

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139344.2 - phase: 0
(1591 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
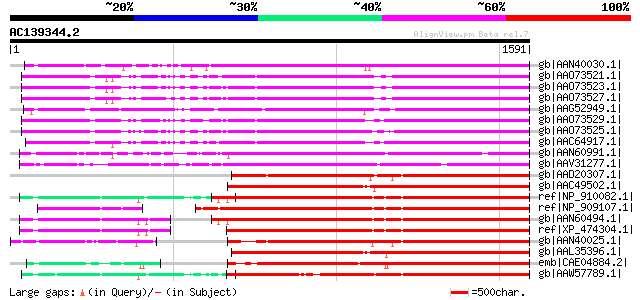
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN40030.1| putative gag-pol polyprotein [Zea mays] 976 0.0
gb|AAO73521.1| gag-pol polyprotein [Glycine max] 969 0.0
gb|AAO73523.1| gag-pol polyprotein [Glycine max] 969 0.0
gb|AAO73527.1| gag-pol polyprotein [Glycine max] 966 0.0
gb|AAG52949.1| gag/pol polyprotein [Arabidopsis thaliana] 965 0.0
gb|AAO73529.1| gag-pol polyprotein [Glycine max] 955 0.0
gb|AAO73525.1| gag-pol polyprotein [Glycine max] 943 0.0
gb|AAC64917.1| gag-pol polyprotein [Glycine max] 939 0.0
gb|AAN60991.1| Putative Zea mays retrotransposon Opie-2 [Oryza s... 921 0.0
gb|AAV31277.1| putative polyprotein [Oryza sativa (japonica cult... 903 0.0
gb|AAD20307.1| copia-type pol polyprotein [Zea mays] 881 0.0
gb|AAC49502.1| Pol [Zea mays] gi|7489803|pir||T04112 pol protein... 880 0.0
ref|NP_910082.1| putative gag-pol polyprotein [Oryza sativa (jap... 867 0.0
ref|NP_909107.1| unnamed protein product [Oryza sativa (japonica... 863 0.0
gb|AAN60494.1| Putative Zea mays retrotransposon Opie-2 [Oryza s... 863 0.0
ref|XP_474304.1| OSJNBb0004A17.2 [Oryza sativa (japonica cultiva... 862 0.0
gb|AAN40025.1| putative gag-pol polyprotein [Zea mays] 847 0.0
gb|AAL35396.1| Opie2a pol [Zea mays] 841 0.0
emb|CAE04884.2| OSJNBa0042I15.6 [Oryza sativa (japonica cultivar... 813 0.0
gb|AAW57789.1| putative polyprotein [Oryza sativa (japonica cult... 791 0.0
>gb|AAN40030.1| putative gag-pol polyprotein [Zea mays]
Length = 2319
Score = 976 bits (2522), Expect = 0.0
Identities = 611/1629 (37%), Positives = 886/1629 (53%), Gaps = 151/1629 (9%)
Query: 44 WKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIV 103
W M S ++G+ LW+I+ G+ EE TP + ++ + II
Sbjct: 138 WADKMKSHLIGVHPSLWEIVNVGMYKPAQGEE--------MTPEMMQEVHRNAQAVSIIK 189
Query: 104 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYELFRMKDDESIEEM 163
S+ EY K+ + A+ ++ L + EG K + L + + ES++ +
Sbjct: 190 GSLCPEEYRKVQGREDARDIWNILRMSHEGDPKAKRHRVEALESELARYDWTKGESLQSL 249
Query: 164 YSRFQTLVSGLQILKKSYVASDHVSKI-LRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSS 222
+ R LV+ +++L + V+++ +R+ + + I + D ++ L +
Sbjct: 250 FDRLMVLVNKIRVLGSEDWSDSKVTRLFMRAYKEKDKSLARMIRDRDDYEDMTPHQLFAK 309
Query: 223 LKVHEMSLNEHETSKKSKSIALPSKGKTS-KSSKAYKASESEEESPDGDSDEDQSVKMAM 281
++ HE +E K S AL + + + K SK +KA + E S D DS D+ M +
Sbjct: 310 IQQHE---SEEAPIKTRDSHALITNEQDNLKKSKDHKAKKVVETSSDEDSSSDEDTAMFI 366
Query: 282 LSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKFKGKSKK 341
+ KKF+ K ++ +K ++ C+ C + GHFIADCP+ ++++ K + KK
Sbjct: 367 ---------KTFKKFVRKNDKFQ--RKGKKRACYECGQTGHFIADCPNKKEQEAKKEYKK 415
Query: 342 SSFNSS-------KFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMGLVATVSSEAV 394
F K +K + + W D S S++EE +VA V+ ++
Sbjct: 416 DKFKKGGKTKGYFKKKKYGQAHIGEEWNSDDESSSSEEEE----------VVANVAIQST 465
Query: 395 SEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLE 454
S A+ + +++ Y IP + K ++LF + D D K+ +++
Sbjct: 466 SSAQLFTNLQDDSY--IPTCLMAKGDK--VTLFSN------DFSNDDDDDQIAMKNKMIK 515
Query: 455 LKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKHEIALDDFIMAGIDRS-KVA 513
E L G+N+I+ KL EK DK A +D ++ +R+ ++
Sbjct: 516 ----EFGLNGYNVIT------------KLMEKLDKRKATLD--AQEDLLILEKERNLELQ 557
Query: 514 SMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVP-----------EGTNAV 562
+I++ K I K+ +L + + ++ + S V + NA
Sbjct: 558 ELIHN------KDIEVINLKTSIDNLANEKNALESSMLSLNVQNQELQVQLENCKNINAP 611
Query: 563 TVV-QSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKI------------LKRSEPV 609
T+V +SK ++ + K +K + + + S ++K+ LK+ EP
Sbjct: 612 TLVFESKSSSNDNSCKHCAK-YHASCCLTNHARKHSPQVKVEEILKRCSSNDGLKKVEPK 670
Query: 610 HQNLIKPESK---IPKQKDQKNKAATASEKTIPKGVKPKVLNDQ--KPLSIHPKVCLRAR 664
+++L KP + + ++N + PK ++ L D + S + K
Sbjct: 671 YKSL-KPNNGRRGLGFNSSKENPSTVHKGWRSPKFIEGTTLYDALGRIHSSNDKSSQVYS 729
Query: 665 EKQRSWYLDSGCSRHMTGEKALFLTLTM-KDGGEVKFGGNQTGKIIGTGTIGNSSI-SIN 722
SW LDSGC+ HMTGEK +F TL + ++ E+ FG + K+IG G I S S++
Sbjct: 730 SGGSSWVLDSGCTNHMTGEKDMFHTLQLTQEAQEIVFGDSGKSKVIGIGKIPISDQQSLS 789
Query: 723 NVWLVDGLKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDL 782
NV LVD L +NLLS+SQ C Y+ FS + ++ ++D S+ F G+ +Y ++F+
Sbjct: 790 NVLLVDSLSYNLLSVSQLCGMGYNCLFSDVDVKILRREDSSVAFTGRLKGKLYLVDFTTS 849
Query: 783 ADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGK 842
CL++ +DK W+WH+RL H R ++K+ K + GL N+ + D +CGACQ GK
Sbjct: 850 KVTPETCLVAKSDKGWLWHRRLAHVGMRNLAKLQKDNHIIGLTNVVFEKDRVCGACQAGK 909
Query: 843 IVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKD 902
+SK++V+T RPLELLH+DLFGPV S+ GSKYGLVIVDD+SR+TWV F+ K
Sbjct: 910 QHGVPHQSKNVVTTKRPLELLHMDLFGPVAYISIGGSKYGLVIVDDFSRFTWVFFLSDKG 969
Query: 903 YACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNG 962
E+ F + Q+E ELKI KVRSD+G EF+N E F + GI HEFS P TPQQNG
Sbjct: 970 ETQEILKKFMRRAQNEFELKIKKVRSDNGTEFKNTGVEEFLGEEGIKHEFSVPYTPQQNG 1029
Query: 963 VVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRP 1022
VVERKNRTL E ARTM+ E +FWAEAVNT+C+ NR+Y+ + +KTAYEL G +P
Sbjct: 1030 VVERKNRTLIEAARTMLDEYKTPDNFWAEAVNTACHAINRLYLHKIYKKTAYELLTGNKP 1089
Query: 1023 NISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVK 1082
+ YF FGC C+ILN K KF + G LGY+ + YRV+N+ T VE ++ V
Sbjct: 1090 KVDYFRVFGCKCFILNKKVKSSKFAPRVDEGFLLGYASNAHGYRVFNNSTGLVEIAIDVT 1149
Query: 1083 FDDREP-------------------------GSKTSEQSESNAGT-------------TD 1104
FD+ G ++ + GT
Sbjct: 1150 FDESNGSQGHVSNDTVGNEKLPCESIKKLAIGEVRPQERDDEEGTLWMTNEVVDVGARVV 1209
Query: 1105 SEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSPIVQNESASEDFQDNTQQVI 1164
S++ S PS S E+ E E + P+ Q + E Q + V
Sbjct: 1210 SDNVSTQANPSTSSHPNHEENHQRMPTVVEDEQENIDGEVPLDQVTNEEEQIQRH-PSVP 1268
Query: 1165 QPKFKH--KSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWIL 1222
P+ H + HP + I+GS TRS +S +EP VEEAL D WI
Sbjct: 1269 HPRVHHTIQRDHPVDNILGSIRRGVTTRSRLANFCEFYSFVSSLEPLKVEEALDDPDWIT 1328
Query: 1223 AMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQEG 1282
AMQEELN F RN+VW LV +P ++N+I TKWVFRNK +E G VT+NKARLVAQGY+Q EG
Sbjct: 1329 AMQEELNNFTRNEVWSLVQRP-KQNVIGTKWVFRNKQDENGVVTKNKARLVAQGYTQVEG 1387
Query: 1283 INYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQPPGFEDLK 1342
+++ ET+APVARLE+IR+L++YA NH LYQMDVKS FLNG ++E VYV+QPPGFED K
Sbjct: 1388 LDFGETYAPVARLESIRILIAYATNHDFKLYQMDVKSAFLNGPLQERVYVEQPPGFEDPK 1447
Query: 1343 HPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYV 1402
P+HVY L K+LYGLKQAPRAWYD L +FLIKN F G+ D+TLF R + ++ + QIYV
Sbjct: 1448 KPNHVYLLHKALYGLKQAPRAWYDCLKDFLIKNGFTIGKADSTLFTRKVDNELFVCQIYV 1507
Query: 1403 DDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLK 1462
DDIIFGSTN C+EFSK+M + FEMSMMGELK+FLG Q+ Q KEG ++ QTKYT+++LK
Sbjct: 1508 DDIIFGSTNEKFCEEFSKVMTNRFEMSMMGELKYFLGFQVKQLKEGTFLCQTKYTQDMLK 1567
Query: 1463 KFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCAR 1522
KF +E K TPM L + G VDQKLYR MIGSLLYL ASRPDI+ SVC+CAR
Sbjct: 1568 KFGMEKAKHAKTPMPSNGHLDLNEEGKPVDQKLYRSMIGSLLYLCASRPDIMLSVCMCAR 1627
Query: 1523 FQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNC 1582
FQ++P++ HL AVKRI RYL T NLGL Y K + L+G+ D+DYAG +++RKST+G C
Sbjct: 1628 FQANPKDCHLVAVKRILRYLVHTQNLGLWYPKGSFFDLLGYSDSDYAGCKVDRKSTTGTC 1687
Query: 1583 QFLGENLIS 1591
QFLG +L+S
Sbjct: 1688 QFLGRSLVS 1696
>gb|AAO73521.1| gag-pol polyprotein [Glycine max]
Length = 1574
Score = 969 bits (2506), Expect = 0.0
Identities = 597/1583 (37%), Positives = 856/1583 (53%), Gaps = 178/1583 (11%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG D T + +L
Sbjct: 17 DGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKEEDEL 76
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ VK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKISRLQLLATKFEN 136
Query: 152 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ KKSK++A S E EE+ D ++
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-AEKKSKNLAFVSN------------DEGEEDEYDLNT 243
Query: 272 DEDQSVKMAMLSNKLEYLARKQKK----------FLSKRGSYKNSKKEDQKG-------C 314
DE + + +L + + + K F ++GS K KK D K C
Sbjct: 244 DEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGS-KYQKKSDVKPSHSKGIQC 302
Query: 315 FNCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH IA+CP K+ KG S S D +SE SD
Sbjct: 303 HGCEGYGHIIAECPTHLKKHRKGLSVCQS-------------------DTESEQESD--- 340
Query: 375 ADDDAKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
+D D A G+ T +ED ++ S+I EL S ++L E +
Sbjct: 341 SDRDVNALTGIFET------------AEDSSDTDSEITFDELATSYRKLCIKSEKILQQE 388
Query: 435 TDLKEKYVDLMKQQKSTLLELKASEEELKG-FNLISTTYEDRLKSLCQKLQEKCDKGSGN 493
LK+ DL ++++ E+ ELKG +++ E+ KS+ + +KGS
Sbjct: 389 AQLKKVIADLEAEKEAHKEEIS----ELKGEVGFLNSKLENMTKSI-----KMLNKGSDT 439
Query: 494 KHEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLK 551
LD+ ++ G KN G +G+G++ + + ++
Sbjct: 440 -----LDEVLLLG--------------KNAGNQRGLGFNPKSAGRTTM------------ 468
Query: 552 STFVP-EGTNAVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVH 610
+ FVP + T+ Q + G Q K KS K + +
Sbjct: 469 TEFVPAKNRTGATMSQHRSRHHGMQQK--------------KSKRKKWRCHYCGK----- 509
Query: 611 QNLIKPES-KIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRS 669
IKP + ++ + +PK K +S+ LRA K+
Sbjct: 510 YGHIKPFCYHLHPHHGTQSSNSRKKMMWVPK---------HKAVSLVVHTSLRASAKE-D 559
Query: 670 WYLDSGCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVD 728
WYLDSGCSRHMTG K L + V FG GKIIG G + + + S+N V LV
Sbjct: 560 WYLDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVK 619
Query: 729 GLKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVV 788
GL NL+SISQ CD ++V F+K+ C LV + + KG R ++ +
Sbjct: 620 GLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSKDNCYLWTPQETSYSST 678
Query: 789 CLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSF 848
CL S D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK S
Sbjct: 679 CLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSH 738
Query: 849 KSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVF 908
+ +TSR LELLH+DL GP+ SL G +Y V+VDD+SR+TWVKFI+ K EVF
Sbjct: 739 QKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVKFIREKSETFEVF 798
Query: 909 SSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKN 968
++Q EK+ I ++RSDHG EFEN FC GI HEFS+ TPQQNG+VERKN
Sbjct: 799 KELSLRLQREKDCVIKRIRSDHGREFENSRLTEFCTSEGITHEFSAAITPQQNGIVERKN 858
Query: 969 RTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFH 1028
