

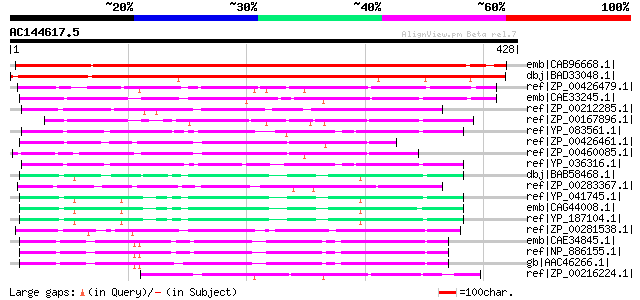
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144617.5 + phase: 0
(428 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB96668.1| putative protein [Arabidopsis thaliana] gi|26449... 549 e-155
dbj|BAD33048.1| monooxygenase-like [Oryza sativa (japonica culti... 460 e-128
ref|ZP_00426479.1| Flavoprotein monooxygenase [Burkholderia viet... 122 2e-26
emb|CAE33245.1| putative monooxygenase [Bordetella bronchiseptic... 107 9e-22
ref|ZP_00212285.1| COG0654: 2-polyprenyl-6-methoxyphenol hydroxy... 97 1e-18
ref|ZP_00167896.1| COG0654: 2-polyprenyl-6-methoxyphenol hydroxy... 97 1e-18
ref|YP_083561.1| probable FAD-dependent monooxygenase [Bacillus ... 96 2e-18
ref|ZP_00426461.1| Flavoprotein monooxygenase [Burkholderia viet... 96 2e-18
ref|ZP_00460085.1| Flavoprotein monooxygenase [Burkholderia ceno... 94 6e-18
ref|YP_036316.1| possible FAD-dependent monooxygenase [Bacillus ... 94 8e-18
dbj|BAB58468.1| similar to monooxygenase [Staphylococcus aureus ... 92 3e-17
ref|ZP_00283367.1| COG0654: 2-polyprenyl-6-methoxyphenol hydroxy... 92 4e-17
ref|YP_041745.1| putative monooxygenase [Staphylococcus aureus s... 91 5e-17
emb|CAG44008.1| putative monooxygenase [Staphylococcus aureus su... 91 5e-17
ref|YP_187104.1| monooxygenase family protein [Staphylococcus au... 91 7e-17
ref|ZP_00281538.1| COG0654: 2-polyprenyl-6-methoxyphenol hydroxy... 89 2e-16
emb|CAE34845.1| putative hydroxylase [Bordetella bronchiseptica ... 89 2e-16
ref|NP_886155.1| putative hydroxylase [Bordetella parapertussis ... 87 1e-15
gb|AAC46266.1| unknown [Bordetella pertussis] 86 2e-15
ref|ZP_00216224.1| COG0654: 2-polyprenyl-6-methoxyphenol hydroxy... 82 3e-14
>emb|CAB96668.1| putative protein [Arabidopsis thaliana] gi|26449436|dbj|BAC41845.1|
unknown protein [Arabidopsis thaliana]
gi|28950969|gb|AAO63408.1| At5g11330 [Arabidopsis
thaliana] gi|15239022|ref|NP_196694.1| monooxygenase
family protein [Arabidopsis thaliana]
Length = 408
Score = 549 bits (1415), Expect = e-155
Identities = 275/414 (66%), Positives = 321/414 (77%), Gaps = 10/414 (2%)
Query: 6 RKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW 65
+K KA+IVGGSI G+S AH+L LA WDVLVLEK++ PP GSPTGAGLGL+ ++QII+SW
Sbjct: 4 KKGKAIIVGGSIAGLSCAHSLTLACWDVLVLEKSSEPPVGSPTGAGLGLDPQARQIIKSW 63
Query: 66 ISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQ 125
++ P Q L T PL+IDQN TDSEKKV R LTRDE +FRAA+W D+ +LY +LP
Sbjct: 64 LTTP-QLLDEITLPLSIDQNQTTDSEKKVTRILTRDEDFDFRAAYWSDIQSLLYNALPQT 122
Query: 126 IFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKL 185
+FLWGH FLSF ++ ++ V VK VV+T E +EI GDLL+AADGCLSSIR+ +LPDFKL
Sbjct: 123 MFLWGHKFLSFTMSQDESNVKVKTLVVETQETVEIEGDLLIAADGCLSSIRKTFLPDFKL 182
Query: 186 RYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKKLNWI 245
RYSGYCAWRGV DFS ENSET+ GIKK YPDLGKCLYFDL THSV YEL NKKLNWI
Sbjct: 183 RYSGYCAWRGVFDFSGNENSETVSGIKKVYPDLGKCLYFDLGEQTHSVFYELFNKKLNWI 242
Query: 246 WYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNFIYDSDPL 305
WYVNQPEP++K SVT KV+ +MI KMHQEAE IWIPELA++M ETK+PFLN IYDSDPL
Sbjct: 243 WYVNQPEPDLKSNSVTLKVSQEMINKMHQEAETIWIPELARLMSETKDPFLNVIYDSDPL 302
Query: 306 EKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTRL 365
E+IFW N+VLVG+AAHPTTPH LRSTNM+ILDA VLGKC+E G E + LEEYQ RL
Sbjct: 303 ERIFWGNIVLVGDAAHPTTPHGLRSTNMSILDAEVLGKCLENCGPENVSLGLEEYQRIRL 362
Query: 366 PVTSKQVLHARRLGRIKQGLVLPDRDPFDPKSARQEECQEIILTNTPFFNDAPL 419
PV S+QVL+ARRLGRIKQGL D D E+ N PFF+ APL
Sbjct: 363 PVVSEQVLYARRLGRIKQGL---DHDGIGDGVFGLEQ------RNMPFFSCAPL 407
>dbj|BAD33048.1| monooxygenase-like [Oryza sativa (japonica cultivar-group)]
gi|50725454|dbj|BAD32925.1| monooxygenase-like [Oryza
sativa (japonica cultivar-group)]
Length = 431
Score = 460 bits (1184), Expect = e-128
Identities = 234/434 (53%), Positives = 303/434 (68%), Gaps = 22/434 (5%)
Query: 1 MVGEGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQ 60
MVG K AV+VGGS+ G++ AHA+ AGW+ +V+EK +P GS TGAGLGL++ S +
Sbjct: 1 MVG---KRVAVVVGGSVAGLACAHAVAEAGWEAVVVEKAAAPGAGSGTGAGLGLDAQSME 57
Query: 61 IIQSWISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYE 120
+ WI P L T PL +D N TD E K RTLTRDE FRAAHWGDLH L+E
Sbjct: 58 TLARWI---PGRLDAATLPLAVDLNRATDGETKAGRTLTRDEGFGFRAAHWGDLHRRLHE 114
