

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146308.4 + phase: 0
(357 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
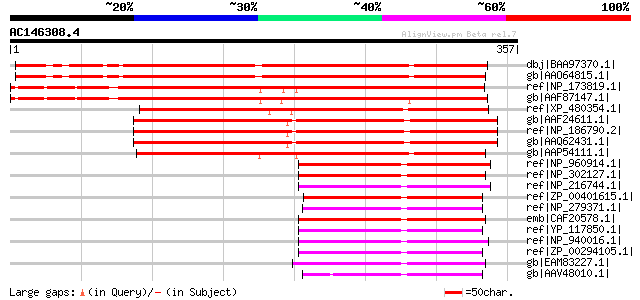
dbj|BAA97370.1| unnamed protein product [Arabidopsis thaliana] 310 6e-83
gb|AAO64815.1| At5g51080 [Arabidopsis thaliana] gi|30695999|ref|... 307 3e-82
ref|NP_173819.1| RNase H domain-containing protein [Arabidopsis ... 291 2e-77
gb|AAF87147.1| T23E23.24 [Arabidopsis thaliana] 290 5e-77
ref|XP_480354.1| putative RNase H domain-containing protein [Ory... 280 5e-74
gb|AAF24611.1| putative RNase H [Arabidopsis thaliana] 259 1e-67
ref|NP_186790.2| RNase H domain-containing protein [Arabidopsis ... 259 1e-67
gb|AAQ62431.1| At3g01410 [Arabidopsis thaliana] gi|51970934|dbj|... 258 3e-67
gb|AAP54111.1| putative RNase [Oryza sativa (japonica cultivar-g... 250 4e-65
ref|NP_960914.1| hypothetical protein MAP1980c [Mycobacterium av... 98 3e-19
ref|NP_302127.1| hypothetical protein ML1637 [Mycobacterium lepr... 98 4e-19
ref|NP_216744.1| hypothetical protein Rv2228c [Mycobacterium tub... 96 2e-18
ref|ZP_00401615.1| RNase H [Anaeromyxobacter dehalogenans 2CP-C]... 94 6e-18
ref|NP_279371.1| hypothetical protein VNG0255C [Halobacterium sp... 92 3e-17
emb|CAF20578.1| Ribonuclease HI [Corynebacterium glutamicum ATCC... 91 7e-17
ref|YP_117850.1| putative phosphoglycerate mutase [Nocardia farc... 90 1e-16
ref|NP_940016.1| hypothetical protein DIP1678 [Corynebacterium d... 89 2e-16
ref|ZP_00294105.1| COG0406: Fructose-2,6-bisphosphatase [Thermob... 89 3e-16
gb|EAM83227.1| Phosphoglycerate/bisphosphoglycerate mutase:RNase... 88 3e-16
gb|AAV48010.1| ribonuclease H-like protein [Haloarcula marismort... 86 1e-15
>dbj|BAA97370.1| unnamed protein product [Arabidopsis thaliana]
Length = 316
Score = 310 bits (793), Expect = 6e-83
Identities = 170/332 (51%), Positives = 223/332 (66%), Gaps = 19/332 (5%)
Query: 5 FSHLSSFTSTILARTTQFVANRSLHTTPHTFSFPVKSHHSLLTSFHSQFVLTTVTRCYST 64
FS S+ S +L R + +V + P F + SL +S V + CYS+
Sbjct: 4 FSRARSYISLVLFRKSSYVTSH----IPWNQCF----YTSLKSSLKPASVSVSSVHCYSS 55
Query: 65 RKLRKTGSSKLHKVEAETTMNQEQNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPP 124
R KT SK+ K + +++ E DAF+VVRKGD+VGIY L D QAQVGSSV DPP
Sbjct: 56 RS--KTAKSKMSK--SSVSVSDSDKEKDAFFVVRKGDIVGIYKDLIDCQAQVGSSVYDPP 111
Query: 125 VSVYKGYSMSNETEEYLLSHGLKNALYTIRASDLKEDLFGTLVPCPFQDPSSTQGTTSNA 184
VSVYKGYS+ +TEE L + GLK LY RA DLKED+FG L PC FQD Q +++
Sbjct: 112 VSVYKGYSLLKDTEECLSTVGLKKPLYVFRALDLKEDMFGALTPCLFQD----QLPSASM 167
Query: 185 DSSKKRALEVLEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATN 244
K LE + +TCI+EFDGASKGNPG +GA A+L+++DG+LI+++R+G+GIATN
Sbjct: 168 SVEKLAELEPSADTSYETCIIEFDGASKGNPGLSGAAAVLKTEDGSLIFKMRQGLGIATN 227
Query: 245 NVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKEL 304
