

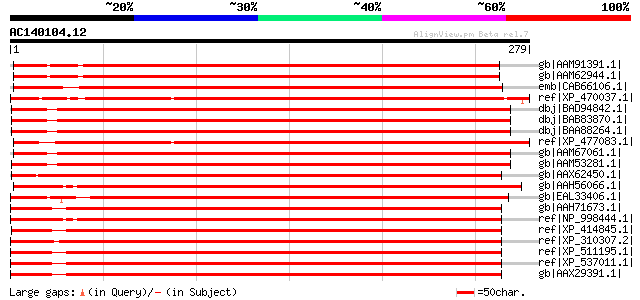
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140104.12 - phase: 0
(279 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM91391.1| At2g41840/T11A7.6 [Arabidopsis thaliana] gi|23350... 472 e-132
gb|AAM62944.1| 40S ribosomal protein S2 [Arabidopsis thaliana] 469 e-131
emb|CAB66106.1| 40S ribosomal protein S2 homolog [Arabidopsis th... 465 e-130
ref|XP_470037.1| putative 40S ribosomal protein S2 [Oryza sativa... 462 e-129
dbj|BAD94842.1| ribosomal protein S2 [Arabidopsis thaliana] gi|2... 459 e-128
dbj|BAB83870.1| ribosomal protein S2 [Arabidopsis thaliana] gi|6... 459 e-128
dbj|BAA88264.1| RF12 [Arabidopsis thaliana] gi|25294520|pir||T52... 459 e-128
ref|XP_477083.1| putative 40S ribosomal protein S2 [Oryza sativa... 458 e-128
gb|AAM67061.1| ribosomal protein S2, putative [Arabidopsis thali... 457 e-127
gb|AAM53281.1| ribosomal protein S2, putative [Arabidopsis thali... 457 e-127
gb|AAX62450.1| ribosomal protein S2 [Lysiphlebus testaceipes] 397 e-109
gb|AAH56066.1| Sop-prov protein [Xenopus laevis] 397 e-109
gb|EAL33406.1| GA19229-PA [Drosophila pseudoobscura] 395 e-109
gb|AAH71673.1| Ribosomal protein S2 [Homo sapiens] 394 e-108
ref|NP_998444.1| hypothetical protein LOC406565 [Danio rerio] gi... 392 e-108
ref|XP_414845.1| PREDICTED: similar to 40S ribosomal protein S2 ... 392 e-108
ref|XP_310307.2| ENSANGP00000015322 [Anopheles gambiae str. PEST... 392 e-108
ref|XP_511195.1| PREDICTED: hypothetical protein XP_511195 [Pan ... 392 e-108
ref|XP_537011.1| PREDICTED: similar to 40S ribosomal protein S2 ... 392 e-108
gb|AAX29391.1| ribosomal protein S2 [synthetic construct] 392 e-108
>gb|AAM91391.1| At2g41840/T11A7.6 [Arabidopsis thaliana] gi|2335095|gb|AAC02764.1|
40S ribosomal protein S2 [Arabidopsis thaliana]
gi|15081715|gb|AAK82512.1| At2g41840/T11A7.6
[Arabidopsis thaliana] gi|3915847|sp|P49688|RS2_ARATH
40S ribosomal protein S2 gi|15227443|ref|NP_181715.1|
40S ribosomal protein S2 (RPS2C) [Arabidopsis thaliana]
Length = 285
Score = 472 bits (1214), Expect = e-132
Identities = 233/261 (89%), Positives = 243/261 (92%), Gaps = 3/261 (1%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
ERGG+RG FG GFGGRGG RG RGGRG GRGGRR GGR EEEKWVPVTKLGRLV G
Sbjct: 11 ERGGERGDFGRGFGGRGG-RGDRGGRGRGGRGGRR--GGRASEEEKWVPVTKLGRLVAAG 67
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
I+ +EQIYLHSLP+KE+QIID L+GPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD
Sbjct: 68 HIKQIEQIYLHSLPVKEYQIIDMLIGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDG 127
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSVTV
Sbjct: 128 NGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 187
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFLTP
Sbjct: 188 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFLTP 247
Query: 243 EFWKETRFSKSPFQEYTDLLA 263
EFWKETRFS+SP+QE+TD LA
Sbjct: 248 EFWKETRFSRSPYQEHTDFLA 268
>gb|AAM62944.1| 40S ribosomal protein S2 [Arabidopsis thaliana]
Length = 285
Score = 469 bits (1207), Expect = e-131
Identities = 232/261 (88%), Positives = 242/261 (91%), Gaps = 3/261 (1%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
ERGG+RG FG GFGGRGG RG RGGRG GRGGRR GGR EEEKWVPVTKLGR V G
Sbjct: 11 ERGGERGDFGRGFGGRGG-RGDRGGRGRGGRGGRR--GGRASEEEKWVPVTKLGRHVAAG 67
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
I+ +EQIYLHSLP+KE+QIID L+GPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD
Sbjct: 68 HIKQIEQIYLHSLPVKEYQIIDMLIGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDG 127
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSVTV
Sbjct: 128 NGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 187
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFLTP
Sbjct: 188 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFLTP 247
Query: 243 EFWKETRFSKSPFQEYTDLLA 263
EFWKETRFS+SP+QE+TD LA
Sbjct: 248 EFWKETRFSRSPYQEHTDFLA 268
>emb|CAB66106.1| 40S ribosomal protein S2 homolog [Arabidopsis thaliana]
gi|30017231|gb|AAP12849.1| At3g57490 [Arabidopsis
thaliana] gi|21536514|gb|AAM60846.1| 40S ribosomal
protein S2 homolog [Arabidopsis thaliana]
gi|15230312|ref|NP_191308.1| 40S ribosomal protein S2
(RPS2D) [Arabidopsis thaliana] gi|11276536|pir||T46185
ribosomal protein S2, cytosolic [similarity] -
Arabidopsis thaliana
Length = 276
Score = 465 bits (1196), Expect = e-130
Identities = 229/263 (87%), Positives = 239/263 (90%), Gaps = 8/263 (3%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
ERGGDRG FG GFGGRGG RGG GRG R GR EEEKWVPVTKLGRLVKEG
Sbjct: 7 ERGGDRGDFGRGFGGRGGGRGGPRGRG--------RRAGRAPEEEKWVPVTKLGRLVKEG 58
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
KI +EQIYLHSLP+KE+QIID LVGP+LKDEVMKIMPVQKQTRAGQRTRFKAF+VVGD+
Sbjct: 59 KITKIEQIYLHSLPVKEYQIIDLLVGPSLKDEVMKIMPVQKQTRAGQRTRFKAFIVVGDS 118
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
NGHVGLGVKCSKEVATAIRGAIILAKLSV+P+RRGYWGNKIGKPHTVPCKVTGKCGSVTV
Sbjct: 119 NGHVGLGVKCSKEVATAIRGAIILAKLSVVPIRRGYWGNKIGKPHTVPCKVTGKCGSVTV 178
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFLTP
Sbjct: 179 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFLTP 238
Query: 243 EFWKETRFSKSPFQEYTDLLAKP 265
EFWKETRFSKSP+QE+TD L P
Sbjct: 239 EFWKETRFSKSPYQEHTDFLLIP 261
>ref|XP_470037.1| putative 40S ribosomal protein S2 [Oryza sativa (japonica
cultivar-group)] gi|30103021|gb|AAP21434.1| putative 40S
ribosomal protein S2 [Oryza sativa (japonica
cultivar-group)]
Length = 274
Score = 462 bits (1188), Expect = e-129
Identities = 237/281 (84%), Positives = 256/281 (90%), Gaps = 9/281 (3%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
MAERGG+RGG GFG RG RGGRG RG R RGGRR G R+EEEKWVPVTKLGRLVK
Sbjct: 1 MAERGGERGGERGGFG-RGFGRGGRGDRGGR-RGGRR---GPRQEEEKWVPVTKLGRLVK 55
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
EG+ +E+IYLHSLP+KEHQI++TLV P LKDEVMKI PVQKQTRAGQRTRFKAFVVVG
Sbjct: 56 EGRFSKIEEIYLHSLPVKEHQIVETLV-PGLKDEVMKITPVQKQTRAGQRTRFKAFVVVG 114
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
DNNGHVGLGVKC+KEVATAIRGAIILAKLSV+PVRRGYWGNKIG+PHTVPCKVTGKCGSV
Sbjct: 115 DNNGHVGLGVKCAKEVATAIRGAIILAKLSVVPVRRGYWGNKIGQPHTVPCKVTGKCGSV 174
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGI+DVFTSSRGSTKTLGNFVKATFDCLMKTYGFL
Sbjct: 175 TVRMVPAPRGSGIVAARVPKKVLQFAGIEDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 234
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKALILEE--ERVEA 279
TP+FW++T+F KSPFQEYTDLLAKPT KAL+++ E VEA
Sbjct: 235 TPDFWRDTKFVKSPFQEYTDLLAKPT-KALMIDAPVENVEA 274
>dbj|BAD94842.1| ribosomal protein S2 [Arabidopsis thaliana]
gi|22137288|gb|AAM91489.1| At1g59359/T4M14_3
[Arabidopsis thaliana] gi|18265374|dbj|BAB84016.1|
ribosomal protein S2 [Arabidopsis thaliana]
gi|18265370|dbj|BAB84012.1| ribosomal protein S2
[Arabidopsis thaliana] gi|18146720|dbj|BAB82426.1|
ribosomal protein S2 [Arabidopsis thaliana]
gi|18086426|gb|AAL57668.1| At1g59359/T4M14_3
[Arabidopsis thaliana] gi|18406353|ref|NP_564740.1| 40S
ribosomal protein S2 (RPS2B) [Arabidopsis thaliana]
gi|18406325|ref|NP_564737.1| 40S ribosomal protein S2,
putative [Arabidopsis thaliana]
gi|22330310|ref|NP_683443.1| 40S ribosomal protein S2,
putative [Arabidopsis thaliana]
gi|14475937|gb|AAK62784.1| ribosomal protein S2,
putative [Arabidopsis thaliana]
gi|14475933|gb|AAK62780.1| ribosomal protein S2,
putative [Arabidopsis thaliana]
Length = 284
Score = 459 bits (1181), Expect = e-128
Identities = 231/269 (85%), Positives = 239/269 (87%), Gaps = 6/269 (2%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
AERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV
Sbjct: 10 AERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVA 64
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 65 DNKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 124
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 125 DGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSV 184
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFL
Sbjct: 185 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFL 244
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKA 269
TPEFWKETRFS+SP+QE+TD L+ A
Sbjct: 245 TPEFWKETRFSRSPYQEHTDFLSTKAVSA 273
>dbj|BAB83870.1| ribosomal protein S2 [Arabidopsis thaliana]
gi|6566272|dbj|BAA88263.1| XW6 [Arabidopsis thaliana]
gi|18377554|gb|AAL66943.1| ribosomal protein S2,
putative [Arabidopsis thaliana]
gi|15217951|ref|NP_176134.1| 40S ribosomal protein S2
(RPS2A) [Arabidopsis thaliana]
gi|14423442|gb|AAK62403.1| ribosomal protein S2,
putative [Arabidopsis thaliana]
gi|12321043|gb|AAG50639.1| ribosomal protein S2,
putative [Arabidopsis thaliana] gi|11276533|pir||T50673
ribosomal protein S2 homolog XW6 [imported] -
Arabidopsis thaliana
Length = 284
Score = 459 bits (1181), Expect = e-128
Identities = 231/269 (85%), Positives = 239/269 (87%), Gaps = 6/269 (2%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
AERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV
Sbjct: 10 AERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVA 64
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 65 DNKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 124
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 125 DGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSV 184
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFL
Sbjct: 185 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFL 244
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKA 269
TPEFWKETRFS+SP+QE+TD L+ A
Sbjct: 245 TPEFWKETRFSRSPYQEHTDFLSTKAVSA 273
>dbj|BAA88264.1| RF12 [Arabidopsis thaliana] gi|25294520|pir||T52466 hypothetical
protein RF12 [imported] - Arabidopsis thaliana
(fragment)
Length = 282
Score = 459 bits (1181), Expect = e-128
Identities = 231/269 (85%), Positives = 239/269 (87%), Gaps = 6/269 (2%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
AERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV
Sbjct: 8 AERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVA 62
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 63 DNKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 122
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 123 DGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSV 182
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFL
Sbjct: 183 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFL 242
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKA 269
TPEFWKETRFS+SP+QE+TD L+ A
Sbjct: 243 TPEFWKETRFSRSPYQEHTDFLSTKAVSA 271
>ref|XP_477083.1| putative 40S ribosomal protein S2 [Oryza sativa (japonica
cultivar-group)] gi|34393314|dbj|BAC83243.1| putative
40S ribosomal protein S2 [Oryza sativa (japonica
cultivar-group)]
Length = 276
Score = 458 bits (1179), Expect = e-128
Identities = 227/277 (81%), Positives = 245/277 (87%), Gaps = 9/277 (3%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
ERGG+RGGFG GFG RGGRGDRGRGGR G R+EEEKWVPVTKLGRLVKE
Sbjct: 9 ERGGERGGFGRGFG--------RGGRGDRGRGGRGGRRGPRQEEEKWVPVTKLGRLVKEN 60
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
KI +E+IYLHSLP+KEHQI++ LV P LKDEVMKI PVQKQTRAGQRTRFKAFVVVGD
Sbjct: 61 KIHKIEEIYLHSLPVKEHQIVEQLV-PGLKDEVMKITPVQKQTRAGQRTRFKAFVVVGDG 119
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
+GHVGLGVKC+KEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSVTV
Sbjct: 120 DGHVGLGVKCAKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 179
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
RMVPAPRGSGIVAA VPKKVLQFAGI+DVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP
Sbjct: 180 RMVPAPRGSGIVAAHVPKKVLQFAGIEDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 239
Query: 243 EFWKETRFSKSPFQEYTDLLAKPTAKALILEEERVEA 279
+FW+ETRF K+PFQEYTDLLA+P + E++EA
Sbjct: 240 DFWRETRFIKTPFQEYTDLLARPKGLVIEAPAEKIEA 276
>gb|AAM67061.1| ribosomal protein S2, putative [Arabidopsis thaliana]
Length = 284
Score = 457 bits (1177), Expect = e-127
Identities = 230/268 (85%), Positives = 238/268 (87%), Gaps = 6/268 (2%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVKE 61
ERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV +
Sbjct: 11 ERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVAD 65
Query: 62 GKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD 121
KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD
Sbjct: 66 NKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD 125
Query: 122 NNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVT 181
NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSVT
Sbjct: 126 GNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSVT 185
Query: 182 VRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLT 241
VRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFLT
Sbjct: 186 VRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFLT 245
Query: 242 PEFWKETRFSKSPFQEYTDLLAKPTAKA 269
PEFWKETRFS+SP+QE+TD L+ A
Sbjct: 246 PEFWKETRFSRSPYQEHTDFLSTKAVSA 273
>gb|AAM53281.1| ribosomal protein S2, putative [Arabidopsis thaliana]
Length = 284
Score = 457 bits (1175), Expect = e-127
Identities = 230/269 (85%), Positives = 238/269 (87%), Gaps = 6/269 (2%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
AERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV
Sbjct: 10 AERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVA 64
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 65 DNKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 124
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 125 DGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSV 184
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLG FVKATFDCL KTYGFL
Sbjct: 185 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGKFVKATFDCLQKTYGFL 244
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKA 269
TPEFWKETRFS+SP+QE+TD L+ A
Sbjct: 245 TPEFWKETRFSRSPYQEHTDFLSTKAVSA 273
>gb|AAX62450.1| ribosomal protein S2 [Lysiphlebus testaceipes]
Length = 285
Score = 397 bits (1021), Expect = e-109
Identities = 192/264 (72%), Positives = 228/264 (85%), Gaps = 2/264 (0%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGG-RGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
A GG RGGFGS GG GG+RGG RGGRG RGRG R G +E++++WVPVTKLGRLV+
Sbjct: 8 ATTGGFRGGFGSR-GGAGGERGGSRGGRGGRGRGRGRGRGRGKEDQKEWVPVTKLGRLVR 66
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+GKIR+LE IYL SLPIKE +IID +GP+LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 67 DGKIRTLEDIYLFSLPIKEFEIIDQFIGPSLKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 126
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D+NGH+GLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 127 DSNGHIGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSV 186
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG+GIV+A VPKK+LQ AGI+D +TS+RGST TLGNF KAT+ + KTY +L
Sbjct: 187 QVRLIPAPRGTGIVSAPVPKKLLQMAGIEDCYTSARGSTCTLGNFAKATYAAIAKTYAYL 246
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WK+ +++P+QE+ D L+K
Sbjct: 247 TPDLWKDMPLTETPYQEHADWLSK 270
>gb|AAH56066.1| Sop-prov protein [Xenopus laevis]
Length = 281
Score = 397 bits (1019), Expect = e-109
