + N + N +S++ S ++ S ++G
Sbjct: 731 ESSHNESSSQGSNNESSNNDSNNQNSNNEPSNNESSNQSSNNLSSNNESSNNESSNQGSN 790
Query: 647 EHDISSD 653
S+
Sbjct: 791 NESSHSE 797
Score = 35.8 bits (81), Expect = 4.5
Identities = 31/160 (19%), Positives = 63/160 (39%), Gaps = 14/160 (8%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
S T S+E+NNS+ SN D + Q S + ++ S + + QG
Sbjct: 490 SGQTSSSESNNSQ-----------SNEDSNHQSSGNHPTNDSSNQGSGSHSNNHSSNQGS 538
Query: 559 QGSHHHGGDE---RGTDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSS 615
+ + ++ E+ H+ST+ E S + Q + +N +S
Sbjct: 539 SSGQDSSSETLKIQNSNNESSDDHSSTNNESSSQGSNNESSNNESSNQGSNNESSHNETS 598
Query: 616 RTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHDISSDDK 655
++ +++ + S + S +ES G ++ SS ++
Sbjct: 599 NQGSNNESSHNESSSQGSNNESAHNESSNQGSNNESSHNE 638
Score = 35.4 bits (80), Expect = 5.9
Identities = 33/181 (18%), Positives = 70/181 (38%), Gaps = 19/181 (10%)
Query: 480 SPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDK 539
SP + + P PP + S T + ++NS S + + +N +E +H +
Sbjct: 458 SPPLTKKPTSPPTQPP--IQSPTEESSSHNSESSSGQTSSSESNNSQSNEDSNHQS---- 511
Query: 540 SKVVHDATSRDYEQRKQGHQGSHHHGGDE---RGTDQENHRSHASTSPERHRSRSRSHEH 596
S ++ +QGS H + +G+ S + + + S
Sbjct: 512 --------SGNHPTNDSSNQGSGSHSNNHSSNQGSSSGQDSSSETLKIQNSNNESSDDHS 563
Query: 597 RVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPS--ESRGVGEHDISSDD 654
++E + +N+ + N++ N TS+ S ++ S ES G ++ S+ +
Sbjct: 564 STNNESSSQGSNNESSNNESSNQGSNNESSHNETSNQGSNNESSHNESSSQGSNNESAHN 623
Query: 655 K 655
+
Sbjct: 624 E 624
>ref|XP_643819.1| hypothetical protein DDB0203095 [Dictyostelium discoideum]
gi|60471924|gb|EAL69878.1| hypothetical protein
DDB0203095 [Dictyostelium discoideum]
Length = 1213
Score = 55.5 bits (132), Expect = 6e-06
Identities = 53/241 (21%), Positives = 101/241 (40%), Gaps = 12/241 (4%)
Query: 418 QHSHHQETQKEKTREELLAEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIKLAAG 477
Q S++ + E + E S+ N+S+ Q L EY E +
Sbjct: 775 QSSNNLSSNNESSNNE---SSNQGSNNESSHSESSNNESSNQ---SLNSEYNNEPSQGSN 828
Query: 478 VKSPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYS 537
+S N S + + SSH ++E++N+ SH S + +N Q +NYS
Sbjct: 829 SESS-NESSNQSLNSESNNDSSSHGSNSESSNNESSSHGSNNESSNN--ESSSQDSSNYS 885
Query: 538 DKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQ--ENHRSHASTSPERHRSRSRSHE 595
+ ++ +S + +QGS++ + + Q N SH S + + S ++E
Sbjct: 886 SNNGSSNNESSSQGSNNESSNQGSNNESSNNESSSQGSNNESSHNEPSNQSSNNESSNNE 945
Query: 596 HRVHHEKQDYSGRKKYNN-SSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHDISSDD 654