Query: 121 SLPPQI-FLWGHIFLSFHVANE---KGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIR 176
+LP + LWGH F+SF +A E G V+V A+V++TGE +E+ GDLLVAADGC S+IR
Sbjct: 115 ALPAGVTVLWGHQFVSFEMAPEGDGDGGVVVTARVLRTGETVEVAGDLLVAADGCTSAIR 174
Query: 177 RKYLPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYE 236
R++LP+ KLRYSGYCAWRGV DFS E T+ I++AYP+LG CLYFDLA TH+VLYE
Sbjct: 175 RRFLPELKLRYSGYCAWRGVFDFSGKEGCTTMVDIRRAYPELGNCLYFDLAYKTHAVLYE 234
Query: 237 LPNKKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFL 296
LP +LNW+WY+N EPE+ G+SVT KV + +M +EAE++W PELA+++ ET EPF+
Sbjct: 235 LPKNRLNWLWYINGDEPELMGSSVTMKVNEATVSEMKEEAERVWCPELARLIGETAEPFV 294
Query: 297 NFIYDSDPLEKIFW--DNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWG----- 349
N IYD++PL + W V LVG+AAHPTTPH LRSTNM+ILDA VLG C+ +WG
Sbjct: 295 NVIYDAEPLPGLSWAGGRVALVGDAAHPTTPHGLRSTNMSILDARVLGCCLARWGGDGNA 354
Query: 350 --TEMLESALEEYQLTRLPVTSKQVLHARRLGRIKQGLVL---PDRDPFDPKSARQEECQ 404
T AL EY+ R PV + QVLHARRLGR+KQGL + D + FD ++A +EE
Sbjct: 355 EPTSTPRRALAEYEAARRPVVAAQVLHARRLGRLKQGLGMGSAGDGEGFDARTATEEEIS 414
Query: 405 EIILTNTPFFNDAP 418
++ ++ P+F+ AP
Sbjct: 415 QLRQSSMPYFSGAP 428
>ref|ZP_00426479.1| Flavoprotein monooxygenase [Burkholderia vietnamiensis G4]
gi|67530124|gb|EAM26971.1| Flavoprotein monooxygenase
[Burkholderia vietnamiensis G4]
Length = 408
Score = 122 bits (306), Expect = 2e-26
Identities = 115/428 (26%), Positives = 190/428 (43%), Gaps = 56/428 (13%)
Query: 7 KPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWI 66
K A IVGGS+GG+ +A+ L+ GWDV V E+ P S GAG+ +
Sbjct: 8 KRNAAIVGGSLGGLFAANLLLRNGWDVDVFERV--PDELSGRGAGI-------------V 52
Query: 67 SKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRA------AHWGDLHGVLYE 120
+ P F + ID ++ + E ++ TL RD SL W ++ VL
Sbjct: 53 THPELFDVLRAAGVRIDASIGVNVEARI--TLARDGSLASERPLPQTLTAWSKMYHVLRA 110
Query: 121 SLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYL 180
+LP Q + G + ++ V + G V+ DL++AADG S+IR +L
Sbjct: 111 ALPDQHYHAGAVVADVADGPDRASV-----TLADGSVVH--ADLVIAADGLRSAIRETFL 163
Query: 181 PDFKLRYSGYCAWRGVLDFSEIENS--ETIKGIKKAY--PDLGKCLYFDLASGTHSVLYE 236
PD +L+Y+GY AWRG++D E+ ++ T+ G K A+ P + L + +A +S +
Sbjct: 164 PDARLQYAGYVAWRGLVDERELSDTTHATLFG-KFAFGLPPREQILGYPVAGRHNST--Q 220
Query: 237 LPNKKLNWIWY-------------VNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPE 283
++ N++WY + + G T + D++ M A + P+
Sbjct: 221 PGERRYNFVWYRATREDTDLPNLLTDATGKQWAGGIPPTLIRPDVLAAMEDAAHALLAPQ 280
Query: 284 LAKVMKETKEPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGK 343
A+V+ +P IYD + + + +D + L+G+AA PH DA L +
Sbjct: 281 FAEVVSRAAQPLFQPIYDLE-VPHMTFDRIALLGDAAFVARPHCGMGVTKAAGDAMALVQ 339
Query: 344 CIEKWGTEMLESALEEYQLTRLPVTSKQVLHARRLGRIKQGLVLPDRDPFDPKSARQEEC 403
+ + AL Y TR + V HAR LG Q + D + + A +
Sbjct: 340 ALGAHADTL--DALHAYSDTRTAFGAAIVQHARHLGAYMQAQLQNDT---EREMAERYRT 394
Query: 404 QEIILTNT 411
EI++ +T
Sbjct: 395 PEIVMRDT 402
>emb|CAE33245.1| putative monooxygenase [Bordetella bronchiseptica RB50]
gi|33601729|ref|NP_889289.1| putative monooxygenase
[Bordetella bronchiseptica RB50]
Length = 402
Score = 107 bits (266), Expect = 9e-22
Identities = 105/420 (25%), Positives = 179/420 (42%), Gaps = 44/420 (10%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWISK 68
KA ++GGS+GG+ +A+ L+ AGWDV V E+ G GAG+ + +++
Sbjct: 3 KAAVIGGSLGGLFAANLLLRAGWDVHVHERVHDELDGR--GAGIVTHPELLDVLRRIGVA 60
Query: 69 PPQFLH-NTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQIF 127
P + + +T+D + + + + + LT WG L +L P + +
Sbjct: 61 PTESIGIEVARRVTLDLDGSVLASRALPQLLTA----------WGRLFSILRTRFPAERY 110
Query: 128 LWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRY 187
G S + +++ + DL+VAADG S++R+ LP + Y
Sbjct: 111 HRGQALESLEQDERRA-------LLRFADGARAEVDLVVAADGLRSTLRQALLPQARPAY 163
Query: 188 SGYCAWRGVLD---FSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKKLNW 244
+GY AWRG++D SE E P + + + +A GT S + ++ N+
Sbjct: 164 AGYVAWRGLVDEASLSERTRRELCPYFAFGLPAHEQMIAYPVA-GTGSGAQDAA-RRFNF 221
Query: 245 IWYVNQPEPEVKGTSVTTK-------------VTSDMIQKMHQEAEKIWIPELAKVMKET 291
+WY E VT + +++ + Q A ++ P+ +V+ +T
Sbjct: 222 VWYRPADEHTTFRDMVTDAQGQAWMDGIPPPLIRPEIVAQARQAAAQVLAPQFGEVVAKT 281
Query: 292 KEPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTE 351
F I+D + ++ V L+G+AA PH DA L + GT
Sbjct: 282 ANLFFQPIFDLES-PRLALGRVALLGDAAFVARPHCGMGVTKAAGDARALAYALA--GTP 338
