

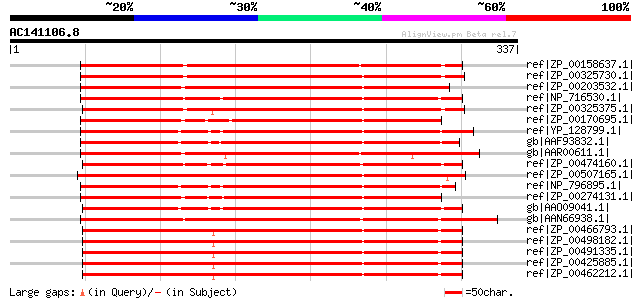
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141106.8 - phase: 0
(337 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|ZP_00158637.1| COG0667: Predicted oxidoreductases (related t... 285 2e-75
ref|ZP_00325730.1| COG0667: Predicted oxidoreductases (related t... 275 1e-72
ref|ZP_00203532.1| COG0667: Predicted oxidoreductases (related t... 271 3e-71
ref|NP_716530.1| oxidoreductase, aldo/keto reductase family [She... 259 1e-67
ref|ZP_00325375.1| COG0667: Predicted oxidoreductases (related t... 257 4e-67
ref|ZP_00170695.1| COG0667: Predicted oxidoreductases (related t... 251 2e-65
ref|YP_128799.1| putative oxidoreductase Tas, aldo/keto reductas... 250 4e-65
gb|AAF93832.1| oxidoreductase Tas, aldo/keto reductase family [V... 249 6e-65
gb|AAR00611.1| putative aldo/keto reductase family protein [Oryz... 246 7e-64
ref|ZP_00474160.1| Aldo/keto reductase [Chromohalobacter salexig... 245 2e-63
ref|ZP_00507165.1| Aldo/keto reductase [Polaromonas sp. JS666] g... 244 3e-63
ref|NP_796895.1| oxidoreductase Tas, aldo/keto reductase family ... 242 1e-62
ref|ZP_00274131.1| COG0667: Predicted oxidoreductases (related t... 241 2e-62
gb|AAO09041.1| Oxidoreductase Tas, aldo/keto reductase family [V... 241 3e-62
gb|AAN66938.1| oxidoreductase, aldo/keto reductase family [Pseud... 236 7e-61
ref|ZP_00466793.1| COG0667: Predicted oxidoreductases (related t... 234 2e-60
ref|ZP_00498182.1| COG0667: Predicted oxidoreductases (related t... 234 3e-60
ref|ZP_00491335.1| COG0667: Predicted oxidoreductases (related t... 234 3e-60
ref|ZP_00425885.1| Aldo/keto reductase [Burkholderia vietnamiens... 234 4e-60
ref|ZP_00462212.1| Aldo/keto reductase [Burkholderia cenocepacia... 234 4e-60
>ref|ZP_00158637.1| COG0667: Predicted oxidoreductases (related to aryl-alcohol
dehydrogenases) [Anabaena variabilis ATCC 29413]
Length = 345
Score = 285 bits (728), Expect = 2e-75
Identities = 138/254 (54%), Positives = 184/254 (72%), Gaps = 5/254 (1%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
+L VS +CLGTMT+G+QNT+ ++ Q LD A G+NF D+AEMYPVP A+T+G++E Y
Sbjct: 10 DLQVSDICLGTMTYGQQNTIEEAHQQLDYAIAQGVNFIDAAEMYPVPTSAETYGLTETYI 69
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDL 167
G W+K++ R++L+IATK+AGP W+R G K++D NI QA+D+SL R+Q DYIDL
Sbjct: 70 GEWLKNQQ--REQLIIATKIAGPGRGFKWVRDGAKAIDRNNIKQAVDDSLQRLQTDYIDL 127
Query: 168 YQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMK 227
YQIHWPDRYVP FG+T +DP Q +I I EQL+ + +N GKIRYIGLSNETP+G+ +
Sbjct: 128 YQIHWPDRYVPRFGQTVFDPTQVGETIPITEQLEVFADVINAGKIRYIGLSNETPWGVAQ 187
Query: 228 FIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFS 287
F A K PK+VS+QN+Y+LL R FD A+AE + E+I LLAYSPL G L+GKY +
Sbjct: 188 FSHAA-KQLGLPKVVSIQNAYNLLNRNFDGALAETVYYENIPLLAYSPLGFGYLTGKYLN 246
Query: 288 HGNGPADARLNLFK 301
P AR+ LF+
Sbjct: 247 --GKPEKARVTLFE 258
>ref|ZP_00325730.1| COG0667: Predicted oxidoreductases (related to aryl-alcohol
dehydrogenases) [Trichodesmium erythraeum IMS101]
Length = 345
Score = 275 bits (703), Expect = 1e-72
Identities = 137/255 (53%), Positives = 180/255 (69%), Gaps = 5/255 (1%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
NLNVS +CLGTMT+G QNT+ ++ Q L+ A GINF D+AEMYPVP RA T G +EEY
Sbjct: 10 NLNVSEICLGTMTYGLQNTIEEAHQQLNYAVAEGINFIDTAEMYPVPSRADTQGKTEEYI 69
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDL 167
G W+ + RD+L+IATKV GPS ++TWIRG + ++ NI QAI++SL +Q DYIDL
