

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140027.12 + phase: 0 /pseudo
(526 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
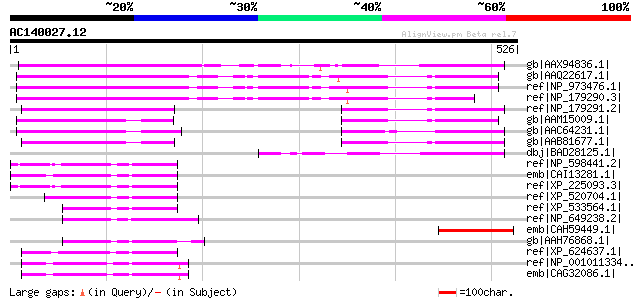
gb|AAX94836.1| Major Facilitator Superfamily, putative [Oryza sa... 296 1e-78
gb|AAQ22617.1| At2g16980 [Arabidopsis thaliana] 186 2e-45
ref|NP_973476.1| expressed protein [Arabidopsis thaliana] 171 4e-41
ref|NP_179290.3| expressed protein [Arabidopsis thaliana] 145 3e-33
ref|NP_179291.2| expressed protein [Arabidopsis thaliana] 114 1e-23
gb|AAM15009.1| putative tetracycline transporter protein [Arabid... 103 1e-20
gb|AAC64231.1| putative tetracycline transporter protein [Arabid... 92 5e-17
gb|AAB81677.1| putative tetracycline transporter protein [Arabid... 90 2e-16
dbj|BAD28125.1| tetracycline transporter protein-like [Oryza sat... 89 3e-16
ref|NP_598441.2| hypothetical protein LOC66631 [Mus musculus] gi... 65 7e-09
emb|CAI13281.1| hippocampus abundant transcript-like 1 [Homo sap... 64 1e-08
ref|XP_225093.3| PREDICTED: similar to RIKEN cDNA 5730414C17 [Ra... 63 2e-08
ref|XP_520704.1| PREDICTED: similar to RIKEN cDNA 5730414C17 [Pa... 62 3e-08
ref|XP_533564.1| PREDICTED: similar to RIKEN cDNA 5730414C17 [Ca... 62 6e-08
ref|NP_649238.2| CG5078-PA [Drosophila melanogaster] gi|45446045... 60 1e-07
emb|CAH59449.1| tetracycline transporter-like protein 1 [Plantag... 60 2e-07
gb|AAH76868.1| LOC445835 protein [Xenopus laevis] 58 8e-07
ref|XP_624637.1| PREDICTED: similar to ENSANGP00000009556 [Apis ... 58 8e-07
ref|NP_001011334.1| hypothetical protein LOC496797 [Xenopus trop... 57 1e-06
emb|CAG32086.1| hypothetical protein [Gallus gallus] 57 1e-06
>gb|AAX94836.1| Major Facilitator Superfamily, putative [Oryza sativa (japonica
cultivar-group)]
Length = 1143
Score = 296 bits (757), Expect = 1e-78
Identities = 207/509 (40%), Positives = 267/509 (51%), Gaps = 105/509 (20%)
Query: 10 LKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQQS-SCSKAIYINGLQQTITGIFKM 68
LKPL HLLL L ++W+AE MTV VLVDVTT ALCP +C +AIY+ GL QT+ GIF+
Sbjct: 704 LKPLMHLLLGLVMYWVAEEMTVPVLVDVTTRALCPGADIACPEAIYLTGLHQTVGGIFRA 763
Query: 69 AVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSI 128
L+GQL+DE+GRKPLLLLT STSIIP+ +LA N+SK VY + +LRT S +I QG+I
Sbjct: 764 VGYTLMGQLADEYGRKPLLLLTASTSIIPYGVLACNKSKIAVYIFLILRTLSFMIGQGTI 823
Query: 129 FCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYIFVILNYGLLLSGF 188
++V Y ADVV SKR F ITG+ SA+H + N F+RFLP+ +IF + L+ S
Sbjct: 824 TSLAVTYTADVVDPSKRAFAFGCITGILSASHALGNGFSRFLPERWIFQVSVALLISSVI 883
Query: 189 NNTIDLLSTLYAFLPS*NCETGSRKEPRVRLLY*I*GMPQK**SSGCLSHVN*HQSFFE- 247
I L+ TL S + E S VRL + +S E
Sbjct: 884 YMKISLVETLQR-ASSGSFEHMSFSSLVVRL------------------PLRRWESIKEN 924
Query: 248 LTCIHES*IFPTLRGVALVSFFYKLGMTGIHSVLLQIFSLLCHYYLTNFF**IP*YNRQS 307
+ I S TL + +SFFY+LGM GI VL+ YYL
Sbjct: 925 INIIRRS---ETLSRITYISFFYELGMIGISDVLM--------YYL-------------- 959
Query: 308 HNSYAVLFESCFWF---QQKSVLRTSDDGRNWFHLFSARFQIVLLPILNPLVGEKVILCS 364
+S F F Q +L G +FS QI++LP++ VGEK +LC
Sbjct: 960 --------KSVFGFDKNQFSEILMVVGIG----SIFS---QILVLPVIINTVGEKGVLCV 1004
Query: 365 ALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLAENFITYISIMKVIEFNPLWQ 424
+LAS+AYA L GLAW+ W