Query: 352 MLESALEEYQLTRLPVTSKQVLHARRLGRIKQGLVLPDRDPFDPKSARQEECQEIILTNT 411
+ +AL+ Y+ RLP V HAR LGR Q + RD + + A + E I+ T
Sbjct: 339 DIGAALQAYEDERLPFGKFIVGHARDLGRYMQAQL---RDDAEREMAERYRTPEAIMRET 395
>ref|ZP_00212285.1| COG0654: 2-polyprenyl-6-methoxyphenol hydroxylase and related
FAD-dependent oxidoreductases [Burkholderia cepacia
R18194]
Length = 408
Score = 97.1 bits (240), Expect = 1e-18
Identities = 99/368 (26%), Positives = 159/368 (42%), Gaps = 34/368 (9%)
Query: 11 VIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWISKPP 70
++VG IGG+++A AL AG V VLE++ + GAGL + + + +++
Sbjct: 10 IVVGAGIGGLAAALALRRAGHRVTVLEQSRA---FREVGAGLQVVPNATRALRALGVLDR 66
Query: 71 QFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWG----DLHGVLYESL---- 122
Q L TI + D + L + F A +W DLH L +++
Sbjct: 67 QRLAAVAPTATIRRRW-QDGSLLGSFPLGNEVEAAFNAPYWNAHRADLHAALLDAVRDPY 125
Query: 123 ---PPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKY 179
PP L G G +V A + DL+VAADG S++R
Sbjct: 126 TAGPPVAVLGGIAVRGLDACGPDGATLVDAAGTRWR------ADLVVAADGIHSTLRHAL 179
Query: 180 LPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPN 239
L D YSG A+R ++D I+ + I K + L G H++ Y + +
Sbjct: 180 LGDDAPHYSGDDAYRALIDVDAIDPQSPVFEIVKEPQ-----VTIWLGPGRHAIHYWVRD 234
Query: 240 KKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKI-WIPELAKVMKETKEPFLNF 298
++L + + V G T + S + EAE W P L +++ +
Sbjct: 235 RRLLNLVVI------VPGDGSTRESWSSKGDRATLEAELDGWDPRLVGLIRCASDLSRWS 288
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGT-EMLESAL 357
++D PL + WD+V L+G+A HP P+ + + DA VLG+C+ + E L +AL
Sbjct: 289 LHDRQPLTRWVWDSVCLLGDACHPMLPYQSQGAAQALEDAVVLGRCVTGLASKEALGAAL 348
Query: 358 EEYQLTRL 365
EYQ R+
Sbjct: 349 VEYQRLRI 356
>ref|ZP_00167896.1| COG0654: 2-polyprenyl-6-methoxyphenol hydroxylase and related
FAD-dependent oxidoreductases [Ralstonia eutropha
JMP134]
Length = 382
Score = 96.7 bits (239), Expect = 1e-18
Identities = 99/387 (25%), Positives = 168/387 (42%), Gaps = 58/387 (14%)
Query: 30 GWDVLVLEKTTSPPTGSPTGAGLGLN-SLSQQIIQSWISKPPQFLHNTTHPLTIDQNLVT 88
GWDV + E+T TG GAG+ + L + + + ++ +T+ ++ T
Sbjct: 6 GWDVTIFERTPETLTGR--GAGIVTHPELFEALAAAGVAIDDSIGVRIRTRVTLSRDGST 63
Query: 89 DSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQIFLWGHIFLSFHVANEKGPVIVK 148
S++++ +TLT WG ++ VL GH F + G +
Sbjct: 64 VSDREMPQTLTA----------WGKMYDVL-----------GHAFSGEY---RTGATVTD 99
Query: 149 AK------VVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRYSGYCAWRGVLDFSEI 202
VVK + DL++AADG S +R + +P L Y+GY AWRG++D ++I
Sbjct: 100 VDSRSDHAVVKLEDGSTHRADLVIAADGFRSGVRERLVPSAALEYAGYIAWRGLVDEAKI 159
Query: 203 ENSETIKGIKKAY----PDLGKCLYFDLASGTHSVLYELPNKKLNWIWYVNQPE----PE 254
+ ET I + P + L + +A +S ++ N++WY E P+
Sbjct: 160 -SRETHAAIFDKFAFCLPPHEQILGYPVAGDGNST--TPGQRRYNFVWYRATREHDELPD 216
Query: 255 VKGTSVTTKV----------TSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNFIYDSDP 304
+ T T KV +D++ M A P+ A+++ T +P I+D
Sbjct: 217 LL-TDATGKVWNNGIPPALIRADVLADMESAAADRLAPQFAEIVSSTAQPLFQPIFDL-T 274
Query: 305 LEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTR 364
+ ++ + + L+G+AA PH DA L + + G E +E+ALE Y R
Sbjct: 275 VPRMAFGRIALLGDAAFVARPHCGMGVTKAAGDAMALVRALA--GHESVETALEAYSAER 332
Query: 365 LPVTSKQVLHARRLGRIKQGLVLPDRD 391
+ V V HAR LG Q + ++D
Sbjct: 333 VQVGQAVVEHARHLGAYMQAQLKSEQD 359
>ref|YP_083561.1| probable FAD-dependent monooxygenase [Bacillus cereus E33L]
gi|51976737|gb|AAU18287.1| probable FAD-dependent
monooxygenase [Bacillus cereus E33L]
Length = 377
Score = 96.3 bits (238), Expect = 2e-18
Identities = 96/376 (25%), Positives = 159/376 (41%), Gaps = 40/376 (10%)
Query: 11 VIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW-ISKP 69
+I+GG I G+ +A +L G DV V +K T P GAG+ + + Q ++ + ISK
Sbjct: 5 IIIGGGIAGLCAAISLQKIGLDVKVYDKNTEPTVA---GAGIIIAPNAMQALELYGISKK 61
Query: 70 PQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQIFLW 129
+ N ++ NLV++ N+ + + H DLH +L L W
Sbjct: 62 IKKFGNESNGF----NLVSEKGTTFNKLIIPTCYPKMYSIHRKDLHQLLLSELKEDTVKW 117
Query: 130 GHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRYSG 189
G + NE + + V + G E +G++L+AADG S IR++ RY+G
Sbjct: 118 GKECVKIE-QNEANALKI---VFQDGS--EALGNILIAADGIHSVIRKQVTQGDNYRYAG 171
Query: 190 YCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHS--VLYELPNKKLNWIWY 247
