

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139344.7 + phase: 1 /pseudo
(478 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
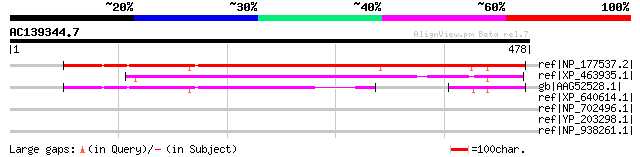
ref|NP_177537.2| expressed protein [Arabidopsis thaliana] 359 1e-97
ref|XP_463935.1| hypothetical protein [Oryza sativa (japonica cu... 276 1e-72
gb|AAG52528.1| unknown protein; 56038-53215 [Arabidopsis thalian... 169 2e-40
ref|XP_640614.1| hypothetical protein DDB0204642 [Dictyostelium ... 41 0.094
ref|NP_702496.1| hypothetical protein PF14_0607 [Plasmodium falc... 36 2.3
ref|YP_203298.1| orf292 [Rhizopus oryzae] gi|57338995|gb|AAW4946... 36 2.3
ref|NP_938261.1| hypothetical protein D3112p54 [Bacteriophage D3... 35 4.0
>ref|NP_177537.2| expressed protein [Arabidopsis thaliana]
Length = 803
Score = 359 bits (921), Expect = 1e-97
Identities = 205/438 (46%), Positives = 283/438 (63%), Gaps = 16/438 (3%)
Query: 50 ESFKENYAPFVVFMSGIGVLRVTDRYASSTGMKVDVLTRMRTSAIVRVEALVSDLVSRTL 109
E+ KE YA F VFM+ GV+R + SS +++ +++R SA R+E + LVS
Sbjct: 264 ETSKEKYAVFAVFMAAAGVVRASTAGFSSGAQSLEI-SKLRNSAEKRIEFVAQILVSNG- 321
Query: 110 RFRNLGNDLQDRVLLQCVTLGMTRTISFSSHSSLFVCLGLSLLTRILPLPRLYES---VF 166
L ++ LL+C + + R S SS + L +CL +LLT++ PL ++YES F
Sbjct: 322 NVVTLPTTQREGPLLKCFAIALARCGSVSSSAPLLLCLTSALLTQVFPLGQIYESFCNAF 381
Query: 167 ELSPSSGGLKVNEIKEHPDNILFKEAGAVTGIFCNLYVLADEENKNIVENLIWEYYRDIY 226
P G ++ ++EH ++LFKE+GA++G FCN Y A EENK IVEN+IW++ +++Y
Sbjct: 382 GKEPI--GPRLIWVREHLSDVLFKESGAISGAFCNQYSSASEENKYIVENMIWDFCQNLY 439
Query: 227 FGHRKVVMDLKGKEDELLTNFEKTAESAFLMVVVFALSVTKHKLSSTFAQEIQTEISLKI 286
HR++ M L G ED LL + EK AES+FLMVVVFAL+VTK L ++E + S+KI
Sbjct: 440 LQHRQIAMLLCGIEDTLLGDIEKIAESSFLMVVVFALAVTKQWLKPIVSKERKMVTSVKI 499
Query: 287 LVSLSCVEYFRHVRLPEYMETIRKVTAIVKKNENACTFFVNSIPSYGDLTNGPD---QKT 343
LVS SCVEYFRH+RLPEYMETIR+V + V++N+ C FV SIP+Y LTN D Q+
Sbjct: 500 LVSFSCVEYFRHIRLPEYMETIREVISCVQENDAPCVSFVESIPAYDSLTNPKDLFTQRI 559
Query: 344 KYFWSKDEVQTARVLLYLRVIPTLIECLRGPVFGDMVAPTMFLYMEHPNGKVARASHSVF 403
KY WS+D+VQT+R+L YLRVIPT I L F +VA TMFLY+ HPN KVA+ASH++
Sbjct: 560 KYEWSRDDVQTSRILFYLRVIPTCIGRLSASAFRGVVASTMFLYIGHPNRKVAQASHTLL 619
Query: 404 TAFISMGKESEKIDSLIEGEA---RFLSLSIIILPNTL---GMASGVVGMVRHLPAGSPT 457
AF+S KESE+ + E ++ S+ + P G+ASGV +V+HLPAGSP
Sbjct: 620 AAFLSSAKESEEDERTQFKEQLVFYYMQRSLEVYPEITPFEGLASGVATLVQHLPAGSPA 679
Query: 458 TFYCIHSLVEKANQLCSE 475
FY +HSLVEKA+ +E
