

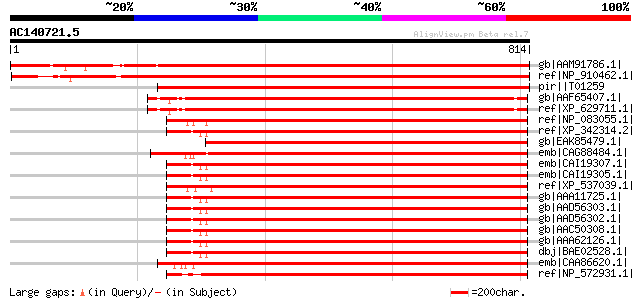
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.5 - phase: 0
(814 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM91786.1| putative AMP deaminase [Arabidopsis thaliana] gi|... 1297 0.0
ref|NP_910462.1| putative AMP deaminase [Oryza sativa (japonica ... 1221 0.0
pir||T01259 AMP deaminase homolog F16M14.21 - Arabidopsis thaliana 1114 0.0
gb|AAF65407.1| AMP deaminase [Dictyostelium discoideum] 708 0.0
ref|XP_629711.1| AMP deaminase [Dictyostelium discoideum] gi|604... 708 0.0
ref|NP_083055.1| adenosine monophosphate deaminase 2 (isoform L)... 647 0.0
ref|XP_342314.2| PREDICTED: similar to adenosine monophosphate d... 647 0.0
gb|EAK85479.1| hypothetical protein UM04622.1 [Ustilago maydis 5... 647 0.0
emb|CAG88484.1| unnamed protein product [Debaryomyces hansenii C... 646 0.0
emb|CAI19307.1| adenosine monophosphate deaminase 2 (isoform L) ... 644 0.0
emb|CAI19305.1| adenosine monophosphate deaminase 2 (isoform L) ... 644 0.0
ref|XP_537039.1| PREDICTED: similar to AMP deaminase 2 (AMP deam... 644 0.0
gb|AAA11725.1| AMP deaminase isoform L [Homo sapiens] gi|345738|... 644 0.0
gb|AAD56303.1| AMP deaminase isoform L [Homo sapiens] 644 0.0
gb|AAD56302.1| AMP deaminase isoform L [Homo sapiens] gi|1264437... 644 0.0
gb|AAC50308.1| AMP deaminase 644 0.0
gb|AAA62126.1| AMP deaminase isoform L splicing variant 644 0.0
dbj|BAE02528.1| unnamed protein product [Macaca fascicularis] 642 0.0
emb|CAA86620.1| AMD1 [Saccharomyces cerevisiae] gi|6323606|ref|N... 637 0.0
ref|NP_572931.1| CG32626-PC, isoform C [Drosophila melanogaster]... 637 0.0
>gb|AAM91786.1| putative AMP deaminase [Arabidopsis thaliana]
gi|15810525|gb|AAL07150.1| putative AMP deaminase
[Arabidopsis thaliana] gi|20196986|gb|AAC27176.2|
putative AMP deaminase [Arabidopsis thaliana]
gi|30687456|ref|NP_850294.1| AMP deaminase, putative /
myoadenylate deaminase, putative [Arabidopsis thaliana]
gi|18404701|ref|NP_565886.1| AMP deaminase, putative /
myoadenylate deaminase, putative [Arabidopsis thaliana]
Length = 839
Score = 1297 bits (3356), Expect = 0.0
Identities = 652/832 (78%), Positives = 711/832 (85%), Gaps = 32/832 (3%)
Query: 1 MDAHAVHLAMAALFGASIVAVSAYYMHRKTLTELLEFA--RTVEPEGDSDGGE---RRRG 55
M+ + LA+AALFGAS VAVS ++MH K L +LE R P+GD RRR
Sbjct: 1 MEPNIYQLALAALFGASFVAVSGFFMHFKALNLVLERGKERKENPDGDEPQNPTLVRRRS 60
Query: 56 GSKRRNGGGGGYRRGSGSLPDVTAIAGGVEG--------NGLMHDEGIPVGLPRLQTLRE 107
+R+ Y R SLPD T G G NG ++ + IP GLPRL T E
Sbjct: 61 QVRRKVNDQ--YGRSPASLPDATPFTDGGGGGGGDTGRSNGHVYVDEIPPGLPRLHTPSE 118
Query: 108 GKSANNGSFK----RNIIRPTSPKSPVASASAFESVEGSDDEDNLTDTKH-DTTYLHTNG 162
G+++ +G+ + +RP SPKSPVASASAFESVE SDD+DNLT+++ D +YL NG
Sbjct: 119 GRASVHGASSIRKTGSFVRPISPKSPVASASAFESVEESDDDDNLTNSEGLDASYLQANG 178
Query: 163 NVGGEGKNPYETLPNHVNTNGEQMAITASSMIRSHSISGDLHGVQPDPIAADILRKEPEQ 222
+ +P + N EQ+++ ASSMIRSHS+SGDLHGVQPDPIAADILRKEPEQ
Sbjct: 179 D---------NEMP--ADANEEQISMAASSMIRSHSVSGDLHGVQPDPIAADILRKEPEQ 227
Query: 223 EIFARLRITPMEAPSPDEIESYVILQECLEMRKRYIFKEAVAPWEKEVISDPSTPKPNLE 282
E F RL + P+E P+ DE+E+Y LQECLE+RKRY+F+E VAPWEKEVISDPSTPKPN E
Sbjct: 228 ETFVRLNV-PLEVPTSDEVEAYKCLQECLELRKRYVFQETVAPWEKEVISDPSTPKPNTE 286
Query: 283 PFFYAPEGKSDHYFEMQDGVIHVYPNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRT 342
PF + P+GKSDH FEMQDGV+HV+ NK++ E+LFPVADAT FFTDLH +L+VIAAGNIRT
Sbjct: 287 PFAHYPQGKSDHCFEMQDGVVHVFANKDAKEDLFPVADATAFFTDLHHVLKVIAAGNIRT 346
Query: 343 LCHHRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRF 402
LCH RL LLEQKFNLHLMLNAD+EFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRF
Sbjct: 347 LCHRRLVLLEQKFNLHLMLNADKEFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRF 406
Query: 403 IKSKLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLK 462
IKSKLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLK
Sbjct: 407 IKSKLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLK 466
Query: 463 YNPCGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQ 522
YNPCGQSRLREIFLKQDNLIQGRFLGE+TKQVFSDLEASKYQMAEYRISIYGRK SEWDQ
Sbjct: 467 YNPCGQSRLREIFLKQDNLIQGRFLGEITKQVFSDLEASKYQMAEYRISIYGRKMSEWDQ 526
Query: 523 LASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQ 582
LASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQN+LDNIFIPLFE TVDPDSHPQ
Sbjct: 527 LASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNILDNIFIPLFEATVDPDSHPQ 586
Query: 583 LHVFLKQVVGLDLVDDESKPERRPTKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLR 642
LHVFLKQVVG DLVDDESKPERRPTKHMPTPAQWTN FNPAFSYYVYYCYANLY LNKLR
Sbjct: 587 LHVFLKQVVGFDLVDDESKPERRPTKHMPTPAQWTNAFNPAFSYYVYYCYANLYVLNKLR 646
Query: 643 ESKGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSP 702
ESKGMTTI RPH+GEAGDIDHLAATFLT H+IAHGINLRKSPVLQYLYYLAQIGLAMSP
Sbjct: 647 ESKGMTTITLRPHSGEAGDIDHLAATFLTCHSIAHGINLRKSPVLQYLYYLAQIGLAMSP 706
Query: 703 LSNNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDL 762
LSNNSLFLDYHRNP PVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLS+CDL
Sbjct: 707 LSNNSLFLDYHRNPFPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSACDL 766
Query: 763 CEIARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFRDTV 814
CEIARNSVYQSGFSHALKSHWIGK+YYKRGP+GNDIH+TNVPHIR+EFRDT+
Sbjct: 767 CEIARNSVYQSGFSHALKSHWIGKDYYKRGPDGNDIHKTNVPHIRVEFRDTI 818
>ref|NP_910462.1| putative AMP deaminase [Oryza sativa (japonica cultivar-group)]
gi|51963676|ref|XP_506591.1| PREDICTED P0034A04.129 gene
product [Oryza sativa (japonica cultivar-group)]
gi|29837186|dbj|BAC75568.1| putative AMP deaminase
[Oryza sativa (japonica cultivar-group)]
Length = 815
Score = 1221 bits (3159), Expect = 0.0
Identities = 600/820 (73%), Positives = 688/820 (83%), Gaps = 39/820 (4%)
Query: 4 HAVHLAMAALFGASIVAVSAYYMHRKTLTELLEFARTVEPEGDSDGGERRRGGSKRRNGG 63
+A+HLA+A L GAS A SAYYMHRKTL +LL FAR+++ +
Sbjct: 5 YALHLAVATLLGASFAAASAYYMHRKTLDQLLRFARSLDRD------------------- 45
Query: 64 GGGYRRGSGSLPDVTAIAGGVEGNGLMHDEG----IPVGLPRLQTLREGKSA-NNGSFKR 118
+RR + L D A + HD IP GLP L T REGK + S KR
Sbjct: 46 ---HRRRNRHLLD--ADDDDDDDPPRDHDRRTTLPIPPGLPPLHTGREGKPIISPASTKR 100
Query: 119 --NIIRPTSPKSPVASASAFESVEGSDDEDN--LTDTKHDTTYLHTNGNVGGEGKNPYET 174
++RPT+P+SPV + SAFE++E SDD+D D K++ L TNG +G +
Sbjct: 101 VGPLVRPTTPRSPVPTVSAFETIEDSDDDDENIAPDAKNNAVSLLTNGTIGSD------P 154
Query: 175 LPNHVNTNGEQMAITASSMIRSHSISGDLHGVQPDPIAADILRKEPEQEIFARLRITPME 234
LP + NG+ + +++MIRS S +G LHG Q +P+AADILRKEPE E F+R+ IT +E
Sbjct: 155 LPGKASQNGDTKPVPSTNMIRSQSATGSLHGAQHNPVAADILRKEPEHETFSRINITAVE 214
Query: 235 APSPDEIESYVILQECLEMRKRYIFKEAVAPWEKEVISDPSTPKPNLEPFFYAPEGKSDH 294