RTLQE AR M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P++ +FH
Sbjct: 859 RTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFH 918
Query: 1029 QFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREP 1088
FG CYIL ++ +K D K+ GIFLGYS S+AYRV+NS T+ V ES++V DD P
Sbjct: 919 IFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLSP 978
Query: 1089 GSKTSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSPIVQ 1148
K + + + DA++S + +++ S+S + S I Q
Sbjct: 979 ARKKDVEEDVRTSGDNVADAAKSGENAEN-------------------SDSATDESNINQ 1019
Query: 1149 NESASEDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEP 1208
+ S + + HP+ELIIG + TRS + S +S IEP
Sbjct: 1020 PDKRSST-------------RIQKMHPKELIIGDPNRGVTTRSREVEIVSNSCFVSKIEP 1066
Query: 1209 KTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRN 1268
K V+EAL+D+ WI AMQEEL QF+RN+VW+LVP+P N+I TKW+F+NK NE+G +TRN
Sbjct: 1067 KNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRN 1126
Query: 1269 KARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEE 1328
KARLVAQGY+Q EG+++ ETFAPVARLE+IRLLL A LYQMDVKS FLNG + E
Sbjct: 1127 KARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNE 1186
Query: 1329 EVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFR 1388
EVYV+QP GF D HPDHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF
Sbjct: 1187 EVYVEQPKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFV 1246
Query: 1389 RTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEG 1448
+ ++++I QIYVDDI+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++
Sbjct: 1247 KQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDS 1306
Query: 1449 VYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTA 1508
+++ Q++Y K ++KKF +E+ TP LSK++ GT VDQ LYR MIGSLLYLTA
Sbjct: 1307 IFLSQSRYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTA 1366
Query: 1509 SRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADY 1568
SRPDI ++V +CAR+Q++P+ SHLT VKRI +Y+ GT++ G++Y + L+G+CDAD+
Sbjct: 1367 SRPDITYAVGVCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADW 1426
Query: 1569 AGDRIERKSTSGNCQFLGENLIS 1591
AG +RKSTSG C +LG NLIS
Sbjct: 1427 AGSADDRKSTSGGCFYLGNNLIS 1449
>gb|AAO73523.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 969 bits (2505), Expect = 0.0
Identities = 596/1582 (37%), Positives = 855/1582 (53%), Gaps = 174/1582 (10%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG D T + +L
Sbjct: 17 DGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKEEDEL 76
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK L + EG+ VK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDACEILKSTHEGTSKVKMSRLQLLATKFEN 136
Query: 152 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ KKSK++A S E EE+ D D+
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-AEKKSKNLAFVSN------------DEGEEDEYDLDT 243
Query: 272 DEDQSVKMAMLSNKLEYLARKQKK----------FLSKRGSYKNSKKEDQKG-------C 314
DE + + +L + + + K F ++GS K K+ D K C
Sbjct: 244 DEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGS-KYQKRSDVKPSHSKGIQC 302
Query: 315 FNCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH IA+CP K+ KG S S D +SE SD
Sbjct: 303 HGCEGYGHIIAECPTHLKKHRKGLSVCQS-------------------DTESEQESD--- 340
Query: 375 ADDDAKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
+D D A +G+ T +ED ++ S+I EL S ++L E +
Sbjct: 341 SDRDVNALIGIFET------------AEDSSDTDSEITFDELAASYRKLCIKSEKILQQE 388
Query: 435 TDLKEKYVDLMKQQKSTLLELKASEEELKG-FNLISTTYEDRLKSLCQKLQEKCDKGSGN 493
LK+ DL ++++ E+ ELKG +++ E+ KS+ + +KGS
Sbjct: 389 AQLKKVIADLEAEKEAHKEEIS----ELKGEVGFLNSKLENMTKSI-----KMLNKGSDT 439
Query: 494 KHEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLK 551
LD+ ++ G KN G +G+G++ + + ++
Sbjct: 440 -----LDEVLLLG--------------KNAGNQRGLGFNPKSAGRTTM------------ 468
Query: 552 STFVP-EGTNAVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVH 610
+ FVP + T+ Q + G Q K KS K + +
Sbjct: 469 TEFVPAKNRTGATMSQHRSRHHGMQQK--------------KSKRKKWRCHYCGK----- 509
Query: 611 QNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRSW 670
IKP ++ S K + K K ++ L +H + A+E W
Sbjct: 510 YGHIKPFCYHLHGHPHHGTQSSNSRKKMMWVPKHKAVS----LVVHTSLRASAKE---DW 562
Query: 671 YLDSGCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDG 729
YLDSGCSRHMTG K L + V FG GKIIG G + + + S+N V LV G
Sbjct: 563 YLDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKG 622
Query: 730 LKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVC 789
L NL+SISQ CD ++V F+K+ C LV + + KG R ++ + C
Sbjct: 623 LTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSKDNCYLWTPQETSYSSTC 681
Query: 790 LLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFK 849
L S D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK S +
Sbjct: 682 LSSKEDEVRIWHQRFGHLHLRGMKKILDKSAVRGIPNLKIEEGRICGECQIGKQVKMSHQ 741
Query: 850 SKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFS 909
+TSR LELLH+DL GP+ SL G +Y V+VDD+SR+TWV FI+ K EVF
Sbjct: 742 KLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSGTFEVFK 801
Query: 910 SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNR 969
++Q EK+ I ++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNR
Sbjct: 802 KLSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQNGIVERKNR 861
Query: 970 TLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQ 1029
TLQE AR M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P++ +FH
Sbjct: 862 TLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFHI 921
Query: 1030 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPG 1089
FG CYIL ++ +K D K+ GIFLGYS S+AYRV+NS T+ V ES++V DD P
Sbjct: 922 FGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLSPA 981
Query: 1090 SKTSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSPIVQN 1149
K + + + DA++S + +++ S+S + S I Q
Sbjct: 982 RKKDVEEDVRTSGDNVADAAKSGENAEN-------------------SDSATDESNINQP 1022
Query: 1150 ESASEDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPK 1209
+ S + + HP+ELIIG + TRS + S +S IEPK
Sbjct: 1023 DKRSST-------------RIQKMHPKELIIGDPNRGVTTRSREVEIVSNSCFVSKIEPK 1069
Query: 1210 TVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNK 1269
V+EAL+D+ WI AMQEEL QF+RN+VW+LVP+P N+I TKW+F+NK NE+G +TRNK
Sbjct: 1070 NVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNK 1129
Query: 1270 ARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEE 1329
ARLVAQGY+Q EG+++ ETFAPVARLE+IRLLL A LYQMDVKS FLNG + EE
Sbjct: 1130 ARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEE 1189
Query: 1330 VYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRR 1389
VYV+QP GF D HPDHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF +
Sbjct: 1190 VYVEQPKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVK 1249
Query: 1390 TLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGV 1449
++++I QIYVDDI+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++ +
Sbjct: 1250 QDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSI 1309
Query: 1450 YVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTAS 1509
++ Q++Y K ++KKF +E+ TP LSK++ GT VDQK YR MIGSLLYLTAS
Sbjct: 1310 FLSQSRYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQKPYRSMIGSLLYLTAS 1369
Query: 1510 RPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYA 1569
RPDI ++V +CAR+Q++P+ SHL VKRI +Y+ GT++ G++Y L+G+CDAD+A
Sbjct: 1370 RPDITYAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSSSMLVGYCDADWA 1429
Query: 1570 GDRIERKSTSGNCQFLGENLIS 1591
G +RKSTSG C +LG NLIS
Sbjct: 1430 GSADDRKSTSGGCFYLGNNLIS 1451
>gb|AAO73527.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 966 bits (2497), Expect = 0.0
Identities = 594/1581 (37%), Positives = 850/1581 (53%), Gaps = 172/1581 (10%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG D T + +L
Sbjct: 17 DGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKEEDEL 76
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ VK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKMSRLQLLATKFEN 136
Query: 152 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ KKSK++A S E EE+ D D+
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-AEKKSKNLAFVSN------------DEGEEDEYDLDT 243
Query: 272 DEDQSVKMAMLSNKLEYLARKQKK----------FLSKRGSYKNSKKEDQKG-------C 314
DE + + +L + + + K F ++GS K K+ D K C
Sbjct: 244 DEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGS-KYQKRSDVKPSHSKGIQC 302
Query: 315 FNCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH IA+CP K+ KG S S D +SE SD
Sbjct: 303 HGCEGYGHIIAECPTHLKKHRKGLSVCQS-------------------DTESEQESD--- 340
Query: 375 ADDDAKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
+D D A G+ T +ED ++ S+I EL S ++L E +
Sbjct: 341 SDRDVNALTGIFET------------AEDSSDTDSEITFDELAASYRKLCIKSEKILQQE 388
Query: 435 TDLKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNK 494
LK+ DL ++++ E+ + E+ N T + +K L NK
Sbjct: 389 AQLKKVIADLEAEKEAHEEEISELKGEVGFLNSKLETMKKSIKML-------------NK 435
Query: 495 HEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLKS 552
LD+ ++ G KN G +G+G++ + + ++ +
Sbjct: 436 GSDTLDEVLLLG--------------KNAGNQRGLGFNPKFAGRTTM------------T 469
Query: 553 TFVP-EGTNAVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQ 611
FVP + T+ Q G+Q K KS K + +
Sbjct: 470 EFVPAKNRTGTTMSQHLSRHHGTQQK--------------KSKRKKWRCHYCGK-----Y 510
Query: 612 NLIKPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRSWY 671
IKP ++ S K + K K ++ L +H + A+E WY
Sbjct: 511 GHIKPFCYHLHGHPHHGTQSSNSRKKMMWVPKHKAVS----LVVHTSLRASAKE---DWY 563
Query: 672 LDSGCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDGL 730
LDSGCSRHMTG K L + V FG GKIIG G + + + S+N V LV GL
Sbjct: 564 LDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKGL 623
Query: 731 KHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCL 790
NL+SISQ CD ++V F+K+ C LV + + KG R ++ + CL
Sbjct: 624 TANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSKDNCYLWTPQETSYSSTCL 682
Query: 791 LSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKS 850
S D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK S +
Sbjct: 683 SSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSHQK 742
Query: 851 KDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSS 910
+TSR LELLH+DL GP+ SL G +Y V+VDD+SR+TWV FI+ K EVF
Sbjct: 743 LRHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSETFEVFKE 802
Query: 911 FCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRT 970
++Q EK+ I ++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNRT
Sbjct: 803 LSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQNGIVERKNRT 862
Query: 971 LQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQF 1030