Identities = 196/273 (71%), Positives = 227/273 (82%), Gaps = 3/273 (1%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
+ GG+RGGF GFGG G RG GRG RGRG R A G + ++++WVPVTKLGRLVK+
Sbjct: 4 DAGGNRGGFRGGFGGGGRGRGRGRGRG-RGRG--RGARGGKADDKEWVPVTKLGRLVKDM 60
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
KI+SLE+IYL SLPIKE +IID +G TLKDEV+KIMPVQKQTRAGQRTRFKAFV +GD
Sbjct: 61 KIKSLEEIYLFSLPIKESEIIDFFLGSTLKDEVLKIMPVQKQTRAGQRTRFKAFVAIGDY 120
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV V
Sbjct: 121 NGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSVLV 180
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
R++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +LTP
Sbjct: 181 RLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYLTP 240
Query: 243 EFWKETRFSKSPFQEYTDLLAKPTAKALILEEE 275
+ WKET F+KSP+QEYTD LAK + + E
Sbjct: 241 DLWKETVFAKSPYQEYTDHLAKTHTRVSVQRTE 273
>gb|EAL33406.1| GA19229-PA [Drosophila pseudoobscura]
Length = 268
Score = 395 bits (1015), Expect = e-109
Identities = 193/271 (71%), Positives = 227/271 (83%), Gaps = 11/271 (4%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRG---GRGDRGRGGRRRAGGRREEEEKWVPVTKLGR 57
MA+ RGGF GFG RGG RGGRG GRG RGRGG+ E+ ++WVPVTKLGR
Sbjct: 1 MADAAPARGGFRGGFGSRGG-RGGRGRGRGRGARGRGGK-------EDSKEWVPVTKLGR 52
Query: 58 LVKEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFV 117
LV+EGKI+SLE+IYL+SLPIKE +IID +G L+DEV+KIMPVQKQTRAGQRTRFKAFV
Sbjct: 53 LVREGKIKSLEEIYLYSLPIKEFEIIDFFLGSALRDEVLKIMPVQKQTRAGQRTRFKAFV 112
Query: 118 VVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKC 177
+GDNNGH+GLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKC
Sbjct: 113 AIGDNNGHIGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKC 172
Query: 178 GSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTY 237
GSV+VR++PAPRG+GIV+A VPKK+L AGI+D +TS+RGST TLGNF KAT+ + KTY
Sbjct: 173 GSVSVRLIPAPRGTGIVSAPVPKKLLTMAGIEDCYTSARGSTGTLGNFAKATYAAIAKTY 232
Query: 238 GFLTPEFWKETRFSKSPFQEYTDLLAKPTAK 268
+LTP+ WKE +P+Q Y+D L+KPTA+
Sbjct: 233 AYLTPDLWKEMPLGSTPYQAYSDFLSKPTAR 263
>gb|AAH71673.1| Ribosomal protein S2 [Homo sapiens]
Length = 293
Score = 394 bits (1011), Expect = e-108
Identities = 193/264 (73%), Positives = 222/264 (83%), Gaps = 7/264 (2%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
M RGG RGGFGSG GRG RG RGRG R A G + E+++W+PVTKLGRLVK
Sbjct: 19 MGNRGGFRGGFGSGIRGRGRGRG-------RGRGRGRGARGGKAEDKEWMPVTKLGRLVK 71
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 72 DMKIKSLEEIYLFSLPIKESEIIDFFLGASLKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 131
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 132 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSV 191
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 192 LVRLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 251
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD LAK
Sbjct: 252 TPDLWKETVFTKSPYQEFTDHLAK 275
>ref|NP_998444.1| hypothetical protein LOC406565 [Danio rerio]
gi|45768675|gb|AAH67645.1| Zgc:85824 [Danio rerio]
Length = 280
Score = 392 bits (1008), Expect = e-108
Identities = 195/264 (73%), Positives = 223/264 (83%), Gaps = 3/264 (1%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
MA+ G RGGF GFG G RG GRG RGRG R A G + E+++WVPVTKLGRLVK
Sbjct: 1 MADDAGGRGGFRGGFGTGGRGRGRGRGRG-RGRG--RGARGGKSEDKEWVPVTKLGRLVK 57
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL+SLPIKE +IID +G LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 58 DMKIKSLEEIYLYSLPIKESEIIDFFLGSALKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 117
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS+IPVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 118 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIIPVRRGYWGNKIGKPHTVPCKVTGRCGSV 177
