

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144617.11 + phase: 0
(688 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
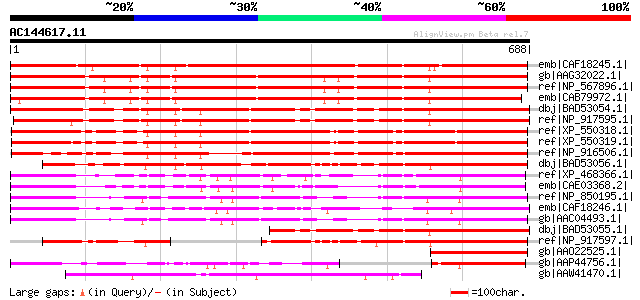
Score E
Sequences producing significant alignments: (bits) Value
emb|CAF18245.1| STYLOSA protein [Antirrhinum majus] 1026 0.0
gb|AAG32022.1| LEUNIG [Arabidopsis thaliana] 983 0.0
ref|NP_567896.1| WD-40 repeat family protein (LEUNIG) [Arabidops... 980 0.0
emb|CAB79972.1| putative protein [Arabidopsis thaliana] gi|49144... 955 0.0
dbj|BAD53054.1| putative LEUNIG [Oryza sativa (japonica cultivar... 823 0.0
ref|NP_917595.1| putative flower development regulator, LEUNI pr... 798 0.0
ref|XP_550318.1| putative transcriptional corepressor LEUNIG [Or... 764 0.0
ref|XP_550319.1| putative transcriptional corepressor LEUNIG [Or... 756 0.0
ref|NP_916506.1| P0013F10.18 [Oryza sativa (japonica cultivar-gr... 599 e-170
dbj|BAD53056.1| putative LEUNIG [Oryza sativa (japonica cultivar... 574 e-162
ref|XP_468366.1| putative LEUNIG [Oryza sativa (japonica cultiva... 455 e-126
emb|CAE03368.2| OSJNBb0065L13.11 [Oryza sativa (japonica cultiva... 440 e-122
ref|NP_850195.1| WD-40 repeat family protein [Arabidopsis thaliana] 391 e-107
emb|CAF18246.1| STY-L protein [Antirrhinum majus] 390 e-107
gb|AAC04493.1| expressed protein [Arabidopsis thaliana] gi|13605... 386 e-105
dbj|BAD53055.1| LEUNIG-like [Oryza sativa (japonica cultivar-gro... 383 e-104
ref|NP_917597.1| putative flower development regulator, LEUNI pr... 313 2e-83
gb|AAO22525.1| leunig [Brassica rapa subsp. pekinensis] 209 2e-52
gb|AAP44756.1| putative WD repeat protein [Oryza sativa (japonic... 174 7e-42
gb|AAW41470.1| hypothetical protein CNB00360 [Cryptococcus neofo... 144 1e-32
>emb|CAF18245.1| STYLOSA protein [Antirrhinum majus]
Length = 915
Score = 1026 bits (2652), Expect = 0.0
Identities = 547/719 (76%), Positives = 595/719 (82%), Gaps = 44/719 (6%)
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW
Sbjct: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
Query: 61 DIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQ---------QQQ 111
DIFIARTNEKHSEVAASYIETQL+KARE QQQQQQPQ Q P QQQQ Q+Q
Sbjct: 61 DIFIARTNEKHSEVAASYIETQLMKARE--QQQQQQPQQSQQPQQQQQQLHMQQLLLQRQ 118
Query: 112 QHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLL 171
Q Q QQ Q+QHQQQQQQQQQQQQQQQQ QQQ Q QQQQQQQQQQQQQQ + R LL
Sbjct: 119 AQQQQQQQQQHQQQHQQQQQQQQQQQQQQQQHQQQQQQQQQQQQQQQQQQQQQQRREGLL 178
Query: 172 NGGGANGLVG-------NPSTANAIATKMYEERLKLPL-QRDSLEDAAMKQRFGD---QL 220
N G ANG+VG NP TANA+ATKMYEE+LKLP+ QR+S++DAA KQRFGD QL
Sbjct: 179 N-GTANGIVGNDPLMRQNPGTANALATKMYEEKLKLPVSQRESMDDAAFKQRFGDNAGQL 237
Query: 221 LDPNHASILKSSAATGQPSGQVLHGAAGAMSPQVQARSQQLPGSTLDIKTEINPVLNPRA 280
LDPNH+SILKS+AA GQPSGQVLHG+AG MSPQVQARSQQ PG T DIK+E+NP+LNPRA
Sbjct: 238 LDPNHSSILKSAAA-GQPSGQVLHGSAGGMSPQVQARSQQFPGPTQDIKSEMNPILNPRA 296
Query: 281 AGPEGSLMAVPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKPFIQSPQPFHQLPMLTQQ 340
AGPEGSL+ +PGSNQGGNNLTLKGWPLTG + LRSGLLQQ K F+Q PQPFHQL ML+ Q
Sbjct: 297 AGPEGSLIGIPGSNQGGNNLTLKGWPLTGFDQLRSGLLQQPKSFMQGPQPFHQLQMLSPQ 356
Query: 341 HQQQLMLAQQNLASPSASD-DSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGG 399
HQQQLMLAQQNL SPSASD +SRRLRMLLN R++ + KDGLSN VGDV N+GSPLQ G
Sbjct: 357 HQQQLMLAQQNLTSPSASDVESRRLRMLLNNRSLSMGKDGLSNSVGDVGPNIGSPLQPGC 416
Query: 400 PPFPRGDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVG 459
PR DP+MLMKLK+AQLQ QQQQ N Q Q QQ H+LS QQ QS+NHN+ QQD +
Sbjct: 417 AVLPRADPEMLMKLKIAQLQQQQQQQQNSNQTQQQQHHTLSGQQPQSSNHNL-QQDKMM- 474
Query: 460 GGGGSVTGDGSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPST 519
G S G+GSMSNSFRGNDQ SK+Q GRKRKQPVSSSGPANSSGTANTAGPSPSSAPST
Sbjct: 475 -GTSSAAGEGSMSNSFRGNDQASKNQTGRKRKQPVSSSGPANSSGTANTAGPSPSSAPST 533
Query: 520 PSGSTHTPGDVISMPSLPHNGSSSKPLMMFSTDGN---TSPSNQL--------ADVDRFV 568
P STHTPGDV+SMP+LPH+GSSSKPLMMF D N TSPSNQL AD+DRFV
Sbjct: 534 P--STHTPGDVMSMPALPHSGSSSKPLMMFGADNNATLTSPSNQLWDDKDLVPADMDRFV 591
Query: 569 ADGSLDDNVESFLSHDDTDPRDPVGR-MDVSKGFTFSELNSVRASTNKVVCSHFSSDGKL 627
D ++DNVESFLS+DD DPRD VGR MDVSKGFTF+E++ VRAS +KVVC HFS DGKL
Sbjct: 592 DD--VEDNVESFLSNDDADPRDAVGRCMDVSKGFTFTEVSYVRASASKVVCCHFSPDGKL 649
Query: 628 LASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDN 686
LASGGHDKK VLWYTD+LK K TLEEHS LITDVRFSPSM RLATSS+DKTVRVWD DN
Sbjct: 650 LASGGHDKKAVLWYTDTLKPKTTLEEHSSLITDVRFSPSMARLATSSFDKTVRVWDADN 708
>gb|AAG32022.1| LEUNIG [Arabidopsis thaliana]
Length = 931
Score = 983 bits (2542), Expect = 0.0
Identities = 525/735 (71%), Positives = 588/735 (79%), Gaps = 60/735 (8%)
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
MSQTNWEADKMLDVYIHDYLVKRDLKA+AQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW
Sbjct: 1 MSQTNWEADKMLDVYIHDYLVKRDLKATAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
Query: 61 DIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQM 120
DIFIARTNEKHSEVAASYIETQ+IKAREQQ QQ Q PQ QQQQQQQQ Q+QMQQ+
Sbjct: 61 DIFIARTNEKHSEVAASYIETQMIKAREQQLQQSQHPQVS----QQQQQQQQQQIQMQQL 116
Query: 121 LLQR-------------QHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQ--------- 158
LLQR HQQQQQQQQQQQQQQQQQQQQ Q+Q QQQQQ
Sbjct: 117 LLQRAQQQQQQQQQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQ 176
Query: 159 ---QQQQQGRDRAHLLNGGGANGLVG---------NPSTANAIATKMYEERLKLPLQRDS 206
QQQ Q RD +HL N G ANGLVG NP + +++A+K YEER+K+P QR+S
Sbjct: 177 PTPQQQPQRRDGSHLAN-GSANGLVGNNSEPVMRQNPGSGSSLASKAYEERVKMPTQRES 235
Query: 207 LEDAAMKQRFGD---QLLDPNHASILKSSAATGQPSGQVLHGAAGAMSPQVQARSQQLPG 263
L++AAMK RFGD QLLDP+HASILKS+AA+GQP+GQVLH +G MSPQVQ R+QQLPG
Sbjct: 236 LDEAAMK-RFGDNVGQLLDPSHASILKSAAASGQPAGQVLHSTSGGMSPQVQTRNQQLPG 294
Query: 264 STLDIKTEINPVLNPRAAGPEGSLMAVPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKP 323
S +DIK+EINPVL PR A PEGSL+ +PGSNQG NNLTLKGWPLTG + LRSGLLQQQKP
Sbjct: 295 SAVDIKSEINPVLTPRTAVPEGSLIGIPGSNQGSNNLTLKGWPLTGFDQLRSGLLQQQKP 354
Query: 324 FIQSPQPFHQLPMLTQQHQQQLMLAQQNLASPSASDDSRRLRMLLNPRNMGVSKDGLSNP 383
F+QS Q FHQL MLT QHQQQLMLAQQNL S S S+++RRL+MLLN R+M + KDGL +
Sbjct: 355 FMQS-QSFHQLNMLTPQHQQQLMLAQQNLNSQSVSEENRRLKMLLNNRSMSLGKDGLGSS 413
Query: 384 VGDVVSNVGSPLQAGGPPFPRGDPDMLMKLKLA----QLQHQQQQNANPQQQQLQ----Q 435
VGDV+ NVGS LQ GG PRGD DML+KLK+A Q QHQQQ NP Q Q Q
Sbjct: 414 VGDVLPNVGSSLQPGGSLLPRGDTDMLLKLKMALLQQQQQHQQQGGGNPPQPQPQPQPLN 473
Query: 436 QHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDGSMSNSFRGNDQVSKSQPGRKRKQPVS 495
Q +L+N Q QS+NH++HQQ+ GGGGS+T DGS+SNSFRGN+QV K+Q GRKRKQPVS
Sbjct: 474 QLALTNPQPQSSNHSIHQQEKL--GGGGSITMDGSISNSFRGNEQVLKNQSGRKRKQPVS 531
Query: 496 SSGPANSSGTANTAGPSPSSAPSTPSGSTHTPGDVISMPSLPHNGSSSKPLMMFSTDGN- 554
SSGPANSSGTANTAGPSPSSAPSTP STHTPGDVISMP+LPH+G SSK +MMF T+G
Sbjct: 532 SSGPANSSGTANTAGPSPSSAPSTP--STHTPGDVISMPNLPHSGGSSKSMMMFGTEGTG 589
Query: 555 --TSPSNQLADVDRFVADGSLDDNVESFLSHDDTDPRDPVGR-MDVSKGFTFSELNSVRA 611
TSPSNQLAD+DRFV DGSLDDNVESFLS +D D RD V R MDVSKGFTF+E+NSVRA
Sbjct: 590 TLTSPSNQLADMDRFVEDGSLDDNVESFLSQEDGDQRDAVTRCMDVSKGFTFTEVNSVRA 649
Query: 612 STNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLA 671