LQE AR M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P++ +FH F
Sbjct: 863 LQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFHIF 922
Query: 1031 GCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPGS 1090
G CYIL ++ +K D K+ GIFLGYS S+AYRV+NS T+ V ES++V DD P
Sbjct: 923 GSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLSPAR 982
Query: 1091 KTSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSPIVQNE 1150
K + + + DA++S + +++ S+S + S I Q +
Sbjct: 983 KKDVEEDVRTLGDNVADAAKSGENAEN-------------------SDSATDESNINQPD 1023
Query: 1151 SASEDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKT 1210
S + + HP+ELIIG + TRS + S +S IEPK
Sbjct: 1024 KRSST-------------RIQKMHPKELIIGDPNRGVTTRSREVEIVSNSCFVSKIEPKN 1070
Query: 1211 VEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNKA 1270
V+EAL+D+ WI AMQEEL QF+RN+VW+LVP+P N+I TKW+F+NK NE+G +TRNKA
Sbjct: 1071 VKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKA 1130
Query: 1271 RLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEEV 1330
RLVAQGY+Q EG+++ ETFAPVARLE+IRLLL A LYQMDVKS FLNG + EEV
Sbjct: 1131 RLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEV 1190
Query: 1331 YVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRT 1390
YV+QP GF D HPDHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF +
Sbjct: 1191 YVEQPKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQ 1250
Query: 1391 LKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVY 1450
++++I QIYVDDI+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++ ++
Sbjct: 1251 DAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIF 1310
Query: 1451 VHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASR 1510
+ Q++Y K ++KKF +E+ TP LSK++ GT VDQ LYR MIGSLLYLTASR
Sbjct: 1311 LSQSRYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASR 1370
Query: 1511 PDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAG 1570
PDI ++V +CAR+Q++P+ SHLT VKRI +Y+ GT++ G++Y + L+G+CDAD+AG
Sbjct: 1371 PDITYAVGVCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAG 1430
Query: 1571 DRIERKSTSGNCQFLGENLIS 1591
+RKSTSG C +LG NLIS
Sbjct: 1431 SADDRKSTSGGCFYLGNNLIS 1451
>gb|AAG52949.1| gag/pol polyprotein [Arabidopsis thaliana]
Length = 1643
Score = 965 bits (2494), Expect = 0.0
Identities = 588/1589 (37%), Positives = 856/1589 (53%), Gaps = 127/1589 (7%)
Query: 41 FSWWKTNMYSFIMGLDEELWDIL-----------EDGVDDLDLDEEGAAIDRRIHTPAQK 89
+ WK M + I GL +E W E+G D L +++ T A++
Sbjct: 21 YGHWKVKMRALIRGLGKEAWIATSVGWKAPVVKGENGEDVLKTEDQW--------TDAEE 72
Query: 90 KLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQY 149
+ + +I S+ + ++ ++ + +AK + L +EG+ +VK ++ ML Q+
Sbjct: 73 AKATANSRALSLIFNSVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQF 132
Query: 150 ELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAK 209
E M + E+IEE + + S L K Y V K+LR LPSR+ K TA+ +
Sbjct: 133 ENLTMDESENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSL 192
Query: 210 DLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEE--ESP 267
D +T+ E++V L+ +E+ + KG SK +SE E E
Sbjct: 193 DTDTIDFEEVVGMLQAYELEITS-------------GKGGYSKGVALAVSSEKNEIQELK 239
Query: 268 DGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADC 327
D S ++ AM + AR Q + + K + C C+ GH A+C
Sbjct: 240 DSMSMMAKNFSRAMKRVEKRGFARNQGSDRDRDRDRDRNSKRSEIQCHECQGYGHIKAEC 299
Query: 328 PDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMGLVA 387
P L+++ K S+ +KF KS +S+S SD E++++D K + V
Sbjct: 300 PSLKRKDLKC-SECRGIGHTKFDCIGSKSKPDRSYIAESDSDSDDEDSEEDVKGFVSFVG 358
Query: 388 TVSSEAVSEAESDSE---DENEVYSKIPRQELVDSLKELLSLFEH---RTNELTDLKEKY 441
+ + VS SDSE ++ E+ + +D E L+E+ + E E+
Sbjct: 359 IIEDDNVSSDSSDSEVGCEKEEISADDESDVEMDVDGEFRKLYENWLVLSKEKVIWLEEK 418
Query: 442 VDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDK-GSGNKHEIALD 500
V + +Q + EL + + L + E++ + L Q L + K NK LD
Sbjct: 419 VKVQEQIEQLKGELAVANQIKSEMILKYSAKEEKNRELSQDLSDTRKKIHMLNKGTKDLD 478
Query: 501 DFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTN 560
+ AG +V + G+GY S K+ FV
Sbjct: 479 SILAAG----RVGKSNF--------GLGYHGGGSST--------------KTNFVRSKAA 512
Query: 561 AVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIKPESKI 620
A T QS + + K +N ++ + R + + + +++
Sbjct: 513 APTQSQSVFRSKSNSVPARRKYQNQN-HYHSQRTVTGYECYYCGRHGHIQRYCYRYAARL 571
Query: 621 PKQKDQKNKAATASEKTIP-KGVKPKVLNDQKPLSIHPKVCLRAREKQRSWYLDSGCSRH 679
K K Q K P +G K+ ++ L H A ++ WY DSG SRH
Sbjct: 572 SKLKRQG--------KLYPHQGRNSKMYVRREDLYCHVAYTSIAEGVKKPWYFDSGASRH 623
Query: 680 MTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSIS-INNVWLVDGLKHNLLSIS 738
MTG +A + V FGG G+I G G + + + NV+ V+GL NL+S+S
Sbjct: 624 MTGSQANLNNYSSVKESNVMFGGGAKGRIKGKGDLTETEKPHLTNVYFVEGLTANLISVS 683
Query: 739 QFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKW 798
Q CD V+F+K C N+ +++ + N Y + ++ +CL + +
Sbjct: 684 QLCDEGLTVSFNKVKCWATNERNQNTLTGVRTGNNCY------MWEEPKICLRAEKEDPV 737
Query: 799 VWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSR 858
+WH+RLGH N R +SK+ ++V+G+P + + +CGAC +GK ++ K + + T++
Sbjct: 738 LWHQRLGHMNARSMSKLVNKEMVRGVPELKHIEKIVCGACNQGKQIRVQHKRVEGIQTTQ 797
Query: 859 PLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSE 918
L+L+H+DL GP+ T S+ G +Y V+VDD+SR+ WV+FI+ K F Q+++E
Sbjct: 798 VLDLIHMDLMGPMQTESIAGKRYVFVLVDDFSRYAWVRFIREKSETANSFKILALQLKNE 857
Query: 919 KELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTM 978
K++ I ++RSD GGEF NE F FCE GI H++S+PRTPQ NGVVERKNRTLQEMAR M
Sbjct: 858 KKMGIKQIRSDRGGEFMNEAFNSFCESQGIFHQYSAPRTPQSNGVVERKNRTLQEMARAM 917
Query: 979 IHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILN 1038
IH N + + FWAEA++T+CY+ NR+Y+R +KT YE++KG++PN+SYF FGC CYI+N
Sbjct: 918 IHGNGVPEKFWAEAISTACYVINRVYVRLGSDKTPYEIWKGKKPNLSYFRVFGCVCYIMN 977
Query: 1039 TKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDD------------- 1085
KD L KFD++++ G FLGY+ S AYRV+N + +EESM+V FDD
Sbjct: 978 DKDQLGKFDSRSEEGFFLGYATNSLAYRVWNKQRGKIEESMNVVFDDGSMPELQIIVRNR 1037
Query: 1086 REPGSKTSEQ--SESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAES 1143
EP + S E N D+ D ++S + SD E P A+ + + S
Sbjct: 1038 NEPQTSISNNHGEERNDNQFDNGDINKSGEESDEE-------------VPPAQVHRDHAS 1084
Query: 1144 SPIVQNESASEDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIG-L 1202
I+ + S + Q Q + G K R S EE +
Sbjct: 1085 KDIIGDPSGERVTRGVKQDYRQ-------------LAGIKQKHRVMASFACFEEIMFSCF 1131
Query: 1203 LSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQ 1262
+SI+EPK V+EAL D WILAM+EEL +F R+ VWDLVP+P Q N+I TKW+F+NK +E
Sbjct: 1132 VSIVEPKNVKEALEDHFWILAMEEELEEFSRHQVWDLVPRPPQVNVIGTKWIFKNKFDEV 1191
Query: 1263 GEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFL 1322
G +TRNKARLVAQGY+Q EG+++ ETFAPVARLE IR LL A G L+QMDVK FL
Sbjct: 1192 GNITRNKARLVAQGYTQVEGLDFDETFAPVARLECIRFLLGTACGMGFKLHQMDVKCAFL 1251
Query: 1323 NGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQV 1382
NG+IEEEVYV+QP GFE+L+ P++VYKLKK+LYGLKQAPRAWY+RL+ FLI + RG V
Sbjct: 1252 NGIIEEEVYVEQPKGFENLEFPEYVYKLKKALYGLKQAPRAWYERLTTFLIVQGYTRGSV 1311
Query: 1383 DTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQI 1442
D TLF + I+I+QIYVDDI+FG T+ L K F K M EF MSM+GELK+FLG+QI
Sbjct: 1312 DKTLFVKNDVHGIIIIQIYVDDIVFGGTSDKLVKTFVKTMTTEFRMSMVGELKYFLGLQI 1371
Query: 1443 NQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGS 1502
NQ+ EG+ + Q+ Y + L+K+F + K TPM T L K++ G VD+KLYRGMIGS
Sbjct: 1372 NQTDEGITISQSTYAQNLVKRFGMCSSKPAPTPMSTTTKLFKDEKGVKVDEKLYRGMIGS 1431
Query: 1503 LLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIG 1562
LLYLTA+RPD+ SV LCAR+QS+P+ SHL AVKRI +Y+ GT N GL Y + L+G
Sbjct: 1432 LLYLTATRPDLCLSVGLCARYQSNPKASHLLAVKRIIKYVSGTINYGLNYTRDTSLVLVG 1491
Query: 1563 FCDADYAGDRIERKSTSGNCQFLGENLIS 1591
+CDAD+ G+ +R+ST+G FLG NLIS
Sbjct: 1492 YCDADWGGNLDDRRSTTGGVFFLGSNLIS 1520
>gb|AAO73529.1| gag-pol polyprotein [Glycine max]
Length = 1577
Score = 955 bits (2469), Expect = 0.0
Identities = 589/1578 (37%), Positives = 855/1578 (53%), Gaps = 165/1578 (10%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG + T + +L
Sbjct: 17 DGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDEL 76
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ VK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLATKFEN 136
Query: 152 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMTILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ T KKSK++A S E EE+ D D+
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-TEKKSKNLAFVSN------------DEGEEDEYDLDT 243
Query: 272 DEDQSVKMAMLSNK----LEYLARKQKKFLS------KRGSYKNSKKEDQ----KG--CF 315
DE + + L + L + R+QK + ++GS K +++ KG C
Sbjct: 244 DEGLTNAVVFLGKQFNKVLNRMDRRQKPHVRNISLDIRKGSEYQRKSDEKPSHSKGIQCR 303
Query: 316 NCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEA 375
C+ GH A+CP K++ KG S S +D +SE SD +
Sbjct: 304 GCEGYGHIKAECPTHLKKQRKGLSVCRS------------------DDTESEQESD---S 342
Query: 376 DDDAKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELT 435
D D A G + +ED ++ S+I EL +EL E +
Sbjct: 343 DRDVNALTGRFES------------AEDSSDTDSEITFDELAIFYRELCIKSEKILQQEA 390
Query: 436 DLKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKH 495
LK+ +L ++++ E+ + E+ GF +++ E+ KS+ + +KGS
Sbjct: 391 QLKKVIANLEAEKEAHEEEISKLKGEV-GF--LNSKLENMTKSI-----KMLNKGSD--- 439
Query: 496 EIALDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFV 555
LD+ + G KV + +G+G++ + + ++ + FV
Sbjct: 440 --MLDZVLQLG---KKVGNQ---------RGLGFNHKSAGRTTM------------TEFV 473
Query: 556 P-EGTNAVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLI 614
P + + T+ Q + G+Q K SK + + K I
Sbjct: 474 PAKNSTGATMSQHRSRHHGTQQK-RSKRKKWRCHYCGK------------------YGHI 514
Query: 615 KPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRSWYLDS 674
KP ++S + + K K+++ L +H + A+E WYLDS
Sbjct: 515 KPFCYHLHGHPHHGTQGSSSGRKMMWVPKHKIVS----LVVHTSLRASAKE---DWYLDS 567
Query: 675 GCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDGLKHN 733
GCSRHMTG K + + V FG GKI G G + + + S+N V LV GL N
Sbjct: 568 GCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLVHEGLPSLNKVLLVKGLTVN 627
Query: 734 LLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSM 793
L+SISQ CD ++V F+K+ C LV + + KG R ++ + + CL S
Sbjct: 628 LISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSKDNCYLWTPQESSHSSTCLFSK 686
Query: 794 NDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDI 853
D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK S +
Sbjct: 687 EDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLQH 746
Query: 854 VSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCT 913