N AEY +ILG++HA++KG+T I ++ DSKLVCMQMK G WKV +E LS L+K AK+L
Sbjct: 228 NAAEYHGLILGLKHAIEKGYTKIKVKTDSKLVCMQMK---GQWKVNHEVLSKLHKEAKQL 284
Query: 305 KDKFVSFKISHVLRDLNSEADAQANLAVNLAG 336
DK +SF+ISHVLR LNS+AD QAN+A L+G
Sbjct: 285 SDKCLSFEISHVLRSLNSDADEQANMAARLSG 316
>gb|AAO64815.1| At5g51080 [Arabidopsis thaliana] gi|30695999|ref|NP_199921.2| RNase
H domain-containing protein [Arabidopsis thaliana]
Length = 322
Score = 307 bits (787), Expect = 3e-82
Identities = 169/331 (51%), Positives = 222/331 (67%), Gaps = 19/331 (5%)
Query: 5 FSHLSSFTSTILARTTQFVANRSLHTTPHTFSFPVKSHHSLLTSFHSQFVLTTVTRCYST 64
FS S+ S +L R + +V + P F + SL +S V + CYS+
Sbjct: 4 FSRARSYISLVLFRKSSYVTSH----IPWNQCF----YTSLKSSLKPASVSVSSVHCYSS 55
Query: 65 RKLRKTGSSKLHKVEAETTMNQEQNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPP 124
R KT SK+ K + +++ E DAF+VVRKGD+VGIY L D QAQVGSSV DPP
Sbjct: 56 RS--KTAKSKMSK--SSVSVSDSDKEKDAFFVVRKGDIVGIYKDLIDCQAQVGSSVYDPP 111
Query: 125 VSVYKGYSMSNETEEYLLSHGLKNALYTIRASDLKEDLFGTLVPCPFQDPSSTQGTTSNA 184
VSVYKGYS+ +TEE L + GLK LY RA DLKED+FG L PC FQD Q +++
Sbjct: 112 VSVYKGYSLLKDTEECLSTVGLKKPLYVFRALDLKEDMFGALTPCLFQD----QLPSASM 167
Query: 185 DSSKKRALEVLEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATN 244
K LE + +TCI+EFDGASKGNPG +GA A+L+++DG+LI+++R+G+GIATN
Sbjct: 168 SVEKLAELEPSADTSYETCIIEFDGASKGNPGLSGAAAVLKTEDGSLIFKMRQGLGIATN 227
Query: 245 NVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKEL 304
N AEY +ILG++HA++KG+T I ++ DSKLVCMQMK G WKV +E LS L+K AK+L
Sbjct: 228 NAAEYHGLILGLKHAIEKGYTKIKVKTDSKLVCMQMK---GQWKVNHEVLSKLHKEAKQL 284
Query: 305 KDKFVSFKISHVLRDLNSEADAQANLAVNLA 335
DK +SF+ISHVLR LNS+AD QAN+A L+
Sbjct: 285 SDKCLSFEISHVLRSLNSDADEQANMAARLS 315
>ref|NP_173819.1| RNase H domain-containing protein [Arabidopsis thaliana]
Length = 535
Score = 291 bits (745), Expect = 2e-77
Identities = 173/358 (48%), Positives = 232/358 (64%), Gaps = 34/358 (9%)
Query: 1 MNALFSHLSSFTSTILARTTQFVANRSLHTTPHTFSFPVKSHHSLLTSFHSQFVLTTVTR 60
MN L SH S+ + L + + +V+ S+ F P KS + + S F + +V
Sbjct: 1 MNCL-SHARSYIALGLLKRSSYVS--SIPWNECFFYMPSKSCLKPV-AVSSVFGICSVHS 56
Query: 61 CYSTRKLRKTGSSKLHKVEAETTMNQEQNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSV 120
S K K+ K+ + T ++ E DAF+VVRKGDV+GIY L+D QAQVGSSV
Sbjct: 57 YSSRSKAVKS------KMLSSTVVSAVDKEKDAFFVVRKGDVIGIYKDLSDCQAQVGSSV 110
Query: 121 CDPPVSVYKGYSMSNETEEYLLSHGLKNALYTIRASDLKEDLFGTLVPCPFQDPS----- 175
D PVSVYKGYS+ +TEEYL S GLK LY++RASDLK+D+FG L PC FQ+P+
Sbjct: 111 FDLPVSVYKGYSLPKDTEEYLSSVGLKKPLYSLRASDLKDDMFGALTPCLFQEPAPCTVK 170
Query: 176 ----STQGTTSNADSSKKRA---------LEVLEQNNV------KTCIVEFDGASKGNPG 216
T T + D K + LE L + +TC +EFDGASKGNPG
Sbjct: 171 VSEDETTSETKSKDDKKDQLPSASISYDPLEKLSKVEPSAYISDETCFIEFDGASKGNPG 230
Query: 217 KAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLV 276