PSPDEIE+Y +LQ+CLE+R++Y+F+E VAPWEKE+I+DPSTPKPN PF+Y + K++H
Sbjct: 215 TPSPDEIEAYKVLQKCLELREKYMFREEVAPWEKEIITDPSTPKPNPNPFYYEQQTKTEH 274
Query: 295 YFEMQDGVIHVYPNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCHHRLNLLEQK 354
+FEM DGVIHVYPNK++ E ++PVADATTFFTD+H ILRV+AAG+IRT+C+ RLNLLEQK
Sbjct: 275 HFEMVDGVIHVYPNKDAKERIYPVADATTFFTDMHYILRVLAAGDIRTVCYKRLNLLEQK 334
Query: 355 FNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLRKEPDEV 414
FNLHLM+NADRE LAQK+APHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLRKEPDEV
Sbjct: 335 FNLHLMVNADRELLAQKAAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLRKEPDEV 394
Query: 415 VIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQSRLREI 474
VIFRDGTYLTL+EVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQSRLREI
Sbjct: 395 VIFRDGTYLTLKEVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQSRLREI 454
Query: 475 FLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLASWIVNNDLYS 534
FLKQDNLIQGRFL ELTK+VFSDLEASKYQMAEYRISIYGRK+SEWDQ+ASWIVNN+LYS
Sbjct: 455 FLKQDNLIQGRFLAELTKEVFSDLEASKYQMAEYRISIYGRKKSEWDQMASWIVNNELYS 514
Query: 535 ENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHVFLKQVVGLD 594
ENVVWLIQ+PR+YN+Y++MG + SFQN+LDNIF+PLFEVTVDP SHPQLHVFL+QVVGLD
Sbjct: 515 ENVVWLIQIPRIYNVYREMGTINSFQNLLDNIFLPLFEVTVDPASHPQLHVFLQQVVGLD 574
Query: 595 LVDDESKPERRPTKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRESKGMTTIKFRP 654
LVDDESKPERRPTKHMPTP QWTNVFNPA++YYVYYCYANLYTLNKLRESKGMTTIK RP
Sbjct: 575 LVDDESKPERRPTKHMPTPEQWTNVFNPAYAYYVYYCYANLYTLNKLRESKGMTTIKLRP 634
Query: 655 HAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNNSLFLDYHR 714
H GEAGDIDHLAA FLT+HNIAHG+NL+KSPVLQYLYYLAQIGLAMSPLSNNSLF+DYHR
Sbjct: 635 HCGEAGDIDHLAAAFLTSHNIAHGVNLKKSPVLQYLYYLAQIGLAMSPLSNNSLFIDYHR 694
Query: 715 NPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCEIARNSVYQSG 774
NP P FFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAAS+WKLSSCDLCEIARNSVYQSG
Sbjct: 695 NPFPTFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASLWKLSSCDLCEIARNSVYQSG 754
Query: 775 FSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFRDTV 814
FSH LKSHWIG+ YYKRG +GNDIH+TNVPHIR+EFR T+
Sbjct: 755 FSHRLKSHWIGRNYYKRGHDGNDIHQTNVPHIRIEFRHTI 794
>pir||T01259 AMP deaminase homolog F16M14.21 - Arabidopsis thaliana
Length = 600
Score = 1114 bits (2882), Expect = 0.0
Identities = 532/582 (91%), Positives = 558/582 (95%)
Query: 233 MEAPSPDEIESYVILQECLEMRKRYIFKEAVAPWEKEVISDPSTPKPNLEPFFYAPEGKS 292
+E P+ DE+E+Y LQECLE+RKRY+F+E VAPWEKEVISDPSTPKPN EPF + P+GKS
Sbjct: 4 LEVPTSDEVEAYKCLQECLELRKRYVFQETVAPWEKEVISDPSTPKPNTEPFAHYPQGKS 63
Query: 293 DHYFEMQDGVIHVYPNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCHHRLNLLE 352
DH FEMQDGV+HV+ NK++ E+LFPVADAT FFTDLH +L+VIAAGNIRTLCH RL LLE
Sbjct: 64 DHCFEMQDGVVHVFANKDAKEDLFPVADATAFFTDLHHVLKVIAAGNIRTLCHRRLVLLE 123
Query: 353 QKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLRKEPD 412
QKFNLHLMLNAD+EFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLRKEPD
Sbjct: 124 QKFNLHLMLNADKEFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLRKEPD 183
Query: 413 EVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQSRLR 472
EVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQSRLR
Sbjct: 184 EVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQSRLR 243
Query: 473 EIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLASWIVNNDL 532
EIFLKQDNLIQGRFLGE+TKQVFSDLEASKYQMAEYRISIYGRK SEWDQLASWIVNNDL
Sbjct: 244 EIFLKQDNLIQGRFLGEITKQVFSDLEASKYQMAEYRISIYGRKMSEWDQLASWIVNNDL 303
Query: 533 YSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHVFLKQVVG 592
YSENVVWLIQLPRLYNIYKDMGIVTSFQN+LDNIFIPLFE TVDPDSHPQLHVFLKQVVG
Sbjct: 304 YSENVVWLIQLPRLYNIYKDMGIVTSFQNILDNIFIPLFEATVDPDSHPQLHVFLKQVVG 363
Query: 593 LDLVDDESKPERRPTKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRESKGMTTIKF 652
DLVDDESKPERRPTKHMPTPAQWTN FNPAFSYYVYYCYANLY LNKLRESKGMTTI
Sbjct: 364 FDLVDDESKPERRPTKHMPTPAQWTNAFNPAFSYYVYYCYANLYVLNKLRESKGMTTITL 423
Query: 653 RPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNNSLFLDY 712
RPH+GEAGDIDHLAATFLT H+IAHGINLRKSPVLQYLYYLAQIGLAMSPLSNNSLFLDY
Sbjct: 424 RPHSGEAGDIDHLAATFLTCHSIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNNSLFLDY 483
Query: 713 HRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCEIARNSVYQ 772
HRNP PVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLS+CDLCEIARNSVYQ
Sbjct: 484 HRNPFPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSACDLCEIARNSVYQ 543
Query: 773 SGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFRDTV 814
SGFSHALKSHWIGK+YYKRGP+GNDIH+TNVPHIR+EFRDTV
Sbjct: 544 SGFSHALKSHWIGKDYYKRGPDGNDIHKTNVPHIRVEFRDTV 585
>gb|AAF65407.1| AMP deaminase [Dictyostelium discoideum]
Length = 743
Score = 708 bits (1828), Expect = 0.0
Identities = 349/607 (57%), Positives = 451/607 (73%), Gaps = 21/607 (3%)
Query: 216 LRKEPEQEIFARLRITPMEAPSPDEIESYVILQE----CLEMRKRYIFKEAVAPWEKEVI 271
+ ++ + ++F R+ +T S EIE Y + E + +R++Y+F + W+ +
Sbjct: 1 MARKDQTQLFQRIILTN---ESESEIEEYAEVAEQLLDAINLREKYVFHPKI--WKADA- 54
Query: 272 SDPSTPKPNLEPFFY--APEGKSDHYFEMQDGVIHVYPNKN---SNEELFPVADA-TTFF 325
P KP PF + ++H F+ +GV VY N+ SN+ LF V +++
Sbjct: 55 --PVGEKPPYSPFESDESTNCATEHMFKEVNGVYFVYSNETDMKSNKALFSVPHTLASYY 112
Query: 326 TDLHQILRVIAAGNIRTLCHHRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVD 385
D++ ++ + + G +T RL LLE KFN+H +LN E QK+APHRDFYNVRKVD
Sbjct: 113 KDINNLMMLSSYGPAKTFTFKRLQLLESKFNMHTLLNDSLELFQQKTAPHRDFYNVRKVD 172
Query: 386 THVHHSACMNQKHLLRFIKSKLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLD 445
THVHHS+ MNQKHLL+FIK KL++ P+E+VIFRD YLTL EVF+SL+L +L+VD LD
Sbjct: 173 THVHHSSSMNQKHLLKFIKRKLKENPNEIVIFRDDKYLTLAEVFKSLNLDVDELSVDTLD 232
Query: 446 VHADKSTFHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQM 505
VHAD +TFHRFDKFNLKYNPCGQSRLREIFLK DNLI+G++L E++K+VF+DLE+SKYQ
Sbjct: 233 VHADNNTFHRFDKFNLKYNPCGQSRLREIFLKTDNLIKGKYLAEISKEVFTDLESSKYQC 292
Query: 506 AEYRISIYGRKQSEWDQLASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDN 565
AEYR+SIYGRK SEWD LASWIV+NDL+S V WLIQ+PRLY++Y++ T+FQ+ L+N
Sbjct: 293 AEYRLSIYGRKMSEWDTLASWIVDNDLFSTKVRWLIQVPRLYDVYRETS-TTTFQDFLNN 351
Query: 566 IFIPLFEVTVDPDSHPQLHVFLKQVVGLDLVDDESKPERRPTKHMPTPAQWTNVFNPAFS 625
+F PLFEVT DP SHP+LH+FL+QVVG+D VDDESK E++ T+ P P +W++ NP ++
Sbjct: 352 VFHPLFEVTKDPSSHPKLHLFLQQVVGIDCVDDESKFEKKFTEKFPVPGEWSSEHNPPYT 411
Query: 626 YYVYYCYANLYTLNKLRESKGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSP 685
YY+YY YANLYTLN+ RE KG+ + RPH+GEAG++DH+ A F AH I HGINLRK+P
Sbjct: 412 YYLYYLYANLYTLNQFREEKGLNILTLRPHSGEAGEVDHMGAAFYLAHGINHGINLRKTP 471