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 178 LVRLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 237
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD LAK
Sbjct: 238 TPDLWKETVFTKSPYQEFTDHLAK 261
>ref|XP_414845.1| PREDICTED: similar to 40S ribosomal protein S2 [Gallus gallus]
Length = 286
Score = 392 bits (1008), Expect = e-108
Identities = 192/263 (73%), Positives = 222/263 (84%), Gaps = 7/263 (2%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKE 61
A RGG RGGFG+G GRG RG RGRG R A G + E+++W+PVTKLGRLVK+
Sbjct: 13 AARGGFRGGFGTGLRGRGRGRG-------RGRGRGRGARGGKAEDKEWIPVTKLGRLVKD 65
Query: 62 GKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD 121
KI+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +GD
Sbjct: 66 MKIKSLEEIYLFSLPIKESEIIDFFLGSSLKDEVLKIMPVQKQTRAGQRTRFKAFVAIGD 125
Query: 122 NNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVT 181
NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 126 YNGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSVL 185
Query: 182 VRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLT 241
VR++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +LT
Sbjct: 186 VRLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYLT 245
Query: 242 PEFWKETRFSKSPFQEYTDLLAK 264
P+ WKET F+KSP+QE+TD LAK
Sbjct: 246 PDLWKETVFTKSPYQEFTDHLAK 268
>ref|XP_310307.2| ENSANGP00000015322 [Anopheles gambiae str. PEST]
gi|55243894|gb|EAA06099.2| ENSANGP00000015322 [Anopheles
gambiae str. PEST]
Length = 271
Score = 392 bits (1007), Expect = e-108
Identities = 190/265 (71%), Positives = 224/265 (83%), Gaps = 3/265 (1%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRG-GRRRAGGRREEEEKWVPVTKLGRLV 59
MA+ RGGF GFG RGG RGG GRG RGRG GR R G +E+ ++WVPVTKLGRLV
Sbjct: 1 MADAAPARGGFRGGFGSRGGARGG--GRGGRGRGRGRGRGRGGKEDSKEWVPVTKLGRLV 58
Query: 60 KEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVV 119
K+GKIR+LE+IYL+SLPIKE +IID + LKDEV+KIMPVQKQTRAGQRTRFKAFV +
Sbjct: 59 KDGKIRTLEEIYLYSLPIKEFEIIDFFLNRLLKDEVLKIMPVQKQTRAGQRTRFKAFVAI 118
Query: 120 GDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGS 179
GD+NGH+GLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIG+PHTVPCKV+GKCGS
Sbjct: 119 GDSNGHIGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGRPHTVPCKVSGKCGS 178
Query: 180 VTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGF 239
V VR++PAPRG+GIV+A VPKK+LQ AGIDD +T++RGST TLGNF KAT+ + KTY F
Sbjct: 179 VLVRLIPAPRGTGIVSAPVPKKLLQMAGIDDCYTTTRGSTTTLGNFAKATYAAIAKTYAF 238
Query: 240 LTPEFWKETRFSKSPFQEYTDLLAK 264
LTP+ WK+ +K+P+QE+ D L K
Sbjct: 239 LTPDLWKDLPLNKTPYQEFADFLDK 263
>ref|XP_511195.1| PREDICTED: hypothetical protein XP_511195 [Pan troglodytes]
gi|60656433|gb|AAX32780.1| ribosomal protein S2
[synthetic construct] gi|50369371|gb|AAH75830.1|
Ribosomal protein S2 [Homo sapiens]
gi|49522753|gb|AAH73966.1| Ribosomal protein S2 [Homo
sapiens] gi|48734991|gb|AAH71923.1| Ribosomal protein S2
[Homo sapiens] gi|48734765|gb|AAH71924.1| Ribosomal
protein S2 [Homo sapiens] gi|48734763|gb|AAH71922.1|
Ribosomal protein S2 [Homo sapiens]
gi|42542643|gb|AAH66321.1| Ribosomal protein S2 [Homo
sapiens] gi|17512041|gb|AAH18993.1| Ribosomal protein S2
[Homo sapiens] gi|15214457|gb|AAH12354.1| Ribosomal
protein S2 [Homo sapiens] gi|14603435|gb|AAH10165.1|
Ribosomal protein S2 [Homo sapiens]
gi|16740591|gb|AAH16178.1| Ribosomal protein S2 [Homo
sapiens] gi|16306879|gb|AAH06559.1| Ribosomal protein S2
[Homo sapiens] gi|19343921|gb|AAH25677.1| Ribosomal
protein S2 [Homo sapiens] gi|12804729|gb|AAH01795.1|
Ribosomal protein S2 [Homo sapiens]
gi|16877410|gb|AAH16951.1| Ribosomal protein S2 [Homo
sapiens] gi|14250786|gb|AAH08862.1| Ribosomal protein S2
[Homo sapiens] gi|18203799|gb|AAH21545.1| Ribosomal