Sbjct: 1005 GILASVAYAVLYGLAWSYW----------------------------------------- 1023
Query: 425 VPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIAGANSISGLLSPIVMSPLTSL 484
VPYL S G+IY+L KPATY IIS S++QGKAQ FI+ S + LL+P+ MSPLTS
Sbjct: 1024 VPYLTSSLGVIYVLVKPATYAIISGEVDSSDQGKAQGFISTVKSTAVLLAPLFMSPLTSY 1083
Query: 485 FLSSDAPFECKGFSIICASVCMVSYLFLS 513
F+S APF CKGFS + A + L +S
Sbjct: 1084 FISEQAPFNCKGFSFLVAGFFLAISLGIS 1112
>gb|AAQ22617.1| At2g16980 [Arabidopsis thaliana]
Length = 461
Score = 186 bits (472), Expect = 2e-45
Identities = 146/506 (28%), Positives = 232/506 (44%), Gaps = 84/506 (16%)
Query: 8 FELKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQ-QSSCSKAIYINGLQQTITGIF 66
F L L HLL+ + + +AE + V+ DVT A+C SCS A+Y+ G+QQ G+
Sbjct: 4 FRLGELRHLLVTVFLSGLAEYLIRPVMTDVTVAAVCSGLDDSCSLAVYLTGVQQVTVGMG 63
Query: 67 KMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQG 126
M ++P++G LSD +G K +L L + S++P A+L + + F YA+YV++T ++ QG
Sbjct: 64 TMVMMPVIGNLSDRYGIKAMLTLPMCLSVLPPAILGYRRDTNFFYAFYVIKTLFDMVCQG 123
Query: 127 SIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYIFVILNYGLLLS 186
+I C++ AYVA VH +KR+++F + G+SS + V A++ ARFL F + L +
Sbjct: 124 TIDCLANAYVAKNVHGTKRISMFGILAGVSSISGVCASLSARFLSIASTFQVAAISLFIG 183
Query: 187 GFNNTIDLLSTLYAFLPS*NCETGSRKEPRVRLLY*I*GMPQK**SSGCLSHVN*HQSFF 246
L + FL + E S GC SH H
Sbjct: 184 --------LVYMRVFLKERLQDADDDDEAD---------------SGGCRSHQEVHNG-G 219
Query: 247 ELTCIHES*IFPTLRGVALVSFFYKLGMTGIHSV--LLQIFSLLCHYYLTNFF**IP*YN 304
+L + E P LR + + + V L+ ++L + FF
Sbjct: 220 DLKMLTE----PILRDAPTKTHVFNSKYSPWKDVVSLINNSTILIQALVVTFFATFSESG 275
Query: 305 RQSHNSYAVLFESCFWFQQKSVLRTSDDGRNWFHL---FSARFQIVLLPILNPLVGEKVI 361
R S Y L ES FW QQ +D F L + Q+ +LP L+ +GE+ +
Sbjct: 276 RGSALMY--LSESSFWVQQ-------NDFAELFLLVTIIGSISQLFILPTLSSTIGERKV 326
Query: 362 LCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLAENFITYISIMKVIEFNP 421
L + LL A +AW+PWV +T +
Sbjct: 327 LSTGLLMEFFNATCLSVAWSPWVPYAMTMLVP---------------------------- 358
Query: 422 LWQVPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIAGANSISGLLSPIVMSPL 481
G ++++ P+ GI S+ S+ QGK Q I+G + + +++P V SPL
Sbjct: 359 -----------GAMFVM--PSVCGIASRQVGSSEQGKVQGCISGVRAFAQVVAPFVYSPL 405
Query: 482 TSLFLSSDAPFECKGFSIICASVCMV 507
T+LFLS +APF GFSI+C ++ ++
Sbjct: 406 TALFLSENAPFYFPGFSILCIAISLM 431
>ref|NP_973476.1| expressed protein [Arabidopsis thaliana]
Length = 461
Score = 171 bits (434), Expect = 4e-41
Identities = 140/506 (27%), Positives = 228/506 (44%), Gaps = 84/506 (16%)
Query: 8 FELKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQ-QSSCSKAIYINGLQQTITGIF 66
F L L HLL+ + + +AE + V+ DVT A+C SCS A+Y+ G+QQ G+
Sbjct: 4 FRLGELRHLLVTVFLSGLAEYLIRPVMTDVTVAAVCSGLDDSCSLAVYLTGVQQVTVGMG 63
Query: 67 KMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQG 126
M ++P++G LSD +G K +L L + S++P A+L + + F YA+YV++T ++ QG
Sbjct: 64 TMVMMPVIGNLSDRYGIKAMLTLPMCLSVLPPAILGYRRDTNFFYAFYVIKTLFDMVCQG 123
Query: 127 SIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYIFVILNYGLLLS 186
+I C++ AYVA VH +KR+++F + G+SS + V A++ ARFL F + L +
Sbjct: 124 TIDCLANAYVAKNVHGTKRISMFGILAGVSSISGVCASLSARFLSIASTFQVAAISLFIG 183
Query: 187 GFNNTIDLLSTLYAFLPS*NCETGSRKEPRVRLLY*I*GMPQK**SSGCLSHVN*HQSFF 246