Query: 686 VLQYLYYLAQIGLAMSPLSNNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLV 745
VLQYLYYL QIG+AMSPLSNNSLFL Y+RNP P FF RGLNVS+STDDPLQ H TKEPL+
Sbjct: 472 VLQYLYYLTQIGIAMSPLSNNSLFLTYNRNPFPAFFARGLNVSISTDDPLQFHYTKEPLM 531
Query: 746 EEYSIAASVWKLSSCDLCEIARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPH 805
EEYSIA VW+LS CD+CEIARNSV QSGF H +KSHW+G +Y G GNDI +TN+
Sbjct: 532 EEYSIATQVWRLSVCDICEIARNSVLQSGFEHNVKSHWLGPDYANSG--GNDIKKTNISD 589
Query: 806 IRLEFRD 812
IR+ FR+
Sbjct: 590 IRVCFRN 596
>ref|XP_629711.1| AMP deaminase [Dictyostelium discoideum] gi|60463062|gb|EAL61257.1|
AMP deaminase [Dictyostelium discoideum]
Length = 790
Score = 708 bits (1828), Expect = 0.0
Identities = 349/607 (57%), Positives = 451/607 (73%), Gaps = 21/607 (3%)
Query: 216 LRKEPEQEIFARLRITPMEAPSPDEIESYVILQE----CLEMRKRYIFKEAVAPWEKEVI 271
+ ++ + ++F R+ +T S EIE Y + E + +R++Y+F + W+ +
Sbjct: 48 MARKDQTQLFQRIILTN---ESESEIEEYAEVAEQLLDAINLREKYVFHPKI--WKADA- 101
Query: 272 SDPSTPKPNLEPFFY--APEGKSDHYFEMQDGVIHVYPNKN---SNEELFPVADA-TTFF 325
P KP PF + ++H F+ +GV VY N+ SN+ LF V +++
Sbjct: 102 --PVGEKPPYSPFESDESTNCATEHMFKEVNGVYFVYSNETDMKSNKALFSVPHTLASYY 159
Query: 326 TDLHQILRVIAAGNIRTLCHHRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVD 385
D++ ++ + + G +T RL LLE KFN+H +LN E QK+APHRDFYNVRKVD
Sbjct: 160 KDINNLMMLSSYGPAKTFTFKRLQLLESKFNMHTLLNDSLELFQQKTAPHRDFYNVRKVD 219
Query: 386 THVHHSACMNQKHLLRFIKSKLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLD 445
THVHHS+ MNQKHLL+FIK KL++ P+E+VIFRD YLTL EVF+SL+L +L+VD LD
Sbjct: 220 THVHHSSSMNQKHLLKFIKRKLKENPNEIVIFRDDKYLTLAEVFKSLNLDVDELSVDTLD 279
Query: 446 VHADKSTFHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQM 505
VHAD +TFHRFDKFNLKYNPCGQSRLREIFLK DNLI+G++L E++K+VF+DLE+SKYQ
Sbjct: 280 VHADNNTFHRFDKFNLKYNPCGQSRLREIFLKTDNLIKGKYLAEISKEVFTDLESSKYQC 339
Query: 506 AEYRISIYGRKQSEWDQLASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDN 565
AEYR+SIYGRK SEWD LASWIV+NDL+S V WLIQ+PRLY++Y++ T+FQ+ L+N
Sbjct: 340 AEYRLSIYGRKMSEWDTLASWIVDNDLFSTKVRWLIQVPRLYDVYRETS-TTTFQDFLNN 398
Query: 566 IFIPLFEVTVDPDSHPQLHVFLKQVVGLDLVDDESKPERRPTKHMPTPAQWTNVFNPAFS 625
+F PLFEVT DP SHP+LH+FL+QVVG+D VDDESK E++ T+ P P +W++ NP ++
Sbjct: 399 VFHPLFEVTKDPSSHPKLHLFLQQVVGIDCVDDESKFEKKFTEKFPVPGEWSSEHNPPYT 458
Query: 626 YYVYYCYANLYTLNKLRESKGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSP 685
YY+YY YANLYTLN+ RE KG+ + RPH+GEAG++DH+ A F AH I HGINLRK+P
Sbjct: 459 YYLYYLYANLYTLNQFREEKGLNILTLRPHSGEAGEVDHMGAAFYLAHGINHGINLRKTP 518
Query: 686 VLQYLYYLAQIGLAMSPLSNNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLV 745
VLQYLYYL QIG+AMSPLSNNSLFL Y+RNP P FF RGLNVS+STDDPLQ H TKEPL+
Sbjct: 519 VLQYLYYLTQIGIAMSPLSNNSLFLTYNRNPFPAFFARGLNVSISTDDPLQFHYTKEPLM 578
Query: 746 EEYSIAASVWKLSSCDLCEIARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPH 805
EEYSIA VW+LS CD+CEIARNSV QSGF H +KSHW+G +Y G GNDI +TN+
Sbjct: 579 EEYSIATQVWRLSVCDICEIARNSVLQSGFEHNVKSHWLGPDYANSG--GNDIKKTNISD 636
Query: 806 IRLEFRD 812
IR+ FR+
Sbjct: 637 IRVCFRN 643
>ref|NP_083055.1| adenosine monophosphate deaminase 2 (isoform L) [Mus musculus]
gi|29145073|gb|AAH49119.1| Adenosine monophosphate
deaminase 2 (isoform L) [Mus musculus]
gi|12836179|dbj|BAB23540.1| unnamed protein product [Mus
musculus]
Length = 798
Score = 647 bits (1669), Expect = 0.0
Identities = 322/585 (55%), Positives = 400/585 (68%), Gaps = 20/585 (3%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTP--------KPNLEPFFY------APEGK 291
LQ +RY+ + A P E P TP P LE Y A G
Sbjct: 179 LQSFCPTTRRYLQQLAEKPLETRTYEQSPDTPVSADAPVHPPALEQHPYEHCEPSAMPGD 238
Query: 292 SDHYFEMQDGVIHVY----PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCHHR 347
M GV+HVY P+++ E P D F D++ ++ +I G I++ C+ R
Sbjct: 239 LGLGLRMVRGVVHVYTRRDPDEHCPEVELPYPDLQEFVADVNVLMALIINGPIKSFCYRR 298
Query: 348 LNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKL 407
L L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK +
Sbjct: 299 LQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKRAM 358
Query: 408 RKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCG 467
++ +E+V G TLREVFES++LT YDL+VD LDVHAD++TFHRFDKFN KYNP G
Sbjct: 359 KRHLEEIVHVEQGREQTLREVFESMNLTAYDLSVDTLDVHADRNTFHRFDKFNAKYNPIG 418
Query: 468 QSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLASWI 527
+S LREIF+K DN I G++ + K+V +DLE SKYQ AE R+SIYGR + EWD+LA W
Sbjct: 419 ESVLREIFIKTDNKISGKYFAHIIKEVMADLEESKYQNAELRLSIYGRSRDEWDKLARWA 478
Query: 528 VNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHVFL 587
VN+ ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+FL
Sbjct: 479 VNHKVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHLFL 538
Query: 588 KQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRESKG 646
+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR +G
Sbjct: 539 EHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQRG 598
Query: 647 MTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNN 706
T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLSNN
Sbjct: 599 FHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLSNN 658
Query: 707 SLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCEIA 766
SLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE+A
Sbjct: 659 SLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCELA 718
Query: 767 RNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
RNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 719 RNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 763
>ref|XP_342314.2| PREDICTED: similar to adenosine monophosphate deaminase 2 (isoform
L) [Rattus norvegicus]
Length = 824
Score = 647 bits (1668), Expect = 0.0
Identities = 321/587 (54%), Positives = 399/587 (67%), Gaps = 24/587 (4%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTPKPNLEPFFYAPEGKSDHYFE-------- 297
LQ +RY+ + A P E P TP P P H +E
Sbjct: 205 LQSFCPTTRRYLQQLAEKPLETRTYEQSPDTPVSADAPVH--PPALEQHPYEHCEPSTMP 262
Query: 298 --------MQDGVIHVY----PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCH 345
M GV+HVY P+++ E P D F D++ ++ +I G I++ C+
Sbjct: 263 GDLGLGLRMVRGVVHVYTRRDPDEHCPEVELPYPDLQEFVADVNVLMALIINGPIKSFCY 322
Query: 346 HRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKS 405
RL L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK
Sbjct: 323 RRLQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKR 382
Query: 406 KLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNP 465
+++ +E+V G TLREVFES++LT YDL+VD LDVHAD++TFHRFDKFN KYNP
Sbjct: 383 AMKRHLEEIVHVEQGREQTLREVFESMNLTAYDLSVDTLDVHADRNTFHRFDKFNAKYNP 442