+GA A+L+++DG+LI RVR+G+GIATNN AEY A+ILG+++A++KG+ +I ++GDSKLV
Sbjct: 231 LSGAAAVLKTEDGSLICRVRQGLGIATNNAAEYHALILGLKYAIEKGYKNIKVKGDSKLV 290
Query: 277 CMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNL 334
CMQ +QI G WKV +E L+ L+K AK L +K VSF+ISHVLR+LN++AD QANLAV L
Sbjct: 291 CMQKQQIKGQWKVNHEVLAKLHKEAKLLCNKCVSFEISHVLRNLNADADEQANLAVRL 348
>gb|AAF87147.1| T23E23.24 [Arabidopsis thaliana]
Length = 360
Score = 290 bits (742), Expect = 5e-77
Identities = 173/370 (46%), Positives = 236/370 (63%), Gaps = 44/370 (11%)
Query: 1 MNALFSHLSSFTSTILARTTQFVANRSLHTTPHTFSFPVKSHHSLLTSFHSQFVLTTVTR 60
MN L SH S+ + L + + +V+ S+ F P KS + + S F + +V
Sbjct: 1 MNCL-SHARSYIALGLLKRSSYVS--SIPWNECFFYMPSKSCLKPV-AVSSVFGICSVHS 56
Query: 61 CYSTRKLRKTGSSKLHKVEAETTMNQEQNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSV 120
S K K+ K+ + T ++ E DAF+VVRKGDV+GIY L+D QAQVGSSV
Sbjct: 57 YSSRSKAVKS------KMLSSTVVSAVDKEKDAFFVVRKGDVIGIYKDLSDCQAQVGSSV 110
Query: 121 CDPPVSVYKGYSMSNETEEYLLSHGLKNALYTIRASDLKEDLFGTLVPCPFQDPS----- 175
D PVSVYKGYS+ +TEEYL S GLK LY++RASDLK+D+FG L PC FQ+P+
Sbjct: 111 FDLPVSVYKGYSLPKDTEEYLSSVGLKKPLYSLRASDLKDDMFGALTPCLFQEPAPCTVK 170
Query: 176 ----STQGTTSNADSSKKR------ALEVLEQNNVKTCIVEFDGASKGNPGKAGAGAILR 225
T T + D K + + + LE+ + +TC +EFDGASKGNPG +GA A+L+
Sbjct: 171 VSEDETTSETKSKDDKKDQLPSASISYDPLEKLSKETCFIEFDGASKGNPGLSGAAAVLK 230
Query: 226 SKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQM----- 280
++DG+LI RVR+G+GIATNN AEY A+ILG+++A++KG+ +I ++GDSKLVCMQ+
Sbjct: 231 TEDGSLICRVRQGLGIATNNAAEYHALILGLKYAIEKGYKNIKVKGDSKLVCMQVSLMNH 290
Query: 281 --------------KQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADA 326
+QI G WKV +E L+ L+K AK L +K VSF+ISHVLR+LN++AD
Sbjct: 291 IRFYLLTFSTLSEKQQIKGQWKVNHEVLAKLHKEAKLLCNKCVSFEISHVLRNLNADADE 350
Query: 327 QANLAVNLAG 336
QANLAV L G
Sbjct: 351 QANLAVRLPG 360
>ref|XP_480354.1| putative RNase H domain-containing protein [Oryza sativa (japonica
cultivar-group)] gi|38636983|dbj|BAD03243.1| putative
RNase H domain-containing protein [Oryza sativa
(japonica cultivar-group)] gi|38636806|dbj|BAD03047.1|
putative RNase H domain-containing protein [Oryza sativa
(japonica cultivar-group)]
Length = 351
Score = 280 bits (716), Expect = 5e-74
Identities = 141/255 (55%), Positives = 187/255 (73%), Gaps = 12/255 (4%)
Query: 92 DAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPPVSVYKGYSMSNETEEYLLSHGLKNALY 151
+ FYVVRKGDV+GIY +L+D QAQV +SVCDP V+VYKGYS+ ETEEYL + GL+N LY
Sbjct: 86 EPFYVVRKGDVIGIYKSLSDCQAQVSNSVCDPSVTVYKGYSLRKETEEYLAARGLRNPLY 145
Query: 152 TIRASDLKEDLFGTLVPCPFQDPSSTQGTT-----SNADSSKKRALEVLEQ----NNVKT 202
+I A+D +++LF LVPCPFQ P T +T K+ +V EQ N+ +
Sbjct: 146 SINAADARDELFDDLVPCPFQQPDGTGTSTLKRPLEMETGPSKKQPKVSEQEPLPNSSLS 205
Query: 203 CIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKK 262