+ + S K NN SS + Q +++ ++S + S +ES G ++ SS++
Sbjct: 946 SSNNESSNNESSSKGSNNESSNNESSNQGSNNESSNNESNNESSNNESSSQGSNNESSNN 1005
Query: 655 K 655
+
Sbjct: 1006 E 1006
Score = 45.4 bits (106), Expect = 0.006
Identities = 26/159 (16%), Positives = 72/159 (44%), Gaps = 2/159 (1%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
S+H S+ ++ E SH+ + SN + +S S+ ++++++ +
Sbjct: 627 SAHNESSNQGSNNESSHNESSNQGSNNESSNNESSNQGSNNESSHNESSNQGSNNESSNN 686
Query: 559 QGSHHHGGDERGTDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTK 618
+ S+ +E ++ +++ + S S S+ H+E + + +++ +
Sbjct: 687 ESSNQGSNNESSHNETSNQGSNNESSHNESSSQGSNNESAHNESSNQGSNNESSHNESSS 746
Query: 619 DRWQNDTHKNHTSDSFSRSDPS--ESRGVGEHDISSDDK 655
N++ N +++ S ++PS ES +++SS+++
Sbjct: 747 QGSNNESSNNDSNNQNSNNEPSNNESSNQSSNNLSSNNE 785
Score = 45.1 bits (105), Expect = 0.007
Identities = 33/159 (20%), Positives = 72/159 (44%), Gaps = 5/159 (3%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSH-TNYSDKSKVVHDATSRDYEQRKQG 557
S++ S + +N+ SH S + S ++E SH +N S+ ++ +S + +
Sbjct: 1022 SNNESSNQGSNNESSSHGSNNESSSKGSNNESSSHGSNNESSSQGSNNESSNNESSNQNS 1081
Query: 558 HQGSHHHGGDERGTDQENHRSHASTSPERHR-SRSRSHEHRVHHE---KQDYSGRKKYNN 613
+ S ++ +G++ E+ + +S+ + S + S+ ++E K++ ++
Sbjct: 1082 NNESSNNESSSKGSNNESSNNESSSQGSNNESSNNESNNQNSNNESSHKEESESNSHQSS 1141
Query: 614 SSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHDISS 652
+ +KD+ N N +S+S S SE E SS
Sbjct: 1142 ETESKDQVSNSGSMNQSSESSESSSLSEQSSAFESSSSS 1180
Score = 42.7 bits (99), Expect = 0.037
Identities = 37/167 (22%), Positives = 67/167 (39%), Gaps = 13/167 (7%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
S++ S +++N+ +++S SN + + SH S+ S S + E +
Sbjct: 768 SNNESSNQSSNNLSSNNESSNNESSNQGSNNESSH---SESSNNESSNQSLNSEYNNEPS 824
Query: 559 QGSHHHGGDERGTDQENHRSHASTSPERHRSRSRSHEHRVH----------HEKQDYSGR 608
QGS+ +E N S+ +S S S ++E H QD S
Sbjct: 825 QGSNSESSNESSNQSLNSESNNDSSSHGSNSESSNNESSSHGSNNESSNNESSSQDSSNY 884
Query: 609 KKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHDISSDDK 655
N SS + Q +++ S + S +ES G ++ SS ++
Sbjct: 885 SSNNGSSNNESSSQGSNNESSNQGSNNESSNNESSSQGSNNESSHNE 931
Score = 42.7 bits (99), Expect = 0.037
Identities = 27/158 (17%), Positives = 70/158 (44%), Gaps = 2/158 (1%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
SSH ++ ++ E SH+ + SN + +S S+ ++++++ +
Sbjct: 599 SSHNETSNQGSNNESSHNESSSQGSNNESAHNESSNQGSNNESSHNESSNQGSNNESSNN 658
Query: 559 QGSHHHGGDERGTDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTK 618
+ S+ +E ++ +++ + S S S+ H+E + + +++ +
Sbjct: 659 ESSNQGSNNESSHNESSNQGSNNESSNNESSNQGSNNESSHNETSNQGSNNESSHNESSS 718