Sbjct: 70 GEWLVKQQ--RDKLIIATKVTGPSPRITWIRGENRKVNRANIQQAIEDSLRALQTDYIDL 127
Query: 168 YQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMK 227
YQIHWPDRYVP+FG +YDP ++ S I EQL+ + + GKIRY+G+SNET +GL +
Sbjct: 128 YQIHWPDRYVPLFGAPDYDPNNEWDSTPIAEQLEVFAELIKAGKIRYLGVSNETAWGLCE 187
Query: 228 FIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFS 287
F +AEK PKIVS+QN++SL+ R F +AE C ++ L+AYSPLA GIL+GKY
Sbjct: 188 FCHLAEK-LGLPKIVSIQNAFSLVNRVFHINIAEACRFNNVGLMAYSPLAFGILTGKYLQ 246
Query: 288 HGNGPADARLNLFKG 302
P ++RL F G
Sbjct: 247 --GVPENSRLAFFPG 259
>ref|ZP_00203532.1| COG0667: Predicted oxidoreductases (related to aryl-alcohol
dehydrogenases) [Dechloromonas aromatica RCB]
Length = 348
Score = 271 bits (692), Expect = 3e-71
Identities = 134/245 (54%), Positives = 175/245 (70%), Gaps = 3/245 (1%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
+ V +CLGTMTFGEQ + S + LD A GINF D+AEMY VP RA+T G SE
Sbjct: 13 HFEVPEVCLGTMTFGEQTSESDAHAQLDFALANGINFIDTAEMYAVPPRAETCGASETIV 72
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDL 167
GHW+K + RD+++IATKVAGPS + WIR GP +LD TNI AID SL R+Q DY+DL
Sbjct: 73 GHWLKRQ--ARDKVLIATKVAGPSRNLNWIRNGPPALDRTNIRAAIDGSLQRLQTDYVDL 130
Query: 168 YQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMK 227
YQ+HWP+R PMFG+ ++DP ++ I EQL+AL V +GKIR+IG+SNE P+G+M+
Sbjct: 131 YQLHWPERNQPMFGQWQFDPDKERDGTPIREQLEALGELVREGKIRHIGVSNEHPWGIMQ 190
Query: 228 FIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFS 287
F ++A++ P IVS QNSYSLL RTF++ +AE CH+E + LLAYSPLA G L+GKY +
Sbjct: 191 FTRLADE-LGLPHIVSTQNSYSLLNRTFETGLAEVCHRERVGLLAYSPLAFGHLTGKYLN 249
Query: 288 HGNGP 292
+ P
Sbjct: 250 QPDAP 254
>ref|NP_716530.1| oxidoreductase, aldo/keto reductase family [Shewanella oneidensis
MR-1] gi|24346485|gb|AAN53975.1| oxidoreductase,
aldo/keto reductase family [Shewanella oneidensis MR-1]
Length = 346
Score = 259 bits (661), Expect = 1e-67
Identities = 137/254 (53%), Positives = 177/254 (68%), Gaps = 4/254 (1%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
NL VS++CLGTMT+GEQNT +++F LD A +GINF D+AEMYPVP + +T G +E
Sbjct: 10 NLEVSKICLGTMTWGEQNTQAEAFAQLDYAIGSGINFIDTAEMYPVPPKPETQGETERIL 69
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDL 167
G +IK R RD LVIATK+A P G+ +IR +LD NI QA+D SL R+Q+D IDL
Sbjct: 70 GQYIKARG-NRDDLVIATKIAAPGGKSDYIRKN-MALDWNNIHQAVDTSLERLQIDTIDL 127
Query: 168 YQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMK 227
YQ+HWPDR FGE YD + I E L+AL+ + GK+RYIG+SNETP+GLMK
Sbjct: 128 YQVHWPDRNTNFFGELFYDEQEVEQQTPILETLEALAEVIRQGKVRYIGVSNETPWGLMK 187
Query: 228 FIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFS 287
++Q+AEK P+IV++QN Y+LL R+F+ M+E H+E + LLAYSPLA G LSGKY
Sbjct: 188 YLQLAEK-HGLPRIVTVQNPYNLLNRSFEVGMSEISHREELPLLAYSPLAFGALSGKY-C 245
Query: 288 HGNGPADARLNLFK 301
+ P ARL LFK
Sbjct: 246 NNQWPEGARLTLFK 259
>ref|ZP_00325375.1| COG0667: Predicted oxidoreductases (related to aryl-alcohol
dehydrogenases) [Trichodesmium erythraeum IMS101]
Length = 348
Score = 257 bits (656), Expect = 4e-67
Identities = 129/257 (50%), Positives = 177/257 (68%), Gaps = 8/257 (3%)
Query: 49 LNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFG 108
L VS +CLGTM +G+QNTL ++ +LLD A+ GINF D+AEMYP P A+T G +EEY G
Sbjct: 11 LQVSEICLGTMNYGKQNTLDEAQKLLDYAFSQGINFIDTAEMYPAPTCAKTLGKTEEYIG 70
Query: 109 HWIKHRNIPRDRLVIATKVAGPSGQ---MTWIRGGPKSLDATNISQAIDNSLLRMQLDYI 165
W+ + RD++ IATKV G + + WIR G ++ N+ +AID SL R+Q DY+
Sbjct: 71 KWLSKKQ--RDKVTIATKVCGRPNETLSLNWIREGKSCINDVNVQKAIDGSLSRLQTDYV 128