C++EFDGASKGNPGKAGAGA++R DG +I ++REG+GIATNN AEYRA+ILG+ +A KK
Sbjct: 206 CLLEFDGASKGNPGKAGAGAVIRRLDGTVIAQLREGLGIATNNAAEYRALILGLTYAAKK 265
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
GF I QGDSKLVC Q+ +W+ +++ ++ L K KE+K +F +F+I+HVLR+ N+
Sbjct: 266 GFKYIRAQGDSKLVC---NQVSDVWRARHDTMADLCKRVKEIKGRFHTFQINHVLREFNT 322
Query: 323 EADAQANLAVNLAGE 337
+ADAQANLAV L GE
Sbjct: 323 DADAQANLAVELPGE 337
>gb|AAF24611.1| putative RNase H [Arabidopsis thaliana]
Length = 290
Score = 259 bits (661), Expect = 1e-67
Identities = 134/292 (45%), Positives = 185/292 (62%), Gaps = 41/292 (14%)
Query: 88 QNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPPVSVYKGYSMSNETEEYLLSHGLK 147
++E DAFY+VRKGD++G+Y +L++ Q Q GSSV P +SVYKGY E+ L S G+K
Sbjct: 2 EDEKDAFYIVRKGDIIGVYRSLSECQGQAGSSVSHPAMSVYKGYGWPKGAEDLLSSFGIK 61
Query: 148 NALYTIRASDLKEDLFGTLVPCPFQDPSSTQGTTSNADSSKKRALEV------------- 194
NAL+++ AS +K+D FG L+PCP Q PSS+QG + N S KR ++
Sbjct: 62 NALFSVNASHVKDDAFGKLIPCPVQQPSSSQGESLNKSSPSKRLQDMGSGESGSFSPSPP 121
Query: 195 -----------------------LEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNL 231
+ QN+ +C +EFDGASKGNPGKAGAGA+LR+ D ++
Sbjct: 122 QKQLKIENDMLRRIPSSLLTRTPIRQND--SCTIEFDGASKGNPGKAGAGAVLRASDNSV 179
Query: 232 IYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKN 291
++ +REGVG ATNNVAEYRA++LG+R AL KGF ++ + GDS LVCM Q+ G WK +
Sbjct: 180 LFYLREGVGNATNNVAEYRALLLGLRSALDKGFKNVHVLGDSMLVCM---QVQGAWKTNH 236
Query: 292 ENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNLAGEAFKIIN 343
++ L K AKEL + F +F I H+ R+ NSEAD QAN A+ LA ++I+
Sbjct: 237 PKMAELCKQAKELMNSFKTFDIKHIAREKNSEADKQANSAIFLADGQTQVIS 288
>ref|NP_186790.2| RNase H domain-containing protein [Arabidopsis thaliana]
Length = 294
Score = 259 bits (661), Expect = 1e-67
Identities = 134/292 (45%), Positives = 185/292 (62%), Gaps = 41/292 (14%)
Query: 88 QNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPPVSVYKGYSMSNETEEYLLSHGLK 147
++E DAFY+VRKGD++G+Y +L++ Q Q GSSV P +SVYKGY E+ L S G+K
Sbjct: 6 EDEKDAFYIVRKGDIIGVYRSLSECQGQAGSSVSHPAMSVYKGYGWPKGAEDLLSSFGIK 65
Query: 148 NALYTIRASDLKEDLFGTLVPCPFQDPSSTQGTTSNADSSKKRALEV------------- 194
NAL+++ AS +K+D FG L+PCP Q PSS+QG + N S KR ++
Sbjct: 66 NALFSVNASHVKDDAFGKLIPCPVQQPSSSQGESLNKSSPSKRLQDMGSGESGSFSPSPP 125
Query: 195 -----------------------LEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNL 231
+ QN+ +C +EFDGASKGNPGKAGAGA+LR+ D ++
Sbjct: 126 QKQLKIENDMLRRIPSSLLTRTPIRQND--SCTIEFDGASKGNPGKAGAGAVLRASDNSV 183
Query: 232 IYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKN 291
++ +REGVG ATNNVAEYRA++LG+R AL KGF ++ + GDS LVCM Q+ G WK +
Sbjct: 184 LFYLREGVGNATNNVAEYRALLLGLRSALDKGFKNVHVLGDSMLVCM---QVQGAWKTNH 240
Query: 292 ENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNLAGEAFKIIN 343
++ L K AKEL + F +F I H+ R+ NSEAD QAN A+ LA ++I+