ST KV C HFSSDGK+LAS GHDKK VLWYTD++K K TLEEH+ +ITD+RFSPS RLA
Sbjct: 650 STTKVTCCHFSSDGKMLASAGHDKKAVLWYTDTMKPKTTLEEHTAMITDIRFSPSQLRLA 709
Query: 672 TSSYDKTVRVWDVDN 686
TSS+DKTVRVWD DN
Sbjct: 710 TSSFDKTVRVWDADN 724
>ref|NP_567896.1| WD-40 repeat family protein (LEUNIG) [Arabidopsis thaliana]
gi|30580400|sp|Q9FUY2|LEUNG_ARATH Transcriptional
corepressor LEUNIG
Length = 931
Score = 980 bits (2533), Expect = 0.0
Identities = 524/735 (71%), Positives = 588/735 (79%), Gaps = 60/735 (8%)
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
MSQTNWEADKMLDVYIHDYLVKRDLKA+AQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW
Sbjct: 1 MSQTNWEADKMLDVYIHDYLVKRDLKATAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
Query: 61 DIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQM 120
DIFIARTNEKHSEVAASYIETQ+IKAREQQ QQ Q PQ QQQQQQQQ Q+QMQQ+
Sbjct: 61 DIFIARTNEKHSEVAASYIETQMIKAREQQLQQSQHPQVS----QQQQQQQQQQIQMQQL 116
Query: 121 LLQR-------------QHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQ--------- 158
LLQR HQQQQQQQQQQQQQQQQQQQQ Q+Q QQQQQ
Sbjct: 117 LLQRAQQQQQQQQQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQ 176
Query: 159 ---QQQQQGRDRAHLLNGGGANGLVG---------NPSTANAIATKMYEERLKLPLQRDS 206
QQQ Q RD +HL N G ANGLVG NP + +++A+K YEER+K+P QR+S
Sbjct: 177 PTPQQQPQRRDGSHLAN-GSANGLVGNNSEPVMRQNPGSGSSLASKAYEERVKMPTQRES 235
Query: 207 LEDAAMKQRFGD---QLLDPNHASILKSSAATGQPSGQVLHGAAGAMSPQVQARSQQLPG 263
L++AAMK RFGD QLLDP+HASILKS+AA+GQP+GQVLH +G MSPQVQ R+QQLPG
Sbjct: 236 LDEAAMK-RFGDNVGQLLDPSHASILKSAAASGQPAGQVLHSTSGGMSPQVQTRNQQLPG 294
Query: 264 STLDIKTEINPVLNPRAAGPEGSLMAVPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKP 323
S +DIK+EINPVL PR A PEGSL+ +PGSNQG NNLTLKGWPLTG + LRSGLLQQQKP
Sbjct: 295 SAVDIKSEINPVLTPRTAVPEGSLIGIPGSNQGSNNLTLKGWPLTGFDQLRSGLLQQQKP 354
Query: 324 FIQSPQPFHQLPMLTQQHQQQLMLAQQNLASPSASDDSRRLRMLLNPRNMGVSKDGLSNP 383
F+QS Q FHQL MLT QHQQQLMLAQQNL S S S+++RRL+MLLN R+M + KDGL +
Sbjct: 355 FMQS-QSFHQLNMLTPQHQQQLMLAQQNLNSQSVSEENRRLKMLLNNRSMTLGKDGLGSS 413
Query: 384 VGDVVSNVGSPLQAGGPPFPRGDPDMLMKLKLA----QLQHQQQQNANPQQQQLQ----Q 435
VGDV+ NVGS LQ GG PRGD DML+KLK+A Q Q+QQQ NP Q Q Q
Sbjct: 414 VGDVLPNVGSSLQPGGSLLPRGDTDMLLKLKMALLQQQQQNQQQGGGNPPQPQPQPQPLN 473
Query: 436 QHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDGSMSNSFRGNDQVSKSQPGRKRKQPVS 495
Q +L+N Q QS+NH++HQQ+ GGGGS+T DGS+SNSFRGN+QV K+Q GRKRKQPVS
Sbjct: 474 QLALTNPQPQSSNHSIHQQEKL--GGGGSITMDGSISNSFRGNEQVLKNQSGRKRKQPVS 531
Query: 496 SSGPANSSGTANTAGPSPSSAPSTPSGSTHTPGDVISMPSLPHNGSSSKPLMMFSTDGN- 554
SSGPANSSGTANTAGPSPSSAPSTP STHTPGDVISMP+LPH+G SSK +MMF T+G
Sbjct: 532 SSGPANSSGTANTAGPSPSSAPSTP--STHTPGDVISMPNLPHSGGSSKSMMMFGTEGTG 589
Query: 555 --TSPSNQLADVDRFVADGSLDDNVESFLSHDDTDPRDPVGR-MDVSKGFTFSELNSVRA 611
TSPSNQLAD+DRFV DGSLDDNVESFLS +D D RD V R MDVSKGFTF+E+NSVRA
Sbjct: 590 TLTSPSNQLADMDRFVEDGSLDDNVESFLSQEDGDQRDAVTRCMDVSKGFTFTEVNSVRA 649
Query: 612 STNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLA 671
ST KV C HFSSDGK+LAS GHDKK VLWYTD++K K TLEEH+ +ITD+RFSPS RLA
Sbjct: 650 STTKVTCCHFSSDGKMLASAGHDKKAVLWYTDTMKPKTTLEEHTAMITDIRFSPSQLRLA 709
Query: 672 TSSYDKTVRVWDVDN 686
TSS+DKTVRVWD DN
Sbjct: 710 TSSFDKTVRVWDADN 724
>emb|CAB79972.1| putative protein [Arabidopsis thaliana] gi|4914452|emb|CAB43692.1|
putative protein [Arabidopsis thaliana]
gi|7486768|pir||T08588 hypothetical protein L23H3.30 -
Arabidopsis thaliana
Length = 930
Score = 955 bits (2469), Expect = 0.0
Identities = 516/734 (70%), Positives = 581/734 (78%), Gaps = 67/734 (9%)
Query: 1 MSQTNWEADKM-------LDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLF 53
MSQTNWEADK+ LDVYIHDYLVKRDLKA+AQAFQAEGKVSSDPVAIDAPGGFLF
Sbjct: 1 MSQTNWEADKIQIFECNRLDVYIHDYLVKRDLKATAQAFQAEGKVSSDPVAIDAPGGFLF 60
Query: 54 EWWSVFWDIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQH 113
EWWSVFWDIFIARTNEKHSEVAASYIETQ+IKAREQQ QQ Q PQ QQQQQQQQ
Sbjct: 61 EWWSVFWDIFIARTNEKHSEVAASYIETQMIKAREQQLQQSQHPQVS----QQQQQQQQQ 116
Query: 114 QMQMQQMLLQR-------------QHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQ-- 158
Q+QMQQ+LLQR HQQQQQQQQQQQQQQQQQQQQ Q+Q QQQQQ
Sbjct: 117 QIQMQQLLLQRAQQQQQQQQQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQPPSQQQQQQS 176
Query: 159 ----------QQQQQGRDRAHLLNGGGANGLVG---------NPSTANAIATKMYEERLK 199
QQQ Q RD +HL N G ANGLVG NP + +++A+K YEER+K
Sbjct: 177 TPQHQQQPTPQQQPQRRDGSHLAN-GSANGLVGNNSEPVMRQNPGSGSSLASKAYEERVK 235
Query: 200 LPLQRDSLEDAAMKQRFGD---QLLDPNHASILKSSAATGQPSGQVLHGAAGAMSPQVQA 256
+P QR+SL++AAMK RFGD QLLDP+HASILKS+AA+GQP+GQVLH +G MSPQVQ
Sbjct: 236 MPTQRESLDEAAMK-RFGDNVGQLLDPSHASILKSAAASGQPAGQVLHSTSGGMSPQVQT 294
Query: 257 RSQQLPGSTLDIKTEINPVLNPRAAGPEGSLMAVPGSNQGGNNLTLKGWPLTGLEHLRSG 316
R+QQLPGS +DIK+EINPVL PR A PEGSL+ +PGSNQG NNLTLKGWPLTG + LRSG
Sbjct: 295 RNQQLPGSAVDIKSEINPVLTPRTAVPEGSLIGIPGSNQGSNNLTLKGWPLTGFDQLRSG 354
Query: 317 LLQQQKPFIQSPQPFHQLPMLTQQHQQQLMLAQQNLASPSASDDSRRLRMLLNPRNMGVS 376
LLQQQKPF+QS Q FHQL MLT QHQQQLMLAQQNL S S S+++RRL+MLLN R+M +
Sbjct: 355 LLQQQKPFMQS-QSFHQLNMLTPQHQQQLMLAQQNLNSQSVSEENRRLKMLLNNRSMTLG 413
Query: 377 KDGLSNPVGDVVSNVGSPLQAGGPPFPRGDPDMLMKLKLA----QLQHQQQQNANPQQQQ 432
KDGL + VGDV+ NVGS LQ GG PRGD DML+KLK+A Q Q+QQQ NP Q Q
Sbjct: 414 KDGLGSSVGDVLPNVGSSLQPGGSLLPRGDTDMLLKLKMALLQQQQQNQQQGGGNPPQPQ 473
Query: 433 LQ----QQHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDGSMSNSFRGNDQVSKSQPGR 488
Q Q +L+N Q QS+NH++HQQ+ GGGGS+T DGS+SNSFRGN+QV K+Q GR
Sbjct: 474 PQPQPLNQLALTNPQPQSSNHSIHQQEKL--GGGGSITMDGSISNSFRGNEQVLKNQSGR 531
Query: 489 KRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGSTHTPGDVISMPSLPHNGSSSKPLMM 548
KRKQPVSSSGPANSSGTANTAGPSPSSAPSTP STHTPGDVISMP+LPH+G SSK +MM
Sbjct: 532 KRKQPVSSSGPANSSGTANTAGPSPSSAPSTP--STHTPGDVISMPNLPHSGGSSKSMMM 589
Query: 549 FSTDGN---TSPSNQLADVDRFVADGSLDDNVESFLSHDDTDPRDPVGR-MDVSKGFTFS 604
F T+G TSPSNQLAD+DRFV DGSLDDNVESFLS +D D RD V R MDVSKGFTF+
Sbjct: 590 FGTEGTGTLTSPSNQLADMDRFVEDGSLDDNVESFLSQEDGDQRDAVTRCMDVSKGFTFT 649
Query: 605 ELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFS 664
E+NSVRAST KV C HFSSDGK+LAS GHDKK VLWYTD++K K TLEEH+ +ITD+RFS
Sbjct: 650 EVNSVRASTTKVTCCHFSSDGKMLASAGHDKKAVLWYTDTMKPKTTLEEHTAMITDIRFS 709
Query: 665 PSMPRLATSSYDKT 678
PS RLATSS+DKT
Sbjct: 710 PSQLRLATSSFDKT 723
>dbj|BAD53054.1| putative LEUNIG [Oryza sativa (japonica cultivar-group)]
gi|53792190|dbj|BAD52823.1| putative LEUNIG [Oryza
sativa (japonica cultivar-group)]
Length = 876
Score = 823 bits (2126), Expect = 0.0
Identities = 465/709 (65%), Positives = 538/709 (75%), Gaps = 64/709 (9%)
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
MSQTNWEADKMLDVYI+DY +KR+L+A+A+AFQAEGKVSSDPVAIDAPGGFLFEWWSVFW
Sbjct: 1 MSQTNWEADKMLDVYIYDYFMKRNLQATAKAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
Query: 61 DIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQM 120
DIFIARTNEKHS+VAASYIETQ +KAREQQQQQQQQP QQ+QQQ QH +QMQQM
Sbjct: 61 DIFIARTNEKHSDVAASYIETQHMKAREQQQQQQQQPP------QQRQQQPQH-IQMQQM 113
Query: 121 LLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLV 180
LLQR QQQQQQQQQQQQQ QQQQQQQQQQQQQQ RD HLLN G A+GL
Sbjct: 114 LLQRAAQQQQQQQQQQQQQ----------QQQQQQQQQQQQQQRRDGTHLLN-GTASGLP 162