L + FL + E S GC SH H
Sbjct: 184 --------LVYMRVFLKERLQDADDDDEAD---------------SGGCRSHQEVHNG-G 219
Query: 247 ELTCIHES*IFPTLRGVALVSFFYKLGMTGIHSV--LLQIFSLLCHYYLTNFF**IP*YN 304
+L + E P LR + + + + L+ ++L + FF
Sbjct: 220 DLKMLTE----PILRDAPTKTHVFNSKYSSWKDMVSLINNSTILIQALVVTFFATFSESG 275
Query: 305 RQSHNSYAVLFESCFWFQQKSVLRTSDDGRNWFHLFSARFQIVLL---PILNPLVGEKVI 361
R S Y ++ F F + +D F L + I L P L+ +GE+ +
Sbjct: 276 RGSALMY--FLKARFGFNK-------NDFAELFLLVTIIGSISQLFILPTLSSTIGERKV 326
Query: 362 LCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLAENFITYISIMKVIEFNP 421
L + LL A +AW+PWV +T +
Sbjct: 327 LSTGLLMEFFNATCLSVAWSPWVPYAMTMLVP---------------------------- 358
Query: 422 LWQVPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIAGANSISGLLSPIVMSPL 481
G ++++ P+ GI S+ S+ QGK Q I+G + + +++P V SPL
Sbjct: 359 -----------GAMFVM--PSVCGIASRQVGSSEQGKVQGCISGVRAFAQVVAPFVYSPL 405
Query: 482 TSLFLSSDAPFECKGFSIICASVCMV 507
T+LFLS +APF GFSI+C ++ ++
Sbjct: 406 TALFLSENAPFYFPGFSILCIAISLM 431
>ref|NP_179290.3| expressed protein [Arabidopsis thaliana]
Length = 408
Score = 145 bits (366), Expect = 3e-33
Identities = 128/481 (26%), Positives = 209/481 (42%), Gaps = 84/481 (17%)
Query: 8 FELKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQ-QSSCSKAIYINGLQQTITGIF 66
F L L HLL+ + + +AE + V+ DVT A+C SCS A+Y+ G+QQ G+
Sbjct: 4 FRLGELRHLLVTVFLSGLAEYLIRPVMTDVTVAAVCSGLDDSCSLAVYLTGVQQVTVGMG 63
Query: 67 KMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQG 126
M ++P++G LSD +G K +L L + S++P A+L + + F YA+YV++T ++ QG
Sbjct: 64 TMVMMPVIGNLSDRYGIKAMLTLPMCLSVLPPAILGYRRDTNFFYAFYVIKTLFDMVCQG 123
Query: 127 SIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYIFVILNYGLLLS 186
+I C++ AYVA VH +KR+++F + G+SS + V A++ ARFL F + L +
Sbjct: 124 TIDCLANAYVAKNVHGTKRISMFGILAGVSSISGVCASLSARFLSIASTFQVAAISLFIG 183
Query: 187 GFNNTIDLLSTLYAFLPS*NCETGSRKEPRVRLLY*I*GMPQK**SSGCLSHVN*HQSFF 246
L + FL + E S GC SH H
Sbjct: 184 --------LVYMRVFLKERLQDADDDDEAD---------------SGGCRSHQEVHNG-G 219
Query: 247 ELTCIHES*IFPTLRGVALVSFFYKLGMTGIHSV--LLQIFSLLCHYYLTNFF**IP*YN 304
+L + E P LR + + + + L+ ++L + FF
Sbjct: 220 DLKMLTE----PILRDAPTKTHVFNSKYSSWKDMVSLINNSTILIQALVVTFFATFSESG 275
Query: 305 RQSHNSYAVLFESCFWFQQKSVLRTSDDGRNWFHLFSARFQIVLL---PILNPLVGEKVI 361
R S Y ++ F F + +D F L + I L P L+ +GE+ +
Sbjct: 276 RGSALMY--FLKARFGFNK-------NDFAELFLLVTIIGSISQLFILPTLSSTIGERKV 326
Query: 362 LCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLAENFITYISIMKVIEFNP 421
L + LL A +AW+PWV +T +
Sbjct: 327 LSTGLLMEFFNATCLSVAWSPWVPYAMTMLVP---------------------------- 358
Query: 422 LWQVPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIAGANSISGLLSPIVMSPL 481
G ++++ P+ GI S+ S+ QGK Q I+G + + +++P V SPL
Sbjct: 359 -----------GAMFVM--PSVCGIASRQVGSSEQGKVQGCISGVRAFAQVVAPFVYSPL 405
Query: 482 T 482
T
Sbjct: 406 T 406
>ref|NP_179291.2| expressed protein [Arabidopsis thaliana]
Length = 456
Score = 114 bits (284), Expect = 1e-23
Identities = 55/160 (34%), Positives = 94/160 (58%), Gaps = 1/160 (0%)
Query: 13 LFHLLLPLSIHWIAEAMTVSVLVDVTTTALCP-QQSSCSKAIYINGLQQTITGIFKMAVL 71
L H+L + + A M V V+ DVT A+C SCS A+Y+ G QQ G+ M ++
Sbjct: 8 LRHMLATVFLSAFAGFMVVPVITDVTVAAVCSGPDDSCSLAVYLTGFQQVAIGMGTMIMM 67