+TSR LELLH+DL GP+ SL G +Y V+VDD+SR+TWV FI+ K EVF
Sbjct: 747 QTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSDTFEVFKELSL 806
Query: 914 QIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQE 973
++Q EK+ I ++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNRTLQE
Sbjct: 807 RLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQE 866
Query: 974 MARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCT 1033
AR M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P + +FH FG
Sbjct: 867 AARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKHFHIFGSP 926
Query: 1034 CYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPGSKTS 1093
CYIL ++ +K D K+ GIFLGYS S+AYRV+NS T+ V ES++V DD P K
Sbjct: 927 CYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLTPARK-- 984
Query: 1094 EQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSPIVQNESAS 1153
D E+ + A+ AE+ +N ++
Sbjct: 985 ---------------------KDVEEDVRTSGDNVADTAKSAEN---------AENSDSA 1014
Query: 1154 EDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEE 1213
D + Q +P + + HP+ELIIG + TRS + S +S IEPK V+E
Sbjct: 1015 TDEPNINQPDKRPSIRIQKMHPKELIIGDPNRGVTTRSREIEIVSNSCFVSKIEPKNVKE 1074
Query: 1214 ALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLV 1273
AL+D+ WI AMQEEL QF+RN+VW+LVP+P N+I TKW+F+NK NE+G +TRNKARLV
Sbjct: 1075 ALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLV 1134
Query: 1274 AQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVK 1333
AQGY+Q EG+++ ETFAPVARLE+IRLLL A LYQMDVKS FLNG + EE YV+
Sbjct: 1135 AQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVE 1194
Query: 1334 QPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKK 1393
QP GF D HPDHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF + +
Sbjct: 1195 QPKGFVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAE 1254
Query: 1394 DILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQ 1453
+++I QIYVDDI+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q
Sbjct: 1255 NLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQ 1314
Query: 1454 TKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDI 1513
+KY K ++KKF +E+ TP LSK++ GT VDQ LYR MIGSLLYLTASRPDI
Sbjct: 1315 SKYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDI 1374
Query: 1514 LFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRI 1573
++V +CAR+Q++P+ SHL VKRI +Y+ GT++ G++Y L+G+CDAD+AG
Sbjct: 1375 TYAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSGSMLVGYCDADWAGSAD 1434
Query: 1574 ERKSTSGNCQFLGENLIS 1591
+RKSTSG C +LG NLIS
Sbjct: 1435 DRKSTSGGCFYLGNNLIS 1452
>gb|AAO73525.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 943 bits (2437), Expect = 0.0
Identities = 590/1582 (37%), Positives = 856/1582 (53%), Gaps = 174/1582 (10%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG + T + +L
Sbjct: 17 DGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDEL 76
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAK-AMFASLCANFEGSKNVKEAKALMLVHQYE 150
+ K + + + + ++ + AK A L EG+ VK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDACGEILKTTHEGTSKVKMSRLQLLATKFE 136
Query: 151 LFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKD 210
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D
Sbjct: 137 NLKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQD 196
Query: 211 LNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGD 270
+ + V++L+ SL+ E+ L++ KKSK++A S E EE+ D D
Sbjct: 197 ICNMRVDELIGSLQTFELGLSDRN-EKKSKNLAFVSN------------DEGEEDEYDLD 243
Query: 271 SDEDQSVKMAMLSNK----LEYLARKQKK------FLSKRGSYKNSKKEDQ----KG--C 314
+DE + + +L + L + R+QK F ++GS + K +++ KG C
Sbjct: 244 TDEGLTNAVGLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYHKKSDEKPSHSKGIQC 303
Query: 315 FNCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH A+CP K++ KG S S +D +SE SD
Sbjct: 304 HGCEGYGHIKAECPTHLKKQRKGLSVCRS------------------DDTESEQESD--- 342
Query: 375 ADDDAKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
+D D A G E++ DS D +I EL S ++L E +
Sbjct: 343 SDRDVNALTGRF---------ESDEDSSD-----IEITFDELAISYRKLCIKSEKILQQE 388
Query: 435 TDLKEKYVDLMKQQKSTLLELKASEEELKG-FNLISTTYEDRLKSLCQKLQEKCDKGSGN 493
LK+ +L ++++ E+ ELKG +++ E+ KS+ + +KGS
Sbjct: 389 AQLKKVIANLEAEKEAHEEEIS----ELKGEVGFLNSKLENMTKSI-----KMLNKGSD- 438
Query: 494 KHEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLK 551
LD+ + G KN G +G+G++ + + ++
Sbjct: 439 ----MLDEVLQLG--------------KNVGNQRGLGFNHKSACRITM------------ 468
Query: 552 STFVP-EGTNAVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVH 610
+ FVP + + T+ Q + G+Q K KS K + +
Sbjct: 469 TEFVPAKNSTGATMSQHRSRHHGTQQK--------------KSKRKKWRCHYCGK----- 509
Query: 611 QNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRSW 670
IKP +++S + + K K+++ L +H + A+E W
Sbjct: 510 YGHIKPFCYHLHGHPHHGTQSSSSGRKMMWVPKHKIVS----LVVHTSLRASAKE---DW 562
Query: 671 YLDSGCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDG 729
YLDSGCSRHMTG K + + V FG GKI G G + + + S+N V LV G
Sbjct: 563 YLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLVHDGLPSLNKVLLVKG 622
Query: 730 LKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVC 789
L NL+SISQ CD ++V F+K+ C LV + + KG R ++ + C
Sbjct: 623 LTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSKDNCYLWTPQETSYSSTC 681
Query: 790 LLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFK 849
L S D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK S +
Sbjct: 682 LSSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSHQ 741
Query: 850 SKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFS 909
+TS LELLH+DL GP+ SL G +Y V+VDD+SR+TWV FI+ K EVF
Sbjct: 742 KLQHQTTSMVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSDTFEVFK 801
Query: 910 SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNR 969
++Q EK+ I ++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNR
Sbjct: 802 ELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVERKNR 861
Query: 970 TLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQ 1029
TLQE R M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P + +FH
Sbjct: 862 TLQEATRVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKHFHI 921
Query: 1030 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPG 1089
FG CYIL ++ +K D K+ GIFLGYS S+AYRV+NS T+ V ES++V DD P
Sbjct: 922 FGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLTPA 981
Query: 1090 SKTSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSPIVQN 1149
K + + + D ++S E++ +++ T + + ++ + SP ++
Sbjct: 982 RKKDVEEDVRTSEDNVADTAKS-----------AENAEKSDSTTDEPNINQPDKSPFIRI 1030
Query: 1150 ESASEDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPK 1209
Q +QPK ELIIG + TRS + S +S IEPK
Sbjct: 1031 ------------QKMQPK---------ELIIGDPNRGVTTRSREIEIVSNSCFVSKIEPK 1069
Query: 1210 TVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNK 1269
V+EAL+D+ WI AMQEEL QF+RN+VW+LVP+P N+I TKW+F+NK NE+G +TRNK
Sbjct: 1070 NVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNK 1129
Query: 1270 ARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEE 1329
ARLVAQGY+Q EG+++ ETFAPVARLE+IRLLL A LYQMDVKS FLNG + EE
Sbjct: 1130 ARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEE 1189
Query: 1330 VYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRR 1389
YV+QP GF D H DHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF +
Sbjct: 1190 AYVEQPKGFVDPTHLDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVK 1249
Query: 1390 TLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGV 1449
++++I QIYVDDI+FG + + + F MQ EFEMS++GEL +FLG+Q+ Q ++ +
Sbjct: 1250 QDAENLMIAQIYVDDIVFGGMSNEMLRHFVPQMQSEFEMSLVGELHYFLGLQVKQMEDSI 1309
Query: 1450 YVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTAS 1509
++ Q+KY K ++KKF +E+ TP LSK++ GT VDQ LYR MIGSLLYLTAS
Sbjct: 1310 FLSQSKYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQNLYRSMIGSLLYLTAS 1369
Query: 1510 RPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYA 1569
RPDI F+V +CAR+Q++P+ SHL VKRI +Y+ GT++ G++Y D L+G+CDAD+A
Sbjct: 1370 RPDITFAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWA 1429
Query: 1570 GDRIERKSTSGNCQFLGENLIS 1591
G +RK TSG C +LG NLIS
Sbjct: 1430 GSADDRKCTSGGCFYLGTNLIS 1451
>gb|AAC64917.1| gag-pol polyprotein [Glycine max]
Length = 1550
Score = 939 bits (2428), Expect = 0.0
Identities = 589/1571 (37%), Positives = 844/1571 (53%), Gaps = 173/1571 (11%)
Query: 48 MYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKLYKKHHKIRGII 102
M +F+ LD W + G + LD EG + T + +L + K +
Sbjct: 1 MVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDELALGNSKALNAL 60
Query: 103 VASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYELFRMKDDESIEE 162
+ + + ++ + AK + L EG+ VK ++ +L ++E +MK++E I E
Sbjct: 61 FNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLATKFENLKMKEEECIHE 120
Query: 163 MYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSS 222
+ + + L + V KILRSLP R+ KVTAIEEA+D+ + V++L+ S
Sbjct: 121 FHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQDICNMRVDELIGS 180
Query: 223 LKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAML 282
L+ E+ L++ T KKSK++A S E EE+ D D+DE + + +L
Sbjct: 181 LQTFELGLSDR-TEKKSKNLAFVSN------------DEGEEDEYDLDTDEGLTNAVVLL 227
Query: 283 SNK----LEYLARKQKK------FLSKRGSYKNSKKEDQKG-------CFNCKKPGHFIA 325
+ L + R+QK F ++GS + K+ D+K C C+ GH A
Sbjct: 228 GKQFNKVLNRMDRRQKPHVRNIPFDIRKGS-EYQKRSDEKPSHSKGIQCHGCEGYGHIKA 286
Query: 326 DCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAMGL 385
+CP K++ KG S S +D +SE SD +D D A G
Sbjct: 287 ECPTHLKKQRKGLSVCRS------------------DDTESEQESD---SDRDVNALTGR 325
Query: 386 VATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLM 445
+ +ED ++ S+I EL S +EL E + LK+ +L
Sbjct: 326 FES------------AEDSSDTDSEITFDELAISYRELCIKSEKILQQEAQLKKVIANLE 373
Query: 446 KQQKSTLLELKASEEELKG-FNLISTTYEDRLKSLCQKLQEKCDKGSGNKHEIALDDFIM 504
++++ E+ ELKG +++ E+ KS+ + +KGS LD+ +
Sbjct: 374 AEKEAHEDEIS----ELKGEIGFLNSKLENMTKSI-----KMLNKGSD-----LLDEVLQ 419
Query: 505 AGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVP-EGTNA 561
G KN G +G+G++ + + ++ + FVP + +
Sbjct: 420 LG--------------KNVGNQRGLGFNHKSAGRTTM------------TEFVPAKNSTG 453
Query: 562 VTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIKPESKIP 621
T+ Q + G+Q K KS K + + IKP
Sbjct: 454 ATMSQHRSRHHGTQQK--------------KSKRKKWRCHYCGK-----YGHIKPFCYHL 494
Query: 622 KQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRSWYLDSGCSRHMT 681
+++S G K + K +S+ LRA K+ WYLDSGCSRHMT