Query: 466 CGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLAS 525
G+S LREIF+K DN I G++ + K+V SDLE SKYQ AE R+SIYGR + EWD+LA
Sbjct: 443 IGESVLREIFIKTDNKISGKYFAHIIKEVMSDLEESKYQNAELRLSIYGRSRDEWDKLAR 502
Query: 526 WIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHV 585
W VN+ ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+
Sbjct: 503 WAVNHRVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHL 562
Query: 586 FLKQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRES 644
FL+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR
Sbjct: 563 FLEHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQ 622
Query: 645 KGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLS 704
+G T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLS
Sbjct: 623 RGFHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLS 682
Query: 705 NNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCE 764
NNSLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE
Sbjct: 683 NNSLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCE 742
Query: 765 IARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
+ARNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 743 LARNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 789
>gb|EAK85479.1| hypothetical protein UM04622.1 [Ustilago maydis 521]
gi|49076548|ref|XP_402237.1| hypothetical protein
UM04622.1 [Ustilago maydis 521]
Length = 954
Score = 647 bits (1668), Expect = 0.0
Identities = 307/506 (60%), Positives = 386/506 (75%)
Query: 307 PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCHHRLNLLEQKFNLHLMLNADRE 366
P E LF V +F DL +L VI+ G +++ RL LE K+NL+ +LN RE
Sbjct: 431 PTGLRKEPLFSVPTIREYFKDLDYLLGVISDGPVKSFAWRRLKYLESKWNLYFLLNEYRE 490
Query: 367 FLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLRKEPDEVVIFRDGTYLTLR 426
K PHRDFYNVRKVDTH+HHSA MNQKHLLRFIK+K+++ PD++VI RDG LTL+
Sbjct: 491 LADMKRVPHRDFYNVRKVDTHIHHSASMNQKHLLRFIKAKIKRFPDDIVIHRDGKDLTLQ 550
Query: 427 EVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGRF 486
+VFESL LT YDL++D LD+HA + FHRFDKFNLKYNP G+SRLREIFLK DNLI+GR+
Sbjct: 551 QVFESLKLTAYDLSIDTLDMHAHQDAFHRFDKFNLKYNPMGESRLREIFLKTDNLIKGRY 610
Query: 487 LGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLASWIVNNDLYSENVVWLIQLPRL 546
L ELTK+V +DLE SKYQMAEYR+SIYGR + EWD+LASW+V+N L+S NV WLIQ+PRL
Sbjct: 611 LAELTKEVMADLEQSKYQMAEYRVSIYGRTRGEWDKLASWVVDNSLFSPNVRWLIQVPRL 670
Query: 547 YNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHVFLKQVVGLDLVDDESKPERRP 606
Y++YK G V +F+ ++ N+F PLFEVT +P SHP+LHVFL++VVG DLVDDESKPERR
Sbjct: 671 YDVYKANGTVDNFEQIIRNVFEPLFEVTQNPQSHPKLHVFLQRVVGFDLVDDESKPERRI 730
Query: 607 TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRESKGMTTIKFRPHAGEAGDIDHLA 666
K P P W +P ++Y++YY +AN+ +LN+ R+ +G T RPHAGEAGD DH+A
Sbjct: 731 HKKFPVPKLWDFKDSPPYNYWLYYMFANISSLNQWRKLRGFNTFVLRPHAGEAGDTDHMA 790
Query: 667 ATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNNSLFLDYHRNPLPVFFLRGLN 726
A FLT+ +I+HGI LRK P LQYLYYL QIGLAMSPLSNN+LFL Y RNP P F G+N
Sbjct: 791 AAFLTSQSISHGILLRKVPALQYLYYLKQIGLAMSPLSNNALFLSYDRNPFPNFLKLGMN 850
Query: 727 VSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCEIARNSVYQSGFSHALKSHWIGK 786
VS+STDDPLQ HL+KEPL+EEYS+A ++KL+ D+CE+ARNSV QSG+ +K HW+G
Sbjct: 851 VSISTDDPLQFHLSKEPLLEEYSVATQIYKLTPADMCELARNSVLQSGWEMEIKRHWLGP 910
Query: 787 EYYKRGPNGNDIHRTNVPHIRLEFRD 812
+ GP GN + ++NVP IRL FR+
Sbjct: 911 NFQLPGPRGNVVAKSNVPDIRLRFRE 936
>emb|CAG88484.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50423261|ref|XP_460211.1| unnamed protein product
[Debaryomyces hansenii]
Length = 746
Score = 646 bits (1666), Expect = 0.0
Identities = 329/619 (53%), Positives = 428/619 (68%), Gaps = 29/619 (4%)
Query: 222 QEIFARLRITPMEAPSPDEIESYVILQECLEMRKRYIFKEAVAPWEKEVISD------PS 275
+E R P E PS + IE Y ++ CL++R +Y+ K ++ P
Sbjct: 110 KESETRSNTLPEEHPSEELIEVYQDVKTCLDLRHKYLRTSVQDDEMKNPKNNDGWTIYPP 169
Query: 276 TPKPNL----------------EPFFY----APEGKSDHYFEM-QDGVIHVYPNKNSNEE 314
PKP E F + PE + YF+ ++GV VY + + +E+
Sbjct: 170 APKPTYKSRNKYNQIAHGGNDEEEFDFNKCIIPERNDEFYFKQDEEGVFQVYKS-DCDEK 228
Query: 315 LFPVADATTFFTDLHQILRVIAAGNIRTLCHHRLNLLEQKFNLHLMLNADREFLAQKSAP 374
L + T+F+DL++I ++ + G ++ RL LE K+N++ +LN E K P
Sbjct: 229 LVEIPTLNTYFSDLNKITKISSDGPAKSFAFKRLQYLEAKWNMYYLLNDFEENKHSKRNP 288
Query: 375 HRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLRKEPDEVVIFRDGTYLTLREVFESLDL 434
HRDFYNVRKVDTH+HHSACMNQKHLLRFIK KL+ +PDE VIFRDG L+L +VF+SL L
Sbjct: 289 HRDFYNVRKVDTHIHHSACMNQKHLLRFIKYKLKTQPDEQVIFRDGRILSLAQVFQSLKL 348
Query: 435 TGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGRFLGELTKQV 494
+ YDL++D LD+HA TFHRFDKFNLKYNP G+SRLREIFLK DN I G++L ELTKQV
Sbjct: 349 SAYDLSIDTLDMHAHTDTFHRFDKFNLKYNPIGESRLREIFLKTDNFINGKYLAELTKQV 408
Query: 495 FSDLEASKYQMAEYRISIYGRKQSEWDQLASWIVNNDLYSENVVWLIQLPRLYNIYKDMG 554
DLE+SKYQM E RIS+YGR EWD+LASWIV+N L+S NV WLIQ+PRLY++YK G
Sbjct: 409 MDDLESSKYQMNELRISVYGRSIHEWDKLASWIVDNKLFSHNVRWLIQVPRLYDLYKKNG 468
Query: 555 IVTSFQNMLDNIFIPLFEVTVDPDSHPQLHVFLKQVVGLDLVDDESKPERR-PTKHMPTP 613
VT+F ++L N+F PLFEV++DP SHP+LHVFL++VVG D VDDESK E+ +++ P
Sbjct: 469 NVTTFHDILVNLFKPLFEVSIDPKSHPKLHVFLQRVVGFDSVDDESKLEKSIQSRNYPVA 528
Query: 614 AQWTNVFNPAFSYYVYYCYANLYTLNKLRESKGMTTIKFRPHAGEAGDIDHLAATFLTAH 673
++W NP +SYY+YY YAN+ +LN+LR G T RPH GEAGD +HL + FLT+H
Sbjct: 529 SKWDLPINPPYSYYLYYLYANITSLNQLRSKNGYNTFLLRPHCGEAGDPEHLISAFLTSH 588
Query: 674 NIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNNSLFLDYHRNPLPVFFLRGLNVSLSTDD 733
+I+HGI LRK P +QYLYYL QIGLAMSPLSNN+LFL Y +NP FF +G+NVSLSTDD
Sbjct: 589 SISHGILLRKIPFIQYLYYLDQIGLAMSPLSNNALFLAYDKNPFYSFFQKGMNVSLSTDD 648
Query: 734 PLQIHLTKEPLVEEYSIAASVWKLSSCDLCEIARNSVYQSGFSHALKSHWIGKEYYKRGP 793
PLQ TKEPL+EEYS+AA ++KLS D+CE+A NSV QSG+ A+K HW+GK + G
Sbjct: 649 PLQFSYTKEPLIEEYSVAAQIYKLSGVDMCELACNSVKQSGWEVAIKKHWLGKNFIVGGI 708
Query: 794 NGNDIHRTNVPHIRLEFRD 812
+GNDI +TNVP IR+ +R+
Sbjct: 709 DGNDIEKTNVPDIRVGYRE 727
>emb|CAI19307.1| adenosine monophosphate deaminase 2 (isoform L) [Homo sapiens]
gi|178547|gb|AAA62127.1| AMP deaminase isoform L
gi|55925574|ref|NP_981949.1| adenosine monophosphate
deaminase 2 (isoform L) isoform 3 [Homo sapiens]
Length = 760
Score = 644 bits (1662), Expect = 0.0
Identities = 319/587 (54%), Positives = 399/587 (67%), Gaps = 24/587 (4%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTPKPNLEPFFYAPEGKSDHYFE-------- 297
LQ +RY+ + A P E P TP P P H +E
Sbjct: 140 LQSFCPTTRRYLQQLAEKPLETRTYEQGPDTPVSADAPVH--PPALEQHPYEHCEPSTMP 197
Query: 298 --------MQDGVIHVY----PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCH 345
M GV+HVY P+++ +E P D F D++ ++ +I G I++ C+