Sbjct: 241 PKMAELCKQAKELMNSFKTFDIKHIAREKNSEADKQANSAIFLADGQTQVIS 292
>gb|AAQ62431.1| At3g01410 [Arabidopsis thaliana] gi|51970934|dbj|BAD44159.1|
putative RNase H [Arabidopsis thaliana]
Length = 294
Score = 258 bits (658), Expect = 3e-67
Identities = 133/292 (45%), Positives = 185/292 (62%), Gaps = 41/292 (14%)
Query: 88 QNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPPVSVYKGYSMSNETEEYLLSHGLK 147
++E DAFY+VRKGD++G+Y +L++ Q Q GSSV P +SVYKGY E+ L S G++
Sbjct: 6 EDEKDAFYIVRKGDIIGVYRSLSECQGQAGSSVSHPAMSVYKGYGWPKGAEDLLSSFGIR 65
Query: 148 NALYTIRASDLKEDLFGTLVPCPFQDPSSTQGTTSNADSSKKRALEV------------- 194
NAL+++ AS +K+D FG L+PCP Q PSS+QG + N S KR ++
Sbjct: 66 NALFSVNASHVKDDAFGKLIPCPVQQPSSSQGESLNKSSPSKRLQDMGSGESGSFSPSPP 125
Query: 195 -----------------------LEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNL 231
+ QN+ +C +EFDGASKGNPGKAGAGA+LR+ D ++
Sbjct: 126 QKQLKIENDMLRRIPSSLLTRTPIRQND--SCTIEFDGASKGNPGKAGAGAVLRASDNSV 183
Query: 232 IYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKN 291
++ +REGVG ATNNVAEYRA++LG+R AL KGF ++ + GDS LVCM Q+ G WK +
Sbjct: 184 LFYLREGVGNATNNVAEYRALLLGLRSALDKGFKNVHVLGDSMLVCM---QVQGAWKTNH 240
Query: 292 ENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNLAGEAFKIIN 343
++ L K AKEL + F +F I H+ R+ NSEAD QAN A+ LA ++I+
Sbjct: 241 PKMAELCKQAKELMNSFKTFDIKHIAREKNSEADKQANSAIFLADGQTQVIS 292
>gb|AAP54111.1| putative RNase [Oryza sativa (japonica cultivar-group)]
gi|37535044|ref|NP_921824.1| putative RNase [Oryza
sativa (japonica cultivar-group)]
gi|14140290|gb|AAK54296.1| putative RNase [Oryza sativa
(japonica cultivar-group)]
Length = 289
Score = 250 bits (639), Expect = 4e-65
Identities = 131/279 (46%), Positives = 176/279 (62%), Gaps = 36/279 (12%)
Query: 90 ESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPPVSVYKGYSMSNETEEYLLSHGLKNA 149
E +Y+VRKG++V +Y +L D QAQ+ SSV P S YKG S S E EEYL S GL NA
Sbjct: 3 EGSNYYLVRKGEMVAVYKSLNDCQAQICSSVSGPAASAYKGNSWSREKEEYLSSRGLSNA 62
Query: 150 LYTIRASDLKEDLFGTLVPCPFQDP------------------SSTQGTTSNADSSKKRA 191
Y I A++L+EDLFGTL+PC FQD S +G N S+ +
Sbjct: 63 TYVINAAELREDLFGTLIPCTFQDAVGSGQASAEHYSQRINQGYSVRGQAFNRLESRPSS 122
Query: 192 LEVLEQNNV---------------KTCIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVR 236
NN+ C++ FDGASKGNPGKAGAGA+L ++DG +I R+R
Sbjct: 123 SSHFSPNNLDQSGTVDAQPLSKQYMVCLLHFDGASKGNPGKAGAGAVLMTEDGRVISRLR 182
Query: 237 EGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLST 296
EG+GI TNNVAEYR +ILG+R+A++ GF I + GDS+LVC Q+K G W+ KN+N+
Sbjct: 183 EGLGIVTNNVAEYRGLILGLRYAIRHGFKKIIVYGDSQLVCYQVK---GTWQTKNQNMME 239
Query: 297 LYKVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNLA 335
L K ++LK+ FVSF+I+H+ R+ N+EAD QAN+A+ L+
Sbjct: 240 LCKEVRKLKENFVSFEINHIRREWNAEADRQANIAITLS 278
>ref|NP_960914.1| hypothetical protein MAP1980c [Mycobacterium avium subsp.