Y WRGV + + + F GT+ + LPN ++ W
Sbjct: 172 YTCWRGVTPANNLSLTND----------------FIETWGTNGRFGIVPLPNNEVYWYAL 215
Query: 248 VNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNFIYDSDPLEK 307
+N + K + TT + + H IP + K + + I D P+++
Sbjct: 216 INAKARDPKYKAYTTTDLYNHFKTYHNP-----IPSILKNASDI-DMIHRDIIDITPMKQ 269
Query: 308 IFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTRLPV 367
F +V +G+AAH TP+ + I DA +L +CI+ A EY+ R
Sbjct: 270 FFDKRIVFIGDAAHALTPNLGQGACQAIEDAIILAECIK--NNAHYRQAFLEYEQKRRDR 327
Query: 368 TSKQVLHARRLGRIKQ 383
K A ++G++ Q
Sbjct: 328 IEKISNTAWKVGKMAQ 343
>ref|ZP_00426461.1| Flavoprotein monooxygenase [Burkholderia vietnamiensis G4]
gi|67530106|gb|EAM26953.1| Flavoprotein monooxygenase
[Burkholderia vietnamiensis G4]
Length = 380
Score = 95.9 bits (237), Expect = 2e-18
Identities = 86/333 (25%), Positives = 146/333 (43%), Gaps = 36/333 (10%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNS-LSQQIIQSWIS 67
KAVIVGGS+GG+ +A AL GW V V E++ G G+ L + + ++
Sbjct: 8 KAVIVGGSVGGLFAATALRARGWHVRVFEQSARELDSR--GGGIVLQPPIERAFAFGGVA 65
Query: 68 KPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQIF 127
P ++ + + D++ +V + LT ++ W ++ L +LP +
Sbjct: 66 LPANAGVDSI------ERIYVDAQDRVVQRLTMPQT----QTGWNVIYTALKHALPDDVI 115
Query: 128 LWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRY 187
G F F E+ + V+T DLL+ ADG S++R + LPD + Y
Sbjct: 116 HAGDAFERFQQDGERVVAQFASGRVETA-------DLLIGADGARSTVRAQLLPDVRPAY 168
Query: 188 SGYCAWRGVLDFSEI-ENSETIKGIKKAYPDLGKCLYFD-LASGTHSVLYELPNKKLNWI 245
+GY AWRG++D + E + + + G L+ L G + E +++NW+
Sbjct: 169 AGYVAWRGLVDEHALPEQVLYLLRERFTFQQGGAHLFLTYLVPGADGAI-EPGKRRVNWV 227
Query: 246 WYVNQPEPEVKGTSVTTKVT------------SDMIQKMHQEAEKIWIPELAKVMKETKE 293
WY + +T T D ++ A+++ P LA ++ T
Sbjct: 228 WYRRLASERLPSLFLTRDGTQRDGSLPPGAMRDDNRAELVDAADRMLAPTLATLVDTTPA 287
Query: 294 PFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPH 326
PF IYD +E++ + VL+G+AA PH
Sbjct: 288 PFAQAIYDL-AVERMAFGRTVLLGDAACVVRPH 319
>ref|ZP_00460085.1| Flavoprotein monooxygenase [Burkholderia cenocepacia HI2424]
gi|67660992|ref|ZP_00458324.1| Flavoprotein
monooxygenase [Burkholderia cenocepacia AU 1054]
gi|67103778|gb|EAM20901.1| Flavoprotein monooxygenase
[Burkholderia cenocepacia HI2424]
gi|67091431|gb|EAM09008.1| Flavoprotein monooxygenase
[Burkholderia cenocepacia AU 1054]
Length = 519
Score = 94.4 bits (233), Expect = 6e-18
Identities = 96/357 (26%), Positives = 151/357 (41%), Gaps = 36/357 (10%)
Query: 3 GEGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQII 62
G GRK AVI GGS+GG+ +A AL AGWDV V E+ SP G G+ L Q I
Sbjct: 143 GAGRK--AVIAGGSVGGLFAATALRAAGWDVRVFEQ--SPHALDSRGGGIVL----QPPI 194
Query: 63 QSWISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESL 122
+ + L T +ID+ V ++ V R W ++ L +L
Sbjct: 195 ERAFAFGGVALPRETGVDSIDRIYVDAHDRIVQRFAMPQTQTG-----WNVIYTALKRAL 249
Query: 123 PPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPD 182
P + G F F E+ + +T DLL+ ADG S++R + LPD
Sbjct: 250 PAGVIHAGDAFERFEQDGERIVAHFASGRAETA-------DLLIGADGGRSTVRAQLLPD 302
Query: 183 FKLRYSGYCAWRGVLDFSEI-ENSETIKGIKKAYPDLGKCLYFD-LASGTHSVLYELPNK 240
+ Y+GY AWRG++D + E + + + + L+ L G + E +
Sbjct: 303 VRPTYAGYVAWRGLVDEHALPEQVLVLLRERFTFQQGDRHLFLTYLVPGADGTI-EPGKR 361
Query: 241 KLNWIWY------------VNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVM 288
++NW+WY V + + G+ + D ++ A+++ P LA ++
Sbjct: 362 RVNWVWYRRLASDRMPSLFVARDGTQRDGSLPPGAMRDDHRAELVDAADRMLAPMLAALV 421
Query: 289 KETKEPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCI 345
T PF I D +E++ VL+G+AA PH DA L + +
Sbjct: 422 DATAAPFAQAILDL-AVERMAVGRAVLLGDAACMVRPHTAAGVAKAADDAVGLAEAV 477
>ref|YP_036316.1| possible FAD-dependent monooxygenase [Bacillus thuringiensis
serovar konkukian str. 97-27] gi|49329093|gb|AAT59739.1|
possible FAD-dependent monooxygenase [Bacillus
thuringiensis serovar konkukian str. 97-27]
Length = 377
Score = 94.0 bits (232), Expect = 8e-18
Identities = 93/374 (24%), Positives = 155/374 (40%), Gaps = 36/374 (9%)
Query: 11 VIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW-ISKP 69
+I+GG I G+ +A +L G DV V +K T P GAG+ + + Q ++ + ISK
Sbjct: 5 IIIGGGIAGLCAAISLQKIGLDVKVYDKNTEPTVA---GAGIIIAPNAMQALELYGISKK 61
Query: 70 PQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQIFLW 129