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
DLYQIHWPDRYVP+FG ++Y+P + +I I EQL+ + V GKIRY+G+SNETP+G+
Sbjct: 129 DLYQIHWPDRYVPLFGASDYNPRYERETIPIIEQLEVFADLVKAGKIRYLGISNETPWGV 188
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKY 285
+F +A++ PKIVS+QN+Y+L R F+ +AE CH + L+AYS LA G LSGKY
Sbjct: 189 SEFSHLAQQ-LGLPKIVSIQNAYNLTNRVFEINLAETCHFHRVGLMAYSTLAFGYLSGKY 247
Query: 286 FSHGNGPADARLNLFKG 302
S P ++RL+LF G
Sbjct: 248 LS--KIPKNSRLDLFPG 262
>ref|ZP_00170695.1| COG0667: Predicted oxidoreductases (related to aryl-alcohol
dehydrogenases) [Ralstonia eutropha JMP134]
Length = 352
Score = 251 bits (641), Expect = 2e-65
Identities = 125/240 (52%), Positives = 169/240 (70%), Gaps = 5/240 (2%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
+L VSR+CLGTMTFGEQNT + LD A+ GINF D+AEMYPV RA+T+ +E+
Sbjct: 10 DLEVSRICLGTMTFGEQNTEADGHSQLDYAFSRGINFIDTAEMYPVRPRAETYSRTEQII 69
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDL 167
G W+K + PRDR+V+ATKVAGP G+M WIR G A I A+D SL R+Q DYIDL
Sbjct: 70 GSWLKRQ--PRDRVVLATKVAGP-GRMPWIRNGDDFTPA-GILAAVDASLQRLQTDYIDL 125
Query: 168 YQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMK 227
YQIHWP R P+FG+T +DP + SI QL+ L+ V GK+RY+G+SNETP+G+ +
Sbjct: 126 YQIHWPARNAPIFGQTSFDPALERPCASIQAQLETLAELVQAGKVRYVGVSNETPWGVSE 185
Query: 228 FIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFS 287
F+++AE+ P+I ++QN Y+L+ R+F+ + E C++ +SLL YSPLA G L+GKY S
Sbjct: 186 FVRLAEQ-HGLPRIATIQNPYNLVNRSFEQGLDEACYRTDVSLLVYSPLAFGQLTGKYLS 244
>ref|YP_128799.1| putative oxidoreductase Tas, aldo/keto reductase family
[Photobacterium profundum SS9]
gi|46912202|emb|CAG18997.1| putative oxidoreductase Tas,
aldo/keto reductase family [Photobacterium profundum
SS9]
Length = 344
Score = 250 bits (639), Expect = 4e-65
Identities = 127/261 (48%), Positives = 183/261 (69%), Gaps = 5/261 (1%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
+L VS++CLGTMTFGEQNT S++ LD A+ G+NF D+AEMYPVP + +T G++E Y
Sbjct: 10 SLEVSKICLGTMTFGEQNTESEAHDQLDFAFERGVNFIDTAEMYPVPPKQETQGLTETYI 69
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDL 167
G+W+K ++ RD++++ATKVAGP + +IR +LD +I A++ SL R+Q DYIDL
Sbjct: 70 GNWLK-KSGKRDKVILATKVAGPRS-LPYIRDD-MALDRRHIHDAVETSLNRLQTDYIDL 126
Query: 168 YQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMK 227
YQ+HWP R FG+ Y + + ++I + L+AL+ VN GKIR+IGLSNETP+G+M
Sbjct: 127 YQLHWPQRETNCFGQLNYTYSEDATGVTITDTLEALAELVNAGKIRHIGLSNETPWGVMS 186
Query: 228 FIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFS 287
F+++AEK + P+++S+QN Y+LL R+F+ ++E H E + LLAYSPLA G LSGKY
Sbjct: 187 FLKLAEK-HNLPRVISIQNPYNLLNRSFEVGLSEISHHEGVELLAYSPLAFGALSGKYL- 244
Query: 288 HGNGPADARLNLFKGLLTHCN 308
+G P AR LF+ + N
Sbjct: 245 NGARPDGARCTLFERFSRYFN 265
>gb|AAF93832.1| oxidoreductase Tas, aldo/keto reductase family [Vibrio cholerae O1
biovar eltor str. N16961] gi|15640686|ref|NP_230316.1|
oxidoreductase Tas, aldo/keto reductase family [Vibrio
cholerae O1 biovar eltor str. N16961]
gi|11355749|pir||C82294 oxidoreductase Tas, aldo/keto
reductase family VC0667 [imported] - Vibrio cholerae
(strain N16961 serogroup O1)
Length = 352
Score = 249 bits (637), Expect = 6e-65
Identities = 129/252 (51%), Positives = 178/252 (70%), Gaps = 5/252 (1%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
+L +S++CLGTMTFGEQN+ + +FQ LD A G+NF D+AEMYPVP AQT G +EE+
Sbjct: 18 SLEISKICLGTMTFGEQNSQADAFQQLDYALERGVNFIDTAEMYPVPPTAQTQGKTEEFI 77