Sbjct: 495 HGHPHHGTQSSSS------GRKMMWVPKHKTVSLVVHTSLRASAKE-DWYLDSGCSRHMT 547
Query: 682 GEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDGLKHNLLSISQF 740
G K + + V FG GKI G G + + + S+N V LV GL NL+SISQ
Sbjct: 548 GVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLVHDGLPSLNKVLLVKGLTANLISISQL 607
Query: 741 CDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVW 800
CD ++V F+K+ C LV + + KG R ++ + CL S D+ +W
Sbjct: 608 CDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSKDNCYLWTPQETSYSSTCLFSKEDEVKIW 666
Query: 801 HKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPL 860
H+R GH + R + KI V+G+PN+ +CG CQ GK VK S + +TSR L
Sbjct: 667 HQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSNQKLQHQTTSRVL 726
Query: 861 ELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKE 920
ELLH+DL GP+ SL +Y V+VDD+SR+TWV FI+ K EVF ++Q EK+
Sbjct: 727 ELLHMDLMGPMQVESLGRKRYAYVVVDDFSRFTWVNFIREKSDTFEVFKELSLRLQREKD 786
Query: 921 LKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIH 980
I ++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNRTLQE AR M+H
Sbjct: 787 CVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLH 846
Query: 981 ENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTK 1040
L + WAEA+NT+CYI NR+ +R T YE++KGR+P + +FH G CYIL +
Sbjct: 847 AKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKHFHICGSPCYILADR 906
Query: 1041 DYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPGSKTSEQSESNA 1100
+ +K D K+ GIFLGYS S+AYRV+NS T+ V ES++V DD P K
Sbjct: 907 EQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLTPARK--------- 957
Query: 1101 GTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSPIVQNESASEDFQDNT 1160
D E+ + A+ AE+ +N ++ D +
Sbjct: 958 --------------KDVEEDVRTSGDNVADTAKSAEN---------AENSDSATDEPNIN 994
Query: 1161 QQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGW 1220
Q +P + + HP+ELIIG + TRS + S +S IEPK V+EAL+D+ W
Sbjct: 995 QPDKRPSIRIQKMHPKELIIGDPNRGVTTRSREIEIISNSCFVSKIEPKNVKEALTDEFW 1054
Query: 1221 ILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQ 1280
I AMQEEL QF+RN+VW+LVP+P N+I TKW+F+NK NE+G +TRNKARLVAQGY+Q
Sbjct: 1055 INAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQI 1114
Query: 1281 EGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQPPGFED 1340
EG+++ ETFAP ARLE+IRLLL A LYQMDVKS FLNG + EE YV+QP GF D
Sbjct: 1115 EGVDFDETFAPGARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVD 1174
Query: 1341 LKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQI 1400
HPDHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF + ++++I QI
Sbjct: 1175 PTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQI 1234
Query: 1401 YVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKEL 1460
YVDDI+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q+KY K +
Sbjct: 1235 YVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYAKNI 1294
Query: 1461 LKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLC 1520
+KKF +E+ TP LSK++ GT VDQ LYR MIGSLLYLTASRPDI ++V C
Sbjct: 1295 VKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGGC 1354
Query: 1521 ARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSG 1580
AR+Q++P+ SHL VKRI +Y+ GT++ G++Y D L+G+CDAD+AG +RKST G
Sbjct: 1355 ARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSVDDRKSTFG 1414
Query: 1581 NCQFLGENLIS 1591
C +LG N IS
Sbjct: 1415 GCFYLGTNFIS 1425
>gb|AAN60991.1| Putative Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)] gi|34902324|ref|NP_912508.1| Putative
Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)]
Length = 2011
Score = 921 bits (2381), Expect = 0.0
Identities = 587/1589 (36%), Positives = 839/1589 (51%), Gaps = 161/1589 (10%)
Query: 29 GKAPKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQ 88
GKAP FNG +S WK M + + + +W I++ G AI T
Sbjct: 9 GKAPMFNGT--NYSTWKIKMSTHLKAMSFHIWSIVDVGF----------AITGTPLTEID 56
Query: 89 KKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQ 148
+ K + + ++ S+ + E+ ++S+ TA ++ L E + K+AK L Q
Sbjct: 57 HRNLKLNAQAMNVLFNSLSQEEFDRVSNLETAYEIWNKLAEIHESTSEYKDAKLHFLKIQ 116
Query: 149 YELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEA 208
YE F M ES+ +MY R +V+ L+ L +Y + K+LR+LP ++ VT + +
Sbjct: 117 YETFSMLPHESVNDMYGRLNVIVNDLKGLGANYTNLEVAQKMLRALPEKYETLVTMLINS 176
Query: 209 KDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPD 268
D++ ++ L+ + ++M ++ KK A PSK + ++ S+S+ +
Sbjct: 177 -DMSRMTPASLLGKINTNDM----YKLKKKEMEEASPSKKCIALQAEVEDKSKSKVNEVN 231
Query: 269 GDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKG-------CFNCKKPG 321
D +E+ + +L+ + L ++K+ RGS N ++ + CF C
Sbjct: 232 KDLEEE----IVLLARRFNDLLGRRKE--RGRGSNSNRRRNRRPNKTLSNLRCFEC---- 281
Query: 322 HFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKA 381
D +E + K+ K +++A E A
Sbjct: 282 -------DSNEESSASSGSEEEGGDDASSKKKKMAVVAIKEA-------------PSLFA 321
Query: 382 AMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKY 441
+ L+A S+ S ++S+S+D+ + S EL+S+FE EL EK
Sbjct: 322 PLCLMAKSPSKVTSLSDSESDDDCDDVS----------YDELVSMFE----ELHAYSEKE 367
Query: 442 VDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKHEIALDD 501
+ K K L+ EELK T +RL +KL+E D AL D
Sbjct: 368 IIKFKTLKKDHASLEVLYEELK-------TSHERLTISHEKLKEAHDNLLSTTQHGALID 420
Query: 502 FIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNA 561
+G S CD + D +++
Sbjct: 421 -------------------------VGIS------------CDLLDDSATCHIAHVASSS 443
Query: 562 VTV----VQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIKPE 617
++ + P +S S ++ +L V+ ++ K Q K+ K E + +
Sbjct: 444 ISTSCDDLMDMPNSSSSSC-VSICDASL---VVENNELKEQVAKLNKSLERCFKGKNTLD 499
Query: 618 SKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQR--------- 668
+ +Q+ NK + KG KP + + + K C + RE
Sbjct: 500 KILSEQQCILNKEGLGF--ILKKGKKPSH-RATRFVKSNGKYCSKCREVGHLVNYRTGGS 556
Query: 669 SWYLDSGCSRHMTGEKALFLTLTM--KDGGEVKFGGNQTGKIIGTGTIGNSS-ISINNVW 725
W LDSGC++HMTG++A+F T + + +V FG N GK+IG G I S+ +SI+NV
Sbjct: 557 HWVLDSGCTQHMTGDRAMFTTFEVGGNEQEKVTFGDNSKGKVIGLGKIAISNDLSIDNVS 616
Query: 726 LVDGLKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQ 785
LV L NLLS++Q CD F + + DKS FKG R N+Y +F+
Sbjct: 617 LVKSLNFNLLSVAQICDLVLSCAFFPQEVIVSSLLDKSCVFKGFRYGNLYLGDFNSSEAN 676
Query: 786 KVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVK 845
CL++ W+WH+RL H +SK+SK LV GL ++ + D LC ACQ GK V
Sbjct: 677 LKTCLVAKTSLGWLWHRRLAHVGMNQLSKLSKRDLVVGLKDVKFEKDKLCSACQAGKQVA 736
Query: 846 SSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYAC 905
S +K I+STSRPLELLH++ FGP S+ G+ + LVIVDDYSR+TW+ F+ K
Sbjct: 737 CSHPTKSIMSTSRPLELLHMEFFGPTTYKSIGGNSHCLVIVDDYSRYTWMFFLHDKSIVA 796
Query: 906 EVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVE 965
E+F F + Q+E ++K+RSD+G +F+N E +C+ GI HE S+ +PQQNGVVE
Sbjct: 797 ELFKKFAKRGQNEFNCTLVKIRSDNGSKFKNTNIEDYCDDLGIKHELSATYSPQQNGVVE 856
Query: 966 RKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNIS 1025
KNRTL EMARTM+ E ++ FWAEA+NT+C+ NR+Y+ +L+KT+YEL GR+PN++
Sbjct: 857 MKNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRLYLHRLLKKTSYELIVGRKPNVA 916
Query: 1026 YFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDD 1085
YF FGC CYI L KF+++ G LGY+ SKAYRVYN VEE+ V+FD+
Sbjct: 917 YFRVFGCKCYIYRKGVRLTKFESRCDEGFLLGYASNSKAYRVYNKNKGIVEETADVQFDE 976
Query: 1086 REPGSKTSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSP 1145
GS+ ++ + G A ++ D K +VE P T + S S S
Sbjct: 977 TN-GSQEGHENLDDVGDEGLMRAMKNMSIGD-VKPIEVEDKPSTS-TQDEPSTSATPSQA 1033
Query: 1146 IVQNESASEDFQDNTQQVIQPKFKH---KSSHPEELIIGSKDSPRRTRSHFRQEESLIGL 1202
V+ E ++ Q + P H HP + ++G +TRS
Sbjct: 1034 QVEVE------EEKAQDLPMPPRIHTALSKDHPIDQVLGDISKGVQTRSRVASICEHYSF 1087
Query: 1203 LSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQ 1262
+S +EPK V+EAL D WI AM +ELN F RN VW LV + N+I TKWVFRNK +E
Sbjct: 1088 VSCLEPKHVDEALCDPDWINAMHKELNNFARNKVWTLVERLRDHNVIGTKWVFRNKQDEN 1147
Query: 1263 GEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFL 1322
G V RNKAR VAQG++Q EG+++ ETFAPV RLE I +LL++A I L+QMDVKS FL
Sbjct: 1148 GLVVRNKARFVAQGFTQVEGLDFGETFAPVTRLEAICILLAFASCFNIKLFQMDVKSAFL 1207
Query: 1323 NGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQV 1382
NG I E V+V+QPPGFED K+P+HVYKL K+LYGLKQAPRAWY+RL +FL+ DF+ G+V
Sbjct: 1208 NGEIAELVFVEQPPGFEDPKYPNHVYKLSKALYGLKQAPRAWYERLRDFLLSKDFKIGKV 1267
Query: 1383 DTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQI 1442
DTTLF + + D + QIYVDDIIFG TN CKEF +M EFEMSM+GEL FF G+QI
Sbjct: 1268 DTTLFTKIIGDDFFVCQIYVDDIIFGCTNEVFCKEFGDMMSREFEMSMIGELSFFHGLQI 1327
Query: 1443 NQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGS 1502
Q K+G F LED K + TPM L ++ G VD KLYR MIGS
Sbjct: 1328 KQLKDGT--------------FGLEDAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGS 1373
Query: 1503 LLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIG 1562
LLYLTASRPDI+FSVC+CARFQ+ P+E HL AVKRI RYLK ++ +GL Y K +KL+G
Sbjct: 1374 LLYLTASRPDIMFSVCMCARFQAAPKECHLVAVKRILRYLKHSSTIGLWYPKGAKFKLVG 1433
Query: 1563 FCDADYAGDRIERKSTSGNCQFLGENLIS 1591
+ D+DYAG +++RKSTSG+CQ LG +L+S
Sbjct: 1434 YSDSDYAGCKVDRKSTSGSCQMLGRSLVS 1462
>gb|AAV31277.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1577
Score = 903 bits (2333), Expect = 0.0
Identities = 580/1583 (36%), Positives = 821/1583 (51%), Gaps = 160/1583 (10%)
Query: 29 GKAPKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDG--VDDLDLDEEGAAIDRR---I 83
GKAP FNG +S WK M + + + +W I++ G + L E ID R +
Sbjct: 9 GKAPMFNGT--NYSTWKIKMSTHLKAMSFHIWSIVDVGFAITGTPLME----IDHRNLQL 62
Query: 84 HTPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKAL 143
+ A L+ S+ + E+ ++S+ TA ++ L EG+ K+AK
Sbjct: 63 NAQAMNALFN-----------SLSQEEFDRVSNLETAYEIWNKLAEIHEGTSEYKDAKLH 111
Query: 144 MLVHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVT 203
L QYE F M ES+ +MY R +V+ L+ L +Y + K+LR+LP ++ VT
Sbjct: 112 FLKIQYETFSMLPHESVNDMYGRLNVIVNDLKGLGANYTDLEVAQKMLRALPEKYETLVT 171
Query: 204 AIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESE 263
+ + D++ ++ L+ + ++M +GK S S++
Sbjct: 172 MLINS-DMSRMTPASLLGKINTNDMR---------------KERGKGSNSNR-------- 207
Query: 264 EESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHF 323
R + + +K CF C + GHF
Sbjct: 208 ------------------------------------RRNRRPNKTLSNLRCFECGEKGHF 231
Query: 324 IADCPDLQKEKFKGKSKKSS-FNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAA 382
+ CP + K KKS + K K+ K + A E+ DS S
Sbjct: 232 ASKCPSKDDDGDKSSKKKSGGYKLMKKLKKEGKKIEAFIEEWDSNEESSPHPGPRKKMVM 291
Query: 383 MGLVATVSSEAVSEAESDSEDENEV-YSKIPRQELVD-----SLKELLSLFEHRTNELTD 436
M + + V + K P E D S EL+S+FE EL