paratuberculosis str. k10] gi|41396433|gb|AAS04297.1|
hypothetical protein MAP1980c [Mycobacterium avium
subsp. paratuberculosis str. k10]
Length = 377
Score = 98.2 bits (243), Expect = 3e-19
Identities = 55/136 (40%), Positives = 84/136 (61%), Gaps = 4/136 (2%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKD-GNLIYRVREGVGIATNNVAEYRAMILGMRHALKK 262
++E DG S+GNPG AG GA++ + D G ++ ++ +G ATNNVAEYR ++ G+ ALK
Sbjct: 4 VIEADGGSRGNPGPAGYGAVVWTPDRGTVLAENKQAIGRATNNVAEYRGLLAGLGDALKL 63
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
G T + DSKL+ ++Q+ G WKVK+ +L L+ A+ L +F + + R+ NS
Sbjct: 64 GATEAEVYLDSKLL---VEQMSGRWKVKHPDLIELHAQARSLAARFDRISYTWIPRERNS 120
Query: 323 EADAQANLAVNLAGEA 338
AD AN A++ A A
Sbjct: 121 HADRLANEAMDAAAAA 136
>ref|NP_302127.1| hypothetical protein ML1637 [Mycobacterium leprae TN]
gi|13093416|emb|CAC30588.1| conserved hypothetical
protein [Mycobacterium leprae]
gi|3150239|emb|CAA19219.1| hypothetical protein
MLCB1243.38 [Mycobacterium leprae]
gi|11356567|pir||T44721 hypothetical protein MLCB1243.38
[imported] - Mycobacterium leprae
Length = 371
Score = 97.8 bits (242), Expect = 4e-19
Identities = 57/133 (42%), Positives = 81/133 (60%), Gaps = 4/133 (3%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKDGNLIY-RVREGVGIATNNVAEYRAMILGMRHALKK 262
I+E DG S+GNPG AG GA++ D + + ++ +G ATNNVAEYRA+I G+ A+K
Sbjct: 4 IIEADGGSRGNPGPAGYGAVVWIADRSAVLTETKQAIGRATNNVAEYRALIAGLDDAVKM 63
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
G T + DSKLV ++Q+ G WKVK+ +L LY A+ L + S + + R NS
Sbjct: 64 GATEAEVLMDSKLV---VEQMSGRWKVKHPDLIELYVHAQTLASRLASVSYTWIPRTRNS 120
Query: 323 EADAQANLAVNLA 335
AD AN A++ A
Sbjct: 121 RADRLANEAMDAA 133
>ref|NP_216744.1| hypothetical protein Rv2228c [Mycobacterium tuberculosis H37Rv]
gi|31793409|ref|NP_855902.1| hypothetical protein
Mb2253c [Mycobacterium bovis AF2122/97]
gi|1261930|emb|CAA94651.1| CONSERVED HYPOTHETICAL
PROTEIN [Mycobacterium tuberculosis H37Rv]
gi|31619001|emb|CAD97106.1| CONSERVED HYPOTHETICAL
PROTEIN [Mycobacterium bovis AF2122/97]
gi|13881979|gb|AAK46573.1| phosphoglycerate mutase
family protein [Mycobacterium tuberculosis CDC1551]
gi|15841722|ref|NP_336759.1| phosphoglycerate mutase
family protein [Mycobacterium tuberculosis CDC1551]
gi|7477115|pir||H70776 hypothetical protein Rv2228c -
Mycobacterium tuberculosis (strain H37RV)
gi|54042871|sp|P64955|YM28_MYCTU Hypothetical protein
Rv2228c/MT2287 gi|54040461|sp|P64956|YM53_MYCBO
Hypothetical protein Mb2253c
Length = 364
Score = 95.9 bits (237), Expect = 2e-18
Identities = 55/136 (40%), Positives = 82/136 (59%), Gaps = 4/136 (2%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKD-GNLIYRVREGVGIATNNVAEYRAMILGMRHALKK 262
++E DG S+GNPG AG GA++ + D ++ ++ +G ATNNVAEYR +I G+ A+K
Sbjct: 4 VIEADGGSRGNPGPAGYGAVVWTADHSTVLAESKQAIGRATNNVAEYRGLIAGLDDAVKL 63
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
G T + DSKLV ++Q+ G WKVK+ +L LY A+ L +F V R N+
Sbjct: 64 GATEAAVLMDSKLV---VEQMSGRWKVKHPDLLKLYVQAQALASQFRRINYEWVPRARNT 120
Query: 323 EADAQANLAVNLAGEA 338
AD AN A++ A ++
Sbjct: 121 YADRLANDAMDAAAQS 136
>ref|ZP_00401615.1| RNase H [Anaeromyxobacter dehalogenans 2CP-C]
gi|66776663|gb|EAL77773.1| RNase H [Anaeromyxobacter
dehalogenans 2CP-C]
Length = 423
Score = 94.0 bits (232), Expect = 6e-18
Identities = 49/126 (38%), Positives = 83/126 (64%), Gaps = 3/126 (2%)
Query: 208 DGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSI 267
DGA++GNPG AGAGA++ + DG+++ ++ + +G +TNNVAEY +ILG++ A G +
Sbjct: 298 DGAARGNPGPAGAGAVIVNADGHIVAKIGKFLGDSTNNVAEYMGLILGLKRAKAMGIKEL 357
Query: 268 CIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQ 327
+ DS+L+ +KQ+ G + VK ++L L+ A+ L F ++ HV R+ N+ ADA
Sbjct: 358 EVLSDSELM---VKQLAGDYAVKADHLRPLHDEAQALIAGFDRIQVRHVPREENALADAM 414
Query: 328 ANLAVN 333
+N A++
Sbjct: 415 SNRAID 420
>ref|NP_279371.1| hypothetical protein VNG0255C [Halobacterium sp. NRC-1]
gi|10579895|gb|AAG18851.1| Vng0255c [Halobacterium sp.