Query: 619 DRWQNDTHKNHTSDSFSRSDPS--ESRGVGEHDISSDD 654
N++ N +S+ S ++ S ES G ++ SS++
Sbjct: 719 QGSNNESAHNESSNQGSNNESSHNESSSQGSNNESSNN 756
Score = 42.7 bits (99), Expect = 0.037
Identities = 28/155 (18%), Positives = 70/155 (45%), Gaps = 5/155 (3%)
Query: 504 STEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGHQGSHH 563
S++ +N+ +++S +N H + S+ +++S H+ +S + H S +
Sbjct: 577 SSQGSNNESSNNESSNQGSNNESSHNETSNQGSNNESS--HNESSSQGSNNESAHNESSN 634
Query: 564 HGGDERGTDQENHRSHASTSPERHRSRSR-SHEHRVHHEKQDYSGRKKYNNSSRTKDRWQ 622
G + + E+ ++ + S ++ S+ H+E + + +N+ +
Sbjct: 635 QGSNNESSHNESSNQGSNNESSNNESSNQGSNNESSHNESSNQGSNNESSNNESSNQGSN 694
Query: 623 NDTHKNHTSDSFSRSDPS--ESRGVGEHDISSDDK 655
N++ N TS+ S ++ S ES G ++ S+ ++
Sbjct: 695 NESSHNETSNQGSNNESSHNESSSQGSNNESAHNE 729
Score = 41.2 bits (95), Expect = 0.11
Identities = 37/175 (21%), Positives = 76/175 (43%), Gaps = 18/175 (10%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
SSH S+ ++ E +H+ SN + + SH N S +++++ D + +
Sbjct: 711 SSHNESSSQGSNNESAHNES----SNQGSNNESSH-NESSSQGSNNESSNNDSNNQNSNN 765
Query: 559 QGSHHHGGDERGTD-QENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNN---- 613
+ S++ ++ + N+ S + S + + SH ++E + S +YNN
Sbjct: 766 EPSNNESSNQSSNNLSSNNESSNNESSNQGSNNESSHSESSNNESSNQSLNSEYNNEPSQ 825
Query: 614 ------SSRTKDRWQNDTHKNHTSDSFSRSDPS--ESRGVGEHDISSDDKYIKPD 660
S+ + ++ N N +S S S+ S ES G ++ SS+++ D
Sbjct: 826 GSNSESSNESSNQSLNSESNNDSSSHGSNSESSNNESSSHGSNNESSNNESSSQD 880
Score = 40.4 bits (93), Expect = 0.18
Identities = 42/229 (18%), Positives = 86/229 (37%), Gaps = 7/229 (3%)
Query: 416 HQQHSHHQETQKEKTREELLAEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIKLA 475
+ + SH++ + + E E + + K N + G E
Sbjct: 924 NNESSHNEPSNQSSNNESSNNESSNNESSNNESSSKGSNNESSNNESSNQGSNNESSNNE 983
Query: 476 AGVKSPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSHDSPAVTISNPDHHEQQSH-T 534
+ +S N S G SS+ S+ +++ S++ + SN +E SH +
Sbjct: 984 SNNESSNNESSSQGSNNESSNNESSNNESSSQSSNNGSSNNESSNQGSN---NESSSHGS 1040
Query: 535 NYSDKSKVVHDATSRDYEQRKQGHQGSHHHGGDERGTDQENHRSHASTSPERHRSRSRSH 594
N SK ++ +S + QGS++ + + N S+ +S S+ ++
Sbjct: 1041 NNESSSKGSNNESSSHGSNNESSSQGSNNESSNN---ESSNQNSNNESSNNESSSKGSNN 1097
Query: 595 EHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESR 643
E + S + NN S ++ +HK + + +S +ES+
Sbjct: 1098 ESSNNESSSQGSNNESSNNESNNQNSNNESSHKEESESNSHQSSETESK 1146
Score = 40.0 bits (92), Expect = 0.24
Identities = 24/154 (15%), Positives = 61/154 (39%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
SSH ++ ++ E SH+ + SN + +S S+ ++++S+ +