Query: 181 G-------NPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGD---QLLDPNHASILK 230
G N STAN +ATKMYEERLKLP QRD L++ ++KQR+G+ QLLD N A +LK
Sbjct: 163 GNNPLMRQNQSTANVMATKMYEERLKLPSQRDGLDEVSIKQRYGENAGQLLDSNEA-LLK 221
Query: 231 SSAATGQPSGQVLHGAAGAMS---PQVQARSQQLPGSTLDIKTEINPVLNPRAAGPEGSL 287
+S A+GQ SGQ+LHG G +S QVQ+RS Q+PG IKTE+NP+L PR+AGPEGS
Sbjct: 222 AS-ASGQSSGQILHGTVGGLSGSLQQVQSRSPQIPGPAQSIKTEMNPILTPRSAGPEGSF 280
Query: 288 MAVPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKPFIQSPQPF-HQLPMLTQQHQQQLM 346
+ V GSNQ GNNLTLKGWPLTGLE LRSGLL QQK F+Q+ Q Q+ LT Q QQQLM
Sbjct: 281 IGVQGSNQAGNNLTLKGWPLTGLEQLRSGLL-QQKSFVQNQQQLQQQIHFLTPQQQQQLM 339
Query: 347 L-AQQNLASPSASD-DSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGGPPFPR 404
L AQQN+ASP++SD DSRRLRM+LN RN+G + G GD++ N+GSP +G
Sbjct: 340 LQAQQNMASPTSSDVDSRRLRMMLNNRNVGQTNSG-----GDIIPNIGSPSLSG------ 388
Query: 405 GDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGS 464
GD D+L+K K+AQ Q QQ +N QQ QQ ++S+QQSQS+N + Q+ G G+
Sbjct: 389 GDVDILIKKKIAQQQQLLQQQSNSQQHPQLQQPAVSSQQSQSSNQFLQQEK----PGIGT 444
Query: 465 VTGDGSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGST 524
+ DG M NSF G DQ +K KRK+P SSSG ANSSGTANTAGPSPSSAPSTP ST
Sbjct: 445 MPVDGGMPNSFGGVDQTTK-----KRKKPGSSSGRANSSGTANTAGPSPSSAPSTP--ST 497
Query: 525 HTPGDVISMPSLPHNGSSSKPLMMFSTDGN---TSPSNQLADVDRFVADGSLDDNVESFL 581
HTPGD +SMP L NG S+KPL+MF +DG TSP+N LADVDR + DGSLD+NVESFL
Sbjct: 498 HTPGDAMSMPQLQQNGGSAKPLVMFGSDGAGSLTSPANALADVDRLLEDGSLDENVESFL 557
Query: 582 SHDDTDPRDPVGR-MDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLW 640
S DD DPRD +GR MD SKGF F+E+ RAS KV C HFSSDGKLLA+GGHDKKV+LW
Sbjct: 558 SQDDMDPRDSLGRSMDASKGFGFAEVAKARASATKVTCCHFSSDGKLLATGGHDKKVLLW 617
Query: 641 YTD-SLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDNVS 688
T+ +LK ++LEEHS LITDVRFSPSM RLATSS+DKTVRVWD DN S
Sbjct: 618 CTEPALKPTSSLEEHSALITDVRFSPSMSRLATSSFDKTVRVWDADNTS 666
>ref|NP_917595.1| putative flower development regulator, LEUNI protein [Oryza sativa
(japonica cultivar-group)]
Length = 883
Score = 798 bits (2062), Expect = 0.0
Identities = 455/709 (64%), Positives = 531/709 (74%), Gaps = 69/709 (9%)
Query: 6 WEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFWDIFIA 65
W+ ++ LDVYI+DY +KR+L+A+A+AFQAEGKVSSDPVAIDAPGGFLFEWWSVFWDIFIA
Sbjct: 15 WKLEQWLDVYIYDYFMKRNLQATAKAFQAEGKVSSDPVAIDAPGGFLFEWWSVFWDIFIA 74
Query: 66 RTNEKHSEVAASYIE-----TQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQM 120
RTNEKHS+VAASYIE TQ +KAREQQQQQQQQP QQ+QQQ QH +QMQQM
Sbjct: 75 RTNEKHSDVAASYIEIHHKQTQHMKAREQQQQQQQQPP------QQRQQQPQH-IQMQQM 127
Query: 121 LLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLV 180
LLQR QQQQQQQQQQQQQ QQQQQQQQQQQQQQ RD HLLN G A+GL
Sbjct: 128 LLQRAAQQQQQQQQQQQQQ----------QQQQQQQQQQQQQQRRDGTHLLN-GTASGLP 176
Query: 181 G-------NPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGD---QLLDPNHASILK 230
G N STAN +ATKMYEERLKLP QRD L++ ++KQR+G+ QLLD N A +LK
Sbjct: 177 GNNPLMRQNQSTANVMATKMYEERLKLPSQRDGLDEVSIKQRYGENAGQLLDSNEA-LLK 235
Query: 231 SSAATGQPSGQVLHGAAGAMS---PQVQARSQQLPGSTLDIKTEINPVLNPRAAGPEGSL 287
+S A+GQ SGQ+LHG G +S QVQ+RS Q+PG IKTE+NP+L PR+AGPEGS
Sbjct: 236 AS-ASGQSSGQILHGTVGGLSGSLQQVQSRSPQIPGPAQSIKTEMNPILTPRSAGPEGSF 294
Query: 288 MAVPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKPFIQSPQPF-HQLPMLTQQHQQQLM 346
+ V GSNQ GNNLTLKGWPLTGLE LRSGLL QQK F+Q+ Q Q+ LT Q QQQLM
Sbjct: 295 IGVQGSNQAGNNLTLKGWPLTGLEQLRSGLL-QQKSFVQNQQQLQQQIHFLTPQQQQQLM 353
Query: 347 L-AQQNLASPSASD-DSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGGPPFPR 404
L AQQN+ASP++SD DSRRLRM+LN RN+G + G GD++ N+GSP +G
Sbjct: 354 LQAQQNMASPTSSDVDSRRLRMMLNNRNVGQTNSG-----GDIIPNIGSPSLSG------ 402
Query: 405 GDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGS 464
GD D+L+K K+AQ Q QQ +N QQ QQ ++S+QQSQS+N + Q+ G G+
Sbjct: 403 GDVDILIKKKIAQQQQLLQQQSNSQQHPQLQQPAVSSQQSQSSNQFLQQEK----PGIGT 458
Query: 465 VTGDGSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGST 524
+ DG M NSF G DQ +K KRK+P SSSG ANSSGTANTAGPSPSSAPSTP ST
Sbjct: 459 MPVDGGMPNSFGGVDQTTK-----KRKKPGSSSGRANSSGTANTAGPSPSSAPSTP--ST 511
Query: 525 HTPGDVISMPSLPHNGSSSKPLMMFSTDGN---TSPSNQLADVDRFVADGSLDDNVESFL 581
HTPGD +SMP L NG S+KPL+MF +DG TSP+N LADVDR + DGSLD+NVESFL
Sbjct: 512 HTPGDAMSMPQLQQNGGSAKPLVMFGSDGAGSLTSPANALADVDRLLEDGSLDENVESFL 571
Query: 582 SHDDTDPRDPVGR-MDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLW 640
S DD DPRD +GR MD SKGF F+E+ RAS KV C HFSSDGKLLA+GGHDKKV+LW
Sbjct: 572 SQDDMDPRDSLGRSMDASKGFGFAEVAKARASATKVTCCHFSSDGKLLATGGHDKKVLLW 631
Query: 641 YTD-SLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDNVS 688
T+ +LK ++LEEHS LITDVRFSPSM RLATSS+DKTVRVWD DN S
Sbjct: 632 CTEPALKPTSSLEEHSALITDVRFSPSMSRLATSSFDKTVRVWDADNTS 680
>ref|XP_550318.1| putative transcriptional corepressor LEUNIG [Oryza sativa (japonica
cultivar-group)] gi|55295951|dbj|BAD67819.1| putative
transcriptional corepressor LEUNIG [Oryza sativa
(japonica cultivar-group)]
Length = 877
Score = 764 bits (1972), Expect = 0.0
Identities = 431/700 (61%), Positives = 512/700 (72%), Gaps = 52/700 (7%)
Query: 3 QTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFWDI 62
Q+ WEA+KMLDVYIHDYL+KR+L+++A+AFQAEG VSSDPVAIDAPGGFL EWWSVFWDI
Sbjct: 8 QSAWEAEKMLDVYIHDYLLKRNLQSTAKAFQAEGSVSSDPVAIDAPGGFLLEWWSVFWDI 67
Query: 63 FIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLL 122
FIARTNEKHS+VAASYIETQ IKAREQQ Q Q QQ H QQ Q Q+QMQQ+LL
Sbjct: 68 FIARTNEKHSDVAASYIETQSIKAREQQPSQLQ----QQEAHSQQSSQ---QIQMQQLLL 120
Query: 123 QRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLVG- 181
QR QQQQQQQ QQQ QQQ ++QQ+QQQ+ + AH NGLV
Sbjct: 121 QRHAQQQQQQQSQQQPQQQ--------RRQQKQQQRSESSHLPTSAH-------NGLVSA 165
Query: 182 ------NPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGD---QLLDPNHASILKSS 232
+ S A++++ KMYEER+K +QRD+L++A KQRF + QLL+ N +S+LKS
Sbjct: 166 DPPTRQSTSAASSLSAKMYEERVKNSVQRDTLDEAPAKQRFTENIGQLLESNSSSMLKSV 225
Query: 233 AATGQPSGQVLHGAAGAMS---PQVQARSQQLPGSTLDIKTEINPVLNPRAAGPEGSLMA 289
A T Q SGQ+ HG+ G +S QVQAR+QQL ST +IK + N ++ RAAG +GSL+
Sbjct: 226 AITAQASGQIFHGSTGGVSGTLQQVQARNQQLQASTQEIKVDTNAAVHMRAAGADGSLIG 285
Query: 290 VPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKPFIQSPQPFHQLPMLTQQHQQQLMLAQ 349
VPG+N GNNLTLKGWPLTGL+ LRSG L QQK F+QSPQP H L LT Q QQ L+ AQ
Sbjct: 286 VPGANPAGNNLTLKGWPLTGLDQLRSGFL-QQKSFMQSPQPLHHLQFLTPQQQQLLLQAQ 344
Query: 350 QNLASPSASDDSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGGPPFPRGDPDM 409
QN+ S DSRRLRMLL+ RN+ +DG SN +V+ +VG LQ P R + DM
Sbjct: 345 QNMTSSPGEMDSRRLRMLLSSRNIVPGRDGQSNAYTEVIPSVGPSLQNMCSPVQRMETDM 404
Query: 410 LMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDG 469
LMK A QHQQ N QQQL QHSL +QQ +NH QQ+ G GSVT DG
Sbjct: 405 LMKKIAAIQQHQQSSN----QQQL-LQHSLLSQQPPISNHLPGQQEKM---GAGSVTIDG 456
Query: 470 SMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGSTHTPGD 529
S+SNSFRG++QVSK+Q GRKRKQP+SSSGPANSSGT NTA PSS PSTP S+ +PGD
Sbjct: 457 SLSNSFRGSEQVSKNQNGRKRKQPISSSGPANSSGTGNTA--VPSSEPSTP--SSQSPGD 512
Query: 530 VISMPSLPHNGSSSKPLMMF--STDGNT-SPSNQLADVDRFVADGSLDDNVESFLSHDDT 586