+ N + NLV+ N+ + + H DLH +L L W
Sbjct: 62 IKKFGNESDGF----NLVSKKGTTFNKLIIPTCYPKMYSIHRKDLHQLLLSELKEDTVKW 117
Query: 130 GHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRYSG 189
G + E IV + G E +G++L+AADG S +R++ RY+G
Sbjct: 118 GKECVKIEQNEENALKIV----FQDGS--EALGNILIAADGIHSIVRKQVTQGDNYRYAG 171
Query: 190 YCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKKLNWIWYVN 249
Y WRGV + +N + + G+ + LPN ++ W +N
Sbjct: 172 YTCWRGV---TPTKNLSLTNDFIETWGTNGR-----------FGIVPLPNNEVYWYALIN 217
Query: 250 QPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNFIYDSDPLEKIF 309
+ K + TT + + H IP + + + + I D P+++ F
Sbjct: 218 AKARDPKYKAYTTTDLYNHFKTYHNP-----IPSILQNASDI-DMIHRDIIDITPMKQFF 271
Query: 310 WDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTRLPVTS 369
+V +G+AAH TP+ + I DA +L +CI+ A E++ R
Sbjct: 272 DKRIVFIGDAAHALTPNLGQGACQAIEDAIILAECIK--NNAYYRQAFTEFEQKRRDRIE 329
Query: 370 KQVLHARRLGRIKQ 383
K A ++G+I Q
Sbjct: 330 KISNTAWKVGKIAQ 343
>dbj|BAB58468.1| similar to monooxygenase [Staphylococcus aureus subsp. aureus Mu50]
gi|15927886|ref|NP_375419.1| hypothetical protein SA2099
[Staphylococcus aureus subsp. aureus N315]
gi|13702106|dbj|BAB43398.1| SA2099 [Staphylococcus
aureus subsp. aureus N315] gi|25507133|pir||E90029
hypothetical protein SA2099 [imported] - Staphylococcus
aureus (strain N315) gi|15925296|ref|NP_372830.1|
similar to monooxygenase [Staphylococcus aureus subsp.
aureus Mu50]
Length = 374
Score = 92.0 bits (227), Expect = 3e-17
Identities = 93/383 (24%), Positives = 154/383 (39%), Gaps = 53/383 (13%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLG-----LNSLSQQIIQ 63
K I+G IGG+++A L G + V EK S GAG+G L L +
Sbjct: 2 KIAIIGAGIGGLTAAALLQEQGHTIKVFEKNESV---KEIGAGIGIGDNVLKKLGNHDLA 58
Query: 64 SWISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLP 123
I Q L T ++ D ++ + + +LN L ++ +
Sbjct: 59 KGIKNAGQILSTMT--------VLDDKDRLLTTVKLKSNTLNVTLPRQ-TLIDIIKSYVK 109
Query: 124 PQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDF 183
+ H H+ NE V + E DL + ADG S +R+ D
Sbjct: 110 DDVIFTNHEVT--HIDNETDKV-----TIHFAEQESEAFDLCIGADGIHSKVRQSVNADS 162
Query: 184 KLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKKLN 243
K+ Y GY +RG++D ++++ + K +G + L N +
Sbjct: 163 KVLYQGYTCFRGLIDDIDLKHPDCAKEYWGRKGRVG--------------IVPLLNNQAY 208
Query: 244 WIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIP-ELAKVMKETKEP--FLNFIY 300
W +N E K +S K H +A P E+ +++ + E L+ IY
Sbjct: 209 WFITINSKENNHKYSS---------FGKPHLQAYFNHYPNEVREILDKQSETGILLHNIY 259
Query: 301 DSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEY 360
D PL+ + +L+G+AAH TTP+ + + DA VL C + E AL+ Y
Sbjct: 260 DLKPLKSFVYGRTILLGDAAHATTPNMGQGAGQAMEDAIVLVNCFNAYD---FEKALQRY 316
Query: 361 QLTRLPVTSKQVLHARRLGRIKQ 383
R+ T+K + +R++G+I Q
Sbjct: 317 DKIRVKHTAKVIKRSRKIGKIAQ 339
>ref|ZP_00283367.1| COG0654: 2-polyprenyl-6-methoxyphenol hydroxylase and related
FAD-dependent oxidoreductases [Burkholderia fungorum
LB400]
Length = 382
Score = 91.7 bits (226), Expect = 4e-17
Identities = 95/383 (24%), Positives = 161/383 (41%), Gaps = 52/383 (13%)
Query: 7 KPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLN-SLSQQIIQSW 65
KP+AV++GGS+GG+ +A+ L AGWDV V E SP G G+ L + + +
Sbjct: 6 KPRAVVIGGSLGGLLTANTLRAAGWDVDVFE--ASPNQLESRGGGVVLQPDVLDALAFAG 63
Query: 66 ISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQ 125
+ P + L D E +V L + W L+ + +LP
Sbjct: 64 VELPDP------PGVASGDRLYLDREDRVVEQLYMPQM----QTSWSLLYRAMKNALPAS 113
Query: 126 IFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKL 185
G F+ F + E+ IV GE DLLV ADG S++R++ LP+
Sbjct: 114 NLHAGETFVDFRMEGEQ---IVAQFASGRGEQ----ADLLVGADGIRSTLRQRLLPEVVP 166
Query: 186 RYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELP------- 238
+Y+GY AWRG+++ +++ A L + F + Y +P
Sbjct: 167 QYAGYVAWRGLVEEADLP--------LHAADMLRERFAFQQGDRHSGLAYLIPGENDSTV 218
Query: 239 --NKKLNWIWYVNQPEPEV-----------KGTSVTTKVTSDM-IQKMHQEAEKIWIPEL 284
++ NW+WY + + S+ T + I ++ ++A+ P
Sbjct: 219 AGQRRFNWVWYRKYSAARLDELLVDRHGIRRAFSLPPGSTKGIDIVQLRKDAQAWLGPTF 278
Query: 285 AKVMKETKEPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKC 344
++ T +PF+ I D + +F VL+G+AA PH ST +A L
Sbjct: 279 RALVDATDDPFMQPIVDLRSPKMVF-GRAVLLGDAASVPRPHTAGSTAKAAANAHALALA 337
Query: 345 IEK-WGT-EMLESALEEYQLTRL 365
+ W + ++SAL+ ++ +L