Sbjct: 198 GDLGLGLRMVRGVVHVYTRREPDEHCSEVELPYPDLQEFVADVNVLMALIINGPIKSFCY 257
Query: 346 HRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKS 405
RL L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK
Sbjct: 258 RRLQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKR 317
Query: 406 KLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNP 465
+++ +E+V G TLREVFES++LT YDL+VD LDVHAD++TFHRFDKFN KYNP
Sbjct: 318 AMKRHLEEIVHVEQGREQTLREVFESMNLTAYDLSVDTLDVHADRNTFHRFDKFNAKYNP 377
Query: 466 CGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLAS 525
G+S LREIF+K DN + G++ + K+V SDLE SKYQ AE R+SIYGR + EWD+LA
Sbjct: 378 IGESVLREIFIKTDNRVSGKYFAHIIKEVMSDLEESKYQNAELRLSIYGRSRDEWDKLAR 437
Query: 526 WIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHV 585
W V + ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+
Sbjct: 438 WAVMHRVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHL 497
Query: 586 FLKQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRES 644
FL+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR
Sbjct: 498 FLEHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQ 557
Query: 645 KGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLS 704
+G T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLS
Sbjct: 558 RGFHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLS 617
Query: 705 NNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCE 764
NNSLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE
Sbjct: 618 NNSLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCE 677
Query: 765 IARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
+ARNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 678 LARNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 724
>emb|CAI19305.1| adenosine monophosphate deaminase 2 (isoform L) [Homo sapiens]
gi|14043443|gb|AAH07711.1| Adenosine monophosphate
deaminase 2 (isoform L), isoform 2 [Homo sapiens]
gi|49904197|gb|AAH75844.1| Adenosine monophosphate
deaminase 2 (isoform L), isoform 2 [Homo sapiens]
gi|5922016|gb|AAC50309.2| AMP deaminase isoform L [Homo
sapiens] gi|34147621|ref|NP_631895.1| adenosine
monophosphate deaminase 2 (isoform L) isoform 2 [Homo
sapiens]
Length = 798
Score = 644 bits (1662), Expect = 0.0
Identities = 319/587 (54%), Positives = 399/587 (67%), Gaps = 24/587 (4%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTPKPNLEPFFYAPEGKSDHYFE-------- 297
LQ +RY+ + A P E P TP P P H +E
Sbjct: 178 LQSFCPTTRRYLQQLAEKPLETRTYEQGPDTPVSADAPVH--PPALEQHPYEHCEPSTMP 235
Query: 298 --------MQDGVIHVY----PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCH 345
M GV+HVY P+++ +E P D F D++ ++ +I G I++ C+
Sbjct: 236 GDLGLGLRMVRGVVHVYTRREPDEHCSEVELPYPDLQEFVADVNVLMALIINGPIKSFCY 295
Query: 346 HRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKS 405
RL L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK
Sbjct: 296 RRLQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKR 355
Query: 406 KLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNP 465
+++ +E+V G TLREVFES++LT YDL+VD LDVHAD++TFHRFDKFN KYNP
Sbjct: 356 AMKRHLEEIVHVEQGREQTLREVFESMNLTAYDLSVDTLDVHADRNTFHRFDKFNAKYNP 415
Query: 466 CGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLAS 525
G+S LREIF+K DN + G++ + K+V SDLE SKYQ AE R+SIYGR + EWD+LA
Sbjct: 416 IGESVLREIFIKTDNRVSGKYFAHIIKEVMSDLEESKYQNAELRLSIYGRSRDEWDKLAR 475
Query: 526 WIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHV 585
W V + ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+
Sbjct: 476 WAVMHRVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHL 535
Query: 586 FLKQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRES 644
FL+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR
Sbjct: 536 FLEHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQ 595
Query: 645 KGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLS 704
+G T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLS
Sbjct: 596 RGFHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLS 655
Query: 705 NNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCE 764
NNSLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE
Sbjct: 656 NNSLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCE 715
Query: 765 IARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
+ARNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 716 LARNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 762
>ref|XP_537039.1| PREDICTED: similar to AMP deaminase 2 (AMP deaminase isoform L)
[Canis familiaris]
Length = 1273
Score = 644 bits (1662), Expect = 0.0
Identities = 319/585 (54%), Positives = 399/585 (67%), Gaps = 20/585 (3%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTP--------KPNLE--PFFYAPE----GK 291
LQ +RY+ + A P E P TP P LE P+ Y G
Sbjct: 653 LQSFCPTTRRYLQQLAEKPLETRTYEQGPDTPVSADAPVHPPALEQHPYEYCEPSTMPGD 712
Query: 292 SDHYFEMQDGVIHVYPNKNSNEEL----FPVADATTFFTDLHQILRVIAAGNIRTLCHHR 347
M GV+HVY + ++E P D F D++ ++ +I G I++ C+ R
Sbjct: 713 LGLGLRMVQGVVHVYTRREADEHCSEVELPYPDLQEFVADVNVLMALIINGPIKSFCYRR 772
Query: 348 LNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKL 407
L L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK +
Sbjct: 773 LQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKRAM 832
Query: 408 RKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCG 467
++ +E+V G TLREVFES++LT YDL+VD LD+HAD++TFHRFDKFN KYNP G
Sbjct: 833 KRHLEEIVHVEQGREQTLREVFESMNLTAYDLSVDTLDMHADRNTFHRFDKFNAKYNPIG 892
Query: 468 QSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLASWI 527
+S LREIF+K DN + G++ + K+V SDLE SKYQ AE R+SIYGR + EWD+LA W
Sbjct: 893 ESVLREIFIKTDNRVSGKYFAHIIKEVMSDLEESKYQNAELRLSIYGRSRDEWDKLACWA 952
Query: 528 VNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHVFL 587
V + ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+FL
Sbjct: 953 VKHRVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHLFL 1012
Query: 588 KQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRESKG 646
+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR +G
Sbjct: 1013 EHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQRG 1072
Query: 647 MTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNN 706
T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLSNN
Sbjct: 1073 FHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLSNN 1132
Query: 707 SLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCEIA 766
SLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE+A
Sbjct: 1133 SLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCELA 1192