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDL 167
G+W+ ++ R+++V+ATKVAGP + +IR +LD NI QA+D+SL R+Q DYIDL
Sbjct: 78 GNWLA-KSGKREKIVLATKVAGPRN-VPYIRD-KMALDHRNIHQAVDDSLRRLQTDYIDL 134
Query: 168 YQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMK 227
YQ+HWP R FG+ Y + +++ E L+AL+ V GK+RYIG+SNETP+G+M
Sbjct: 135 YQLHWPQRQTNTFGQLNYPYPDKQEEVTLIETLEALNDLVRMGKVRYIGVSNETPWGVMS 194
Query: 228 FIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFS 287
++++AEK P+IVS+QN Y+LL R+F+ +AE H E + LLAYSPLA G LSGKY
Sbjct: 195 YLRLAEK-HELPRIVSIQNPYNLLNRSFEVGLAEISHLEGVKLLAYSPLAFGALSGKYL- 252
Query: 288 HGNGPADARLNL 299
+G PA AR L
Sbjct: 253 NGARPAGARCTL 264
>gb|AAR00611.1| putative aldo/keto reductase family protein [Oryza sativa (japonica
cultivar-group)] gi|50901464|ref|XP_463165.1| putative
aldo/keto reductase family protein [Oryza sativa
(japonica cultivar-group)]
Length = 405
Score = 246 bits (628), Expect = 7e-64
Identities = 127/271 (46%), Positives = 183/271 (66%), Gaps = 9/271 (3%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
+L +S + LGTMTFGEQNT ++ +L ++ G+N D+AEMYPVP R +T G ++ Y
Sbjct: 58 DLVISEVTLGTMTFGEQNTEKEAHDILSYSFDQGVNILDTAEMYPVPPRKETQGRTDLYI 117
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPK--SLDATNISQAIDNSLLRMQLDYI 165
G W++ + PRD++++ATKV+G S + T++R K +DA NI ++++ SL R+ DYI
Sbjct: 118 GSWMQSK--PRDKIILATKVSGYSERSTFLRDNAKVVRVDAANIKESVEKSLSRLSTDYI 175
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
DL QIHWPDRYVP+FGE Y+P + S+ +EQL A +++GK+RYIG+SNET YG+
Sbjct: 176 DLLQIHWPDRYVPLFGEYCYNPTKWRPSVPFEEQLKAFQELIDEGKVRYIGVSNETSYGV 235
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCRT-FDSAMAECCHQE--SISLLAYSPLAMGILS 282
M+F+ A K PKIVS+QNSYSL+ R F+ + E CH ++ LLAYSPLA G+L+
Sbjct: 236 MEFVHAA-KVQGLPKIVSIQNSYSLIVRCHFEVDLVEVCHPNNCNVGLLAYSPLAGGVLT 294
Query: 283 GKYF-SHGNGPADARLNLFKGLLTHCNICFA 312
GKY ++ + +RLNLF G + N A
Sbjct: 295 GKYIDTNPDISKKSRLNLFPGYMERYNASLA 325
>ref|ZP_00474160.1| Aldo/keto reductase [Chromohalobacter salexigens DSM 3043]
gi|67518511|gb|EAM22478.1| Aldo/keto reductase
[Chromohalobacter salexigens DSM 3043]
Length = 343
Score = 245 bits (625), Expect = 2e-63
Identities = 128/253 (50%), Positives = 175/253 (68%), Gaps = 6/253 (2%)
Query: 49 LNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFG 108
++VSRLCLGTMTFGEQN+ +++ LD A AGINF D+AEMYPVP +A+T G +E Y G
Sbjct: 11 MHVSRLCLGTMTFGEQNSEAEAHAQLDRATAAGINFIDTAEMYPVPPKAETQGRTEAYIG 70
Query: 109 HWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLY 168
W+K R RD L++ATK AGP + IRGGP+ L ++ +A+D+SL R+Q DYIDLY
Sbjct: 71 SWLKARG-KRDDLILATKAAGPG--LDHIRGGPR-LTRDHLHRAVDDSLARLQTDYIDLY 126
Query: 169 QIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKF 228
Q+HWPDR FG+ Y + ++ + E L AL V+ GKIR IGLSNETP+G+M+
Sbjct: 127 QLHWPDRNANFFGKLGYQHDESENATPLAESLQALKELVDAGKIRAIGLSNETPWGVMQS 186
Query: 229 IQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFSH 288
+++A+ + P++ S+QN Y+LL R+F+ +AE H+E + LLAYSPLA G L+GKY
Sbjct: 187 LRLAD-TLGVPRVASVQNPYNLLNRSFEVGLAEIAHREDVGLLAYSPLAFGALTGKYLD- 244
Query: 289 GNGPADARLNLFK 301
G P RL LF+
Sbjct: 245 GARPPQGRLTLFE 257
>ref|ZP_00507165.1| Aldo/keto reductase [Polaromonas sp. JS666]
gi|67778853|gb|EAM38473.1| Aldo/keto reductase
[Polaromonas sp. JS666]
Length = 354
Score = 244 bits (622), Expect = 3e-63
Identities = 122/262 (46%), Positives = 174/262 (65%), Gaps = 5/262 (1%)
Query: 46 APNLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEE 105