ISMPSL HN S SK L+++ ST G SPSNQLAD+DRFV DG L+D+V+SFLSHDD
Sbjct: 513 TISMPSLHHNASLSKALVVYGTSTAGTMGSPSNQLADMDRFVEDGCLEDHVDSFLSHDDA 572
Query: 587 DPRDPVGRMDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLK 646
D RD RM+ +KGF F E++SV+ASTNKVVC HFSSDGKLLA+GGHDKKVVLW+ ++LK
Sbjct: 573 DRRDG-SRMESTKGFIFREVSSVQASTNKVVCCHFSSDGKLLATGGHDKKVVLWHAETLK 631
Query: 647 QKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDN 686
QK+ LEEHSLLITDVRFSPS+PRLATSS+DKTVRVWD DN
Sbjct: 632 QKSVLEEHSLLITDVRFSPSIPRLATSSFDKTVRVWDADN 671
>ref|XP_550319.1| putative transcriptional corepressor LEUNIG [Oryza sativa (japonica
cultivar-group)] gi|55295950|dbj|BAD67818.1| putative
transcriptional corepressor LEUNIG [Oryza sativa
(japonica cultivar-group)]
Length = 875
Score = 756 bits (1952), Expect = 0.0
Identities = 429/700 (61%), Positives = 511/700 (72%), Gaps = 54/700 (7%)
Query: 3 QTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFWDI 62
Q+ WEA+KMLDVYIHDYL+KR+L+++A+AFQAEG VSSDPVAIDAPGGFL EWWSVFWDI
Sbjct: 8 QSAWEAEKMLDVYIHDYLLKRNLQSTAKAFQAEGSVSSDPVAIDAPGGFLLEWWSVFWDI 67
Query: 63 FIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLL 122
FIARTNEKHS+VAASYIE+ IKAREQQ Q Q QQ H QQ Q Q+QMQQ+LL
Sbjct: 68 FIARTNEKHSDVAASYIES--IKAREQQPSQLQ----QQEAHSQQSSQ---QIQMQQLLL 118
Query: 123 QRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLVG- 181
QR QQQQQQQ QQQ QQQ ++QQ+QQQ+ + AH NGLV
Sbjct: 119 QRHAQQQQQQQSQQQPQQQ--------RRQQKQQQRSESSHLPTSAH-------NGLVSA 163
Query: 182 ------NPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGD---QLLDPNHASILKSS 232
+ S A++++ KMYEER+K +QRD+L++A KQRF + QLL+ N +S+LKS
Sbjct: 164 DPPTRQSTSAASSLSAKMYEERVKNSVQRDTLDEAPAKQRFTENIGQLLESNSSSMLKSV 223
Query: 233 AATGQPSGQVLHGAAGAMS---PQVQARSQQLPGSTLDIKTEINPVLNPRAAGPEGSLMA 289
A T Q SGQ+ HG+ G +S QVQAR+QQL ST +IK + N ++ RAAG +GSL+
Sbjct: 224 AITAQASGQIFHGSTGGVSGTLQQVQARNQQLQASTQEIKVDTNAAVHMRAAGADGSLIG 283
Query: 290 VPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKPFIQSPQPFHQLPMLTQQHQQQLMLAQ 349
VPG+N GNNLTLKGWPLTGL+ LRSG L QQK F+QSPQP H L LT Q QQ L+ AQ
Sbjct: 284 VPGANPAGNNLTLKGWPLTGLDQLRSGFL-QQKSFMQSPQPLHHLQFLTPQQQQLLLQAQ 342
Query: 350 QNLASPSASDDSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGGPPFPRGDPDM 409
QN+ S DSRRLRMLL+ RN+ +DG SN +V+ +VG LQ P R + DM
Sbjct: 343 QNMTSSPGEMDSRRLRMLLSSRNIVPGRDGQSNAYTEVIPSVGPSLQNMCSPVQRMETDM 402
Query: 410 LMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDG 469
LMK A QHQQ N QQQL QHSL +QQ +NH QQ+ G GSVT DG
Sbjct: 403 LMKKIAAIQQHQQSSN----QQQL-LQHSLLSQQPPISNHLPGQQEKM---GAGSVTIDG 454
Query: 470 SMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGSTHTPGD 529
S+SNSFRG++QVSK+Q GRKRKQP+SSSGPANSSGT NTA PSS PSTP S+ +PGD
Sbjct: 455 SLSNSFRGSEQVSKNQNGRKRKQPISSSGPANSSGTGNTA--VPSSEPSTP--SSQSPGD 510
Query: 530 VISMPSLPHNGSSSKPLMMF--STDGNT-SPSNQLADVDRFVADGSLDDNVESFLSHDDT 586
ISMPSL HN S SK L+++ ST G SPSNQLAD+DRFV DG L+D+V+SFLSHDD
Sbjct: 511 TISMPSLHHNASLSKALVVYGTSTAGTMGSPSNQLADMDRFVEDGCLEDHVDSFLSHDDA 570
Query: 587 DPRDPVGRMDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLK 646
D RD RM+ +KGF F E++SV+ASTNKVVC HFSSDGKLLA+GGHDKKVVLW+ ++LK
Sbjct: 571 DRRDG-SRMESTKGFIFREVSSVQASTNKVVCCHFSSDGKLLATGGHDKKVVLWHAETLK 629
Query: 647 QKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDN 686
QK+ LEEHSLLITDVRFSPS+PRLATSS+DKTVRVWD DN
Sbjct: 630 QKSVLEEHSLLITDVRFSPSIPRLATSSFDKTVRVWDADN 669
>ref|NP_916506.1| P0013F10.18 [Oryza sativa (japonica cultivar-group)]
Length = 816
Score = 599 bits (1545), Expect = e-170
Identities = 370/700 (52%), Positives = 446/700 (62%), Gaps = 110/700 (15%)
Query: 3 QTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFWDI 62
Q+ WEA+KMLDVYIHDYL+KR+L+++A+AFQAEG VSSDPV W + I
Sbjct: 47 QSAWEAEKMLDVYIHDYLLKRNLQSTAKAFQAEGSVSSDPVG----------WLQIDHLI 96
Query: 63 FIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLL 122
+ T +++ TQ IKAREQQ Q QQ Q H QQ QQ +QMQQ+LL
Sbjct: 97 SLISTT-------LTFVHTQSIKAREQQPSQLQQ----QEAHSQQSSQQ---IQMQQLLL 142
Query: 123 QRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLVG- 181
QR QQQQQQQ Q QQ QQQ++QQ+QQQ + +HL NGLV
Sbjct: 143 QRHAQQQQQQQSQ--------------QQPQQQRRQQKQQQRSESSHLPTSAH-NGLVSA 187
Query: 182 ------NPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGD---QLLDPNHASILKSS 232
+ S A++++ KMYEER+K +QRD+L++A KQRF + QLL+ N +S+LKS
Sbjct: 188 DPPTRQSTSAASSLSAKMYEERVKNSVQRDTLDEAPAKQRFTENIGQLLESNSSSMLKSV 247
Query: 233 AATGQPSGQVLHGAAGAMS---PQVQARSQQLPGSTLDIKTEINPVLNPRAAGPEGSLMA 289
A T Q SGQ+ HG+ G +S QVQAR+QQL ST
Sbjct: 248 AITAQASGQIFHGSTGGVSGTLQQVQARNQQLQAST------------------------ 283
Query: 290 VPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKPFIQSPQPFHQLPMLTQQHQQQLMLAQ 349
GL+ LRSG LQQ K F+QSPQP H L LT Q QQ L+ AQ
Sbjct: 284 ------------------QGLDQLRSGFLQQ-KSFMQSPQPLHHLQFLTPQQQQLLLQAQ 324
Query: 350 QNLASPSASDDSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGGPPFPRGDPDM 409
QN+ S DSRRLRMLL+ RN+ +DG SN +V+ +VG LQ P R + DM
Sbjct: 325 QNMTSSPGEMDSRRLRMLLSSRNIVPGRDGQSNAYTEVIPSVGPSLQNMCSPVQRMETDM 384
Query: 410 LMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDG 469
LMK A QHQQ N QQQL Q HSL +QQ +NH QQ+ G GSVT DG
Sbjct: 385 LMKKIAAIQQHQQSSN----QQQLLQ-HSLLSQQPPISNHLPGQQEKM---GAGSVTIDG 436
Query: 470 SMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGSTHTPGD 529
S+SNSFRG++QVSK+Q GRKRKQP+SSSGPANSSGT NTA PSS PSTPS + +PGD
Sbjct: 437 SLSNSFRGSEQVSKNQNGRKRKQPISSSGPANSSGTGNTA--VPSSEPSTPS--SQSPGD 492
Query: 530 VISMPSLPHNGSSSKPLMMF--STDGNT-SPSNQLADVDRFVADGSLDDNVESFLSHDDT 586
ISMPSL HN S SK L+++ ST G SPSNQLAD+DRFV DG L+D+V+SFLSHDD
Sbjct: 493 TISMPSLHHNASLSKALVVYGTSTAGTMGSPSNQLADMDRFVEDGCLEDHVDSFLSHDDA 552
Query: 587 DPRDPVGRMDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLK 646
D RD GF F E++SV+ASTNKVVC HFSSDGKLLA+GGHDKKVVLW+ ++LK
Sbjct: 553 DRRDGSRMESTKAGFIFREVSSVQASTNKVVCCHFSSDGKLLATGGHDKKVVLWHAETLK 612
Query: 647 QKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDN 686
QK+ LEEHSLLITDVRFSPS+PRLATSS+DKTVRVWD DN
Sbjct: 613 QKSVLEEHSLLITDVRFSPSIPRLATSSFDKTVRVWDADN 652
>dbj|BAD53056.1| putative LEUNIG [Oryza sativa (japonica cultivar-group)]
gi|53792192|dbj|BAD52825.1| putative LEUNIG [Oryza
sativa (japonica cultivar-group)]
Length = 774
Score = 574 bits (1480), Expect = e-162
Identities = 354/663 (53%), Positives = 428/663 (64%), Gaps = 81/663 (12%)
Query: 44 AIDAPGGFLFEWWSVFWDIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSP 103
AIDAPGGFLFEWWS+FWDIFIA+T+ +HS+VA SYIETQ KA
Sbjct: 6 AIDAPGGFLFEWWSIFWDIFIAQTDREHSDVATSYIETQQAKAE---------------- 49
Query: 104 HQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQ 163
HQ+QQQQQ H +QHQ QQ Q QQ Q+ QQQQ QQQQQQQQQQQQ
Sbjct: 50 HQKQQQQQYHH---------QQHQHQQIQMQQMLLQRAAQQQQ----QQQQQQQQQQQQL 96
Query: 164 GRDRAHLLNG--GGANG----LVGNPSTANAIATKMYEERLKLPLQRDSLEDAAMK--QR 215
RD +HL NG G +G + NP+ ANA+A K+YEERLKLP QRDSL++A++K QR
Sbjct: 97 HRDGSHLFNGITSGFSGNDLLMRHNPAIANAMAVKIYEERLKLPSQRDSLDEASIKLQQR 156
Query: 216 FGD---QLLDPNHASILKSSAATGQPSGQVLHGAAGAMSP---QVQARSQQLPGSTLDIK 269
+G+ Q+LDPN AS+LK+ A GQ SG +L G G +S QVQARS +LP +IK