Sbjct: 680 IFYSVHSLVEKASTFSTE 697
>ref|XP_463935.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|41053021|dbj|BAD07952.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 749
Score = 276 bits (706), Expect = 1e-72
Identities = 158/385 (41%), Positives = 228/385 (59%), Gaps = 31/385 (8%)
Query: 107 RTLRFRNL-----GNDLQDR-VLLQCVTLGMTRTISFSSHSSLFVCLGLSLLTRILPLPR 160
RT+RF G + DR +LLQCV LG+T+ + H S+ C+ ++LL +LPLP
Sbjct: 295 RTIRFAAEKAVLEGKHVDDRRLLLQCVALGLTQCGQVTPHESVLRCVCMALLEELLPLPD 354
Query: 161 LYE-SVFELSPSSGGLKVNEIKEHPDNILFKEAGAVTGIFCNLYVLADEENKNIVENLIW 219
L + SV +S + N +K+H D++LFKEAG V GI CN Y A ++ K VE +W
Sbjct: 355 LLKMSVQCPDGNSPEIVKNRVKQHLDSVLFKEAGPVAGILCNQYSFASDKAKTSVETCVW 414
Query: 220 EYYRDIYFGHRKVVMDLKGKEDELLTNFEKTAESAFLMVVVFALSVTKHKLSSTFAQEIQ 279
EY + +Y R V+ +GK+D+L+T+ EK AE+AFLMVVVF+ VTKH+L++ ++ Q
Sbjct: 415 EYAQVLYCHLRAAVILHQGKQDDLITDIEKIAEAAFLMVVVFSAEVTKHRLNAKSSEGFQ 474
Query: 280 TEISLKILVSLSCVEYFRHVRLPEYMETIRKVTAIVKKNENACTFFVNSIPSYGDLTNGP 339
++++KILVS SC+E+ R +RLPEY E +R+ + ++N F+ SIPSY +LTN
Sbjct: 475 PDVAVKILVSFSCLEHLRRLRLPEYTEAVRRAVLVNQENAAVAALFIESIPSYAELTNLL 534
Query: 340 D-QKTKYFWSKDEVQTARVLLYLRVIPTLIECLRGPVFGDMVAPTMFLYMEHPNGKVARA 398
T+Y W D VQT+R+L YLR+ +C + Y++H N KV RA
Sbjct: 535 TLDGTRYIWHGDVVQTSRILFYLRIFDIKKKCPYHKI---------VRYIQHSNEKVTRA 585
Query: 399 SHSVFTAFISMGKESEKIDSLIEGEARFLSLSIIILPNTL----------GMASGVVGMV 448
SHSV +F+S G +++ D + E L+ + TL G+ASGV +
Sbjct: 586 SHSVVVSFLSSGNDTDPDDRMALKE----QLAFYYIKRTLEAYPGVTPFEGLASGVAALA 641
Query: 449 RHLPAGSPTTFYCIHSLVEKANQLC 473
RHLPAGSP T +CIH+LV KA LC
Sbjct: 642 RHLPAGSPATLFCIHNLVVKAKDLC 666
>gb|AAG52528.1| unknown protein; 56038-53215 [Arabidopsis thaliana]
gi|25373334|pir||D96767 unknown protein F2P9.16
[imported] - Arabidopsis thaliana
Length = 699
Score = 169 bits (428), Expect = 2e-40
Identities = 109/291 (37%), Positives = 158/291 (53%), Gaps = 43/291 (14%)
Query: 50 ESFKENYAPFVVFMSGIGVLRVTDRYASSTGMKVDVLTRMRTSAIVRVEALVSDLVSRTL 109
E+ KE YA F VFM+ GV+R + SS +++ +++R SA R+E + LVS
Sbjct: 264 ETSKEKYAVFAVFMAAAGVVRASTAGFSSGAQSLEI-SKLRNSAEKRIEFVAQILVSNG- 321
Query: 110 RFRNLGNDLQDRVLLQCVTLGMTRTISFSSHSSLFVCLGLSLLTRILPLPRLYES---VF 166
L ++ LL+C + + R S SS + L +CL +LLT++ PL ++YES F
Sbjct: 322 NVVTLPTTQREGPLLKCFAIALARCGSVSSSAPLLLCLTSALLTQVFPLGQIYESFCNAF 381
Query: 167 ELSPSSGGLKVNEIKEHPDNILFKEAGAVTGIFCNLYVLADEENKNIVENLIWEYYRDIY 226