A +L V+ +C+GTMTFGEQ + +LD ++ G+NF D+AEMY VP RA+T+G +E
Sbjct: 8 ASDLQVTPICMGTMTFGEQVDEVTAHAILDRSFERGVNFLDTAEMYSVPARAETFGATET 67
Query: 106 YFGHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYI 165
G W R +LV+ATKVAGP+ M+WIR G L A +I+QA +NSL R++ D I
Sbjct: 68 IIGSWFAKNPAARSKLVLATKVAGPARGMSWIREGSPDLTAADITQACNNSLKRLKTDVI 127
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
DLYQIHWP R+VP FG ++P + + SI +QL+AL V GK+R IGLSNETPYG+
Sbjct: 128 DLYQIHWPARHVPAFGMLYFEPAKDQAVTSIHQQLEALGGLVKAGKVRAIGLSNETPYGV 187
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKY 285
+F+++AE+ P++ ++QN + L+ RT ++ + E H+ +SLLAYSPLA G+L+GKY
Sbjct: 188 HEFVRLAEQ-HGLPRVATVQNPFCLVNRTVENGLDETMHRLGVSLLAYSPLAFGLLTGKY 246
Query: 286 FSHG----NGPADARLNLFKGL 303
G P DAR+ F+ +
Sbjct: 247 DESGTEGPRAPKDARIGKFESV 268
>ref|NP_796895.1| oxidoreductase Tas, aldo/keto reductase family [Vibrio
parahaemolyticus RIMD 2210633]
gi|28805499|dbj|BAC58779.1| oxidoreductase Tas,
aldo/keto reductase family [Vibrio parahaemolyticus RIMD
2210633]
Length = 352
Score = 242 bits (618), Expect = 1e-62
Identities = 124/249 (49%), Positives = 174/249 (69%), Gaps = 5/249 (2%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
+L +S++CLGTMTFGEQN ++F+ LD A G+NF D+AEMYPVP +A+T G++E+Y
Sbjct: 18 SLEISKICLGTMTFGEQNIEQEAFEQLDFAVDHGVNFIDTAEMYPVPPKAETQGLTEQYI 77
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDL 167
G+W+ ++ R+++V+ATKVAGP + IR SL+ NI AID+SL R+Q DY+DL
Sbjct: 78 GNWLT-KSGKREKVVLATKVAGPRN-VPHIREN-MSLNRRNIHDAIDSSLQRLQTDYVDL 134
Query: 168 YQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMK 227
YQ+HWP R FG+ Y + +++ E L+AL+ V GK+RYIG+SNETP+G+M
Sbjct: 135 YQLHWPQRQTNCFGQLNYPYPDKQEEVTLIETLEALTELVKMGKVRYIGVSNETPWGVMS 194
Query: 228 FIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFS 287
+++AEK P+IVS+QN Y+LL R+F+ ++E H E + LLAYSPLA G LSGKY
Sbjct: 195 LLRLAEK-HDLPRIVSIQNPYNLLNRSFEVGLSEISHYEGVQLLAYSPLAFGCLSGKYL- 252
Query: 288 HGNGPADAR 296
HG P AR
Sbjct: 253 HGAKPEGAR 261
>ref|ZP_00274131.1| COG0667: Predicted oxidoreductases (related to aryl-alcohol
dehydrogenases) [Ralstonia metallidurans CH34]
Length = 350
Score = 241 bits (616), Expect = 2e-62
Identities = 120/240 (50%), Positives = 169/240 (70%), Gaps = 5/240 (2%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
+L VSR+CLGTMTFGEQNT ++ LD A+ GINF D+AEMYPV RA+T+G +E
Sbjct: 8 DLEVSRICLGTMTFGEQNTEAEGHSQLDYAFSRGINFIDTAEMYPVKPRAETYGSTERII 67
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDL 167
G W+K + PRDR+V+A+KVAGP+ + WIR G L +I A+++SL R+Q DYIDL
Sbjct: 68 GTWLKKQ--PRDRVVLASKVAGPA-RNPWIRNGG-DLTPDSIRAAVEDSLKRLQTDYIDL 123
Query: 168 YQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMK 227
YQIHWP R P+FG+ ++P + +I+ QL+A+ V GKIRY+G+SNETP+G+ +
Sbjct: 124 YQIHWPARNAPIFGQKRFNPEAERQCATIEAQLEAMGDLVRAGKIRYVGVSNETPWGVTE 183
Query: 228 FIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFS 287
F+ VAE+ P+I ++QN Y+L+ R+F+ + E C + +SLLAYSPL G L+GKY +
Sbjct: 184 FVNVAER-HGLPRIATIQNPYNLVNRSFEQGLDETCFRTEVSLLAYSPLGFGQLTGKYLT 242
>gb|AAO09041.1| Oxidoreductase Tas, aldo/keto reductase family [Vibrio vulnificus
CMCP6] gi|37678856|ref|NP_933465.1| oxidoreductase Tas,
aldo/keto reductase family [Vibrio vulnificus YJ016]
gi|37197597|dbj|BAC93436.1| oxidoreductase Tas,
aldo/keto reductase family [Vibrio vulnificus YJ016]
gi|27363986|ref|NP_759514.1| Oxidoreductase Tas,
aldo/keto reductase family [Vibrio vulnificus CMCP6]
Length = 344