Query: 767 RNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
RNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 1193 RNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 1237
>gb|AAA11725.1| AMP deaminase isoform L [Homo sapiens] gi|345738|pir||A44313 AMP
deaminase (EC 3.5.4.6) isoform L - human
Length = 760
Score = 644 bits (1662), Expect = 0.0
Identities = 319/587 (54%), Positives = 399/587 (67%), Gaps = 24/587 (4%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTPKPNLEPFFYAPEGKSDHYFE-------- 297
LQ +RY+ + A P E P TP P P H +E
Sbjct: 140 LQSFCPTTRRYLQQLAEKPLETRTYEQGPDTPVSADAPVH--PPALEQHPYEHCEPSTMP 197
Query: 298 --------MQDGVIHVY----PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCH 345
M GV+HVY P+++ +E P D F D++ ++ +I G I++ C+
Sbjct: 198 GDLGLGLRMVRGVVHVYTRREPDEHCSEVELPYPDLQEFVADVNVLMALIINGPIKSFCY 257
Query: 346 HRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKS 405
RL L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK
Sbjct: 258 RRLQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKR 317
Query: 406 KLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNP 465
+++ +E+V G TLREVFES++LT YDL+VD LDVHAD++TFHRFDKFN KYNP
Sbjct: 318 AMKRHLEEIVHVEQGREQTLREVFESMNLTAYDLSVDTLDVHADRNTFHRFDKFNAKYNP 377
Query: 466 CGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLAS 525
G+S LREIF+K DN + G++ + K+V SDLE SKYQ AE R+SIYGR + EWD+LA
Sbjct: 378 IGESVLREIFIKTDNRVSGKYFAHIIKEVMSDLEESKYQNAELRLSIYGRSRDEWDKLAR 437
Query: 526 WIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHV 585
W V + ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+
Sbjct: 438 WAVMHRVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHL 497
Query: 586 FLKQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRES 644
FL+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR
Sbjct: 498 FLEHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQ 557
Query: 645 KGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLS 704
+G T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLS
Sbjct: 558 RGFHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLS 617
Query: 705 NNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCE 764
NNSLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE
Sbjct: 618 NNSLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCE 677
Query: 765 IARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
+ARNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 678 LARNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 724
>gb|AAD56303.1| AMP deaminase isoform L [Homo sapiens]
Length = 804
Score = 644 bits (1662), Expect = 0.0
Identities = 319/587 (54%), Positives = 399/587 (67%), Gaps = 24/587 (4%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTPKPNLEPFFYAPEGKSDHYFE-------- 297
LQ +RY+ + A P E P TP P P H +E
Sbjct: 184 LQSFCPTTRRYLQQLAEKPLETRTYEQGPDTPVSADAPVH--PPALEQHPYEHCEPSTMP 241
Query: 298 --------MQDGVIHVY----PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCH 345
M GV+HVY P+++ +E P D F D++ ++ +I G I++ C+
Sbjct: 242 GDLGLGLRMVRGVVHVYTRREPDEHCSEVELPYPDLQEFVADVNVLMALIINGPIKSFCY 301
Query: 346 HRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKS 405
RL L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK
Sbjct: 302 RRLQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKR 361
Query: 406 KLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNP 465
+++ +E+V G TLREVFES++LT YDL+VD LDVHAD++TFHRFDKFN KYNP
Sbjct: 362 AMKRHLEEIVHVEQGREQTLREVFESMNLTAYDLSVDTLDVHADRNTFHRFDKFNAKYNP 421
Query: 466 CGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLAS 525
G+S LREIF+K DN + G++ + K+V SDLE SKYQ AE R+SIYGR + EWD+LA
Sbjct: 422 IGESVLREIFIKTDNRVSGKYFAHIIKEVMSDLEESKYQNAELRLSIYGRSRDEWDKLAR 481
Query: 526 WIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHV 585
W V + ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+
Sbjct: 482 WAVMHRVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHL 541
Query: 586 FLKQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRES 644
FL+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR
Sbjct: 542 FLEHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQ 601
Query: 645 KGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLS 704
+G T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLS
Sbjct: 602 RGFHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLS 661
Query: 705 NNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCE 764
NNSLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE
Sbjct: 662 NNSLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCE 721
Query: 765 IARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
+ARNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 722 LARNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 768
>gb|AAD56302.1| AMP deaminase isoform L [Homo sapiens]
gi|12644375|sp|Q01433|AMPD2_HUMAN AMP deaminase 2 (AMP
deaminase isoform L) gi|21264318|ref|NP_004028.3|
adenosine monophosphate deaminase 2 (isoform L) isoform
1 [Homo sapiens]
Length = 879
Score = 644 bits (1662), Expect = 0.0
Identities = 319/587 (54%), Positives = 399/587 (67%), Gaps = 24/587 (4%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTPKPNLEPFFYAPEGKSDHYFE-------- 297
LQ +RY+ + A P E P TP P P H +E
Sbjct: 259 LQSFCPTTRRYLQQLAEKPLETRTYEQGPDTPVSADAPVH--PPALEQHPYEHCEPSTMP 316
Query: 298 --------MQDGVIHVY----PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCH 345
M GV+HVY P+++ +E P D F D++ ++ +I G I++ C+
Sbjct: 317 GDLGLGLRMVRGVVHVYTRREPDEHCSEVELPYPDLQEFVADVNVLMALIINGPIKSFCY 376
Query: 346 HRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKS 405
RL L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK
Sbjct: 377 RRLQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKR 436
Query: 406 KLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNP 465
+++ +E+V G TLREVFES++LT YDL+VD LDVHAD++TFHRFDKFN KYNP
Sbjct: 437 AMKRHLEEIVHVEQGREQTLREVFESMNLTAYDLSVDTLDVHADRNTFHRFDKFNAKYNP 496
Query: 466 CGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLAS 525
G+S LREIF+K DN + G++ + K+V SDLE SKYQ AE R+SIYGR + EWD+LA
Sbjct: 497 IGESVLREIFIKTDNRVSGKYFAHIIKEVMSDLEESKYQNAELRLSIYGRSRDEWDKLAR 556
Query: 526 WIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHV 585
W V + ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+
Sbjct: 557 WAVMHRVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHL 616
Query: 586 FLKQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRES 644
FL+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR
Sbjct: 617 FLEHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQ 676