Sbjct: 157 YGEKYGQVLDPNQASLLKA-ATCGQSSGPILPGGIGDLSSTLQQVQARSPRLPIPEQNIK 215
Query: 270 TEINPVLNPRAAGPEGSLMAVPGSNQGGNNLTLKGWPLTGLEHLRSGLLQQQKPFIQSPQ 329
INP+L R +GSL+ + GSN GG N LKGW L QKP +QSPQ
Sbjct: 216 IRINPILTNRDVISDGSLLGLQGSNHGGRNFMLKGWSL------------MQKPLLQSPQ 263
Query: 330 PFHQLPMLTQQHQQQLMLAQQNLASPSASD-DSRRLRMLLNPRNMGVSKDG-LSNPVGDV 387
F QL LT Q QQ L+ QN+AS A+D ++RRL ML N +NM + DG ++N G +
Sbjct: 264 QFQQLQFLTPQ-QQLLLQTHQNMASLPANDVETRRLWMLHNNKNMAIHLDGQINNNSGHI 322
Query: 388 VSNVGSPLQAGGPPFPRGDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSA 447
+ N+GSP Q GG R DML+ K+A LQ QQQ + QQQQLQQ ++S+QQ+QS
Sbjct: 323 IPNIGSPDQIGGS---RNKIDMLIA-KIAHLQQLQQQG-HSQQQQLQQS-TISHQQAQSL 376
Query: 448 NHNMHQQDNKVGGGGGSVTGDGSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTAN 507
N HQQ +G DGS+ NSF ++ SK KRK+ VSSS ANSSGT+N
Sbjct: 377 NQLHHQQAQSIGS-----MLDGSIPNSFGLANRASK-----KRKKIVSSSERANSSGTSN 426
Query: 508 TAGPSPSSAPSTPSGSTHTPGDVISMPSLPHNGSSSKPLMMFSTDGNTS---PSNQLADV 564
G S SSAPSTP THTP D +SMP L +NG SK L MF D S P+N L DV
Sbjct: 427 NVGSSSSSAPSTPF--THTPRDEMSMPQLKYNGGKSKSLSMFGYDDTKSLISPTNPLGDV 484
Query: 565 DRFVADGSLDDNVESFLSHDDTDPRDPVGR-MDVSKGFTFSELNSVRASTNKVVCSHFSS 623
D+ DGSLD+NVESFLS +D DP++ +G MD SKGF F E+ RASTNKV C HFSS
Sbjct: 485 DQLQEDGSLDENVESFLSQEDMDPQETMGHCMDASKGFGFIEVAKARASTNKVDCCHFSS 544
Query: 624 DGKLLASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWD 683
DGKLLA+GGHDKKVVLW+TD L KA EEHS++ITDVRFS M RLATSS+DKT+RVWD
Sbjct: 545 DGKLLATGGHDKKVVLWFTDDLNIKAIFEEHSMIITDVRFSSIMTRLATSSFDKTIRVWD 604
Query: 684 VDN 686
+N
Sbjct: 605 ANN 607
>ref|XP_468366.1| putative LEUNIG [Oryza sativa (japonica cultivar-group)]
gi|47848544|dbj|BAD22396.1| putative LEUNIG [Oryza
sativa (japonica cultivar-group)]
gi|47847864|dbj|BAD21657.1| putative LEUNIG [Oryza
sativa (japonica cultivar-group)]
Length = 802
Score = 455 bits (1170), Expect = e-126
Identities = 302/717 (42%), Positives = 402/717 (55%), Gaps = 156/717 (21%)
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
M+Q+NWEADKMLDVYI+DYL+KR+L+++A+AF AEGKV++DPVAIDAPGGFLFEWWSVFW
Sbjct: 1 MAQSNWEADKMLDVYIYDYLLKRNLQSTAKAFMAEGKVAADPVAIDAPGGFLFEWWSVFW 60
Query: 61 DIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQM 120
DIFIARTNEKHSEVAA+Y+E Q IKARE HQ QMQ
Sbjct: 61 DIFIARTNEKHSEVAAAYLEAQQIKARE------------------------HQQQMQ-- 94
Query: 121 LLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLV 180
QQ Q Q +H Q Q+ G +N ++G++
Sbjct: 95 ------------------MQQLQLIQHRHAQLQRTNASHPSLNGP-----INTLNSDGIL 131
Query: 181 GNPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGDQLLDPNHASILKSSAATGQPSG 240
G+ STA+ +A KMYEERLK P DS G QLLD + ++LKS+A +G
Sbjct: 132 GH-STASVLAAKMYEERLKHPQSLDS---------EGSQLLDASRMALLKSAAT--NHAG 179
Query: 241 QVLHGAAGAMS---PQVQARSQQLPGSTLDIKTEINP-------VLNPRAAGPEGSLMAV 290
Q++ G G +S Q+Q+R+QQ T+DIK+E N ++P + +G +
Sbjct: 180 QLVPGTPGNVSTTLQQIQSRNQQ----TMDIKSEGNMGVAQRSLPMDPSSLYGQGIIQPK 235
Query: 291 P-----GSNQGGNNLTLKGWPLTGLEHLRSGL-LQQQKPFIQSPQPFHQLPMLTQQHQQQ 344
P G NQG + L LKGWPLTG++ LR L Q QKPF+ + F +++ Q QQQ
Sbjct: 236 PGLGGAGLNQGVSGLPLKGWPLTGIDQLRPNLGAQMQKPFLSTQSQFQ---LMSPQQQQQ 292
Query: 345 LML---AQQNLASPSASDDSRRLRMLLNPRN-MGVSKDGLSNPVGDVVSN---VGSPLQA 397
+ AQ NL++ S D L+PR +++ GL+ G + SP+Q+
Sbjct: 293 FLAQAQAQGNLSNSSNYGD-------LDPRRYTALTRGGLNGKDGQPAGTDGCISSPMQS 345
Query: 398 GGPPFPRGDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNK 457
P R D + L+K+ QQ ++ Q Q+ QQQ S QQ QS M Q +
Sbjct: 346 SSPKV-RSDQEYLIKV---------QQTSSQQPQEQQQQQSQQQQQQQSQQQQMQQSN-- 393
Query: 458 VGGGGGSVTGDGSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAP 517
RKRKQP +SSG ANS+GT NT GPS +S P
Sbjct: 394 ------------------------------RKRKQP-TSSGAANSTGTGNTVGPSTNSPP 422
Query: 518 STPSGSTHTPGDVISMPSLPHNGSSSKPLMMFSTDGN--TSPSNQLADVDRFVADGSLDD 575
STP STHTPGD + MP + K L+M+ DG S SNQ+ D++ F GSL+D
Sbjct: 423 STP--STHTPGDGLGMPG--NMRHVPKNLVMYGADGTGLASSSNQMDDLEPFGDVGSLED 478
Query: 576 NVESFLSHDDTDPRDPVGRMD---------VSKGFTFSELNSVRASTNKVVCSHFSSDGK 626
NVESFL++DD D RD + SKGFTF+E+N +R + +KVVC HFSSDGK
Sbjct: 479 NVESFLANDDGDARDIFAALKRSPAEPNPAASKGFTFNEVNCLRTNNSKVVCCHFSSDGK 538
Query: 627 LLASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWD 683
+LAS GH+KK VLW D+ + + T EEHSL+ITDVRF P+ +LATSS+D+T+++W+
Sbjct: 539 ILASAGHEKKAVLWNMDTFQSQYTSEEHSLIITDVRFRPNSSQLATSSFDRTIKLWN 595
>emb|CAE03368.2| OSJNBb0065L13.11 [Oryza sativa (japonica cultivar-group)]
gi|50926376|ref|XP_473135.1| OSJNBb0065L13.11 [Oryza
sativa (japonica cultivar-group)]
Length = 793
Score = 440 bits (1132), Expect = e-122
Identities = 301/712 (42%), Positives = 392/712 (54%), Gaps = 155/712 (21%)
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
M+Q+NWEADKMLDVYI+DYL+KR+L+A+A++F AEGKVS+DPVAIDAPGGFLFEWWSVFW
Sbjct: 1 MAQSNWEADKMLDVYIYDYLLKRNLQATAKSFMAEGKVSADPVAIDAPGGFLFEWWSVFW 60
Query: 61 DIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQM 120
DIFIARTNEKHSE+AA+Y+E Q KARE
Sbjct: 61 DIFIARTNEKHSEIAAAYLEAQQTKARE-------------------------------- 88
Query: 121 LLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLV 180
HQQQ Q QQ Q QQ+ H Q Q+ G ++G ++G++
Sbjct: 89 -----HQQQMQMQQLQLIQQR-------HAQLQRTNATHPSLNGP-----ISGLNSDGIL 131
Query: 181 GNPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGDQLLDPNHASILKSSAATGQPSG 240
G PSTA+ +A KMYEERLK DS G QLLD + ++LKS A+ SG
Sbjct: 132 G-PSTASVLAAKMYEERLKHSHPMDS---------DGSQLLDASRLALLKS--ASTNHSG 179
Query: 241 QVLHGAAGAMSP---QVQARSQQLPGSTLDIKTEINPV-------LNPRAAGPEGSLMAV 290
Q + G G++S Q+QAR+QQ +DIK+E N ++P + +G +
Sbjct: 180 QSVPGTPGSVSTTLQQIQARNQQ----NIDIKSEGNMSVAQRSMPMDPSSLYGQGIIQPK 235
Query: 291 PGS-----NQGGNNLTLKGWPLTGLEHLRSGLL-QQQKPFIQSPQPFHQLPMLTQQHQQQ 344
PG NQG + L LKGWPLTG++ LR L Q QKPF+ + F +++ Q QQQ
Sbjct: 236 PGLGGGVLNQGVSGLPLKGWPLTGIDQLRPNLGGQMQKPFLSTQSQFQ---LMSPQQQQQ 292
Query: 345 LML---AQQNLASPSASDDSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGGPP 401
+ AQ NL + + D R+ R++ KDG P G + SP+Q+ P
Sbjct: 293 FLAQAQAQGNLGNSTNLGDMDPRRLSALTRSVLNGKDG--QPAG-TDGCITSPMQSSSPK 349
Query: 402 FPRGDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGG 461
R D + LMK Q Q Q QQ N QQQQ QQ
Sbjct: 350 V-RPDQEYLMKTSSQQTQEQLQQQHNQQQQQQTQQ------------------------- 383
Query: 462 GGSVTGDGSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPS 521
GN RKRKQP +SSG ANS+GTANT GPS +S PSTPS
Sbjct: 384 ---------------GN---------RKRKQP-TSSGAANSTGTANTVGPSTNSPPSTPS 418
Query: 522 GSTHTPGDVISMPSLPHNGSSSKPLMMFSTDGNTSPSNQ-LADVDRFVADGSLDDNVESF 580
THTPGD + M + K LMM+ +G PS+ L D+++F GSLDDNVESF
Sbjct: 419 --THTPGDGLGMTGNMRH--VPKNLMMYGVEGTGLPSSSNLDDLEQFGDMGSLDDNVESF 474
Query: 581 LSHDDTDPRDPVGRMD---------VSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASG 631
L++ D D RD + SKGFTFSE+N R + +KVVC HFSSDGK+LAS
Sbjct: 475 LANGDGDARDIFAAPEKSPAEPNPVASKGFTFSEVNCWRTNNSKVVCCHFSSDGKILASA 534
Query: 632 GHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWD 683
GH+KK VLW ++ + + T EEH+++ITDVRF P+ +LATSS+D+T+++W+
Sbjct: 535 GHEKKAVLWNMETFQTQYTAEEHAVIITDVRFRPNSNQLATSSFDRTIKLWN 586
>ref|NP_850195.1| WD-40 repeat family protein [Arabidopsis thaliana]