Score = 241 bits (614), Expect = 3e-62
Identities = 123/253 (48%), Positives = 175/253 (68%), Gaps = 5/253 (1%)
Query: 49 LNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFG 108
L +S+LCLGTMTFGEQN+ +F+ LD A GINF D+AEMYPVP +AQ+ G++E++ G
Sbjct: 11 LEISKLCLGTMTFGEQNSQQDAFEQLDYAVAQGINFIDTAEMYPVPPKAQSQGLTEQFIG 70
Query: 109 HWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLY 168
+W+ ++ R++++IATK+AGP + +IR SL+ +I AID+SL R+Q DY+DLY
Sbjct: 71 NWLS-KSGKREKVLIATKIAGPRN-VPYIRDN-MSLNRRHIHTAIDDSLTRLQTDYVDLY 127
Query: 169 QIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKF 228
Q+HWP R FG+ Y +++ E L+AL+ V GK+RYIG+SNETP+G+M
Sbjct: 128 QLHWPQRQTNCFGQLNYPYPDAQEEVTLIETLEALNELVKAGKVRYIGVSNETPWGVMTL 187
Query: 229 IQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFSH 288
+++AEK P+IVS+QN Y+LL R+F+ ++E H E + LLAYSP+A G LSGKY
Sbjct: 188 LRLAEK-HDLPRIVSIQNPYNLLNRSFEVGLSEISHYEGVQLLAYSPMAFGTLSGKYLD- 245
Query: 289 GNGPADARLNLFK 301
G P AR LF+
Sbjct: 246 GARPKGARCTLFE 258
>gb|AAN66938.1| oxidoreductase, aldo/keto reductase family [Pseudomonas putida
KT2440] gi|26988049|ref|NP_743474.1| oxidoreductase,
aldo/keto reductase family [Pseudomonas putida KT2440]
Length = 346
Score = 236 bits (602), Expect = 7e-61
Identities = 127/277 (45%), Positives = 175/277 (62%), Gaps = 3/277 (1%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
+LNVS LCLGTMT+GEQN +++++ + A AG+NF D+AEMYPVP R +T+ +E
Sbjct: 10 DLNVSALCLGTMTWGEQNIQAEAYEQIARAKAAGVNFIDTAEMYPVPPRPETYAATERII 69
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDL 167
G+W + RD ++A+KVAGP ++ IR G + +I A++ SL R+Q D IDL
Sbjct: 70 GNWFRDHG-DRDAWILASKVAGPGNGISHIRDGQLKHNRQHIVAALEESLKRLQTDRIDL 128
Query: 168 YQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMK 227
YQ+HWP+R FG+ Y + Q ++E L+ L V GKIR+IGLSNETP+G MK
Sbjct: 129 YQLHWPERSTNFFGKLGYQHLPQDHFTPLEETLEVLDEQVRAGKIRHIGLSNETPWGTMK 188
Query: 228 FIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFS 287
F+Q+AE S +P+ VS+QN Y+LL R+F+ +AE +E LLAYSPLA G+LSGKY
Sbjct: 189 FLQLAE-SRGWPRAVSIQNPYNLLNRSFEVGLAEVAIREQCGLLAYSPLAFGMLSGKY-E 246
Query: 288 HGNGPADARLNLFKGLLTHCNICFAFMCFHPFQHKRE 324
+G P +ARL LF + N C Q RE
Sbjct: 247 NGARPDNARLTLFSRFARYSNPQTVAACSRYVQLARE 283
>ref|ZP_00466793.1| COG0667: Predicted oxidoreductases (related to aryl-alcohol
dehydrogenases) [Burkholderia pseudomallei 1655]
gi|67652335|ref|ZP_00449755.1| COG0667: Predicted
oxidoreductases (related to aryl-alcohol dehydrogenases)
[Burkholderia mallei SAVP1]
gi|67647983|ref|ZP_00446218.1| COG0667: Predicted
oxidoreductases (related to aryl-alcohol dehydrogenases)
[Burkholderia mallei NCTC 10247]
gi|67640012|ref|ZP_00438835.1| COG0667: Predicted
oxidoreductases (related to aryl-alcohol dehydrogenases)
[Burkholderia mallei GB8 horse 4]
gi|67636602|ref|ZP_00435547.1| COG0667: Predicted
oxidoreductases (related to aryl-alcohol dehydrogenases)
[Burkholderia mallei 10399]
gi|67630624|ref|ZP_00430481.1| COG0667: Predicted
oxidoreductases (related to aryl-alcohol dehydrogenases)
[Burkholderia mallei 10229] gi|53723473|ref|YP_102901.1|
oxidoreductase, aldo/keto reductase family [Burkholderia
mallei ATCC 23344] gi|52426896|gb|AAU47489.1|
oxidoreductase, aldo/keto reductase family [Burkholderia
mallei ATCC 23344]
Length = 350
Score = 234 bits (598), Expect = 2e-60
Identities = 123/256 (48%), Positives = 166/256 (64%), Gaps = 5/256 (1%)
Query: 49 LNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFG 108