P G ++ ++EH ++LFKE+GA++G FCN Y A EENK IVEN+IW++ +++Y
Sbjct: 382 GKEPI--GPRLIWVREHLSDVLFKESGAISGAFCNQYSSASEENKYIVENMIWDFCQNLY 439
Query: 227 FGHRKVVMDLKGKEDELLTNFEKTAESAFLMVVVFALSVTKHKLSSTFAQEIQTEISLKI 286
HR++ M L G ED LL + EK AES+FLMVVVFAL+VTK L ++E + E
Sbjct: 440 LQHRQIAMLLCGIEDTLLGDIEKIAESSFLMVVVFALAVTKQWLKPIVSKERKME----- 494
Query: 287 LVSLSCVEYFRHVRLPEYMETIRKVTAIVKKNENACTFFVNSIPSYGDLTN 337
N+ C FV SIP+Y LTN
Sbjct: 495 -------------------------------NDAPCVSFVESIPAYDSLTN 514
Score = 56.6 bits (135), Expect = 2e-06
Identities = 33/77 (42%), Positives = 46/77 (58%), Gaps = 6/77 (7%)
Query: 405 AFISMGKESEKIDSLIEGEAR---FLSLSIIILPNTL---GMASGVVGMVRHLPAGSPTT 458
AF+S KESE+ + E ++ S+ + P G+ASGV +V+HLPAGSP
Sbjct: 517 AFLSSAKESEEDERTQFKEQLVFYYMQRSLEVYPEITPFEGLASGVATLVQHLPAGSPAI 576
Query: 459 FYCIHSLVEKANQLCSE 475
FY +HSLVEKA+ +E
Sbjct: 577 FYSVHSLVEKASTFSTE 593
>ref|XP_640614.1| hypothetical protein DDB0204642 [Dictyostelium discoideum]
gi|60468630|gb|EAL66633.1| hypothetical protein
DDB0204642 [Dictyostelium discoideum]
Length = 1329
Score = 40.8 bits (94), Expect = 0.094
Identities = 59/314 (18%), Positives = 125/314 (39%), Gaps = 46/314 (14%)
Query: 177 VNEIKEHPDNILFKEAG----AVTGIFCNLYVLADEENKNIVE----------NLIWEYY 222
+ +I+ H LF ++G+ NL +D +N +V N W Y
Sbjct: 859 IKKIESHKTTPLFLSIPPFIQTISGLVLNL---SDNDNNKLVLLKLCDFTYSLNENWSNY 915
Query: 223 RDIYFGHRKVVMDLKGKEDELLTNFEKTAESAFLMVVVFALSVTKHKLSSTFAQEIQTEI 282
++ F + + K + +F + +LS+ ++SS I + +
Sbjct: 916 GNVEFPSPPIPKSM----------LRKALDPSF-QAIFQSLSLLFFEISSILTP-IDSVV 963
Query: 283 SLKILVSLSCVEYFRHVRLPEYMETIRKVTAIVKKNENACTFFVNSIPSYGDLTNGPDQK 342
+++ L+ + V+L ++IR + V +E A ++ +P Y +
Sbjct: 964 GIELFCKLTFIGSDDDVQL--IGKSIRTLAKNVLVSECAIIRLIDKMPLYYQVPKD---- 1017
Query: 343 TKYFWSKDEVQTARVLLYLRVIPTLIECLRGPVFGDMVAPTMFLYMEHPNGKVARASHSV 402
D++ +V++Y + +LI + + D V P +F YME+PN K+ SHS+
Sbjct: 1018 -------DKIACLQVVIYFIALDSLITSVPHNLIVDHVVPNLFEYMEYPNSKLNIQSHSI 1070
Query: 403 FTAFISMGKESEKIDSLIEGEARFLSLSIIILPNTLGMASGVVGMVRHLPAGSPTTFYCI 462
++ I+ +L ++ P ++S MV + +PT +
Sbjct: 1071 LAKMFAI----PNFHLSIKMVPMYLKSALKSYPKFTKVSSLYEVMVSIVENNAPTNPIIL 1126
Query: 463 HSLVEKANQLCSEV 476
+S+ +N + + +
Sbjct: 1127 YSIKTISNSIINHM 1140
>ref|NP_702496.1| hypothetical protein PF14_0607 [Plasmodium falciparum 3D7]
gi|23497681|gb|AAN37220.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 1068
Score = 36.2 bits (82), Expect = 2.3