Sbjct: 697 SSHNETSNQGSNNESSHNESSSQGSNNESAHNESSNQGSNNESSHNESSSQGSNNESSNN 756
Query: 559 QGSHHHGGDERGTDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTK 618
++ + +E ++ +++S + S S + S ++E +++
Sbjct: 757 DSNNQNSNNEPSNNESSNQSSNNLSSNNESSNNESSNQGSNNESSHSESSNNESSNQSLN 816
Query: 619 DRWQNDTHKNHTSDSFSRSDPSESRGVGEHDISS 652
+ N+ + S+S + S +D SS
Sbjct: 817 SEYNNEPSQGSNSESSNESSNQSLNSESNNDSSS 850
Score = 39.7 bits (91), Expect = 0.31
Identities = 41/247 (16%), Positives = 96/247 (38%), Gaps = 14/247 (5%)
Query: 416 HQQHSHHQETQKEKTREELLAEERDYKRRRMSYRGKKRNQSTIQVMRDLIGEYMEEIKLA 475
++ H T E + + E + + + + T + + E
Sbjct: 563 NESSDDHSSTNNESSSQGSNNESSNNESSNQGSNNESSHNETSNQGSNNESSHNESSSQG 622
Query: 476 AGVKSPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSHDSPAVTISNPD--HHE--QQ 531
+ +S N S + G SSH S+ ++ E S++ + SN + H+E Q
Sbjct: 623 SNNESAHNESSNQGSNNE-----SSHNESSNQGSNNESSNNESSNQGSNNESSHNESSNQ 677
Query: 532 SHTNYSDKSKVVHDATSRDYEQRKQGHQGSH----HHGGDERGTDQEN-HRSHASTSPER 586
N S ++ + ++ + + +QGS+ H+ +G++ E+ H ++
Sbjct: 678 GSNNESSNNESSNQGSNNESSHNETSNQGSNNESSHNESSSQGSNNESAHNESSNQGSNN 737
Query: 587 HRSRSRSHEHRVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPSESRGVG 646
S + S ++E + + +N+ + + N + N +S++ S ++ S ++G
Sbjct: 738 ESSHNESSSQGSNNESSNNDSNNQNSNNEPSNNESSNQSSNNLSSNNESSNNESSNQGSN 797
Query: 647 EHDISSD 653
S+
Sbjct: 798 NESSHSE 804
Score = 35.8 bits (81), Expect = 4.5
Identities = 31/160 (19%), Positives = 63/160 (39%), Gaps = 14/160 (8%)
Query: 499 SSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDKSKVVHDATSRDYEQRKQGH 558
S T S+E+NNS+ SN D + Q S + ++ S + + QG
Sbjct: 497 SGQTSSSESNNSQ-----------SNEDSNHQSSGNHPTNDSSNQGSGSHSNNHSSNQGS 545
Query: 559 QGSHHHGGDE---RGTDQENHRSHASTSPERHRSRSRSHEHRVHHEKQDYSGRKKYNNSS 615
+ + ++ E+ H+ST+ E S + Q + +N +S
Sbjct: 546 SSGQDSSSETLKIQNSNNESSDDHSSTNNESSSQGSNNESSNNESSNQGSNNESSHNETS 605
Query: 616 RTKDRWQNDTHKNHTSDSFSRSDPSESRGVGEHDISSDDK 655
++ +++ + S + S +ES G ++ SS ++
Sbjct: 606 NQGSNNESSHNESSSQGSNNESAHNESSNQGSNNESSHNE 645
Score = 35.4 bits (80), Expect = 5.9
Identities = 33/181 (18%), Positives = 70/181 (38%), Gaps = 19/181 (10%)
Query: 480 SPVNVSEDSGMPPPPPKLLSSHTISTEANNSREVSHDSPAVTISNPDHHEQQSHTNYSDK 539
SP + + P PP + S T + ++NS S + + +N +E +H +
Sbjct: 465 SPPLTKKPTSPPTQPP--IQSPTEESSSHNSESSSGQTSSSESNNSQSNEDSNHQS---- 518
Query: 540 SKVVHDATSRDYEQRKQGHQGSHHHGGDE---RGTDQENHRSHASTSPERHRSRSRSHEH 596
S ++ +QGS H + +G+ S + + + S
Sbjct: 519 --------SGNHPTNDSSNQGSGSHSNNHSSNQGSSSGQDSSSETLKIQNSNNESSDDHS 570