Query: 72 PLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCI 131
P++G LSD +G K +L L + SI+P +L + + +F Y +Y+ + + ++ +G++ C+
Sbjct: 68 PVIGNLSDRYGIKTILTLPMCLSIVPPVILGYRRDIKFFYVFYISKILTSMVCEGTVDCL 127
Query: 132 SVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLP 171
+ AYVA +H S R++ F + G+ + A + + ARFLP
Sbjct: 128 AYAYVAVNIHGSTRISAFGILAGIKTIAGLFGTLVARFLP 167
Score = 68.9 bits (167), Expect = 4e-10
Identities = 45/169 (26%), Positives = 72/169 (41%), Gaps = 41/169 (24%)
Query: 345 QIVLLPILNPLVGEKVILCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLA 404
Q+ +LP +GE +L + L + ++WAPWV + T ++
Sbjct: 306 QLFVLPRFASAIGECKLLSTGLFMEFINMAIVSISWAPWVPYLTTVFVP----------- 354
Query: 405 ENFITYISIMKVIEFNPLWQVPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIA 464
G ++++ P+ GI S+ QGK Q I+
Sbjct: 355 ----------------------------GALFVM--PSVCGIASRQVGPGEQGKVQGCIS 384
Query: 465 GANSISGLLSPIVMSPLTSLFLSSDAPFECKGFSIICASVCMVSYLFLS 513
G S +++P V SPLT+LFLS +APF GFS++C S+ + F S
Sbjct: 385 GVRSFGKVVAPFVFSPLTALFLSKNAPFYFPGFSLLCISLSSLIGFFQS 433
>gb|AAM15009.1| putative tetracycline transporter protein [Arabidopsis thaliana]
gi|6598356|gb|AAF18594.1| putative tetracycline
transporter protein [Arabidopsis thaliana]
gi|25411765|pir||F84546 probable tetracycline
transporter protein [imported] - Arabidopsis thaliana
Length = 415
Score = 103 bits (258), Expect = 1e-20
Identities = 58/164 (35%), Positives = 94/164 (56%), Gaps = 15/164 (9%)
Query: 8 FELKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQ-QSSCSKAIYINGLQQTITGIF 66
F L L HLL+ + + +AE + V+ DVT A+C SCS A+Y+ G+QQ G+
Sbjct: 4 FRLGELRHLLVTVFLSGLAEYLIRPVMTDVTVAAVCSGLDDSCSLAVYLTGVQQVTVGMG 63
Query: 67 KMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQG 126
M ++P++G LSD +G K +L L + S++P A+L + + F YA+YV++T
Sbjct: 64 TMVMMPVIGNLSDRYGIKAMLTLPMCLSVLPPAILGYRRDTNFFYAFYVIKTLFD----- 118
Query: 127 SIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFL 170
+A VH +KR+++F + G+SS + V A++ ARFL
Sbjct: 119 ---------MAKNVHGTKRISMFGILAGVSSISGVCASLSARFL 153
Score = 77.0 bits (188), Expect = 1e-12
Identities = 46/163 (28%), Positives = 77/163 (47%), Gaps = 41/163 (25%)
Query: 345 QIVLLPILNPLVGEKVILCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLA 404
Q+ +LP L+ +GE+ +L + LL A +AW+PWV +T +
Sbjct: 264 QLFILPTLSSTIGERKVLSTGLLMEFFNATCLSVAWSPWVPYAMTMLVP----------- 312
Query: 405 ENFITYISIMKVIEFNPLWQVPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIA 464
G ++++ P+ GI S+ S+ QGK Q I+
Sbjct: 313 ----------------------------GAMFVM--PSVCGIASRQVGSSEQGKVQGCIS 342
Query: 465 GANSISGLLSPIVMSPLTSLFLSSDAPFECKGFSIICASVCMV 507
G + + +++P V SPLT+LFLS +APF GFSI+C ++ ++
Sbjct: 343 GVRAFAQVVAPFVYSPLTALFLSENAPFYFPGFSILCIAISLM 385
>gb|AAC64231.1| putative tetracycline transporter protein [Arabidopsis thaliana]
gi|25411763|pir||E84546 probable tetracycline
transporter protein [imported] - Arabidopsis thaliana
gi|15227337|ref|NP_179289.1| expressed protein
[Arabidopsis thaliana]
Length = 414
Score = 91.7 bits (226), Expect = 5e-17
Identities = 55/172 (31%), Positives = 89/172 (50%), Gaps = 15/172 (8%)
Query: 8 FELKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQ-QSSCSKAIYINGLQQTITGIF 66