Query: 645 KGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLS 704
+G T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLS
Sbjct: 677 RGFHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLS 736
Query: 705 NNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCE 764
NNSLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE
Sbjct: 737 NNSLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCE 796
Query: 765 IARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
+ARNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 797 LARNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 843
>gb|AAC50308.1| AMP deaminase
Length = 624
Score = 644 bits (1662), Expect = 0.0
Identities = 319/587 (54%), Positives = 399/587 (67%), Gaps = 24/587 (4%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTPKPNLEPFFYAPEGKSDHYFE-------- 297
LQ +RY+ + A P E P TP P P H +E
Sbjct: 4 LQSFCPTTRRYLQQLAEKPLETRTYEQGPDTPVSADAPVH--PPALEQHPYEHCEPSTMP 61
Query: 298 --------MQDGVIHVY----PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCH 345
M GV+HVY P+++ +E P D F D++ ++ +I G I++ C+
Sbjct: 62 GDLGLGLRMVRGVVHVYTRREPDEHCSEVELPYPDLQEFVADVNVLMALIINGPIKSFCY 121
Query: 346 HRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKS 405
RL L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK
Sbjct: 122 RRLQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKR 181
Query: 406 KLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNP 465
+++ +E+V G TLREVFES++LT YDL+VD LDVHAD++TFHRFDKFN KYNP
Sbjct: 182 AMKRHLEEIVHVEQGREQTLREVFESMNLTAYDLSVDTLDVHADRNTFHRFDKFNAKYNP 241
Query: 466 CGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLAS 525
G+S LREIF+K DN + G++ + K+V SDLE SKYQ AE R+SIYGR + EWD+LA
Sbjct: 242 IGESVLREIFIKTDNRVSGKYFAHIIKEVMSDLEESKYQNAELRLSIYGRSRDEWDKLAR 301
Query: 526 WIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHV 585
W V + ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+
Sbjct: 302 WAVMHRVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHL 361
Query: 586 FLKQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRES 644
FL+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR
Sbjct: 362 FLEHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQ 421
Query: 645 KGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLS 704
+G T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLS
Sbjct: 422 RGFHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLS 481
Query: 705 NNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCE 764
NNSLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE
Sbjct: 482 NNSLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCE 541
Query: 765 IARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
+ARNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 542 LARNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 588
>gb|AAA62126.1| AMP deaminase isoform L splicing variant
Length = 753
Score = 644 bits (1662), Expect = 0.0
Identities = 319/587 (54%), Positives = 399/587 (67%), Gaps = 24/587 (4%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTPKPNLEPFFYAPEGKSDHYFE-------- 297
LQ +RY+ + A P E P TP P P H +E
Sbjct: 133 LQSFCPTTRRYLQQLAEKPLETRTYEQGPDTPVSADAPVH--PPALEQHPYEHCEPSTMP 190
Query: 298 --------MQDGVIHVY----PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCH 345
M GV+HVY P+++ +E P D F D++ ++ +I G I++ C+
Sbjct: 191 GDLGLGLRMVRGVVHVYTRREPDEHCSEVELPYPDLQEFVADVNVLMALIINGPIKSFCY 250
Query: 346 HRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKS 405
RL L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK
Sbjct: 251 RRLQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKR 310
Query: 406 KLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNP 465
+++ +E+V G TLREVFES++LT YDL+VD LDVHAD++TFHRFDKFN KYNP
Sbjct: 311 AMKRHLEEIVHVEQGREQTLREVFESMNLTAYDLSVDTLDVHADRNTFHRFDKFNAKYNP 370
Query: 466 CGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLAS 525
G+S LREIF+K DN + G++ + K+V SDLE SKYQ AE R+SIYGR + EWD+LA
Sbjct: 371 IGESVLREIFIKTDNRVSGKYFAHIIKEVMSDLEESKYQNAELRLSIYGRSRDEWDKLAR 430
Query: 526 WIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHV 585
W V + ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+
Sbjct: 431 WAVMHRVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHL 490
Query: 586 FLKQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRES 644
FL+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR
Sbjct: 491 FLEHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQ 550
Query: 645 KGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLS 704
+G T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLS
Sbjct: 551 RGFHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLS 610
Query: 705 NNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCE 764
NNSLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE
Sbjct: 611 NNSLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCE 670
Query: 765 IARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
+ARNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 671 LARNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 717
>dbj|BAE02528.1| unnamed protein product [Macaca fascicularis]
Length = 625
Score = 642 bits (1655), Expect = 0.0
Identities = 318/587 (54%), Positives = 398/587 (67%), Gaps = 24/587 (4%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISD-PSTPKPNLEPFFYAPEGKSDHYFE-------- 297
LQ +RY+ + A P E P TP P P H +E
Sbjct: 5 LQSFCPTTRRYLQQLAEKPLETRTYEQGPDTPVSADAPVH--PPALEQHPYEHCEPSTMP 62
Query: 298 --------MQDGVIHVY----PNKNSNEELFPVADATTFFTDLHQILRVIAAGNIRTLCH 345
M GV+HVY P+++ +E P D F D++ ++ +I G I++ C+
Sbjct: 63 GDLGLGLRMVRGVVHVYTRREPDEHCSEVELPYPDLQEFVADVNVLMALIINGPIKSFCY 122
Query: 346 HRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKS 405
RL L KF +H++LN +E AQK PHRDFYN+RKVDTH+H S+CMNQKHLLRFIK
Sbjct: 123 RRLQYLSSKFQMHVLLNEMKELAAQKKVPHRDFYNIRKVDTHIHASSCMNQKHLLRFIKR 182
Query: 406 KLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNP 465
+++ +E+V G TLREVFES++ T YDL+VD LDVHAD++TFHRFDKFN KYNP
Sbjct: 183 AMKRHLEEIVHVEQGREQTLREVFESMNPTAYDLSVDTLDVHADRNTFHRFDKFNAKYNP 242
Query: 466 CGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLAS 525
G+S LREIF+K DN + G++ + K+V SDLE SKYQ AE R+SIYGR + EWD+LA
Sbjct: 243 IGESVLREIFIKTDNRVSGKYFAHIIKEVMSDLEESKYQNAELRLSIYGRSRDEWDKLAR 302
Query: 526 WIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHV 585
W V + ++S NV WL+Q+PRL+++Y+ G + +FQ ML+NIF+PLFE TV P SHP+LH+
Sbjct: 303 WAVMHRVHSPNVRWLVQVPRLFDVYRTKGQLANFQEMLENIFLPLFEATVHPASHPELHL 362