+ VS + LGTMT+GEQNT S + LD A G+ D+AEMYPVP +AQT G +E Y G
Sbjct: 11 VKVSLIGLGTMTWGEQNTESDAHAQLDYAIDQGVTLIDTAEMYPVPPKAQTQGRTETYLG 70
Query: 109 HWIKHRNIPRDRLVIATKVAGPSGQM---TWIRGGPKSLDATNISQAIDNSLLRMQLDYI 165
W+ R++L IATK+AGP+ Q T IRG D N+++A+D SL R++ DY+
Sbjct: 71 TWLAKHPALREKLTIATKIAGPARQPHNPTHIRGAGNQFDRKNLTEALDGSLARLRTDYV 130
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
DLYQ+HWPDR FG Y + ++ I+E L L V GK+R++G+SNETP+G+
Sbjct: 131 DLYQLHWPDRSTTTFGRPAYPWIDDPYTVPIEETLAVLGDFVKAGKVRHVGVSNETPWGV 190
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKY 285
+F++ AEK P+IVS+QN YSLL RTF++ ++E H+E I LLAYSPLA G LSGKY
Sbjct: 191 AQFLRAAEK-LGLPRIVSIQNPYSLLNRTFENGLSEFSHRERIGLLAYSPLAFGSLSGKY 249
Query: 286 FSHGNGPADARLNLFK 301
G P AR+ F+
Sbjct: 250 -EGGARPEGARITRFE 264
>ref|ZP_00498182.1| COG0667: Predicted oxidoreductases (related to aryl-alcohol
dehydrogenases) [Burkholderia pseudomallei S13]
gi|67714631|ref|ZP_00483992.1| COG0667: Predicted
oxidoreductases (related to aryl-alcohol dehydrogenases)
[Burkholderia pseudomallei 1710b]
gi|67681870|ref|ZP_00476171.1| COG0667: Predicted
oxidoreductases (related to aryl-alcohol dehydrogenases)
[Burkholderia pseudomallei 1710a]
gi|52209875|emb|CAH35847.1| putative oxidoreductase
[Burkholderia pseudomallei K96243]
gi|53719461|ref|YP_108447.1| putative oxidoreductase
[Burkholderia pseudomallei K96243]
Length = 350
Score = 234 bits (597), Expect = 3e-60
Identities = 123/256 (48%), Positives = 166/256 (64%), Gaps = 5/256 (1%)
Query: 49 LNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFG 108
+ VS + LGTMT+GEQNT S + LD A G+ D+AEMYPVP +AQT G +E Y G
Sbjct: 11 VKVSLIGLGTMTWGEQNTESDAHAQLDYAIDQGVTLIDTAEMYPVPPKAQTQGRTETYLG 70
Query: 109 HWIKHRNIPRDRLVIATKVAGPSGQM---TWIRGGPKSLDATNISQAIDNSLLRMQLDYI 165
W+ R++L IATK+AGP+ Q T IRG D N+++A+D SL R++ DY+
Sbjct: 71 TWLAKHPALREKLTIATKIAGPARQPHNPTHIRGAGNQFDRKNLTEALDGSLARLRTDYV 130
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
DLYQ+HWPDR FG Y + ++ I+E L L V GK+R++G+SNETP+G+
Sbjct: 131 DLYQLHWPDRSTTTFGRPAYPWIDDPYTVPIEETLAVLGDFVKAGKVRHVGVSNETPWGV 190
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKY 285
+F++ AEK P+IVS+QN YSLL RTF++ ++E H+E I LLAYSPLA G LSGKY
Sbjct: 191 AQFLRAAEK-LGLPRIVSIQNPYSLLNRTFENGLSEFSHRERIGLLAYSPLAFGWLSGKY 249
Query: 286 FSHGNGPADARLNLFK 301
G P AR+ F+
Sbjct: 250 -EGGARPEGARITRFE 264
>ref|ZP_00491335.1| COG0667: Predicted oxidoreductases (related to aryl-alcohol
dehydrogenases) [Burkholderia pseudomallei Pasteur]
gi|67737718|ref|ZP_00488450.1| COG0667: Predicted
oxidoreductases (related to aryl-alcohol dehydrogenases)
[Burkholderia pseudomallei 668]
Length = 350
Score = 234 bits (597), Expect = 3e-60
Identities = 123/256 (48%), Positives = 166/256 (64%), Gaps = 5/256 (1%)
Query: 49 LNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFG 108
+ VS + LGTMT+GEQNT S + LD A G+ D+AEMYPVP +AQT G +E Y G
Sbjct: 11 VKVSLIGLGTMTWGEQNTESDAHAQLDYAIDQGVTLIDTAEMYPVPPKAQTQGRTETYLG 70
Query: 109 HWIKHRNIPRDRLVIATKVAGPSGQM---TWIRGGPKSLDATNISQAIDNSLLRMQLDYI 165
W+ R++L IATK+AGP+ Q T IRG D N+++A+D SL R++ DY+
Sbjct: 71 TWLAKHPALREKLTIATKIAGPARQPHNPTHIRGAGNQFDRKNLTEALDGSLARLRTDYV 130
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
DLYQ+HWPDR FG Y + ++ I+E L L V GK+R++G+SNETP+G+
Sbjct: 131 DLYQLHWPDRSTTTFGRPAYPWIDDPYTVPIEETLAVLGDFVKAGKVRHVGVSNETPWGV 190
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKY 285
+F++ AEK P+IVS+QN YSLL RTF++ ++E H+E I LLAYSPLA G LSGKY