Length = 785
Score = 391 bits (1004), Expect = e-107
Identities = 285/719 (39%), Positives = 362/719 (49%), Gaps = 171/719 (23%)
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
M+Q+NWEADKMLDVYI+DYLVK+ L +A++F EGKVS DPVAIDAPGGFLFEWWSVFW
Sbjct: 1 MAQSNWEADKMLDVYIYDYLVKKKLHNTAKSFMTEGKVSPDPVAIDAPGGFLFEWWSVFW 60
Query: 61 DIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQM 120
DIFIARTNEKHSE AA+YIE Q KA+E
Sbjct: 61 DIFIARTNEKHSEAAAAYIEAQQGKAKE-------------------------------- 88
Query: 121 LLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGG-----G 175
QQ Q QQ Q +Q Q Q RD H GG G
Sbjct: 89 ----------QQMQ----------------IQQLQMMRQAQMQRRDPNHPSLGGPMNAIG 122
Query: 176 ANGLVGNPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGDQLLDPNHASILKSSAAT 235
+ G++G S A+A+A KMYEER+K P M LD A + ++
Sbjct: 123 SEGMIGQ-SNASALAAKMYEERMKQP--------NPMNSETSQPHLDARMALLKSATNHH 173
Query: 236 GQPSGQVLHGAAGAMSPQVQARSQQLPGSTLDIKTEINPVLNPR-------AAGPEGSLM 288
GQ G A Q+Q+R+QQ +IKTE+N +PR +G L
Sbjct: 174 GQIVQGNHQGGVSAALQQIQSRTQQ----PTEIKTEVNLGTSPRQLPVDPSTVYGQGILQ 229
Query: 289 AVPGS-----NQGGNNLTLKGWPLTGLEHLRSGL--LQQQKPFIQSPQPFHQLPMLTQQH 341
+ PG N G + L LKGWPLTG+E +R GL Q QK F+Q+ F QL QQH
Sbjct: 230 SKPGMGSAGLNPGVSGLPLKGWPLTGIEQMRPGLGGPQVQKSFLQNQSQF-QLSPQQQQH 288
Query: 342 QQQLML-AQQNLA-SPSASDDSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGG 399
Q + AQ N+ SP D R PR KDG N ++GSP+Q+
Sbjct: 289 QMLAQVQAQGNMTNSPMYGGDMDPRRFTGLPRGNLNPKDGQQNAND---GSIGSPMQSSS 345
Query: 400 PPFPRGDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVG 459
P QQ QQQ L +QQSQ N
Sbjct: 346 SKHISMPP--------------------VQQSSSQQQDHLLSQQSQQNN----------- 374
Query: 460 GGGGSVTGDGSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPST 519
RKRK P SSSGPANS+GT NT GPS +S PST
Sbjct: 375 ----------------------------RKRKGP-SSSGPANSTGTGNTVGPS-NSQPST 404
Query: 520 PSGSTHTPGDVISMP-SLPHNGSSSKPLMMFSTD---GNTSPSNQLADVDRFVADGSLDD 575
P STHTP D +++ ++ H S K MM+ +D G S +NQL D+D+F G+L+D
Sbjct: 405 P--STHTPVDGVAIAGNMHHVNSMPKGPMMYGSDGIGGLASSANQLDDMDQFGDVGALED 462
Query: 576 NVESFLSHDDTDPRDPVGRM--------DVSKGFTFSELNSVRASTNKVVCSHFSSDGKL 627
NVESFLS DD D G + + SK F+F+E++ +R S +KV+C FS DGKL
Sbjct: 463 NVESFLSQDDGDGGSLFGTLKRNSSVHTETSKPFSFNEVSCIRKSASKVICCSFSYDGKL 522
Query: 628 LASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDN 686
LAS GHDKKV +W ++L+ ++T EEH+ +ITDVRF P+ +LATSS+DKT+++WD +
Sbjct: 523 LASAGHDKKVFIWNMETLQVESTPEEHAHIITDVRFRPNSTQLATSSFDKTIKIWDASD 581
Score = 35.4 bits (80), Expect = 6.2
Identities = 29/93 (31%), Positives = 47/93 (50%), Gaps = 4/93 (4%)
Query: 590 DPVGRMDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKA 649
D V +S G EL++ + VV D LL GG+ + + LW T K
Sbjct: 696 DAVKLWSLSSGDCIHELSNSGNKFHSVVFHPSYPD--LLVIGGY-QAIELWNTMENKCM- 751
Query: 650 TLEEHSLLITDVRFSPSMPRLATSSYDKTVRVW 682
T+ H +I+ + SPS +A++S+DK+V++W
Sbjct: 752 TVAGHECVISALAQSPSTGVVASASHDKSVKIW 784
>emb|CAF18246.1| STY-L protein [Antirrhinum majus]
Length = 777
Score = 390 bits (1003), Expect = e-107
Identities = 282/714 (39%), Positives = 359/714 (49%), Gaps = 166/714 (23%)
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
M+Q+NWEADKMLDVYIHDYL+KR L SA+AF EGKV++DPVAIDAPGGFLFEWWSVFW
Sbjct: 1 MAQSNWEADKMLDVYIHDYLLKRKLHNSAKAFMTEGKVATDPVAIDAPGGFLFEWWSVFW 60
Query: 61 DIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQM 120
DIFIARTN+KHSE AA+YIETQ IKAREQQQQ MQMQQ+
Sbjct: 61 DIFIARTNDKHSEAAAAYIETQQIKAREQQQQ----------------------MQMQQL 98
Query: 121 LLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLV 180
Q QQ+ Q Q++ P H +N + G++
Sbjct: 99 ------------QLLQQRNAQLQRRDPNHP---------------PLGGPMNSMNSEGMI 131
Query: 181 GNPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGDQLLDPNHASILKSSAATGQPSG 240
G PS A+ +A KMYEER+K P DS L+D N ++LKS++ G
Sbjct: 132 GQPS-ASVLAMKMYEERMKHPHSMDS--------ETSPGLIDANRMALLKSAS---NQQG 179
Query: 241 QVLHGAAGAMSPQVQARSQQLPGSTLDIKTEI-------NPVLNPRAAGPEGSL-----M 288
Q++ G G+MS +Q + Q P DIK E+ + ++P + + L +
Sbjct: 180 QLMQGNTGSMSAALQ-QMQGRPQMANDIKGEVGLGSTQKSLPMDPSSIYGQAILQSKSGL 238
Query: 289 AVPGSNQGGNNLTLKGWPLTGLEHLRSGL-LQQQKPFIQSPQPFHQLPMLTQQHQQQLML 347
G NQG L LKGWPLTG++ LR L LQ QKP +Q+ F +L Q QQ L
Sbjct: 239 GGAGLNQGVTGLPLKGWPLTGIDQLRPSLGLQVQKPNLQTQNQF----LLASQQQQVLAQ 294
Query: 348 AQQNLASPSASDDSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGGPPFPRGDP 407
AQ + + +S PR +KDG P D ++ SP+QA P
Sbjct: 295 AQ----AQGSLGNSPNYGYGGLPRGNSNAKDG-QPPRND--GSICSPVQANSP------- 340
Query: 408 DMLMKLKLAQLQH---QQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGS 464
K+K+AQ+Q QQQ QQQQLQQ + S S N N VG G+
Sbjct: 341 ----KMKMAQMQQASSQQQDQLQQQQQQLQQNNRKRKTHSSSGPANSTGTGNTVGPSPGT 396
Query: 465 VTGDGSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGST 524
P S+ P + TA+
Sbjct: 397 ----------------------------PQSTHTPGDGMATAS----------------- 411
Query: 525 HTPGDVISMPSLPHNGSSSKPLMMFSTD---GNTSPSNQLADVDRFVADGSLDDNVESFL 581
SL H S SK +MM+ D G S +NQL D++ F GSL+DNVESFL
Sbjct: 412 ----------SLQHVNSVSKSMMMYGADAAGGIASSTNQLDDLENFGDVGSLEDNVESFL 461
Query: 582 SHD-------DTDPRDPVGRMDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHD 634
SHD + + SKGF+F E+ +R + NKV C HFSSDGKLLAS GHD
Sbjct: 462 SHDGDGNLYGSLKQTLTEHKTETSKGFSFGEVGCIR-TRNKVTCCHFSSDGKLLASAGHD 520
Query: 635 KKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDNVS 688
KK VLW D+L+ + T EEH LITDVRF P+ +LAT+S+DK+VR+WD N S
Sbjct: 521 KKAVLWNMDTLQTETTPEEHQYLITDVRFRPNSTQLATASFDKSVRLWDAANPS 574
>gb|AAC04493.1| expressed protein [Arabidopsis thaliana] gi|13605815|gb|AAK32893.1|
At2g32700/F24L7.16 [Arabidopsis thaliana]
gi|25090107|gb|AAN72230.1| At2g32700/F24L7.16
[Arabidopsis thaliana] gi|7486097|pir||T00798
hypothetical protein At2g32700 [imported] - Arabidopsis
thaliana gi|30685392|ref|NP_850192.1| WD-40 repeat
family protein [Arabidopsis thaliana]
gi|30685401|ref|NP_850194.1| WD-40 repeat family protein
[Arabidopsis thaliana] gi|30685398|ref|NP_850193.1|
WD-40 repeat family protein [Arabidopsis thaliana]
gi|18403052|ref|NP_565749.1| WD-40 repeat family protein
[Arabidopsis thaliana]
Length = 787
Score = 386 bits (992), Expect = e-105
Identities = 285/721 (39%), Positives = 362/721 (49%), Gaps = 173/721 (23%)
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
M+Q+NWEADKMLDVYI+DYLVK+ L +A++F EGKVS DPVAIDAPGGFLFEWWSVFW
Sbjct: 1 MAQSNWEADKMLDVYIYDYLVKKKLHNTAKSFMTEGKVSPDPVAIDAPGGFLFEWWSVFW 60
Query: 61 DIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQM 120
DIFIARTNEKHSE AA+YIE Q KA+E
Sbjct: 61 DIFIARTNEKHSEAAAAYIEAQQGKAKE-------------------------------- 88
Query: 121 LLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGG-----G 175
QQ Q QQ Q +Q Q Q RD H GG G
Sbjct: 89 ----------QQMQ----------------IQQLQMMRQAQMQRRDPNHPSLGGPMNAIG 122
Query: 176 ANGLVGNPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGDQLLDPNHASILKSSAAT 235
+ G++G S A+A+A KMYEER+K P M LD A + ++
Sbjct: 123 SEGMIGQ-SNASALAAKMYEERMKQP--------NPMNSETSQPHLDARMALLKSATNHH 173
Query: 236 GQPSGQVLHGAAGAMSPQVQARSQQLPGSTLDIKTEINPVLNPR-------AAGPEGSLM 288