Sbjct: 292 MQAPGRRRWPLLPSRRLHHSSLHFVSWQKAPLSESDDDCDDVSYDELVSMFE----ELHA 347
Query: 437 LKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDKG-SGNKH 495
EK + K K L+ EELK T +RL +KL+E D S +H
Sbjct: 348 YSEKEIVKFKALKKDHASLEVLYEELK-------TSHERLTISHEKLKEAHDNLLSTTQH 400
Query: 496 EIALDDFIMAGI-DRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTF 554
+D I + D S + Y S+ + CD + D S+
Sbjct: 401 GALIDVGISCDLLDDSATFHIAYVASS----------------SISTSCDDLVDMSSSS- 443
Query: 555 VPEGTNAVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLI 614
++ V++ + ++ K N +++ K KI +R + L
Sbjct: 444 ---SSSCVSICDASLVVENNELKEQVAKLNKRLERCFKGKNTLDKILSEQRCILNKEGL- 499
Query: 615 KPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRSWYLDS 674
IPK+ + + AT K+ + + + D + + + W LDS
Sbjct: 500 ---GFIPKKGKKPSHRATRFVKSNGRLMMVALFLD---MWVPLYSVVNYHTGGSHWVLDS 553
Query: 675 GCSRHMTGEKALFLTLTM--KDGGEVKFGGNQTGKIIGTGTIGNSS-ISINNVWLVDGLK 731
GC++HMTG++A+F T + + +V FG N GK+IG G I S+ +SI+NV LV L
Sbjct: 554 GCTQHMTGDRAMFTTFEVGRNEQEKVTFGDNSKGKVIGLGKIAISNDLSIDNVSLVKSLN 613
Query: 732 HNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLL 791
NLLS++Q CD F + + DKS FKG R N+Y ++F+ CL+
Sbjct: 614 FNLLSVAQICDLSLSCAFFPQEVIVSSLLDKSCVFKGFRYGNLYLVDFNSSEANLKTCLV 673
Query: 792 SMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSK 851
+ W+WH+RL H +SK SK LV GL ++ + D LC ACQ GK V S +K
Sbjct: 674 AKTSLGWLWHRRLAHVGMNQLSKFSKRDLVMGLKDVKFEKDKLCSACQAGKQVACSHPTK 733
Query: 852 DIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSF 911
I+STS+PLELLH+DLF P S+ G+ + LVIVDDYSR+TWV F+ K ++F F
Sbjct: 734 SIMSTSKPLELLHMDLFDPTTYKSIGGNSHCLVIVDDYSRYTWVFFLHDKSIVADLFKKF 793
Query: 912 CTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTL 971
+ Q+E ++K+RS+ G EF+N E +C+ GI HE + +PQQNGVVERKNRTL
Sbjct: 794 AKRAQNEFSCTLVKIRSNIGSEFKNTNIEDYCDDLGIKHELFATYSPQQNGVVERKNRTL 853
Query: 972 QEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFG 1031
EMARTM+ E ++ FWAEA+NT+C+ NR+Y+ +L+KT+YE+ GR+PNI+YF FG
Sbjct: 854 IEMARTMLDEYGVSDSFWAEAINTACHATNRLYLHRVLKKTSYEVIVGRKPNIAYFRVFG 913
Query: 1032 CTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPGSK 1091
C CYI L KF+++ G LGY+ +SKAYRVYN VEE+ V+FD+ GS+
Sbjct: 914 CKCYIHRKGVRLTKFESRCDEGFLLGYASKSKAYRVYNKNKGIVEETADVQFDETN-GSQ 972
Query: 1092 TSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSPIVQNES 1151
++ + G ++ D K +VE P + S + S+ + +++
Sbjct: 973 EGHENLDDVGDEGLMRVMKNMSIGD-VKPIEVEDKP-------STSTQDEPSTSAMPSQA 1024
Query: 1152 ASEDFQDNTQQVIQPKFKH---KSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEP 1208
E ++ Q+ P H HP + ++G +TRS +S +E
Sbjct: 1025 QVEVEEEKAQEPPMPPRIHTALSKDHPIDQVLGDISKGVQTRSRVTSICEHYSFVSCLER 1084
Query: 1209 KTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRN 1268
K V+EAL D W+ AM EEL F RN VW LV +P N+I TKWVFRNK +E G V RN
Sbjct: 1085 KHVDEALCDPDWMNAMHEELKNFARNKVWTLVERPRDHNVIGTKWVFRNKQDENGLVVRN 1144
Query: 1269 KARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEE 1328
KARLVAQG++Q EG+++ ETFAPVARLE I +LL++A I L+QMDVKS FLN
Sbjct: 1145 KARLVAQGFTQVEGLDFGETFAPVARLEAICILLAFASCFDIKLFQMDVKSAFLN----- 1199
Query: 1329 EVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFR 1388
D K+P+HVYKL K+LYGL+QAPRAWY+RL +FL+ DF+ G+VD TLF
Sbjct: 1200 -----------DTKYPNHVYKLSKALYGLRQAPRAWYERLRDFLLSKDFKIGKVDITLFT 1248
Query: 1389 RTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEG 1448
+ + D + QIYVDDIIFGSTN CKEF +M EFEMSM+GEL FFLG+QI Q K G
Sbjct: 1249 KIIGDDFFVYQIYVDDIIFGSTNEVFCKEFGDMMSREFEMSMIGELSFFLGLQIKQLKNG 1308
Query: 1449 VYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTA 1508
+V QTKY K+LLK+F LED K + TPM L ++ G VD KLYR MIGSLLYLT
Sbjct: 1309 TFVSQTKYIKDLLKRFGLEDAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGSLLYLTV 1368
Query: 1509 SRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADY 1568
SRPDI+FSVC+CARFQ+ P+E HL AVKRI RYLK ++ +GL Y K +KL+G+ D DY
Sbjct: 1369 SRPDIMFSVCMCARFQAAPKECHLVAVKRILRYLKHSSTIGLWYPKGAKFKLVGYSDPDY 1428
Query: 1569 AGDRIERKSTSGNCQFLGENLIS 1591
AG +++RKSTS +CQ LG +L+S
Sbjct: 1429 AGCKVDRKSTSSSCQMLGRSLVS 1451
>gb|AAD20307.1| copia-type pol polyprotein [Zea mays]
Length = 1063
Score = 881 bits (2276), Expect = 0.0
Identities = 473/948 (49%), Positives = 610/948 (63%), Gaps = 48/948 (5%)
Query: 680 MTGEKALFLTLTMKDGGE--VKFGGNQTGKIIGTGTIGNS-SISINNVWLVDGLKHNLLS 736
MTGEK +F + + + FG G + G G I S SI+NV+LVD L +NLLS
Sbjct: 1 MTGEKRMFSSYEKNQDPQRAITFGDGNQGLVKGLGKIAISPDHSISNVFLVDSLDYNLLS 60
Query: 737 ISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDK 796
+SQ C Y+ F+ T+ + D SI FKG +Y ++F D A+ CL++ +
Sbjct: 61 VSQLCQMGYNCLFTDIGVTVFRRSDDSIAFKGVLEGQLYLVDF-DRAELDT-CLIAKTNM 118
Query: 797 KWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVST 856
W+WH+RL H + + K+ K + + GL N+ + D +C ACQ GK V + K+I++T
Sbjct: 119 GWLWHRRLAHVGMKNLHKLLKGEHILGLTNVHFEKDRICSACQAGKQVGTHHPHKNIMTT 178
Query: 857 SRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQ 916
RPLELLH+DLFGP+ S+ GSKY LVIVDDYSR+TWV F++ K E F + Q
Sbjct: 179 DRPLELLHMDLFGPIAYISIGGSKYCLVIVDDYSRFTWVFFLQEKSQTQETLKGFLRRAQ 238
Query: 917 SEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 976
+E L+I K+RSD+G EF+N E F E+ GI HEFSSP TPQQNGVVERKNRTL +MAR
Sbjct: 239 NEFGLRIKKIRSDNGTEFKNSQIESFLEEEGIKHEFSSPYTPQQNGVVERKNRTLLDMAR 298
Query: 977 TMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYI 1036
TM+ E FWAEAVNT+CY NR+Y+ +L+KT+YEL G++PNISYF FG C+I
Sbjct: 299 TMLDEYKTPDRFWAEAVNTACYAINRLYLHRILKKTSYELLTGKKPNISYFRVFGSKCFI 358
Query: 1037 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPGSKTSEQS 1096
L + KF K G LGY ++AYRV+N T VE S V FD+ GS+ +
Sbjct: 359 LIKRGRKSKFAPKTVEGFLLGYDSNTRAYRVFNKSTGLVEVSCDVVFDETN-GSQVEQVD 417
Query: 1097 ESNAGTTDS-------------------EDASESDQPSDSEKYTKVESSPEAEITPEA-- 1135
G + E S DQPS S +++SP + EA
Sbjct: 418 LDEIGEEQAPCIALRNMSIGDVCPKESEEPPSTQDQPSSS-----MQASPPTQNEDEAQN 472
Query: 1136 --ESNSEAESSPIVQNESASEDFQDNTQQVIQPKFKH-------KSSHPEELIIGSKDSP 1186
E N E E N+ + + +P+ H + HP + I+G
Sbjct: 473 DEEQNQEDEPPQDDSNDQGGDTNDQEKEDEEEPRPPHPRVHQAIQRDHPVDTILGDIHKG 532
Query: 1187 RRTRS---HFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKP 1243
TRS HF + S + S IEP VEEAL D W++AMQEELN F RN+VW LVP+P
Sbjct: 533 VTTRSRVAHFCEHYSFV---SSIEPHRVEEALQDSDWVVAMQEELNNFTRNEVWHLVPRP 589
Query: 1244 FQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLS 1303
Q N++ TKWVFRNK +E G VTRNKARLVA+GYSQ EG+++ ET+APVARLE+IR+LL+
Sbjct: 590 NQ-NVVGTKWVFRNKQDEHGVVTRNKARLVAKGYSQVEGLDFGETYAPVARLESIRILLA 648
Query: 1304 YAINHGIILYQMDVKSVFLNGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRA 1363
YA HG LYQMDVKS FLNG I+EEVYV+QPPGFED ++P+HVY+L K+LYGLKQAPRA
Sbjct: 649 YATYHGFKLYQMDVKSAFLNGPIKEEVYVEQPPGFEDSEYPNHVYRLSKALYGLKQAPRA 708
Query: 1364 WYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQ 1423
WY+ L +FLI N F+ G+ D TLF +TL+ D+ + QIYVDDIIFGSTN S C+EFS++M
Sbjct: 709 WYECLRDFLIANGFKVGKADPTLFTKTLENDLFVCQIYVDDIIFGSTNKSTCEEFSRIMT 768
Query: 1424 DEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLS 1483
+FEMSMMGELK+FLG Q+ Q +EG ++ QTKYT+++L KF ++D K + TPM L
Sbjct: 769 QKFEMSMMGELKYFLGFQVKQLQEGTFICQTKYTQDILTKFGMKDAKPIKTPMGTNGHLD 828
Query: 1484 KEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLK 1543
+ G VDQK+YR MIGSLLYL ASRPDI+ SVC+CARFQSDP+ESHLTAVKRI RYL
Sbjct: 829 LDTGGKSVDQKVYRSMIGSLLYLCASRPDIMLSVCMCARFQSDPKESHLTAVKRILRYLA 888
Query: 1544 GTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLIS 1591
T GL Y + + LIG+ DAD+AG +I RKSTSG CQFLG +L+S
Sbjct: 889 YTPKFGLWYPRGSTFDLIGYSDADWAGCKINRKSTSGTCQFLGRSLVS 936
>gb|AAC49502.1| Pol [Zea mays] gi|7489803|pir||T04112 pol protein homolog - maize
retrotransposon Opie-2
Length = 1068
Score = 880 bits (2273), Expect = 0.0
Identities = 462/942 (49%), Positives = 606/942 (64%), Gaps = 28/942 (2%)
Query: 669 SWYLDSGCSRHMTGEKALFLTLTMKDGGE--VKFGGNQTGKIIGTGTIGNSSI-SINNVW 725
SW +DSGC+ HMTGEK +F + + + FG GK+ G G I S+ SI+NV+
Sbjct: 10 SWIIDSGCTNHMTGEKKMFTSYVKNKDSQDSIIFGDGNQGKVKGLGKIAISNEHSISNVF 69
Query: 726 LVDGLKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQ 785
LV+ L +NLLS+SQ C+ Y+ F+ + ++ + D S+ FKG +Y ++F+
Sbjct: 70 LVESLGYNLLSVSQLCNMGYNCLFTNIDVSVFRRCDGSLAFKGVLDGKLYLVDFAKEEAG 129
Query: 786 KVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVK 845
CL++ W+WH+RL H + + K+ K + V GL N+ + D C ACQ GK V
Sbjct: 130 LDACLIAKTSMGWLWHRRLAHVGMKNLHKLLKGEHVIGLTNVQFEKDRPCAACQAGKQVG 189
Query: 846 SSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYAC 905
S +K++++TSRPLE+LH+DLFGPV S+ GSKYGLVIVDD+SR+TWV F++ K
Sbjct: 190 GSHHTKNVMTTSRPLEMLHMDLFGPVAYLSIGGSKYGLVIVDDFSRFTWVFFLQEKSETQ 249
Query: 906 EVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVE 965
F + Q+E ELK+ K+RSD+G EF+N E F E+ GI HEFS+P TPQQNGVVE
Sbjct: 250 GTLKRFLRRAQNEFELKVKKIRSDNGSEFKNLQVEEFLEEEGIKHEFSAPYTPQQNGVVE 309
Query: 966 RKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNIS 1025
RKNRTL +MARTM+ E + FW EAVNT+C+ NR+Y+ +L+ T+YEL G +PN+S
Sbjct: 310 RKNRTLIDMARTMLGEFKTPECFWTEAVNTACHAINRVYLHRILKNTSYELLTGNKPNVS 369
Query: 1026 YFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDD 1085
YF FG CYIL K KF KA G LGY +KAYRV+N + VE S V FD
Sbjct: 370 YFRVFGSKCYILVKKGRNSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSGDVVFD- 428
Query: 1086 REPGSKTSEQSESNAGTTDSEDASESDQPSDS------------EKYTKVESSPEAEITP 1133
E EQ D +D E D P+ + E+ + + SP + P
Sbjct: 429 -ETNGSPREQ------VVDCDDVDEEDIPTAAIRTMAIGEVRPQEQDEREQPSPSTMVHP 481
Query: 1134 EAESNSEAESSPIVQNESASED--FQDNTQQV--IQPKFKHKSSHPEELIIGSKDSPRRT 1189
+ + + + A +D ++ Q Q + + HP + I+G T
Sbjct: 482 PTQDDEQVHQQEVCDQGGAQDDHVLEEEAQPAPPTQVRAMIQRDHPVDQILGDISKGVTT 541
Query: 1190 RSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNII 1249
RS +S IEP VEEAL D W+LAMQEELN F+RN+VW LVP+P Q N++
Sbjct: 542 RSRLVNFCEHNSFVSSIEPFRVEEALLDPDWVLAMQEELNNFKRNEVWTLVPRPKQ-NVV 600
Query: 1250 ETKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHG 1309
TKWVFRNK +E+G VTRNKARLVA+GY+Q G+++ ETFAPVARLE+IR+LL+YA +H
Sbjct: 601 GTKWVFRNKQDERGVVTRNKARLVAKGYAQVAGLDFEETFAPVARLESIRILLAYAAHHS 660
Query: 1310 IILYQMDVKSVFLNGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLS 1369
LYQMDVKS FLNG I+EEVYV+QPPGFED ++PDHV KL K+LYGLKQAPRAWY+ L
Sbjct: 661 FRLYQMDVKSAFLNGPIKEEVYVEQPPGFEDERYPDHVCKLSKALYGLKQAPRAWYECLR 720
Query: 1370 NFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMS 1429
+FLI N F+ G+ D TLF +T D+ + QIYVDDIIFGSTN C+EFS++M +FEMS
Sbjct: 721 DFLIANAFKVGKADPTLFTKTCDGDLFVCQIYVDDIIFGSTNQKSCEEFSRVMTQKFEMS 780
Query: 1430 MMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGT 1489