Query: 597 RVHHEKQDYSGRKKYNNSSRTKDRWQNDTHKNHTSDSFSRSDPS--ESRGVGEHDISSDD 654
++E + +N+ + N++ N TS+ S ++ S ES G ++ S+ +
Sbjct: 571 STNNESSSQGSNNESSNNESSNQGSNNESSHNETSNQGSNNESSHNESSSQGSNNESAHN 630
Query: 655 K 655
+
Sbjct: 631 E 631
>gb|AAG15240.1| ENOD2 [Styphnolobium japonicum]
Length = 269
Score = 55.5 bits (132), Expect = 6e-06
Identities = 33/106 (31%), Positives = 43/106 (40%), Gaps = 17/106 (16%)
Query: 18 PTMNPPPAQLPPPSQLPPP-LSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILS 76
P PPP + PPP PPP + PPP PPP + PPP
Sbjct: 43 PEYQPPPHEKPPPEYQPPPHVKPPPAYQPPPHVKPPPVYQ--------------PPHHEK 88
Query: 77 TTPQPQTPTTNLIPCLFNPNHLIPPPSLF--LHHLRCPSSPRPLPH 120
P+ Q P P + P H + PP ++ HH + P +P PH
Sbjct: 89 PPPEYQPPPHEKPPPEYQPPHHVKPPPVYPPPHHEKPPPVHQPPPH 134
Score = 50.8 bits (120), Expect = 1e-04
Identities = 30/101 (29%), Positives = 41/101 (39%), Gaps = 22/101 (21%)
Query: 22 PPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTPQP 81
PPPA PP ++PPP+ P P + PPP PPP P+
Sbjct: 18 PPPAYPPPHHEIPPPVYPSPHEKPPPEYQPPPH--------------------EKPPPEY 57
Query: 82 QTPTTNLIPCLFNPNHLIPPPSLFL--HHLRCPSSPRPLPH 120
Q P P + P + PP ++ HH + P +P PH
Sbjct: 58 QPPPHVKPPPAYQPPPHVKPPPVYQPPHHEKPPPEYQPPPH 98
Score = 50.1 bits (118), Expect = 2e-04
Identities = 20/36 (55%), Positives = 24/36 (66%), Gaps = 1/36 (2%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPP-QQHPPPLLPPP 52
P + PPPA PPP + PPP+ PPP + PPP PPP
Sbjct: 230 PHVKPPPAYQPPPHEKPPPVYPPPHHEKPPPAYPPP 265
Score = 48.5 bits (114), Expect = 7e-04
Identities = 33/105 (31%), Positives = 44/105 (41%), Gaps = 8/105 (7%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPP-QQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILS 76
P PPP PP PPP+ PPP + PPP+ PPP ++ +
Sbjct: 97 PHEKPPPEYQPPHHVKPPPVYPPPHHEKPPPVHQPPPHEKPPPEYQPPPHVK--PPPVYQ 154
Query: 77 TTPQPQTPTTNLIPCLFNPNHLIPPPSLF-LHHLRCPSSPRPLPH 120
P + P P P+H IPPP HH++ P +P PH
Sbjct: 155 PPPHVKPPPVYQPP----PHHEIPPPEYQPPHHVKPPPVYQPPPH 195
Score = 45.8 bits (107), Expect = 0.004
Identities = 31/102 (30%), Positives = 37/102 (35%), Gaps = 20/102 (19%)
Query: 20 MNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPPLNLNTTLSSLTNLLTFSTQILSTTP 79
+ PPP PPP + PPP+ PP PP + PPP N P
Sbjct: 184 VKPPPVYQPPPHEKPPPVYQPPHHEIPPPVYPPPREN-------------------KPPP 224
Query: 80 QPQTPTTNLIPCLFN-PNHLIPPPSLFLHHLRCPSSPRPLPH 120
+ Q P P + P H PPP H P P PH
Sbjct: 225 EYQPPPHVKPPPAYQPPPHEKPPPVYPPPHHEKPPPAYPPPH 266
Score = 45.4 bits (106), Expect = 0.006
Identities = 18/37 (48%), Positives = 24/37 (64%), Gaps = 1/37 (2%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQH-PPPLLPPPP 53