GQ G A Q+Q+R+QQ +IKTE+N +PR +G L
Sbjct: 174 GQIVQGNHQGGVSAALQQIQSRTQQ----PTEIKTEVNLGTSPRQLPVDPSTVYGQGILQ 229
Query: 289 AVPGS-----NQGGNNLTLKGWPLTGLEHLRSGL--LQQQKPFIQSPQPFHQLPMLTQQH 341
+ PG N G + L LKGWPLTG+E +R GL Q QK F+Q+ F QL QQH
Sbjct: 230 SKPGMGSAGLNPGVSGLPLKGWPLTGIEQMRPGLGGPQVQKSFLQNQSQF-QLSPQQQQH 288
Query: 342 QQQLML-AQQNLA-SPSASDDSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGG 399
Q + AQ N+ SP D R PR KDG N ++GSP+Q+
Sbjct: 289 QMLAQVQAQGNMTNSPMYGGDMDPRRFTGLPRGNLNPKDGQQNAND---GSIGSPMQSSS 345
Query: 400 PPFPRGDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVG 459
P QQ QQQ L +QQSQ N
Sbjct: 346 SKHISMPP--------------------VQQSSSQQQDHLLSQQSQQNN----------- 374
Query: 460 GGGGSVTGDGSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPST 519
RKRK P SSSGPANS+GT NT GPS +S PST
Sbjct: 375 ----------------------------RKRKGP-SSSGPANSTGTGNTVGPS-NSQPST 404
Query: 520 PSGSTHTPGDVISMP-SLPHNGSSSKPLMMFSTD---GNTSPSNQLA--DVDRFVADGSL 573
P STHTP D +++ ++ H S K MM+ +D G S +NQL D+D+F G+L
Sbjct: 405 P--STHTPVDGVAIAGNMHHVNSMPKGPMMYGSDGIGGLASSANQLLQDDMDQFGDVGAL 462
Query: 574 DDNVESFLSHDDTDPRDPVGRM--------DVSKGFTFSELNSVRASTNKVVCSHFSSDG 625
+DNVESFLS DD D G + + SK F+F+E++ +R S +KV+C FS DG
Sbjct: 463 EDNVESFLSQDDGDGGSLFGTLKRNSSVHTETSKPFSFNEVSCIRKSASKVICCSFSYDG 522
Query: 626 KLLASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVD 685
KLLAS GHDKKV +W ++L+ ++T EEH+ +ITDVRF P+ +LATSS+DKT+++WD
Sbjct: 523 KLLASAGHDKKVFIWNMETLQVESTPEEHAHIITDVRFRPNSTQLATSSFDKTIKIWDAS 582
Query: 686 N 686
+
Sbjct: 583 D 583
Score = 35.4 bits (80), Expect = 6.2
Identities = 29/93 (31%), Positives = 47/93 (50%), Gaps = 4/93 (4%)
Query: 590 DPVGRMDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKA 649
D V +S G EL++ + VV D LL GG+ + + LW T K
Sbjct: 698 DAVKLWSLSSGDCIHELSNSGNKFHSVVFHPSYPD--LLVIGGY-QAIELWNTMENKCM- 753
Query: 650 TLEEHSLLITDVRFSPSMPRLATSSYDKTVRVW 682
T+ H +I+ + SPS +A++S+DK+V++W
Sbjct: 754 TVAGHECVISALAQSPSTGVVASASHDKSVKIW 786
>dbj|BAD53055.1| LEUNIG-like [Oryza sativa (japonica cultivar-group)]
gi|53792191|dbj|BAD52824.1| LEUNIG-like [Oryza sativa
(japonica cultivar-group)]
Length = 538
Score = 383 bits (983), Expect = e-104
Identities = 214/350 (61%), Positives = 257/350 (73%), Gaps = 28/350 (8%)
Query: 345 LMLAQQNLASPSASD-DSRRLRMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGGPPFP 403
++ AQQN+ASP++SD DSRRLRM+LN RN+G + G GD++ N+GSP +GG
Sbjct: 1 MLQAQQNMASPTSSDVDSRRLRMMLNNRNVGQTNSG-----GDIIPNIGSPSLSGG---- 51
Query: 404 RGDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGG 463
D D+L+K K+AQ Q QQ +N QQ QQ ++S+QQSQS+N + Q+ G G
Sbjct: 52 --DVDILIKKKIAQQQQLLQQQSNSQQHPQLQQPAVSSQQSQSSNQFLQQEKP----GIG 105
Query: 464 SVTGDGSMSNSFRGNDQVSKSQPGRKRKQPVSSSGPANSSGTANTAGPSPSSAPSTPSGS 523
++ DG M NSF G DQ +K KRK+P SSSG ANSSGTANTAGPSPSSAPSTPS
Sbjct: 106 TMPVDGGMPNSFGGVDQTTK-----KRKKPGSSSGRANSSGTANTAGPSPSSAPSTPS-- 158
Query: 524 THTPGDVISMPSLPHNGSSSKPLMMFSTDGN---TSPSNQLADVDRFVADGSLDDNVESF 580
THTPGD +SMP L NG S+KPL+MF +DG TSP+N LADVDR + DGSLD+NVESF
Sbjct: 159 THTPGDAMSMPQLQQNGGSAKPLVMFGSDGAGSLTSPANALADVDRLLEDGSLDENVESF 218
Query: 581 LSHDDTDPRDPVGR-MDVSKGFTFSELNSVRASTNKVVCSHFSSDGKLLASGGHDKKVVL 639
LS DD DPRD +GR MD SKGF F+E+ RAS KV C HFSSDGKLLA+GGHDKKV+L
Sbjct: 219 LSQDDMDPRDSLGRSMDASKGFGFAEVAKARASATKVTCCHFSSDGKLLATGGHDKKVLL 278
Query: 640 WYTD-SLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTVRVWDVDNVS 688
W T+ +LK ++LEEHS LITDVRFSPSM RLATSS+DKTVRVWD DN S
Sbjct: 279 WCTEPALKPTSSLEEHSALITDVRFSPSMSRLATSSFDKTVRVWDADNTS 328
Score = 36.2 bits (82), Expect = 3.7
Identities = 24/61 (39%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query: 127 QQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQ--QQQQGRDRAHLLNGGGANGLVGNPS 184
QQQQ QQQ QQ Q QQP QQ Q Q QQ++ ++GG N G
Sbjct: 63 QQQQLLQQQSNSQQHPQLQQPAVSSQQSQSSNQFLQQEKPGIGTMPVDGGMPNSFGGVDQ 122
Query: 185 T 185
T
Sbjct: 123 T 123
>ref|NP_917597.1| putative flower development regulator, LEUNI protein [Oryza sativa
(japonica cultivar-group)]
Length = 650
Score = 313 bits (801), Expect = 2e-83
Identities = 191/367 (52%), Positives = 239/367 (65%), Gaps = 32/367 (8%)
Query: 335 PMLTQQHQQQLMLAQQNLASPSASD-DSRRLRMLLNPRNMGVSKDG-LSNPVGDVVSNVG 392
P+LT + +++ +L A+D ++RRL ML N +NM + DG ++N G ++ N+G
Sbjct: 160 PILTNRD----VISDGSLLGLQANDVETRRLWMLHNNKNMAIHLDGQINNNSGHIIPNIG 215
Query: 393 SPLQAGGPPFPRGDPDMLMKLKLAQLQHQQQQNANPQQQQLQQQHSLSNQQSQSANHNMH 452
SP Q GG R DML+ K+A LQ QQQ + QQQQLQQ ++S+QQ+QS N H
Sbjct: 216 SPDQIGGS---RNKIDMLIA-KIAHLQQLQQQG-HSQQQQLQQS-TISHQQAQSLNQLHH 269
Query: 453 QQDNKVGGGGGSVTGDGSMSNSFRGNDQVSKS---------QPGRKRKQPVSSSGPANSS 503
QQ +G DGS+ NSF +++ K Q +KRK+ VSSS ANSS
Sbjct: 270 QQAQSIGS-----MLDGSIPNSFGLANRIGKLHVYNVSLVVQASKKRKKIVSSSERANSS 324
Query: 504 GTANTAGPSPSSAPSTPSGSTHTPGDVISMPSLPHNGSSSKPLMMFSTDGNTS---PSNQ 560
GT+N G S SSAPSTP THTP D +SMP L +NG SK L MF D S P+N
Sbjct: 325 GTSNNVGSSSSSAPSTPF--THTPRDEMSMPQLKYNGGKSKSLSMFGYDDTKSLISPTNP 382
Query: 561 LADVDRFVADGSLDDNVESFLSHDDTDPRDPVGR-MDVSKGFTFSELNSVRASTNKVVCS 619
L DVD+ DGSLD+NVESFLS +D DP++ +G MD SKGF F E+ RASTNKV C
Sbjct: 383 LGDVDQLQEDGSLDENVESFLSQEDMDPQETMGHCMDASKGFGFIEVAKARASTNKVDCC 442
Query: 620 HFSSDGKLLASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLATSSYDKTV 679
HFSSDGKLLA+GGHDKKVVLW+TD L KA EEHS++ITDVRFS M RLATSS+DKT+
Sbjct: 443 HFSSDGKLLATGGHDKKVVLWFTDDLNIKAIFEEHSMIITDVRFSSIMTRLATSSFDKTI 502
Query: 680 RVWDVDN 686
RVWD +N
Sbjct: 503 RVWDANN 509
Score = 176 bits (445), Expect = 3e-42
Identities = 101/176 (57%), Positives = 118/176 (66%), Gaps = 35/176 (19%)
Query: 44 AIDAPGGFLFEWWSVFWDIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSP 103
AIDAPGGFLFEWWS+FWDIFIA+T+ +HS+VA SYIETQ KA Q+QQQQQ
Sbjct: 6 AIDAPGGFLFEWWSIFWDIFIAQTDREHSDVATSYIETQQAKAEHQKQQQQQY------- 58
Query: 104 HQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQ 163
H QQ Q Q Q+QMQQMLLQR QQQQQQQQQQQQQQQQ
Sbjct: 59 HHQQHQHQ--QIQMQQMLLQRAAQQQQQQQQQQQQQQQQLH------------------- 97
Query: 164 GRDRAHLLNG--GGANG----LVGNPSTANAIATKMYEERLKLPLQRDSLEDAAMK 213
RD +HL NG G +G + NP+ ANA+A K+YEERLKLP QRDSL++A++K
Sbjct: 98 -RDGSHLFNGITSGFSGNDLLMRHNPAIANAMAVKIYEERLKLPSQRDSLDEASIK 152
Score = 36.6 bits (83), Expect = 2.8
Identities = 20/43 (46%), Positives = 22/43 (50%)
Query: 120 MLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQ 162
ML+ + QQ QQQ QQQQ QQ HQQ Q Q QQ
Sbjct: 229 MLIAKIAHLQQLQQQGHSQQQQLQQSTISHQQAQSLNQLHHQQ 271
Score = 35.8 bits (81), Expect = 4.8
Identities = 45/158 (28%), Positives = 63/158 (39%), Gaps = 22/158 (13%)
Query: 130 QQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLVGNPSTANAI 189
QQ QQQ QQQQ QQ QQ Q Q QQ + +L+G N AN I
Sbjct: 238 QQLQQQGHSQQQQLQQSTISHQQAQSLNQLHHQQAQSIGSMLDGSIPNSF----GLANRI 293
Query: 190 ATKMYEERLKLPLQRDSLEDAAMKQR----FGDQLLDPNHASILKSSAATGQPSGQVLHG 245
KL + SL A K+R + + + S S+++ PS H
Sbjct: 294 G--------KLHVYNVSLVVQASKKRKKIVSSSERANSSGTSNNVGSSSSSAPSTPFTHT 345
Query: 246 AAGAMS-PQVQARSQQLPGSTL----DIKTEINPVLNP 278
MS PQ++ + ++ D K+ I+P NP
Sbjct: 346 PRDEMSMPQLKYNGGKSKSLSMFGYDDTKSLISPT-NP 382
>gb|AAO22525.1| leunig [Brassica rapa subsp. pekinensis]