Sbjct: 338 LSSTWQSGATIDSALKRWENQQL 360
>ref|YP_041745.1| putative monooxygenase [Staphylococcus aureus subsp. aureus
MRSA252] gi|49242650|emb|CAG41371.1| putative
monooxygenase [Staphylococcus aureus subsp. aureus
MRSA252]
Length = 374
Score = 91.3 bits (225), Expect = 5e-17
Identities = 99/385 (25%), Positives = 156/385 (39%), Gaps = 57/385 (14%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLG-----LNSLSQQIIQ 63
K I+G IGG+++A L G + V EK S GAG+G L L +
Sbjct: 2 KIAIIGAGIGGLTAAALLQEQGHTIKVFEKNESV---KEIGAGIGIGDNVLKKLGNHDLA 58
Query: 64 SWISKPPQFLHNTTHPLTIDQNLVTDSEKK--VNRTLTRDESLNFRAAHWGDLHGVLYES 121
I Q L T D+ L T K +N TL R ++ ++ D
Sbjct: 59 KGIKNAGQILSTMTVLDDKDRPLTTVKLKSNTLNVTLPRQTLIDIIKSYVKD-------- 110
Query: 122 LPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLP 181
IF + H+ NE V + E DL + ADG S +R+
Sbjct: 111 --DAIFTNHEVT---HIDNETDKV-----TIHFAEQESEAFDLCIGADGIHSKVRQSVNA 160
Query: 182 DFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKK 241
D K+ Y GY +RG++D ++++ + K +G + L N +
Sbjct: 161 DSKVLYQGYTCFRGLIDDIDLKHPDCAKEYWGRKGRVG--------------IVPLLNNQ 206
Query: 242 LNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIP-ELAKVMKETKEP--FLNF 298
W +N E K +S K H +A P E+ +++ + E L+
Sbjct: 207 AYWFITINSKENNHKYSS---------FGKPHLQAYFNHYPNEVREILDKQSETGILLHN 257
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALE 358
IYD PL+ + +L+G+AAH TTP+ + + DA VL C + E AL+
Sbjct: 258 IYDLKPLKSFVYGRTILLGDAAHATTPNMGQGAGQAMEDAIVLVNCFNAYD---FEKALQ 314
Query: 359 EYQLTRLPVTSKQVLHARRLGRIKQ 383
Y R+ T+K + +R++G+I Q
Sbjct: 315 RYDKIRVKHTAKVIKRSRKIGKIAQ 339
>emb|CAG44008.1| putative monooxygenase [Staphylococcus aureus subsp. aureus
MSSA476] gi|21205396|dbj|BAB96090.1| MW2225
[Staphylococcus aureus subsp. aureus MW2]
gi|49487088|ref|YP_044309.1| putative monooxygenase
[Staphylococcus aureus subsp. aureus MSSA476]
gi|21283954|ref|NP_647042.1| hypothetical protein MW2225
[Staphylococcus aureus subsp. aureus MW2]
Length = 374
Score = 91.3 bits (225), Expect = 5e-17
Identities = 100/385 (25%), Positives = 157/385 (39%), Gaps = 57/385 (14%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLG-----LNSLSQQIIQ 63
K I+G IGG+++A L G + V EK S GAG+G L L +
Sbjct: 2 KIAIIGAGIGGLTAAALLQEQGHTIKVFEKNESV---KEIGAGIGIGDNVLKKLGNHDLA 58
Query: 64 SWISKPPQFLHNTTHPLTIDQNLVTDSEKK--VNRTLTRDESLNFRAAHWGDLHGVLYES 121
I Q L T D+ L T K +N TL R ++ ++ D
Sbjct: 59 KGIKNAGQILSTMTVLDDKDRLLTTVKLKSNTLNVTLPRQTLIDIIKSYVKD-------- 110
Query: 122 LPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLP 181
IF + H+ NE V + E DL + ADG S +R+
Sbjct: 111 --DAIFTNHEVT---HIDNETDKV-----TIHFAEQESEAFDLCIGADGIHSKVRQSVNA 160
Query: 182 DFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKK 241
D K+ Y GY +RG++D ++++ + K +G + L N +
Sbjct: 161 DSKVLYQGYTCFRGLIDDIDLKHPDCAKEYWGRKGRVG--------------IVPLLNNQ 206
Query: 242 LNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIP-ELAKVMKETKEP--FLNF 298
W +N E K +S K H +A P E+ +++ + E L+
Sbjct: 207 AYWFITINSKENNHKYSS---------FGKPHLQAYFNHYPNEVREILDKQSETGILLHN 257
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALE 358
IYD PL+ + +L+G+AAH TTP+ + + DA VL C + T E AL+
Sbjct: 258 IYDLKPLKSFVYGRTILLGDAAHATTPNMGQGAGQAMEDAIVLVNC---FNTYDFEKALQ 314
Query: 359 EYQLTRLPVTSKQVLHARRLGRIKQ 383
Y R+ T+K + +R++G+I Q
Sbjct: 315 RYDKIRVKHTAKVIKRSRKIGKIAQ 339
>ref|YP_187104.1| monooxygenase family protein [Staphylococcus aureus subsp. aureus
COL] gi|57286423|gb|AAW38517.1| monooxygenase family
protein [Staphylococcus aureus subsp. aureus COL]
Length = 374
Score = 90.9 bits (224), Expect = 7e-17
Identities = 99/385 (25%), Positives = 156/385 (39%), Gaps = 57/385 (14%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLG-----LNSLSQQIIQ 63
K I+G IGG+++A L G + V EK S GAG+G L L +
Sbjct: 2 KIAIIGAGIGGLTAAALLQEQGHTIKVFEKNESV---KEIGAGIGIGDNVLKKLGNHDLA 58
Query: 64 SWISKPPQFLHNTTHPLTIDQNLVTDSEKK--VNRTLTRDESLNFRAAHWGDLHGVLYES 121
I Q L T D+ L T K +N TL R ++ ++ D
Sbjct: 59 KGIKNAGQILSTMTVLDDKDRLLTTVKLKSNTLNVTLPRQTLIDIIKSYVKD-------- 110
Query: 122 LPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLP 181
IF + H+ NE V + E DL + ADG S +R+
Sbjct: 111 --DAIFTNHEVT---HIDNETDKV-----TIHFAEQESEAFDLCIGADGIHSKVRQSVNA 160
Query: 182 DFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKK 241