MMGEL +FLG Q+ Q K+G ++ QTKYT++LLK+F ++D K TPM G
Sbjct: 781 MMGELNYFLGFQVKQLKDGTFISQTKYTQDLLKRFGMKDAKPAKTPMGTDGHTDLNKGGK 840
Query: 1490 VVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLG 1549
VDQK YR MIGSLLYL ASRPDI+ SVC+CARFQSDP+E HL AVKRI RYL T G
Sbjct: 841 SVDQKAYRSMIGSLLYLCASRPDIMLSVCMCARFQSDPKECHLVAVKRILRYLVATPCFG 900
Query: 1550 LLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLIS 1591
L Y K + L+G+ D+DYAG +++RKSTSG CQFLG +L+S
Sbjct: 901 LWYPKGSTFDLVGYSDSDYAGCKVDRKSTSGTCQFLGRSLVS 942
>ref|NP_910082.1| putative gag-pol polyprotein [Oryza sativa (japonica cultivar-group)]
gi|28269414|gb|AAO37957.1| putative gag-pol polyprotein
[Oryza sativa (japonica cultivar-group)]
Length = 1969
Score = 867 bits (2240), Expect = 0.0
Identities = 465/994 (46%), Positives = 620/994 (61%), Gaps = 29/994 (2%)
Query: 618 SKIPKQKDQKNKAATASEKTI-------PKGVKPKVLNDQ-KPLSIHPKVCLRARE---- 665
S PK + K T+ K + PK V+ K Q KP+ C +
Sbjct: 685 SSTPKTSEPKMVPMTSKSKPVELPRPKNPKQVEHKQNQRQTKPVEKTKYECTYCGKAGHL 744
Query: 666 -----KQRSWYLDSGCSRHMTGEKALFLTLTMKDGGE-VKFGGNQTGKIIGTGTIG-NSS 718
+ SW +DSGCSRHMTGE F +LT G E + FG +G+++ GTI N
Sbjct: 745 DFGVGRSNSWLVDSGCSRHMTGEAKWFTSLTRASGDETITFGDASSGRVMAKGTIKVNDK 804
Query: 719 ISINNVWLVDGLKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKIN 778
+ +V LV LK+NLLS+SQ CD +V F K +++ + + F RV V+ N
Sbjct: 805 FMLKDVALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFAN 863
Query: 779 FSDLADQKVVCLL-SMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGA 837
F A CL+ S N + WH+RLGH + +S+IS + L++GLP + D +C
Sbjct: 864 FDSSAPGPSRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVQKDLVCAP 923
Query: 838 CQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKF 897
C+ GK+ SS K +V T P +LLH+D GP S+ G Y LV+VDD+SR++WV F
Sbjct: 924 CRHGKMTSSSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRYSWVYF 983
Query: 898 IKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRT 957
++SK+ F S + E + +RSD+G EF+N FE FC+ G+ H+FSSP
Sbjct: 984 LESKEETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYV 1043
Query: 958 PQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELF 1017
PQQNGVVERKNRTL EMARTM+ E + FW EA++ +C+I NR+++R +L KT YEL
Sbjct: 1044 PQQNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYELR 1103
Query: 1018 KGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEE 1077
GRRP +S+ FGC C++L + + L KF++++ GIFLGY+ S+AYRVY T + E
Sbjct: 1104 FGRRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVE 1162
Query: 1078 SMHVKFDDREPGSKTSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAES 1137
+ V FD+ PG++ + ED+ + D D ++S+P + T +
Sbjct: 1163 TCEVTFDEASPGARPEISGVPDESIFVDEDSDDDD---DDSIPPPLDSTPPVQETGSPST 1219
Query: 1138 NSEAESSPIVQNESASEDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEE 1197
S + +P + SA+E+ T P+ P+ +I G + R RS+
Sbjct: 1220 TSPSGDAPTTSS-SAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYELVNS 1278
Query: 1198 SLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRN 1257
+ + + EPK V ALSD+ W+ AM EEL F+RN VW LV P N+I TKWVF+N
Sbjct: 1279 AFV---ASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKN 1335
Query: 1258 KLNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDV 1317
KL E G + RNKARLVAQG++Q EG+++ ETFAPVARLE IR+LL++A + G L+QMDV
Sbjct: 1336 KLGEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMDV 1395
Query: 1318 KSVFLNGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDF 1377
KS FLNGVIEEEVYVKQPPGFE+ K P+HV+KL+K+LYGLKQAPRAWY+RL FL++N F
Sbjct: 1396 KSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFLLQNGF 1455
Query: 1378 ERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFF 1437
E G VD TLF D L+VQIYVDDIIFG ++ +L +FS +M EFEMSMMGEL FF
Sbjct: 1456 EMGAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFF 1515
Query: 1438 LGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYR 1497
LG+QI Q+KEG++VHQTKY+KELLKKF + DCK + TPM T +L ++ G VDQ+ YR
Sbjct: 1516 LGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREYR 1575
Query: 1498 GMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLD 1557
MIGSLLYLTASRPDI FSVCLCARFQ+ PR SH AVKRIFRY+K T G+ Y S
Sbjct: 1576 SMIGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRIFRYIKSTLEYGIWYSCSSA 1635
Query: 1558 YKLIGFCDADYAGDRIERKSTSGNCQFLGENLIS 1591
+ F DAD+AG +I+RKSTSG C FLG +L+S
Sbjct: 1636 LSVRAFSDADFAGCKIDRKSTSGTCHFLGTSLVS 1669
Score = 119 bits (297), Expect = 1e-24
Identities = 157/684 (22%), Positives = 267/684 (38%), Gaps = 84/684 (12%)
Query: 31 APKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGV---DDLDLDEEGAAIDRRIHTPA 87
A F D F WK M +++ +W+ ++ DD D+ TPA
Sbjct: 7 AKPFVFDGHNFVIWKARMEAYLQSQGHNVWNKVKSPYTVPDDADI------------TPA 54
Query: 88 QKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVH 147
+++ R I+ I E+ ++ +A M+ +LC EG+ +++ +
Sbjct: 55 NMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQFHK 114
Query: 148 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 206
+Y+ F M ESI+ + RF ++S L+ + K + +D+ +L L W KVT+I
Sbjct: 115 EYQRFEMHPGESIDSYFKRFGEIISKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVTSIT 174
Query: 207 EAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEES 266
E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+ +
Sbjct: 175 ESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAFVYGGFSLAA 234
Query: 267 PDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIAD 326
++E + + L ARK + K K + CF C +P H +
Sbjct: 235 LHSVTEEQLE---KIPEDDLALFARKFSRAYKNVRDRKRGKTNEPFVCFECGEPNHKRVN 291
Query: 327 CPDLQKEKFKGKSKKSSFNSSKFRKQIKK----SLMATWEDLD-SESGSDKEEADDDAKA 381
CP L+K+ K +KK K +K + K ++A E++ S+ SD ++ + K
Sbjct: 292 CPKLKKKSDK-TTKKPEGQGRKGKKDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDKD 350
Query: 382 AMGLVATVSSE--------AVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNE 433
G+ ++E A+ + + SE IP SL L N+
Sbjct: 351 FSGMCCLANNEDFINLCLMALEDKDDSSEHPEVCLDDIP------SLDGSLCDDSCSDND 404
Query: 434 LTD---LKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDKG 490
D KE+ LM + K E+LK N RL+S+ + ++ C
Sbjct: 405 SVDDELSKERMAHLMIEISDKYRSSKYKIEKLKSENDGMALEIARLRSMIPE-EDTCSTC 463
Query: 491 SGNKHEI-ALDDFIMA---GIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCI 546
+ EI L D + + G AS ST +G E + KE
Sbjct: 464 ASYLSEINLLKDKLKSCALGAGNPSSASAACSTCYEMKVDMGLLEMELKE---------- 513
Query: 547 KDGLKSTFVPEGTNAVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRS 606
LK FV + + ++ P + ++ + LK TK+D ++
Sbjct: 514 ---LKEKFVHD---RIGRCENCPILTSDNDELRQQVAMLK----TKND----LLESFATK 559
Query: 607 EPVHQNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREK 666
EP+H + + KD KT+ +K D I KV L + +K
Sbjct: 560 EPIHSSCANCAILETELKD---------AKTVIDSIKS---IDSCSSCISLKVDLESAKK 607
Query: 667 QRSWYLDSGCSRHMTGEKALFLTL 690
+ S YL R G+K L + L
Sbjct: 608 ENS-YLQQSLERFAQGKKKLNMIL 630
>ref|NP_909107.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 1410
Score = 863 bits (2230), Expect = 0.0
Identities = 474/1029 (46%), Positives = 635/1029 (61%), Gaps = 20/1029 (1%)
Query: 569 PEASGSQAKITSKPENLKIKVMTKSDPKSQKI-KILKRSEPVHQNLIKPESKIPKQ--KD 625
P+ K T KPE K K+D + I K+L E V + I + ++ KD
Sbjct: 270 PKLKKKSDKTTKKPEGQGRK--GKNDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDKD 327
Query: 626 QKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRSWYLDSGCSRHMTGEKA 685
A+ + + L D+ S HP+ R SW +DSGCSRHMTGE
Sbjct: 328 FSGMCCLANNEDFIN-LCLMALEDKDDSSEHPEDFGVGRSN--SWLVDSGCSRHMTGEAK 384
Query: 686 LFLTLTMKDGGE-VKFGGNQTGKIIGTGTIG-NSSISINNVWLVDGLKHNLLSISQFCDN 743
F +LT G E + FG +G+++ GTI N + +V LV LK+NLLS+SQ CD
Sbjct: 385 WFTSLTRASGDETITFGDASSGRVMAKGTIKVNDKFMLKDVALVSKLKYNLLSVSQLCDE 444
Query: 744 EYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLL-SMNDKKWVWHK 802
+V F K +++ + + F RV V+ NF A CL+ S N + WH+
Sbjct: 445 NLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFDSSAPGPSRCLVASENRDLFFWHR 503
Query: 803 RLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLEL 862
RLGH + +S+IS + L++GLP + D +C C+ GK+ SS K +V T P +L
Sbjct: 504 RLGHIGFDHLSRISGMDLIRGLPKLKAPKDLVCAPCRHGKMTSSSHKPVTMVMTDGPGQL 563
Query: 863 LHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELK 922
LH++ GP S+ G Y LV+VDD+SR++WV F++SK+ F S + E
Sbjct: 564 LHMNTVGPARVQSVGGKWYVLVVVDDFSRYSWVYFLESKEETFGFFQSLARSLALEFPGA 623
Query: 923 ILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHEN 982
+ +RSD+G EF+N FE FC+ G+ H+FSSP PQQNGVVERKNRTL EMARTM+ E
Sbjct: 624 LRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYVPQQNGVVERKNRTLVEMARTMLDEF 683
Query: 983 NLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDY 1042
+ FW EA++ +C+I NR+++R +L KT YEL GRRP +S+ FGC C++L + +
Sbjct: 684 TTPRKFWTEAISAACFISNRVFLRTILHKTPYELRFGRRPKVSHLRVFGCKCFVLKSGN- 742
Query: 1043 LKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPGSKTSEQSESNAGT 1102
L KF++++ GIFLGY+ S+AYRVY T + E+ V FD+ PG++ +
Sbjct: 743 LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVETCEVTFDEASPGARPEISGVLDESI 802
Query: 1103 TDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSPIVQNESASEDFQDNTQQ 1162
ED+ + D D ++S+P + T + S + +P + SA+E+ T
Sbjct: 803 FVDEDSDDDD---DDSIPPPLDSTPPVQETGSPSTTSPSGDAPTTSS-SAAEEIDGGTSG 858
Query: 1163 VIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWIL 1222
P+ P+ +I G + R RS+ + + + EPK V ALSD+ W+
Sbjct: 859 PTAPRHIQNRHPPDSMIGGLGERVTRNRSYDLVNSAFV---ASFEPKNVCHALSDENWVN 915
Query: 1223 AMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQEG 1282
AM EEL F+RN VW LV P N+I TKWVF+NKL E G + RNKARLVAQG++Q EG
Sbjct: 916 AMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKNKLGEDGSIVRNKARLVAQGFTQVEG 975
Query: 1283 INYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQPPGFEDLK 1342
+++ ETFAPVARLE IR+LL++A + G L+QMDVKS FLNGVIEEEVYVKQPPGFE+ K
Sbjct: 976 LDFEETFAPVARLEAIRILLAFAASKGFKLFQMDVKSAFLNGVIEEEVYVKQPPGFENPK 1035
Query: 1343 HPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYV 1402
P+HV+KL K+LYGLKQAPRAWY+RL FL++N FE G VD TLF D L+VQIYV
Sbjct: 1036 FPNHVFKLDKALYGLKQAPRAWYERLKTFLLQNGFEMGAVDKTLFTLHSGIDFLLVQIYV 1095
Query: 1403 DDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLK 1462
DDIIFG ++ +L +FS +M EFEMSMMGEL FFLG+QI Q+KEG++VHQTKY+KELLK
Sbjct: 1096 DDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFFLGLQIKQTKEGIFVHQTKYSKELLK 1155
Query: 1463 KFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCAR 1522
KF + DCK + TPM T +L ++ G VDQ+ YR MIGSLLYLTASRPDI FSVCLCAR
Sbjct: 1156 KFDMADCKPIATPMATTSSLGPDEDGEEVDQREYRSMIGSLLYLTASRPDIHFSVCLCAR 1215