+ L L HLL + + +E + V+ DVT A+C +CS A+Y+ G++Q G+
Sbjct: 4 YRLGELRHLLTTVFLSGFSEFLVKPVMTDVTVAAVCSGLNETCSLAVYLTGVEQVTVGLG 63
Query: 67 KMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQG 126
M ++P++G LSD +G K LL L + SI+P A+LA+ + F YA+Y+ +
Sbjct: 64 TMVMMPVIGNLSDRYGIKTLLTLPMCLSILPPAILAYRRDTNFFYAFYITKILFD----- 118
Query: 127 SIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYIFVI 178
+A V KR+++F + G+ S + V A AR LP IF +
Sbjct: 119 ---------MAKNVCGRKRISMFGVLAGVRSISGVCATFSARLLPIASIFQV 161
Score = 73.9 bits (180), Expect = 1e-11
Identities = 49/169 (28%), Positives = 74/169 (42%), Gaps = 41/169 (24%)
Query: 345 QIVLLPILNPLVGEKVILCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLA 404
Q+ +LP L +GE+ +L + LL A ++W+ WV T L+ + MF+
Sbjct: 264 QLFILPKLVSAIGERRVLSTGLLMDSVNAACLSVSWSAWVPYATTV---LVPVTMFVM-- 318
Query: 405 ENFITYISIMKVIEFNPLWQVPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIA 464
P+ GI S+ QGK Q I+
Sbjct: 319 ------------------------------------PSVCGIASRQVGPGEQGKVQGCIS 342
Query: 465 GANSISGLLSPIVMSPLTSLFLSSDAPFECKGFSIICASVCMVSYLFLS 513
G S SG+++P + SPLT+LFLS APF GFS++C + ++ FLS
Sbjct: 343 GVKSFSGVVAPFIYSPLTALFLSEKAPFYFPGFSLLCVTFSLMIGFFLS 391
>gb|AAB81677.1| putative tetracycline transporter protein [Arabidopsis thaliana]
gi|25411767|pir||G84546 probable tetracycline
transporter protein [imported] - Arabidopsis thaliana
Length = 418
Score = 90.1 bits (222), Expect = 2e-16
Identities = 50/160 (31%), Positives = 83/160 (51%), Gaps = 15/160 (9%)
Query: 13 LFHLLLPLSIHWIAEAMTVSVLVDVTTTALCP-QQSSCSKAIYINGLQQTITGIFKMAVL 71
L H+L + + A M V V+ DVT A+C SCS A+Y+ G QQ G+ M ++
Sbjct: 8 LRHMLATVFLSAFAGFMVVPVITDVTVAAVCSGPDDSCSLAVYLTGFQQVAIGMGTMIMM 67
Query: 72 PLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCI 131
P++G LSD +G K +L L + SI+P +L + + +F Y +Y+ + +
Sbjct: 68 PVIGNLSDRYGIKTILTLPMCLSIVPPVILGYRRDIKFFYVFYISKILTS---------- 117
Query: 132 SVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLP 171
+A +H S R++ F + G+ + A + + ARFLP
Sbjct: 118 ----MAVNIHGSTRISAFGILAGIKTIAGLFGTLVARFLP 153
Score = 68.9 bits (167), Expect = 4e-10
Identities = 45/169 (26%), Positives = 72/169 (41%), Gaps = 41/169 (24%)
Query: 345 QIVLLPILNPLVGEKVILCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLA 404
Q+ +LP +GE +L + L + ++WAPWV + T ++
Sbjct: 268 QLFVLPRFASAIGECKLLSTGLFMEFINMAIVSISWAPWVPYLTTVFVP----------- 316
Query: 405 ENFITYISIMKVIEFNPLWQVPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIA 464
G ++++ P+ GI S+ QGK Q I+
Sbjct: 317 ----------------------------GALFVM--PSVCGIASRQVGPGEQGKVQGCIS 346
Query: 465 GANSISGLLSPIVMSPLTSLFLSSDAPFECKGFSIICASVCMVSYLFLS 513
G S +++P V SPLT+LFLS +APF GFS++C S+ + F S
Sbjct: 347 GVRSFGKVVAPFVFSPLTALFLSKNAPFYFPGFSLLCISLSSLIGFFQS 395
>dbj|BAD28125.1| tetracycline transporter protein-like [Oryza sativa (japonica
cultivar-group)] gi|50251859|dbj|BAD27788.1|
tetracycline transporter protein-like [Oryza sativa
(japonica cultivar-group)]
Length = 211
Score = 89.4 bits (220), Expect = 3e-16
Identities = 69/255 (27%), Positives = 105/255 (41%), Gaps = 75/255 (29%)
Query: 259 TLRGVALVSFFYKLGMTGIHSVLLQIFSLLCHYYLTNFF**IP*YNRQSHNSYAVLFESC 318
TL G A+V+FFY LG G+ + LL YYL F Y++ + ++ +