D K+ Y GY +RG++D ++++ + K +G + L N +
Sbjct: 161 DSKVLYQGYTCFRGLIDDIDLKHPDCAKEYWGRKGRVG--------------IVPLLNNQ 206
Query: 242 LNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIP-ELAKVMKETKEP--FLNF 298
W +N E K +S K H +A P E+ +++ + E L+
Sbjct: 207 AYWFITINSKENNHKYSS---------FGKPHLQAYFNHYPNEVREILDKQSETGILLHN 257
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALE 358
IYD PL+ + +L+G+AAH TTP+ + + DA VL C + E AL+
Sbjct: 258 IYDLKPLKSFVYGRTILLGDAAHATTPNMGQGAGQAMEDAIVLVNCFNAYD---FEKALQ 314
Query: 359 EYQLTRLPVTSKQVLHARRLGRIKQ 383
Y R+ T+K + +R++G+I Q
Sbjct: 315 RYDKIRVKHTAKVIKRSRKIGKIAQ 339
>ref|ZP_00281538.1| COG0654: 2-polyprenyl-6-methoxyphenol hydroxylase and related
FAD-dependent oxidoreductases [Burkholderia fungorum
LB400]
Length = 403
Score = 89.4 bits (220), Expect = 2e-16
Identities = 94/388 (24%), Positives = 165/388 (42%), Gaps = 51/388 (13%)
Query: 6 RKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW 65
RK + ++GG IGG++ AL G+DV + E+T S GAG+ + + +++++
Sbjct: 3 RKLRVGVIGGGIGGLALTAALAQQGFDVRIFERTGS---FGEVGAGIQVTPNAVKVLEAM 59
Query: 66 --------ISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDE-----SLNFRAAHWG 112
++ PQ + + +N D+ K++ R +E + F H
Sbjct: 60 GLGDALRKVAFLPQAI--------VGRNW--DTAKEIFRIPLAEECPRLYNAPFFHVHRA 109
Query: 113 DLHGVLYESLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCL 172
DLH +L + +P + E V + + E DL+V ADG
Sbjct: 110 DLHHLLIDQVPAHAATLATACVDVRQTGETA-------VARFEDGSEFEADLIVGADGVR 162
Query: 173 SSIRRKYLPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHS 232
S++R K + ++G +R V+ + T PD F L +H
Sbjct: 163 STVRSKLFGETAPGFTGNMCFRAVVPVEGDFDFVT--------PDSS----FWLGPKSHV 210
Query: 233 VLYELPNKKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETK 292
V Y + K I VN+ V+ + +++ A + W P L ++ + +
Sbjct: 211 VTYYVRGGKAVNIVAVNETADWVEESWNAPSSREELLA-----AFEGWHPNLIQLFERVE 265
Query: 293 EPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEM 352
F ++D DP+ + L+G+AAHP P + M+I D VL + + G++
Sbjct: 266 SVFKWGLFDRDPMPAWSRGRITLLGDAAHPMLPFLSQGAAMSIEDGYVLARSLTAHGSD- 324
Query: 353 LESALEEYQLTRLPVTSKQVLHARRLGR 380
+ SAL +Y+ RLP TS+ L +R GR
Sbjct: 325 VASALRDYEAERLPRTSRVQLESRERGR 352
>emb|CAE34845.1| putative hydroxylase [Bordetella bronchiseptica RB50]
gi|33603456|ref|NP_891016.1| putative hydroxylase
[Bordetella bronchiseptica RB50]
Length = 406
Score = 89.4 bits (220), Expect = 2e-16
Identities = 93/372 (25%), Positives = 164/372 (44%), Gaps = 44/372 (11%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWISK 68
+ +I G IGG + A AL D +VLE+ P GAG+ L+ ++Q
Sbjct: 2 RVIIAGCGIGGAALAVALEKFKIDHVVLEQA---PRLEEVGAGVQLSPNGVAVLQHL--- 55
Query: 69 PPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESL-----NFRA----AHWGDLHGVLY 119
+H + + + + + + L R+ + +F A AH DL GVL
Sbjct: 56 ---GVHEALSKVAFEPRDLLYRDWQSGQVLMRNPLMPTIKEHFGAPYYHAHRADLLGVLT 112
Query: 120 ESLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKY 179
E L P G + E+ V A + + I GD+LV ADG S +R ++
Sbjct: 113 ERLDPAKLRLGSRIVDI----EQDARQVTATLA---DGTRIQGDILVGADGIHSLVRSRF 165
Query: 180 LPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPN 239
+ + SG AWRG++D + D+ + L +V+Y +
Sbjct: 166 FQADQPQASGCIAWRGIVDADAARHL-----------DISPSAHLWLGPERSAVIYYVSG 214
Query: 240 -KKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNF 298
+K+NWI ++P + S TT V D + + + W ++ +++ T +PF+
Sbjct: 215 GRKINWICIGSRPGDRKESWSATTTV--DEVLREYAG----WNEQVTGLIRLTDKPFVTA 268
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALE 358
+YD PL+ + L+G++AH P++ + ++ DA VL + +++ G + + ALE
Sbjct: 269 LYDRAPLDSWINGRIALLGDSAHAMLPYHAQGAVQSMEDAWVLARTLQQSGGD-IPPALE 327
Query: 359 EYQLTRLPVTSK 370
YQ R T++
Sbjct: 328 RYQSLRKDRTAR 339
>ref|NP_886155.1| putative hydroxylase [Bordetella parapertussis 12822]
gi|33574641|emb|CAE39292.1| putative hydroxylase
[Bordetella parapertussis]
Length = 406
Score = 87.0 bits (214), Expect = 1e-15
Identities = 92/372 (24%), Positives = 163/372 (43%), Gaps = 44/372 (11%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWISK 68
+ +I G IGG + A AL D +VLE+ P GAG+ L+ ++Q
Sbjct: 2 RVIIAGCGIGGAALAVALEKFKIDHVVLEQA---PRLEEVGAGVQLSPNGVAVLQHL--- 55
Query: 69 PPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESL-----NFRA----AHWGDLHGVLY 119
+H + + + + + + L R+ + +F A AH DL GVL