Sbjct: 191 AQFLRAAEK-LGLPRIVSIQNPYSLLNRTFENGLSEFSHRERIGLLAYSPLAFGWLSGKY 249
Query: 286 FSHGNGPADARLNLFK 301
G P AR+ F+
Sbjct: 250 -EGGARPEGARITRFE 264
>ref|ZP_00425885.1| Aldo/keto reductase [Burkholderia vietnamiensis G4]
gi|67530708|gb|EAM27543.1| Aldo/keto reductase
[Burkholderia vietnamiensis G4]
Length = 350
Score = 234 bits (596), Expect = 4e-60
Identities = 120/256 (46%), Positives = 168/256 (64%), Gaps = 5/256 (1%)
Query: 49 LNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFG 108
+ VS + LGTMT+GEQN+ + + +D A G+ D+AEMYPVP +A T G +E+Y G
Sbjct: 11 IEVSLIGLGTMTWGEQNSERDAHEQIDYALAQGVTLIDTAEMYPVPPKADTQGRTEQYIG 70
Query: 109 HWIKHRNIPRDRLVIATKVAGPSGQM---TWIRGGPKSLDATNISQAIDNSLLRMQLDYI 165
W+ R R+V+ATK+AGP+ Q IRG D N+++A+D SL R+Q DY+
Sbjct: 71 TWLAQHRAQRARIVLATKIAGPARQPHNPRHIRGEGNQFDRKNLTEALDGSLKRLQTDYV 130
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
DLYQ+HWPDR FG + Y V ++ I+E L L+ V GK+R IG+SNETP+G+
Sbjct: 131 DLYQLHWPDRSTTTFGRSAYPWVDDAYTVPIEETLGVLAEFVKAGKVRAIGVSNETPWGV 190
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKY 285
+F++ AE+ P+I S+QN YSLL RTF++ ++E H++ I LLAYSPLA G LSGKY
Sbjct: 191 AQFLRAAER-LGLPRIASIQNPYSLLNRTFENGLSEFTHRDGIGLLAYSPLAFGWLSGKY 249
Query: 286 FSHGNGPADARLNLFK 301
+G PA AR+ LF+
Sbjct: 250 -ENGARPAGARITLFE 264
>ref|ZP_00462212.1| Aldo/keto reductase [Burkholderia cenocepacia HI2424]
gi|67658404|ref|ZP_00455776.1| Aldo/keto reductase
[Burkholderia cenocepacia AU 1054]
gi|67101468|gb|EAM18609.1| Aldo/keto reductase
[Burkholderia cenocepacia HI2424]
gi|67093976|gb|EAM11513.1| Aldo/keto reductase
[Burkholderia cenocepacia AU 1054]
Length = 350
Score = 234 bits (596), Expect = 4e-60
Identities = 120/256 (46%), Positives = 167/256 (64%), Gaps = 5/256 (1%)
Query: 49 LNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFG 108
+ VS + LGTMT+GEQN+ + + +D A G+ D+AEMYPVP + T G +E+Y G
Sbjct: 11 IEVSLIGLGTMTWGEQNSERDAHEQIDYALGHGVTLIDAAEMYPVPPKPDTQGRTEQYIG 70
Query: 109 HWIKHRNIPRDRLVIATKVAGPSGQM---TWIRGGPKSLDATNISQAIDNSLLRMQLDYI 165
WI RDR+V+ATK+AGP+ Q IRG D N+++A+D SL R+Q DY+
Sbjct: 71 TWIAQHRAQRDRIVLATKIAGPARQPHNPRHIRGEGNQFDRKNLTEALDGSLKRLQTDYV 130
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
DLYQ+HWPDR FG Y V ++ I+E L L+ V GK+R IG+SNETP+G+
Sbjct: 131 DLYQLHWPDRSTTTFGRPAYPWVDDAYTVPIEETLGVLAEFVKAGKVRAIGVSNETPWGV 190
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKY 285
+F++ AE+ P+I S+QN YSLL RTF++ ++E H++ + LLAYSPLA G LSGKY
Sbjct: 191 AQFLRAAER-LRLPRIASIQNPYSLLNRTFENGLSEFTHRDGVGLLAYSPLAFGWLSGKY 249
Query: 286 FSHGNGPADARLNLFK 301
+G PA AR+ LF+
Sbjct: 250 -ENGARPAGARITLFE 264
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.323 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 567,839,415
Number of Sequences: 2540612
Number of extensions: 23154713
Number of successful extensions: 55655
Number of sequences better than 10.0: 2207
Number of HSP's better than 10.0 without gapping: 1043
Number of HSP's successfully gapped in prelim test: 1164
Number of HSP's that attempted gapping in prelim test: 51194
Number of HSP's gapped (non-prelim): 2273
length of query: 337
length of database: 863,360,394
effective HSP length: 128
effective length of query: 209
effective length of database: 538,162,058
effective search space: 112475870122
effective search space used: 112475870122
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 75 (33.5 bits)
Medicago: description of AC141106.8