Sbjct: 10 TLSGAAIVTFFYSLGEHGLQTALL--------YYLKAQFG----YSKDEFANLLLIAGAA 57
Query: 319 FWFQQKSVLRTSDDGRNWFHLFSARFQIVLLPILNPLVGEKVILCSALLASIAYAWLSGL 378
Q +V+ P+L VGE ++L LL + +L G+
Sbjct: 58 GMLSQLTVM----------------------PVLARFVGEDILLIIGLLGGCTHVFLYGI 95
Query: 379 AWAPWVSKIVTFYINLIQIYMFIWLAENFITYISIMKVIEFNPLWQVPYLGGSFGIIYIL 438
AW+ WV PYL F I+
Sbjct: 96 AWSYWV-----------------------------------------PYLSAVFIILSAF 114
Query: 439 EKPATYGIISKASSSTNQGKAQTFIAGANSISGLLSPIVMSPLTSLFLSSDAPFECKGFS 498
P+ +SK+ S QG AQ I+G +S + +L+P++ +PLT+ LS APF+ KGFS
Sbjct: 115 VHPSIRTNVSKSVGSNEQGIAQGCISGISSFASILAPLIFTPLTAWVLSETAPFKFKGFS 174
Query: 499 IICASVCMVSYLFLS 513
I+CA C + +S
Sbjct: 175 IMCAGFCTLIAFIIS 189
>ref|NP_598441.2| hypothetical protein LOC66631 [Mus musculus]
gi|23271594|gb|AAH33469.1| RIKEN cDNA 5730414C17 [Mus
musculus]
Length = 484
Score = 64.7 bits (156), Expect = 7e-09
Identities = 47/173 (27%), Positives = 78/173 (44%), Gaps = 14/173 (8%)
Query: 2 SWWSGFFELKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQQSSCSKAIYINGLQQT 61
SW GF ++H + + + A + + ++ V PQ + +NGL Q
Sbjct: 14 SWLQGFGH-PSVYHAAFVIFLEFFAWGLLTTPMLTVLHETF-PQHT-----FLMNGLIQG 66
Query: 62 ITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSH 121
+ G+ PL+G LSD GRKP LL T+ + P L+ N + Y+ + + S
Sbjct: 67 VKGLLSFLSAPLIGALSDVWGRKPFLLGTVFFTCFPIPLMRINP-----WWYFGMISVSG 121
Query: 122 IISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNY 174
+ S F + AYVAD E +R + W++ +A+ V + +L NY
Sbjct: 122 VFS--VTFSVIFAYVADFTQEHERSTAYGWVSATFAASLVSSPAIGTYLSANY 172
>emb|CAI13281.1| hippocampus abundant transcript-like 1 [Homo sapiens]
gi|55959067|emb|CAI15197.1| hippocampus abundant
transcript-like 1 [Homo sapiens]
Length = 506
Score = 63.9 bits (154), Expect = 1e-08
Identities = 43/173 (24%), Positives = 78/173 (44%), Gaps = 13/173 (7%)
Query: 2 SWWSGFFELKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQQSSCSKAIYINGLQQT 61
S W F ++H + + + + A + + ++ V ++ +NGL Q
Sbjct: 34 STWLQGFGRPSVYHAAIVIFLEFFAWGLLTTPMLTVL------HETFSQHTFLMNGLIQG 87
Query: 62 ITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSH 121
+ G+ PL+G LSD GRKP LL T+ + P L+ + + Y+ + + S
Sbjct: 88 VKGLLSFLSAPLIGALSDVWGRKPFLLGTVFFTCFPIPLMRISP-----WWYFAMISVSG 142
Query: 122 IISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNY 174
+ S F + AYVADV E +R + W++ +A+ V + +L +Y
Sbjct: 143 VFS--VTFSVIFAYVADVTQEHERSTAYGWVSATFAASLVSSPAIGAYLSASY 193
>ref|XP_225093.3| PREDICTED: similar to RIKEN cDNA 5730414C17 [Rattus norvegicus]
Length = 606
Score = 63.2 bits (152), Expect = 2e-08
Identities = 46/173 (26%), Positives = 78/173 (44%), Gaps = 14/173 (8%)
Query: 2 SWWSGFFELKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQQSSCSKAIYINGLQQT 61
SW GF + ++H + + A + + ++ V PQ + +NGL Q
Sbjct: 136 SWLQGFGQ-PSVYHAAFVIFFEFFAWGLLTTPMLTVLHETF-PQHT-----FLMNGLIQG 188
Query: 62 ITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSH 121
+ G+ PL+G LSD GRKP LL T+ + P L+ + + Y+ + + S
Sbjct: 189 VKGLLSFLSAPLIGALSDVWGRKPFLLGTVFFTCFPIPLMRISP-----WWYFGMISVSG 243
Query: 122 IISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNY 174
+ S F + AYVAD E +R + W++ +A+ V + +L NY
Sbjct: 244 VFS--VTFSVIFAYVADFTQEHERSTAYGWVSATFAASLVSSPAIGTYLSSNY 294