protein S2 [Homo sapiens] gi|23270746|gb|AAH23541.1|
Ribosomal protein S2 [Homo sapiens]
gi|45786137|gb|AAH68051.1| Ribosomal protein S2 [Homo
sapiens] gi|1710756|sp|P15880|RS2_HUMAN 40S ribosomal
protein S2 (S4) (LLRep3 protein)
gi|15055539|ref|NP_002943.2| ribosomal protein S2 [Homo
sapiens]
Length = 293
Score = 392 bits (1007), Expect = e-108
Identities = 192/264 (72%), Positives = 221/264 (82%), Gaps = 7/264 (2%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
M RGG RGGFGSG GRG RG RGRG R A G + E+++W+PVTKLGRLVK
Sbjct: 19 MGNRGGFRGGFGSGIRGRGRGRG-------RGRGRGRGARGGKAEDKEWMPVTKLGRLVK 71
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 72 DMKIKSLEEIYLFSLPIKESEIIDFFLGASLKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 131
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 132 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSV 191
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 192 LVRLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 251
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD L K
Sbjct: 252 TPDLWKETVFTKSPYQEFTDHLVK 275
>ref|XP_537011.1| PREDICTED: similar to 40S ribosomal protein S2 [Canis familiaris]
Length = 331
Score = 392 bits (1007), Expect = e-108
Identities = 192/264 (72%), Positives = 221/264 (82%), Gaps = 7/264 (2%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
M RGG RGGFGSG GRG RG RGRG R A G + E+++W+PVTKLGRLVK
Sbjct: 57 MGGRGGFRGGFGSGIRGRGRGRG-------RGRGRGRGARGGKAEDKEWIPVTKLGRLVK 109
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 110 DMKIKSLEEIYLFSLPIKESEIIDFFLGASLKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 169
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 170 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSV 229
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 230 LVRLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 289
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD L K
Sbjct: 290 TPDLWKETVFTKSPYQEFTDHLVK 313
>gb|AAX29391.1| ribosomal protein S2 [synthetic construct]
Length = 294
Score = 392 bits (1007), Expect = e-108
Identities = 192/264 (72%), Positives = 221/264 (82%), Gaps = 7/264 (2%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
M RGG RGGFGSG GRG RG RGRG R A G + E+++W+PVTKLGRLVK
Sbjct: 19 MGNRGGFRGGFGSGIRGRGRGRG-------RGRGRGRGARGGKAEDKEWMPVTKLGRLVK 71
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI+SLE+IYL SLPIKE +IID +G +LKDEV+KIMPVQKQTRAGQRTRFKAFV +G
Sbjct: 72 DMKIKSLEEIYLFSLPIKESEIIDFFLGASLKDEVLKIMPVQKQTRAGQRTRFKAFVAIG 131
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLS++PVRRGYWGNKIGKPHTVPCKVTG+CGSV
Sbjct: 132 DYNGHVGLGVKCSKEVATAIRGAIILAKLSIVPVRRGYWGNKIGKPHTVPCKVTGRCGSV 191
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
VR++PAPRG+GIV+A VPKK+L AGIDD +TS+RG T TLGNF KATFD + KTY +L
Sbjct: 192 LVRLIPAPRGTGIVSAPVPKKLLMMAGIDDCYTSARGCTATLGNFAKATFDAISKTYSYL 251
Query: 241 TPEFWKETRFSKSPFQEYTDLLAK 264
TP+ WKET F+KSP+QE+TD L K
Sbjct: 252 TPDLWKETVFTKSPYQEFTDHLVK 275
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 506,053,576
Number of Sequences: 2540612
Number of extensions: 24044063
Number of successful extensions: 313653
Number of sequences better than 10.0: 4189
Number of HSP's better than 10.0 without gapping: 2815
Number of HSP's successfully gapped in prelim test: 1489
Number of HSP's that attempted gapping in prelim test: 193627
Number of HSP's gapped (non-prelim): 57118
length of query: 279
length of database: 863,360,394
effective HSP length: 126
effective length of query: 153
effective length of database: 543,243,282
effective search space: 83116222146
effective search space used: 83116222146
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC140104.12