P PPP PP ++PPP+ PPP+++ PPP PPP
Sbjct: 194 PHEKPPPVYQPPHHEIPPPVYPPPRENKPPPEYQPPP 230
Score = 44.3 bits (103), Expect = 0.013
Identities = 18/37 (48%), Positives = 20/37 (53%)
Query: 18 PTMNPPPAQLPPPSQLPPPLSPPPQQHPPPLLPPPPP 54
P PPP PPP+ PPP PP +PPP PPP
Sbjct: 224 PEYQPPPHVKPPPAYQPPPHEKPPPVYPPPHHEKPPP 260
Score = 42.0 bits (97), Expect = 0.063
Identities = 18/42 (42%), Positives = 20/42 (46%), Gaps = 7/42 (16%)
Query: 18 PTMNPPPAQLPPPSQLPPP-------LSPPPQQHPPPLLPPP 52
P PP PPP PPP PPP + PPP+ PPP
Sbjct: 212 PVYPPPRENKPPPEYQPPPHVKPPPAYQPPPHEKPPPVYPPP 253
Score = 40.8 bits (94), Expect = 0.14
Identities = 17/37 (45%), Positives = 21/37 (55%), Gaps = 1/37 (2%)
Query: 18 PTMNPPPAQLPPPSQLPP-PLSPPPQQHPPPLLPPPP 53
P PP ++PPP PP PPP+ PPP + PPP
Sbjct: 200 PVYQPPHHEIPPPVYPPPRENKPPPEYQPPPHVKPPP 236
Score = 38.9 bits (89), Expect = 0.54
Identities = 18/38 (47%), Positives = 21/38 (54%), Gaps = 2/38 (5%)
Query: 18 PTMNPPPA-QLPPPSQLPP-PLSPPPQQHPPPLLPPPP 53
P PPP ++PPP PP + PPP PPP PPP
Sbjct: 163 PVYQPPPHHEIPPPEYQPPHHVKPPPVYQPPPHEKPPP 200
>gb|EAA15226.1| developmental protein DG1037 [Plasmodium yoelii yoelii]
Length = 345
Score = 55.5 bits (132), Expect = 6e-06
Identities = 37/127 (29%), Positives = 54/127 (42%), Gaps = 12/127 (9%)
Query: 39 PPPQQHPPPLLPPPPPL-----NLNTTLSSLTNLLTFSTQILSTTPQPQTPTTNLIPCLF 93
PPP HPPP PPPPP N L TN + I+S T P N
Sbjct: 84 PPPPTHPPP--PPPPPFIPKEKQQNWKLKKKTNNSDINNDIISITTYPANSNNNNNNNNN 141
Query: 94 NPNHLIPPPSLFLHHLRCPSSPRPLPH-----IDHLLTSLSYPKTLQNPTSIHLSSYLDF 148
NP++ I P ++ L PS P P P +H +++++ P ++L + ++
Sbjct: 142 NPHYRINEPHMYNKSLSIPSIPGPTPPYPSNIYNHPVSNINSPPCTNTNIKVNLQTNHNY 201
Query: 149 TNFFYNN 155
N F N
Sbjct: 202 NNNFKKN 208
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.132 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,288,374,179
Number of Sequences: 2540612
Number of extensions: 63691467
Number of successful extensions: 753311
Number of sequences better than 10.0: 7348
Number of HSP's better than 10.0 without gapping: 2204
Number of HSP's successfully gapped in prelim test: 5661
Number of HSP's that attempted gapping in prelim test: 547807
Number of HSP's gapped (non-prelim): 74208
length of query: 661
length of database: 863,360,394
effective HSP length: 135
effective length of query: 526
effective length of database: 520,377,774
effective search space: 273718709124
effective search space used: 273718709124
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 79 (35.0 bits)
Medicago: description of AC146559.13