NRC-1] gi|25409284|pir||G84185 hypothetical protein
Vng0255c [imported] - Halobacterium sp. NRC-1
Length = 199
Score = 91.7 bits (226), Expect = 3e-17
Identities = 54/127 (42%), Positives = 73/127 (56%), Gaps = 3/127 (2%)
Query: 207 FDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTS 266
FDGAS+GNPG A G +L S DG ++ + +G ATNN AEY A+I + A GF
Sbjct: 74 FDGASRGNPGPAAVGWVLVSGDGGIVAEGGDTIGRATNNQAEYDALIAALEAAADFGFDD 133
Query: 267 ICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADA 326
I ++GDS+LV KQ+ G W + +L A+EL F + I+HV R N ADA
Sbjct: 134 IELRGDSQLV---EKQLTGAWDTNDPDLRRKRVRARELLTGFDDWSITHVPRATNERADA 190
Query: 327 QANLAVN 333
AN A++
Sbjct: 191 LANEALD 197
>emb|CAF20578.1| Ribonuclease HI [Corynebacterium glutamicum ATCC 13032]
gi|21325007|dbj|BAB99629.1| Phosphoglycerate
mutase/fructose-2,6-bisphosphatase [Corynebacterium
glutamicum ATCC 13032] gi|19110481|dbj|BAB85788.1| RNase
HI [Corynebacterium glutamicum]
gi|62391077|ref|YP_226479.1| Ribonuclease HI
[Corynebacterium glutamicum ATCC 13032]
gi|19553436|ref|NP_601438.1| phosphoglycerate mutase
[Corynebacterium glutamicum ATCC 13032]
Length = 382
Score = 90.5 bits (223), Expect = 7e-17
Identities = 51/134 (38%), Positives = 84/134 (62%), Gaps = 5/134 (3%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKD-GNLIYRVREGVGI-ATNNVAEYRAMILGMRHALK 261
++E DG S+GNPG AG+G ++ S + ++ + VG ATNNVAEYR ++ G++ A +
Sbjct: 4 VIEADGGSRGNPGVAGSGTVVYSDNKAEVLKEIAYVVGTKATNNVAEYRGLLEGLKAARE 63
Query: 262 KGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLN 321
G TS+ + DSKLV ++Q+ G WK+K+ ++ L AKE+ + S + + R+ N
Sbjct: 64 LGATSVDVYMDSKLV---VEQMSGRWKIKHPDMKVLAIEAKEIASEIGSVSYTWIPREKN 120
Query: 322 SEADAQANLAVNLA 335
ADA +N+A++ A
Sbjct: 121 KRADALSNVAMDAA 134
>ref|YP_117850.1| putative phosphoglycerate mutase [Nocardia farcinica IFM 10152]
gi|54015116|dbj|BAD56486.1| putative phosphoglycerate
mutase [Nocardia farcinica IFM 10152]
Length = 406
Score = 89.7 bits (221), Expect = 1e-16
Identities = 55/131 (41%), Positives = 79/131 (59%), Gaps = 4/131 (3%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKDG-NLIYRVREGVGIATNNVAEYRAMILGMRHALKK 262
IVE DG S+GNPG AG GA++ D ++ RE VGIATNNVAEYR +I G+ A +
Sbjct: 7 IVEADGGSRGNPGPAGYGAVVFDADHVAVLAERRESVGIATNNVAEYRGLIAGLEAAAEL 66
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
G ++ ++ DSKLV ++Q+ G WKVK+ + L A+ L F + + + R N+
Sbjct: 67 GARTVDVRMDSKLV---VEQMSGRWKVKHAAMIPLADRARRLVAGFDAVTFTWIPRAQNA 123
Query: 323 EADAQANLAVN 333
AD AN A++
Sbjct: 124 HADRLANEAMD 134
>ref|NP_940016.1| hypothetical protein DIP1678 [Corynebacterium diphtheriae NCTC
13129] gi|38200511|emb|CAE50207.1| Conserved
hypothetical protein [Corynebacterium diphtheriae]
Length = 377
Score = 89.4 bits (220), Expect = 2e-16
Identities = 51/135 (37%), Positives = 79/135 (57%), Gaps = 4/135 (2%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGV-GIATNNVAEYRAMILGMRHALKK 262
I+E DG S+GNPG AG+G ++ + D + + + V G ATNNVAEY ++ G+R A +
Sbjct: 4 IIEADGGSRGNPGIAGSGTLIYNADRSQVLKEYSYVVGKATNNVAEYHGLLNGLRGAAEL 63
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