Query: 586 FLKQVVGLDLVDDESKPERRP-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRES 644
FL+ V G D VDDESKPE P P W NP ++YY+YY +AN+ LN LR
Sbjct: 363 FLEHVDGFDSVDDESKPENHVFNLESPLPEAWVEEDNPPYAYYLYYTFANMAMLNHLRRQ 422
Query: 645 KGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLS 704
+G T RPH GEAG I HL + F+ A NI+HG+ LRK+PVLQYLYYLAQIG+AMSPLS
Sbjct: 423 RGFHTFVLRPHCGEAGPIHHLVSAFMLAENISHGLLLRKAPVLQYLYYLAQIGIAMSPLS 482
Query: 705 NNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCE 764
NNSLFL YHRNPLP + RGL VSLSTDDPLQ H TKEPL+EEYSIA VWKLSSCD+CE
Sbjct: 483 NNSLFLSYHRNPLPEYLSRGLMVSLSTDDPLQFHFTKEPLMEEYSIATQVWKLSSCDMCE 542
Query: 765 IARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHIRLEFR 811
+ARNSV SGFSH +KSHW+G Y K GP GNDI RTNVP IR+ +R
Sbjct: 543 LARNSVLMSGFSHKVKSHWLGPNYTKEGPEGNDIRRTNVPDIRVGYR 589
>emb|CAA86620.1| AMD1 [Saccharomyces cerevisiae] gi|6323606|ref|NP_013677.1| AMP
deaminase, tetrameric enzyme that catalyzes the
deamination of AMP to form IMP and ammonia; may be
involved in regulation of intracellular adenine
nucleotide pools; Amd1p [Saccharomyces cerevisiae]
gi|1077011|pir||S49744 AMP deaminase (EC 3.5.4.6) -
yeast (Saccharomyces cerevisiae)
gi|1351916|sp|P15274|AMDM_YEAST AMP deaminase
(Myoadenylate deaminase)
Length = 810
Score = 637 bits (1644), Expect = 0.0
Identities = 321/605 (53%), Positives = 413/605 (68%), Gaps = 26/605 (4%)
Query: 233 MEAPSPDEIESYVILQECLEMRKRY----IFKEAVAPWEK------------EVISD--- 273
++ PS + I+ Y + EC +R +Y + + P K SD
Sbjct: 182 LDDPSSELIDLYSKVAECRNLRAKYQTISVQNDDQNPKNKPGWVVYPPPPKPSYNSDTKT 241
Query: 274 --PSTPKPNLEPFFYA----PEGKSDHYFEMQDGVIHVYPNKNSNEELFP-VADATTFFT 326
P T KP+ E F + P D F + D +V +EL + ++
Sbjct: 242 VVPVTNKPDAEVFDFTKCEIPGEDPDWEFTLNDDDSYVVHRSGKTDELIAQIPTLRDYYL 301
Query: 327 DLHQILRVIAAGNIRTLCHHRLNLLEQKFNLHLMLNADREFLAQKSAPHRDFYNVRKVDT 386
DL +++ + + G ++ + RL LE ++NL+ +LN +E K PHRDFYNVRKVDT
Sbjct: 302 DLEKMISISSDGPAKSFAYRRLQYLEARWNLYYLLNEYQETSVSKRNPHRDFYNVRKVDT 361
Query: 387 HVHHSACMNQKHLLRFIKSKLRKEPDEVVIFRDGTYLTLREVFESLDLTGYDLNVDLLDV 446
HVHHSACMNQKHLLRFIK KLR DE VIFRDG LTL EVF SL LTGYDL++D LD+
Sbjct: 362 HVHHSACMNQKHLLRFIKHKLRHSKDEKVIFRDGKLLTLDEVFRSLHLTGYDLSIDTLDM 421
Query: 447 HADKSTFHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGRFLGELTKQVFSDLEASKYQMA 506
HA K TFHRFDKFNLKYNP G+SRLREIFLK +N I+G +L ++TKQV DLE SKYQ
Sbjct: 422 HAHKDTFHRFDKFNLKYNPIGESRLREIFLKTNNYIKGTYLADITKQVIFDLENSKYQNC 481
Query: 507 EYRISIYGRKQSEWDQLASWIVNNDLYSENVVWLIQLPRLYNIYKDMGIVTSFQNMLDNI 566
EYRIS+YGR EWD+LASW+++N + S NV WL+Q+PRLY+IYK GIV SFQ++ N+
Sbjct: 482 EYRISVYGRSLDEWDKLASWVIDNKVISHNVRWLVQIPRLYDIYKKTGIVQSFQDICKNL 541
Query: 567 FIPLFEVTVDPDSHPQLHVFLKQVVGLDLVDDESKPERRPTKHMPTPAQWTNVFNPAFSY 626
F PLFEVT +P SHP+LHVFL++V+G D VDDESK +RR + P P+ W NP +SY
Sbjct: 542 FQPLFEVTKNPQSHPKLHVFLQRVIGFDSVDDESKVDRRFHRKYPKPSLWEAPQNPPYSY 601
Query: 627 YVYYCYANLYTLNKLRESKGMTTIKFRPHAGEAGDIDHLAATFLTAHNIAHGINLRKSPV 686
Y+YY Y+N+ +LN+ R +G T+ RPH GEAGD +HL + +L AH I+HGI LRK P
Sbjct: 602 YLYYLYSNVASLNQWRAKRGFNTLVLRPHCGEAGDPEHLVSAYLLAHGISHGILLRKVPF 661
Query: 687 LQYLYYLAQIGLAMSPLSNNSLFLDYHRNPLPVFFLRGLNVSLSTDDPLQIHLTKEPLVE 746
+QYLYYL Q+G+AMSPLSNN+LFL Y +NP P +F RGLNVSLSTDDPLQ T+EPL+E
Sbjct: 662 VQYLYYLDQVGIAMSPLSNNALFLTYDKNPFPRYFKRGLNVSLSTDDPLQFSYTREPLIE 721
Query: 747 EYSIAASVWKLSSCDLCEIARNSVYQSGFSHALKSHWIGKEYYKRGPNGNDIHRTNVPHI 806
EYS+AA ++KLS+ D+CE+ARNSV QSG+ +K HWIGK++ K G GND+ RTNVP I
Sbjct: 722 EYSVAAQIYKLSNVDMCELARNSVLQSGWEAQIKKHWIGKDFDKSGVEGNDVVRTNVPDI 781
Query: 807 RLEFR 811
R+ +R
Sbjct: 782 RINYR 786
>ref|NP_572931.1| CG32626-PC, isoform C [Drosophila melanogaster]
gi|22832227|gb|AAN09337.1| CG32626-PC, isoform C
[Drosophila melanogaster] gi|15291169|gb|AAK92853.1|
GH10492p [Drosophila melanogaster]
Length = 707
Score = 637 bits (1642), Expect = 0.0
Identities = 307/567 (54%), Positives = 395/567 (69%), Gaps = 20/567 (3%)
Query: 247 LQECLEMRKRYIFKEAVAPWEKEVISDPSTPKPNLEPFFYAPEGKSDHYFEMQDGVIHVY 306
+ E ++ R+ + PW E PN E F P +GV H+Y
Sbjct: 133 VSESADVHLRHSPMKITNPWNVEF--------PNDEDFKIKP----------LNGVFHIY 174
Query: 307 PNKNSNEEL-FPVADATTFFTDLHQILRVIAAGNIRTLCHHRLNLLEQKFNLHLMLNADR 365
N + + E+ + D + F D+ + +IA G +++ C+ RL L K+ +H++LN R
Sbjct: 175 ENDDESSEIKYEYPDMSQFVNDMQVMCNMIADGPLKSFCYRRLCYLSSKYQMHVLLNELR 234
Query: 366 EFLAQKSAPHRDFYNVRKVDTHVHHSACMNQKHLLRFIKSKLRKEPDEVVIFRDGTYLTL 425
E AQK+ PHRDFYN RKVDTH+H ++CMNQKHLLRFIK L+ +EVV +G +TL
Sbjct: 235 ELAAQKAVPHRDFYNTRKVDTHIHAASCMNQKHLLRFIKKTLKNNANEVVTVTNGQQMTL 294
Query: 426 REVFESLDLTGYDLNVDLLDVHADKSTFHRFDKFNLKYNPCGQSRLREIFLKQDNLIQGR 485
+VF+S++LT YDL VD+LDVHAD++TFHRFDKFN KYNP G+SRLRE+FLK DN + G+
Sbjct: 295 AQVFQSMNLTTYDLTVDMLDVHADRNTFHRFDKFNSKYNPIGESRLREVFLKTDNYLNGK 354
Query: 486 FLGELTKQVFSDLEASKYQMAEYRISIYGRKQSEWDQLASWIVNNDLYSENVVWLIQLPR 545
+ ++ K+V DLE SKYQ AE R+SIYG+ EW +LA W ++ND+YS N+ WLIQ+PR
Sbjct: 355 YFAQIIKEVAFDLEESKYQNAELRLSIYGKSPDEWYKLAKWAIDNDVYSSNIRWLIQIPR 414
Query: 546 LYNIYKDMGIVTSFQNMLDNIFIPLFEVTVDPDSHPQLHVFLKQVVGLDLVDDESKPERR 605
L++I+K ++ SFQ +L+NIF+PLFE T P HP+LH FL+ V+G D VDDESKPE
Sbjct: 415 LFDIFKSNKMMKSFQEILNNIFLPLFEATARPSKHPELHRFLQYVIGFDSVDDESKPENP 474
Query: 606 P-TKHMPTPAQWTNVFNPAFSYYVYYCYANLYTLNKLRESKGMTTIKFRPHAGEAGDIDH 664
+P P +WT NP ++YY+YY YAN+ LNK R+S+ M T RPH GEAG + H
Sbjct: 475 LFDNDVPRPEEWTYEENPPYAYYIYYMYANMTVLNKFRQSRNMNTFVLRPHCGEAGPVQH 534
Query: 665 LAATFLTAHNIAHGINLRKSPVLQYLYYLAQIGLAMSPLSNNSLFLDYHRNPLPVFFLRG 724
L FL A NI+HG+ LRK PVLQYLYYL QIG+AMSPLSNNSLFL+YHRNPLP + RG
Sbjct: 535 LVCGFLMAENISHGLLLRKVPVLQYLYYLTQIGIAMSPLSNNSLFLNYHRNPLPEYLARG 594
Query: 725 LNVSLSTDDPLQIHLTKEPLVEEYSIAASVWKLSSCDLCEIARNSVYQSGFSHALKSHWI 784
L +SLSTDDPLQ H TKEPL+EEYSIAA VWKLSSCD+CE+ARNSV SGF HA+K W+
Sbjct: 595 LIISLSTDDPLQFHFTKEPLMEEYSIAAQVWKLSSCDMCELARNSVMMSGFPHAIKQQWL 654
Query: 785 GKEYYKRGPNGNDIHRTNVPHIRLEFR 811
G YY+ G GNDI RTNVP IR+ +R
Sbjct: 655 GPIYYEDGIMGNDITRTNVPEIRVAYR 681
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,494,348,170
Number of Sequences: 2540612
Number of extensions: 70203164
Number of successful extensions: 223733
Number of sequences better than 10.0: 371
Number of HSP's better than 10.0 without gapping: 141
Number of HSP's successfully gapped in prelim test: 238
Number of HSP's that attempted gapping in prelim test: 222484
Number of HSP's gapped (non-prelim): 864
length of query: 814
length of database: 863,360,394
effective HSP length: 136
effective length of query: 678
effective length of database: 517,837,162
effective search space: 351093595836
effective search space used: 351093595836
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC140721.5