Length = 249
Score = 209 bits (533), Expect = 2e-52
Identities = 100/129 (77%), Positives = 115/129 (88%), Gaps = 1/129 (0%)
Query: 559 NQLADVDRFVADGSLDDNVESFLSHDDTDPRDPVGR-MDVSKGFTFSELNSVRASTNKVV 617
NQLAD+DRFV DGSLDDNVESFLSH+D D RD VGR MDVSKGFTF+E+NSVRAST+KV
Sbjct: 1 NQLADMDRFVEDGSLDDNVESFLSHEDGDQRDTVGRCMDVSKGFTFAEVNSVRASTSKVT 60
Query: 618 CSHFSSDGKLLASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLATSSYDK 677
C HFS DGK+LAS GHDKK V+W+TD++K K TLEEH+ +ITDVRFSP++PRLATSS+DK
Sbjct: 61 CCHFSLDGKMLASAGHDKKAVIWHTDTMKPKTTLEEHTAMITDVRFSPNLPRLATSSFDK 120
Query: 678 TVRVWDVDN 686
TVRVWD DN
Sbjct: 121 TVRVWDADN 129
>gb|AAP44756.1| putative WD repeat protein [Oryza sativa (japonica cultivar-group)]
gi|50920301|ref|XP_470511.1| putative WD repeat protein
[Oryza sativa (japonica cultivar-group)]
Length = 684
Score = 174 bits (442), Expect = 7e-42
Identities = 151/487 (31%), Positives = 213/487 (43%), Gaps = 162/487 (33%)
Query: 1 MSQTNWEADKMLDVYIHDYLVKRDLKASAQAFQAEGKVSSDPVAIDAPGGFLFEWWSVFW 60
M+++NWEADKMLDVYI+DYLVKR++ +A+AF EGKV++DPVAIDAPGGFLFEWWS+FW
Sbjct: 1 MARSNWEADKMLDVYIYDYLVKRNMHNTAKAFMTEGKVATDPVAIDAPGGFLFEWWSIFW 60
Query: 61 DIFIARTNEKHSEVAASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQM 120
DIF ART +K QPQPQ P +
Sbjct: 61 DIFDARTRDK-----------------------PPQPQPQPPP------------PIPID 85
Query: 121 LLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLV 180
+ R+ Q + Q QQQQ QQ++ P +
Sbjct: 86 IKSREQQMRLQLLQQQQNAHQQRRDPPLNAAMDAL------------------------- 120
Query: 181 GNPSTANAIATKMYEERLKLPLQRDSLEDAAMKQRFGDQLLDPNHASILKSSAATGQPSG 240
N + +A+KM ++R++ P DS DA+ QLLD N ++LK AT Q +G
Sbjct: 121 -NSDVSAVLASKMMQDRMRNPNPTDS--DAS------HQLLDANRIALLK--PATNQ-TG 168
Query: 241 QVLHGAAGAMS--PQVQARSQQ----------------------LPGSTLDIK------- 269
Q++ GA+ MS Q+ +R+QQ P D+K
Sbjct: 169 QLVQGASVNMSALQQIHSRNQQPVIPFHLLKLHWFMYRLSLLTLCPTIFKDMKGDAAMSQ 228
Query: 270 ----TEINPVLNPRAAGPEGSLMAVPGSNQGGNNLTLKGWPLT------------GLEHL 313
T+ + + P+ L++ G NQG ++ LKGWPLT G++ L
Sbjct: 229 RSMPTDPSTLYGSGMMQPKSGLVST-GLNQGVGSVPLKGWPLTKSLPTSCLLKVPGIDQL 287
Query: 314 RSGLLQQQKPFIQSPQPFHQLPMLTQQHQQQLMLAQQNLASPSASDDSRRLRMLLNPRNM 373
RS L QK + SP F QL+ QQ L + + S +
Sbjct: 288 RSN-LGVQKQLMASPNQF------------QLLSPQQQLIAQAQSQND------------ 322
Query: 374 GVSKDGLSNPVGDVVSNVGSPLQAGGPPFPRGDPDMLMKLKLAQLQH---QQQQNANPQQ 430
++ +GSP +G P + D +MKLK+AQ+Q + QQ
Sbjct: 323 --------------LARMGSPAPSGSPKVRPDESDYMMKLKMAQMQQPSGHRLMELQQQQ 368
Query: 431 QQLQQQH 437
QQLQQ +
Sbjct: 369 QQLQQDN 375
Score = 124 bits (310), Expect = 1e-26
Identities = 66/131 (50%), Positives = 84/131 (63%), Gaps = 10/131 (7%)
Query: 560 QLADVDRFVADGSLDDNVESFLSHDDTDPRDPVGRM-------DVSKGFTFSELNSVRAS 612
Q ++D FV D+NV+SFLS+DD D RD + D KG + SE + R S
Sbjct: 372 QQDNLDSFV---DFDENVDSFLSNDDGDGRDIFASLKKGSSEQDSLKGLSLSEFGNNRTS 428
Query: 613 TNKVVCSHFSSDGKLLASGGHDKKVVLWYTDSLKQKATLEEHSLLITDVRFSPSMPRLAT 672
NKVVC HFS+DGKLLAS GH+KKV LW D+L +EEH+ ITD+RF P+ +LAT
Sbjct: 429 NNKVVCCHFSTDGKLLASAGHEKKVFLWNMDNLNMDTKIEEHTNFITDIRFKPNSTQLAT 488
Query: 673 SSYDKTVRVWD 683
SS D TVR+W+
Sbjct: 489 SSSDGTVRLWN 499
>gb|AAW41470.1| hypothetical protein CNB00360 [Cryptococcus neoformans var.
neoformans JEC21] gi|58262734|ref|XP_568777.1|
hypothetical protein CNB00360 [Cryptococcus neoformans
var. neoformans JEC21]
Length = 936
Score = 144 bits (363), Expect = 1e-32
Identities = 141/497 (28%), Positives = 210/497 (41%), Gaps = 36/497 (7%)
Query: 75 AASYIETQLIKAREQQQQQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQ 134
A Y++ Q+ + ++Q QQQQQQ Q QQ QQQQQQQQ Q Q QQ Q Q QQQQQQQQ
Sbjct: 232 ANQYVQQQVQQIQQQHQQQQQQLQQQQQQQQQQQQQQQQQQQQQQQQRQPQQQQQQQQQQ 291
Query: 135 QQQQQQQQQQQQPQHQQQQQQQQQQQQ----QQGRDRAHLLNGGGANGLVGNPSTA--NA 188
QQQQQQQQQQQQ Q QQQQQQQQQQQQ QQ + + + +G P++
Sbjct: 292 QQQQQQQQQQQQQQQQQQQQQQQQQQQHAMMQQQQQQRFQFAPTSSPAAIGRPASRQDQQ 351
Query: 189 IATKMYEERLKLPLQRDSLEDAAMKQRFGDQLLDPNHASILKSSAATGQPSGQVLHGAAG 248
MY+ + ++ LQ + + M Q + N + ++++ Q S +H A
Sbjct: 352 AQPTMYQPQPQMELQPPAPQHPHMTPTQQQQHMIQNQSVNVQTAQQRAQQSLAEMHAQA- 410
Query: 249 AMSPQVQARSQQLPGSTLDIKTEINPVLNPRAAGPEGSLMAVPGSNQGGNNLTLKGWPLT 308
QVQA++QQ + ++ A + A+ Q + + W
Sbjct: 411 ----QVQAQAQQ---QQQLLHSQQQAQAQAAAQAAQQREQAIQAQQQHIQAVNSQMWEGL 463
Query: 309 GLEHLRSGLLQQQKPFI----QSPQPFHQLPMLTQQHQQQLMLAQQNLASPSASDDSRRL 364
GL H+ G++ + + PQ + H +L L + + + S++
Sbjct: 464 GLGHVNHGVMNYSAQSLGLGGKDPQSMDEEEKQRLIHTYKLTLQENQRKAMMGAQQSQQQ 523
Query: 365 RMLLNPRNMGVSKDGLSNPVGDVVSNVGSPLQAGGPPF-PRGDPDMLMKLKLAQLQHQQQ 423
+ + G N ++ + S G P P G +L+Q Q Q+
Sbjct: 524 TPQMQHQGQGGPGQLHQNQQYTMMPHQQSVHPQGYPDMRPNGTMTRNQSYELSQQQQPQE 583
Query: 424 QN-ANPQQQQLQQQHSLSNQQSQSANHNMHQQDNKVGGGGGSVTGDGS----MSNSFRGN 478
+N A P L Q + S+ SQSA+ NK S +S G
Sbjct: 584 ENKAAPTPGSLNQSTNNSSSASQSASTPTTSTANKPSPPERKRKRKNSEKENKPSSPFGM 643
Query: 479 DQVSKSQPGRKRKQP-VSSSGPANSSGTA---------NTAGPSPSSAPSTPSGSTHTPG 528
Q + Q G+ ++ P ++GP S A TA P P SA +P T
Sbjct: 644 SQFGQGQAGQGQQGPNAGAAGPVTSGANATNDPMRSIPGTAPPRPVSA--SPRNPTRREK 701
Query: 529 DVISMPSLPHNGSSSKP 545
+ LP ++ KP
Sbjct: 702 RNQAKEMLPPRTTTPKP 718
Score = 38.9 bits (89), Expect = 0.56
Identities = 22/49 (44%), Positives = 26/49 (52%)
Query: 109 QQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQ 157
QQQQH +Q Q Q+ Q Q Q Q Q Q Q Q Q + Q QQQ Q+
Sbjct: 14 QQQQHALQQAQARQQQIIQAQAQVQAQAQAQAQAQTRAAQSGQQQANQR 62
Score = 37.4 bits (85), Expect = 1.6
Identities = 22/48 (45%), Positives = 25/48 (51%)
Query: 116 QMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQ 163
Q QQ LQ+ +QQQ Q Q Q Q Q Q Q Q Q + Q QQQ Q
Sbjct: 14 QQQQHALQQAQARQQQIIQAQAQVQAQAQAQAQAQTRAAQSGQQQANQ 61
Score = 36.2 bits (82), Expect = 3.7
Identities = 25/70 (35%), Positives = 28/70 (39%), Gaps = 1/70 (1%)
Query: 92 QQQQQPQPQQSPHQQQQQQQQHQMQMQQMLLQRQHQQQQQQQQQQQQQQQQQQQQPQHQQ 151
QQQQ Q QQQ Q Q Q+Q Q Q Q Q + Q QQQ Q+ P H
Sbjct: 14 QQQQHALQQAQARQQQIIQAQAQVQAQAQ-AQAQAQTRAAQSGQQQANQRVVPPSPHHPS 72
Query: 152 QQQQQQQQQQ 161
Q Q
Sbjct: 73 PSPHMDHQFQ 82
Score = 35.4 bits (80), Expect = 6.2
Identities = 24/54 (44%), Positives = 26/54 (47%)
Query: 131 QQQQQQQQQQQQQQQQPQHQQQQQQQQQQQQQQGRDRAHLLNGGGANGLVGNPS 184
QQQQ QQ Q +QQQ Q Q Q Q Q Q Q + RA AN V PS
Sbjct: 14 QQQQHALQQAQARQQQIIQAQAQVQAQAQAQAQAQTRAAQSGQQQANQRVVPPS 67
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.310 0.126 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,229,503,827
Number of Sequences: 2540612
Number of extensions: 58649509
Number of successful extensions: 2201242
Number of sequences better than 10.0: 19556
Number of HSP's better than 10.0 without gapping: 14500
Number of HSP's successfully gapped in prelim test: 5366
Number of HSP's that attempted gapping in prelim test: 686215
Number of HSP's gapped (non-prelim): 379637
length of query: 688
length of database: 863,360,394
effective HSP length: 135
effective length of query: 553
effective length of database: 520,377,774
effective search space: 287768909022
effective search space used: 287768909022
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 79 (35.0 bits)
Medicago: description of AC144617.11