>ref|XP_520704.1| PREDICTED: similar to RIKEN cDNA 5730414C17 [Pan troglodytes]
Length = 1073
Score = 62.4 bits (150), Expect = 3e-08
Identities = 41/139 (29%), Positives = 68/139 (48%), Gaps = 8/139 (5%)
Query: 37 VTTTALCPQQSSCSKAIYI-NGLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSI 95
+TT L + S+ ++ NGL Q + G+ PL+G LSD GRKP LL T+ +
Sbjct: 89 LTTPVLTVLHETFSQHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKPFLLGTVFFTC 148
Query: 96 IPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGL 155
P L+ + + Y+ + + S + S F + AYVADV E +R + W++
Sbjct: 149 FPIPLMRISP-----WWYFAMISVSGVFS--VTFSVIFAYVADVTQEHERSTAYGWVSAT 201
Query: 156 SSAAHVIANVFARFLPQNY 174
+A+ V + +L +Y
Sbjct: 202 FAASLVSSPAIGAYLSASY 220
>ref|XP_533564.1| PREDICTED: similar to RIKEN cDNA 5730414C17 [Canis familiaris]
Length = 972
Score = 61.6 bits (148), Expect = 6e-08
Identities = 37/120 (30%), Positives = 60/120 (49%), Gaps = 7/120 (5%)
Query: 55 INGLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYY 114
+NGL Q + G+ PL+G LSD GRKP LL T+ + P L+ + + Y+
Sbjct: 547 MNGLIQGVKGLLSFLSAPLIGALSDVWGRKPFLLGTVFFTCFPIPLMRISP-----WWYF 601
Query: 115 VLRTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNY 174
+ + S + S F + AYVADV E +R + W++ +A+ V + +L +Y
Sbjct: 602 AMISVSGVFS--VTFSVIFAYVADVTQEHERSTAYGWVSATFAASLVSSPAIGAYLSASY 659
>ref|NP_649238.2| CG5078-PA [Drosophila melanogaster] gi|45446045|gb|AAF51617.2|
CG5078-PA [Drosophila melanogaster]
Length = 405
Score = 60.5 bits (145), Expect = 1e-07
Identities = 41/143 (28%), Positives = 73/143 (50%), Gaps = 8/143 (5%)
Query: 55 INGLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYY 114
+NGL + GI PL+G LSD +GRK LLL+T+ + +P ++ + + ++
Sbjct: 18 MNGLVMGVKGILSFLSSPLIGALSDIYGRKVLLLITVIFTSLPIPMMTMDN-----WWFF 72
Query: 115 VLRTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNY 174
V+ + S ++ G F ++ AYVADV + +R + ++ +A+ VIA + Y
Sbjct: 73 VISSISGVL--GVSFSVAFAYVADVTTKEERSRSYELVSATFAASLVIAPAMGNLIMDRY 130
Query: 175 -IFVILNYGLLLSGFNNTIDLLS 196
I ++ L+S N LL+
Sbjct: 131 GINTVVLVATLVSTTNVMFVLLA 153
>emb|CAH59449.1| tetracycline transporter-like protein 1 [Plantago major]
Length = 94
Score = 59.7 bits (143), Expect = 2e-07
Identities = 30/77 (38%), Positives = 48/77 (61%)
Query: 446 IISKASSSTNQGKAQTFIAGANSISGLLSPIVMSPLTSLFLSSDAPFECKGFSIICASVC 505
IISK QG AQ + G +S++ +LSP++ SPL+++FLS +APF+ GFSI+C +
Sbjct: 4 IISKNVGPYEQGIAQGCMMGISSLANVLSPLIYSPLSAVFLSDNAPFDFPGFSILCVGLA 63
Query: 506 MVSYLFLSPIHHLVVFF 522
+ LS + ++ F
Sbjct: 64 WLIGFVLSTMIKIIPHF 80
>gb|AAH76868.1| LOC445835 protein [Xenopus laevis]
Length = 626
Score = 57.8 bits (138), Expect = 8e-07
Identities = 41/148 (27%), Positives = 69/148 (45%), Gaps = 20/148 (13%)
Query: 55 INGLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYY 114
+NGL Q + G PL+G LSD +GRK LLLT+ + P L++ + + Y+
Sbjct: 205 MNGLIQGVKGFLSFMCAPLIGALSDVYGRKSFLLLTVFFTCFPIPLMSISP-----WWYF 259
Query: 115 VLRTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNY 174
+ + S S F + AYVAD+ E +R + ++ +A+ V + F+ + Y
Sbjct: 260 AMISVSGAFS--VTFSVIFAYVADITQEHERSTAYGLVSATFAASLVTSPAIGAFISEYY 317
Query: 175 IFVILNYGLLLSGFNNTIDLLSTLYAFL 202
+N + LL+T+ A L
Sbjct: 318 -------------GDNLVVLLATVVALL 332