Sbjct: 56 ---GVHEALSKVAFEPRDLLYRDWQSGQVLMRNPLMPTIKEHFGAPYYHAHRADLLGVLT 112
Query: 120 ESLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKY 179
E L P G + E+ V A + + I GD+LV AD S +R ++
Sbjct: 113 ERLDPAKLRLGSRIVDI----EQDARQVTATLA---DGTRIQGDILVGADSIHSLVRSRF 165
Query: 180 LPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPN 239
+ + SG AWRG++D + D+ + L +V+Y +
Sbjct: 166 FQADQPQASGCIAWRGIVDADAARHL-----------DISPSAHLWLGPERSAVIYYVSG 214
Query: 240 -KKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNF 298
+K+NWI ++P + S TT V D + + + W ++ +++ T +PF+
Sbjct: 215 GRKINWICIGSRPGDRKESWSATTTV--DEVLREYAG----WNEQVTGLIRLTDKPFVTA 268
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALE 358
+YD PL+ + L+G++AH P++ + ++ DA VL + +++ G + + ALE
Sbjct: 269 LYDRAPLDSWINGRIALLGDSAHAMLPYHAQGAVQSMEDAWVLARTLQQSGGD-IPPALE 327
Query: 359 EYQLTRLPVTSK 370
YQ R T++
Sbjct: 328 RYQSLRKDRTAR 339
>gb|AAC46266.1| unknown [Bordetella pertussis]
Length = 406
Score = 86.3 bits (212), Expect = 2e-15
Identities = 89/372 (23%), Positives = 159/372 (41%), Gaps = 44/372 (11%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWISK 68
+ +I G IGG + A AL D +VLE+ P GAG+ L+ ++Q
Sbjct: 2 RVIIAGCGIGGAALAVALEKFKIDHVVLEQA---PRLEEVGAGVQLSPNGVAVLQHL--- 55
Query: 69 PPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESL-----NFRA----AHWGDLHGVLY 119
+H + + + + + + L R+ + +F A AH DL GVL
Sbjct: 56 ---GVHEALSKVAFEPRELLYRDWQSGQVLMRNPLMPTIKEHFGAPYYHAHRADLLGVLT 112
Query: 120 ESLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKY 179
E L P G + + + + GD+LV ADG S +R ++
Sbjct: 113 ERLDPAKLRLGSRIVDIDQD-------ARQVTATLADGTRVQGDILVGADGIHSLVRGRF 165
Query: 180 LPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPN 239
+ + SG AWRG++D + D+ + L +V+Y +
Sbjct: 166 FQADQPQASGCIAWRGIVDADAARHL-----------DISPSAHLWLGPERSAVIYYVSG 214
Query: 240 -KKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNF 298
+K+NWI ++P + S TT V D + + + W + +++ T +PF+
Sbjct: 215 GRKINWICIGSRPGDRKESWSATTTV--DEVLREYAG----WNELVTGLIRLTDKPFVTA 268
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALE 358
+YD PL+ + L+G++AH P++ + ++ DA VL + +++ G + + ALE
Sbjct: 269 LYDRAPLDSWINGRIALLGDSAHAMLPYHAQGAVQSMEDAWVLARTLQQSGGD-IPPALE 327
Query: 359 EYQLTRLPVTSK 370
YQ R T++
Sbjct: 328 RYQSLRKDRTAR 339
>ref|ZP_00216224.1| COG0654: 2-polyprenyl-6-methoxyphenol hydroxylase and related
FAD-dependent oxidoreductases [Burkholderia cepacia
R18194]
Length = 297
Score = 82.0 bits (201), Expect = 3e-14
Identities = 71/302 (23%), Positives = 127/302 (41%), Gaps = 34/302 (11%)
Query: 111 WGDLHGVLYESLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADG 170
W L+ + LP Q+F G F+ F ++ + V+TG DLLV ADG
Sbjct: 8 WNMLYSTMKAHLPAQVFHPGERFVRFEQNGDRITAYFASGRVETG-------DLLVGADG 60
Query: 171 CLSSIRRKYLPDFKLRYSGYCAWRGVLDFSEIENS--ETIKGIKKAYPDLGKCLYFDLAS 228
S++R + Y+GY AWRG++ + + S +KG G + L
Sbjct: 61 ARSAVREQVSAGLSPNYAGYVAWRGLVPEAALPASAAAVLKGTFAFQQGPGHLMLEYLVP 120
Query: 229 GTHSVLYELPNKKLNWIWYVNQPEPEVKGTSVTTK-------------VTSDMIQKMHQE 275
G E ++ NW+WY E T +T + + + + + Q
Sbjct: 121 GEDLSTQE-GQRRWNWVWYRKAAHGEELSTLLTDRSGIRHSFSLPPGALKDEDLTSLTQA 179
Query: 276 AEKIWIPELAKVMKETKEPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTI 335
++ + P +++ TK+PF+ I D ++++ + +L+G+AA PH ST
Sbjct: 180 SKALLAPTFQTLVEATKDPFVQAILDLQ-VKQMVYGRAILLGDAAFVPRPHTAGSTAKAA 238
Query: 336 LDAAVLGKCIEKWGTEMLESALEEYQLTRLPVTSKQVLHARRLGRIKQGLVLPDRDPFDP 395
+A L K + ++ AL +Q +L ++ +G + G+ + DR P
Sbjct: 239 ANALALAKALHS-AESGIDEALSRWQTGQL---------SQGIGMTEWGMNIGDRIMGIP 288
Query: 396 KS 397
+S
Sbjct: 289 RS 290
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 764,545,910
Number of Sequences: 2540612
Number of extensions: 33237445
Number of successful extensions: 99943
Number of sequences better than 10.0: 460
Number of HSP's better than 10.0 without gapping: 213
Number of HSP's successfully gapped in prelim test: 247
Number of HSP's that attempted gapping in prelim test: 99199
Number of HSP's gapped (non-prelim): 710
length of query: 428
length of database: 863,360,394
effective HSP length: 131
effective length of query: 297
effective length of database: 530,540,222
effective search space: 157570445934
effective search space used: 157570445934
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC144617.5