G T + ++ DSKLV ++Q+ G WK+K+ ++ L K++ S + R NS
Sbjct: 64 GATEVSVRMDSKLV---VEQMSGRWKIKHPDMKELALECKKVASSIGSVSYEWIPRSENS 120
Query: 323 EADAQANLAVNLAGE 337
ADA AN A++ E
Sbjct: 121 HADALANKAMDALAE 135
>ref|ZP_00294105.1| COG0406: Fructose-2,6-bisphosphatase [Thermobifida fusca]
Length = 378
Score = 88.6 bits (218), Expect = 3e-16
Identities = 54/132 (40%), Positives = 80/132 (59%), Gaps = 5/132 (3%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKD-GNLIYRVREGVGIATNNVAEYRAMILGMRHALKK 262
++E DGAS+GNPG AG GA +R D G L+ V +G ATNNVAEYR +I G+ A
Sbjct: 6 VIEADGASRGNPGPAGYGARVRDADTGELLAEVASPIGEATNNVAEYRGLIAGLEAAAAI 65
Query: 263 GFT-SICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLN 321
T ++ ++ DSKLV ++Q+ G WK+K+ L L + A+++ S V R+ N
Sbjct: 66 DPTAAVEVRMDSKLV---VEQMSGRWKIKHPALQPLARQARQVAASLASVTYRWVPREQN 122
Query: 322 SEADAQANLAVN 333
++AD AN A++
Sbjct: 123 AQADRLANEALD 134
>gb|EAM83227.1| Phosphoglycerate/bisphosphoglycerate mutase:RNase H [Frankia sp.
CcI3]
Length = 378
Score = 88.2 bits (217), Expect = 3e-16
Identities = 54/138 (39%), Positives = 81/138 (58%), Gaps = 5/138 (3%)
Query: 200 VKTCIVEFDGASKGNPGKAGAGAILRSK-DGNLIYRVREGVGIATNNVAEYRAMILGMRH 258
V+ ++E DG S+GNPG AG GA++R G L+ + +G ATNNVAEY +I G+R
Sbjct: 4 VRRLVIEADGGSRGNPGPAGYGAVVRDAGSGKLLAERADSIGRATNNVAEYSGLIAGLRA 63
Query: 259 ALKKG-FTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVL 317
A + + ++ DSKLV ++Q+ G WKVK+ + L A EL +F + + V
Sbjct: 64 AAEIAPDAELEVRMDSKLV---VEQMSGRWKVKHPAMRPLVAEATELAARFPAVRFQWVP 120
Query: 318 RDLNSEADAQANLAVNLA 335
R N++AD AN A++ A
Sbjct: 121 RARNADADRLANEAMDAA 138
>gb|AAV48010.1| ribonuclease H-like protein [Haloarcula marismortui ATCC 43049]
gi|55379866|ref|YP_137716.1| ribonuclease H-like protein
[Haloarcula marismortui ATCC 43049]
Length = 198
Score = 86.3 bits (212), Expect = 1e-15
Identities = 50/127 (39%), Positives = 73/127 (57%), Gaps = 4/127 (3%)
Query: 207 FDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTS 266
FDGAS+GNPG A G +L + D ++ E +G ATNN AEY+A+I + A GF
Sbjct: 74 FDGASRGNPGPASVGYVLVN-DSGIVTEGGETIGTATNNQAEYKALIRAVEVARDYGFDD 132
Query: 267 ICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADA 326
+ I+GDS+L+ +KQ+ G W + L +EL F ++I HV R++N AD
Sbjct: 133 VHIRGDSELI---VKQVRGEWDTNDPELREHRVRVRELLTDFDDWQIEHVPREINDRADE 189
Query: 327 QANLAVN 333
AN A++
Sbjct: 190 LANDALD 196
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 555,115,646
Number of Sequences: 2540612
Number of extensions: 22117229
Number of successful extensions: 61769
Number of sequences better than 10.0: 782
Number of HSP's better than 10.0 without gapping: 476
Number of HSP's successfully gapped in prelim test: 306
Number of HSP's that attempted gapping in prelim test: 60925
Number of HSP's gapped (non-prelim): 853
length of query: 357
length of database: 863,360,394
effective HSP length: 129
effective length of query: 228
effective length of database: 535,621,446
effective search space: 122121689688
effective search space used: 122121689688
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146308.4