>ref|XP_624637.1| PREDICTED: similar to ENSANGP00000009556 [Apis mellifera]
Length = 518
Score = 57.8 bits (138), Expect = 8e-07
Identities = 39/162 (24%), Positives = 75/162 (46%), Gaps = 13/162 (8%)
Query: 13 LFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQQSSCSKAIYINGLQQTITGIFKMAVLP 72
++H L+ + + + A + ++ V + +NGL I GI P
Sbjct: 36 VYHALVVIFLEFFAWGLLTMPVISVLNITFP------NHTFLMNGLIMGIKGILSFLSAP 89
Query: 73 LLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCIS 132
L+G LSD GRK LL+T++ + P L++ N + ++ + + S + + F +
Sbjct: 90 LIGALSDVWGRKFFLLITVAFTCAPIPLMSIN-----TWWFFAMISISGVFA--CTFSVV 142
Query: 133 VAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNY 174
AYVADV E +R + ++ +A+ VI+ ++ + Y
Sbjct: 143 FAYVADVTEEHQRSPAYGMVSATFAASMVISPALGDYIMKEY 184
>ref|NP_001011334.1| hypothetical protein LOC496797 [Xenopus tropicalis]
gi|56789744|gb|AAH88481.1| Hypothetical LOC496797
[Xenopus tropicalis]
Length = 493
Score = 57.4 bits (137), Expect = 1e-06
Identities = 44/176 (25%), Positives = 81/176 (46%), Gaps = 16/176 (9%)
Query: 13 LFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQQSSCSKAIYINGLQQTITGIFKMAVLP 72
++H ++ + + + A + + ++ V Q+ +NGL + GI P
Sbjct: 37 VYHAVVVIFLEFFAWGLLTTPMLTVL------HQTFPQHTFLMNGLIHGVKGILSFLSAP 90
Query: 73 LLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCIS 132
L+G LSD GRK LLLT+ + P LL K + Y+ + + S + + F +
Sbjct: 91 LIGALSDVWGRKSFLLLTVFFTCAPIPLL-----KISPWWYFAVISMSGVFA--VTFSVI 143
Query: 133 VAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNY---IFVILNYGLLL 185
AYVAD+ E +R + ++ +A+ V + +L + Y + V+L G+ L
Sbjct: 144 FAYVADITQEHERSTAYGLVSATFAASLVTSPAIGAYLSRAYGDTLVVVLASGVAL 199
>emb|CAG32086.1| hypothetical protein [Gallus gallus]
Length = 492
Score = 57.4 bits (137), Expect = 1e-06
Identities = 43/176 (24%), Positives = 81/176 (45%), Gaps = 16/176 (9%)
Query: 13 LFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQQSSCSKAIYINGLQQTITGIFKMAVLP 72
++H ++ + + + A + + ++ V Q+ +NGL + G+ P
Sbjct: 37 VYHAVVVIFLEFFAWGLLTTPMLTVL------HQTFPQHTFLMNGLIHGVKGLLSFLSAP 90
Query: 73 LLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCIS 132
L+G LSD GRK LLLT+ + P L+ K + Y+ + + S + + F +
Sbjct: 91 LIGALSDVWGRKSFLLLTVFFTCAPIPLM-----KISPWWYFAVISMSGVFA--VTFSVI 143
Query: 133 VAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNY---IFVILNYGLLL 185
AYVAD+ E +R + ++ +A+ V + +L Q Y + V+L G+ L
Sbjct: 144 FAYVADITQEHERSTAYGLVSATFAASLVTSPAIGAYLSQAYGDTLVVVLASGVAL 199
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.339 0.147 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 821,635,827
Number of Sequences: 2540612
Number of extensions: 31824384
Number of successful extensions: 135518
Number of sequences better than 10.0: 467
Number of HSP's better than 10.0 without gapping: 163
Number of HSP's successfully gapped in prelim test: 304
Number of HSP's that attempted gapping in prelim test: 134935
Number of HSP's gapped (non-prelim): 739
length of query: 526
length of database: 863,360,394
effective HSP length: 133
effective length of query: 393
effective length of database: 525,458,998
effective search space: 206505386214
effective search space used: 206505386214
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC140027.12