Identities = 32/90 (35%), Positives = 46/90 (50%), Gaps = 9/90 (10%)
Query: 175 LKVNEIKEHPD-NILFKEAGAVTGIFCNLYVLADEENKNIVENLIWEYYRDIYFGHRKVV 233
L+ N IKE D + L K + GI N++ A E K I++N +E FG
Sbjct: 375 LRRNNIKEIFDVDDLVKNIKSFLGIKSNIHE-AFENQKLIIKNCNYES-----FGPELCS 428
Query: 234 MDLKGKEDELLTNFEKTAESAFLMVVVFAL 263
+D K KE +L +EK SAFL +++F L
Sbjct: 429 VDEKAKE--MLWKYEKKKNSAFLFIILFTL 456
>ref|YP_203298.1| orf292 [Rhizopus oryzae] gi|57338995|gb|AAW49465.1| orf292
[Rhizopus oryzae]
Length = 292
Score = 36.2 bits (82), Expect = 2.3
Identities = 32/178 (17%), Positives = 79/178 (43%), Gaps = 2/178 (1%)
Query: 260 VFALS-VTKHKLSSTFAQEIQTEISLKILVSLSCVEYFRHVRLPEYMETIRKVTAIVKKN 318
V ALS + K +S+ + +++ I + + + +R+ E++E ++V I K N
Sbjct: 104 VAALSLIGKFTISAVKSSPASNKLTATIGTATAMGTTYLGIRILEHIENQKRVFTITK-N 162
Query: 319 ENACTFFVNSIPSYGDLTNGPDQKTKYFWSKDEVQTARVLLYLRVIPTLIECLRGPVFGD 378
+ + + P G ++ K + + +L +++I E L V
Sbjct: 163 DKSANLIIEPNPELGGTEKDVEEVIKNIKINSPYEASEILNGIKMILFCTEILSTLVIIS 222
Query: 379 MVAPTMFLYMEHPNGKVARASHSVFTAFISMGKESEKIDSLIEGEARFLSLSIIILPN 436
+ ++F+++++ N + ++ + FI++ +++ I L G ++S I+ N
Sbjct: 223 FIFYSLFIFIKYVNFNKFKINNVICIKFINLVQKASIIYVLFWGGLTLFNISAILYLN 280
>ref|NP_938261.1| hypothetical protein D3112p54 [Bacteriophage D3112]
gi|37595217|gb|AAQ94492.1| hypothetical protein
[Bacteriophage D3112]
Length = 383
Score = 35.4 bits (80), Expect = 4.0
Identities = 24/89 (26%), Positives = 41/89 (45%), Gaps = 12/89 (13%)
Query: 384 MFLYMEHPNGKVARASHSVFTAFISMGKESEKIDSLIEGEARFLSLSIIILPNTLGMASG 443
+ + M P+G +AR +TA ++ G + S ++G R+L ++ + L +A G
Sbjct: 221 LLVAMGRPSGSLARYGVEFYTALVTAGPNATTGSSGLDGTTRYLQMTNV--SRALFIADG 278
Query: 444 ----------VVGMVRHLPAGSPTTFYCI 462
G + +PAG PTT Y I
Sbjct: 279 WSTAVLWVRAETGSLHFMPAGVPTTDYRI 307
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.326 0.140 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 757,249,927
Number of Sequences: 2540612
Number of extensions: 30083048
Number of successful extensions: 93255
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 93238
Number of HSP's gapped (non-prelim): 9
length of query: 478
length of database: 863,360,394
effective HSP length: 132
effective length of query: 346
effective length of database: 527,999,610
effective search space: 182687865060
effective search space used: 182687865060
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC139344.7