Query: 1523 FQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNC 1582
FQ+ PR SH AVKRIFRY+K T G+ Y S + F DAD+AG +I+RKSTSG C
Sbjct: 1216 FQASPRTSHRQAVKRIFRYIKSTLEYGIWYSCSSALSVRAFSDADFAGCKIDRKSTSGTC 1275
Query: 1583 QFLGENLIS 1591
FLG +L+S
Sbjct: 1276 HFLGTSLVS 1284
Score = 111 bits (277), Expect = 2e-22
Identities = 84/333 (25%), Positives = 148/333 (44%), Gaps = 16/333 (4%)
Query: 85 TPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALM 144
TPA +++ R I+ I E+ ++ +A M+ +LC EG+ +++ +
Sbjct: 29 TPANMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQ 88
Query: 145 LVHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVT 203
+Y+ F M ESI+ + RF +VS L+ + K + +D+ +L L W KVT
Sbjct: 89 FHKEYQRFEMHPGESIDSYFKRFGEIVSKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVT 148
Query: 204 AIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESE 263
+I E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+
Sbjct: 149 SITESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAFVCGGFS 208
Query: 264 EESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHF 323
+ ++E + + L ARK + + K K + CF C +P H
Sbjct: 209 LAALHSVTEEQLE---KIPEDDLALFARKFSRAYKNVRNKKRGKTNEPFVCFECGEPNHI 265
Query: 324 IADCPDLQK------EKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADD 377
+CP L+K +K +G+ +K + +K I K L A E S+ SD ++ +
Sbjct: 266 RVNCPKLKKKSDKTTKKPEGQGRKG--KNDLMKKAIHKVLAALEEVQLSDIDSDDDDQEK 323
Query: 378 DAKAAMGLVATVSSEAVSE----AESDSEDENE 406
K G+ ++E A D +D +E
Sbjct: 324 GDKDFSGMCCLANNEDFINLCLMALEDKDDSSE 356
>gb|AAN60494.1| Putative Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)] gi|34902378|ref|NP_912535.1| Putative
Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)]
Length = 2145
Score = 863 bits (2229), Expect = 0.0
Identities = 463/994 (46%), Positives = 619/994 (61%), Gaps = 29/994 (2%)
Query: 618 SKIPKQKDQKNKAATASEKTI-------PKGVKPKVLNDQ-KPLSIHPKVCLRARE---- 665
S PK + K T+ K + PK V+ K Q KP+ C +
Sbjct: 685 SSTPKTSEPKMVPMTSKSKPVELPRPKNPKQVEHKQNQRQTKPVEKTKYECTYCGKAGHL 744
Query: 666 -----KQRSWYLDSGCSRHMTGEKALFLTLTMKDGGE-VKFGGNQTGKIIGTGTIG-NSS 718
+ SW +DSGCSRHMTGE F +LT E + FG +G+++ GTI N
Sbjct: 745 DFGVGRSNSWLVDSGCSRHMTGEAKWFTSLTRASSDETITFGDASSGRVMAKGTIKVNDK 804
Query: 719 ISINNVWLVDGLKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKIN 778
+ +V LV LK+NLLS+SQ CD +V F K +++ + + F RV V+ N
Sbjct: 805 FMLKDVALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFAN 863
Query: 779 FSDLADQKVVCLL-SMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGA 837
F A CL+ S N + WH+RLGH + +S+IS + L++GLP + D +C
Sbjct: 864 FDSSAPGPSRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVPKDLVCAP 923
Query: 838 CQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKF 897
C+ GK+ SS K +V T P +LLH+D GP S+ G Y LV+VDD+SR++WV F
Sbjct: 924 CRHGKMTSSSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRYSWVYF 983
Query: 898 IKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRT 957
++SK+ F S + E + +RSD+G EF+N FE FC+ G+ H+FSSP
Sbjct: 984 LESKEETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYV 1043
Query: 958 PQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELF 1017
PQQNGVVERKNRTL EMARTM+ E + FW EA++ +C+I NR+++R +L KT YEL
Sbjct: 1044 PQQNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYELR 1103
Query: 1018 KGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEE 1077
GRRP +S+ FGC C++L + + L KF++++ GIFLGY+ S+AYRVY T + E
Sbjct: 1104 FGRRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVE 1162
Query: 1078 SMHVKFDDREPGSKTSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAES 1137
+ V FD+ PG++ + ED+ + D D ++S+P + T +
Sbjct: 1163 TCEVTFDEASPGARPEISGVPDESIFVDEDSDDDD---DDSIPPPLDSTPPVQETGSPST 1219
Query: 1138 NSEAESSPIVQNESASEDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEE 1197
S + +P + SA+E+ T P+ P+ +I G + R RS+
Sbjct: 1220 TSPSGDAPTTSS-SAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYELVNS 1278
Query: 1198 SLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRN 1257
+ + + EPK V ALSD+ W+ AM EEL F+RN VW LV P N+I TKWVF+N
Sbjct: 1279 AFV---ASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKN 1335
Query: 1258 KLNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDV 1317
KL E G + RNKARLVAQG++Q EG+++ ETFAPVARLE IR+LL++A + G L+QMDV
Sbjct: 1336 KLGEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMDV 1395
Query: 1318 KSVFLNGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDF 1377
KS FLNGVIEEEVYVKQPPGFE+ K P+HV+KL+K+LYGLKQAPRAWY+RL FL++N F
Sbjct: 1396 KSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFLLQNGF 1455
Query: 1378 ERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFF 1437
E G VD TLF D L+VQIYVDDIIFG ++ +L +FS +M EFEMSMMGEL FF
Sbjct: 1456 EMGAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFF 1515
Query: 1438 LGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYR 1497
LG+QI Q+KEG++VHQTKY+KELLKKF + DCK + TPM T +L ++ G VDQ+ YR
Sbjct: 1516 LGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREYR 1575
Query: 1498 GMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLD 1557
MIGSLLYLTASRPDI FSVCLCARFQ+ PR SH AVKR+FRY+K T G+ Y S
Sbjct: 1576 SMIGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRMFRYIKSTLEYGIWYSCSSA 1635
Query: 1558 YKLIGFCDADYAGDRIERKSTSGNCQFLGENLIS 1591
+ F DAD+AG +I+RKSTSG C FLG +L+S
Sbjct: 1636 LSVRAFSDADFAGCKIDRKSTSGTCHFLGTSLVS 1669
Score = 118 bits (295), Expect = 2e-24
Identities = 114/498 (22%), Positives = 209/498 (41%), Gaps = 53/498 (10%)
Query: 31 APKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGV---DDLDLDEEGAAIDRRIHTPA 87
A F D F WK M +++ +W+ ++ DD D+ TPA
Sbjct: 7 AKPFVFDGHNFVIWKARMEAYLQSQGHNVWNKVKSPYTVPDDADI------------TPA 54
Query: 88 QKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVH 147
+++ R I+ I E+ ++ +A M+ +LC EG+ +++ +
Sbjct: 55 NMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQFHK 114
Query: 148 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 206
+Y+ F M ESI+ + RF ++S L+ + K + +D+ +L L W KVT+I
Sbjct: 115 EYQRFEMHPGESIDSYFKRFGEIISKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVTSIT 174
Query: 207 EAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEES 266
E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+ +
Sbjct: 175 ESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAFVYGGFSLAA 234
Query: 267 PDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIAD 326
++E + + L ARK + K K + CF C +P H +
Sbjct: 235 LHSVTEEQLE---KIPEDDLALFARKFSRAYKNVRDRKRGKTNEPFVCFECGEPNHKRVN 291
Query: 327 CPDLQKEKFKGKSKKSSFNSSKFRKQIKK----SLMATWEDLD-SESGSDKEEADDDAKA 381
CP L+K+ K +KK K +K + K ++A E++ S+ SD ++ + K
Sbjct: 292 CPKLKKKSDK-TTKKPEGQGRKGKKDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDKD 350
Query: 382 AMGLVATVSSE--------AVSEAESDSEDENEVYSKIPRQELV---------DSLKELL 424
G+ ++E A+ + + SE IP + DS+ + L
Sbjct: 351 FSGMCCLANNEDFINLCLMALEDKDDSSEHPEVCLDDIPSLDGSLCDDSCSDNDSVDDEL 410
Query: 425 SLFEHRTNELTDLKEKY------VDLMKQQKSTL-LELKASEEELKGFNLIST--TYEDR 475
S E + + ++ +KY ++ +K + + LE+ + + ST +Y
Sbjct: 411 SK-ERMAHLMIEISDKYRSSKYKIEKLKSENDGMALEIARLRSMIPEEDTCSTCASYLSE 469
Query: 476 LKSLCQKLQEKCDKGSGN 493
+ L KL + C G+GN
Sbjct: 470 INLLKDKL-KSCALGAGN 486
>ref|XP_474304.1| OSJNBb0004A17.2 [Oryza sativa (japonica cultivar-group)]
gi|32488723|emb|CAE03600.1| OSJNBb0004A17.2 [Oryza sativa
(japonica cultivar-group)]
Length = 1877
Score = 862 bits (2228), Expect = 0.0
Identities = 449/929 (48%), Positives = 599/929 (64%), Gaps = 12/929 (1%)
Query: 666 KQRSWYLDSGCSRHMTGEKALFLTLTMKDGGE-VKFGGNQTGKIIGTGTIG-NSSISINN 723
+ SW +DSGCSRHMTGE F +LT E + FG +G+++ GTI N + +
Sbjct: 832 RSNSWLVDSGCSRHMTGEAKWFTSLTRASSDETITFGDASSGRVMAKGTIKVNDKFMLKD 891
Query: 724 VWLVDGLKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLA 783
V LV LK+NLLS+SQ CD +V F K +++ + + F RV V+ NF A
Sbjct: 892 VALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFDSSA 950
Query: 784 DQKVVCLL-SMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGK 842
CL+ S N + WH+RLGH + +S+IS + L++GLP + D +C C+ GK
Sbjct: 951 PGPSRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVPKDLVCAPCRHGK 1010
Query: 843 IVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKD 902
+ SS K +V T P +LLH+D GP S+ G Y LV+VDD+SR++WV F++SK+
Sbjct: 1011 MTSSSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRYSWVYFLESKE 1070
Query: 903 YACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNG 962
F S + E + +RSD+G EF+N FE FC+ G+ H+FSSP PQQNG
Sbjct: 1071 ETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYVPQQNG 1130
Query: 963 VVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRP 1022
VVERKNRTL EMARTM+ E + FW EA++ +C+I NR+++R +L KT YEL GRRP
Sbjct: 1131 VVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYELRFGRRP 1190
Query: 1023 NISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVK 1082
+S+ FGC C++L + + L KF++++ GIFLGY+ S+AYRVY T + E+ V
Sbjct: 1191 KVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVETCEVT 1249
Query: 1083 FDDREPGSKTSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAE 1142
FD+ PG++ + ED+ + D D ++S+P + T + S +
Sbjct: 1250 FDEASPGARPEISGVPDESIFVDEDSDDDD---DDSIPPPLDSTPPVQETGSPSTTSPSG 1306
Query: 1143 SSPIVQNESASEDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGL 1202
+P + SA+E+ T P+ P+ +I G + R RS+ + +
Sbjct: 1307 DAPTTSS-SAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYELVNSAFV-- 1363
Query: 1203 LSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQ 1262
+ EPK V ALSD+ W+ AM EEL F+RN VW LV P N+I TKWVF+NKL E
Sbjct: 1364 -ASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKNKLGED 1422
Query: 1263 GEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFL 1322
G + RNKARLVAQG++Q EG+++ ETFAPVARLE IR+LL++A + G L+QMDVKS FL
Sbjct: 1423 GSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMDVKSAFL 1482
Query: 1323 NGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQV 1382
NGVIEEEVYVKQPPGFE+ K P+HV+KL+K+LYGLKQAPRAWY+RL FL++N FE G V
Sbjct: 1483 NGVIEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFLLQNGFEMGAV 1542
Query: 1383 DTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQI 1442
D TLF D L+VQIYVDDIIFG ++ +L +FS +M EFEMSMMGEL FFLG+QI
Sbjct: 1543 DKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFFLGLQI 1602
Query: 1443 NQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGS 1502
Q+KEG++VHQTKY+KELLKKF + DCK + TPM T +L ++ G VDQ+ YR MIGS
Sbjct: 1603 KQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREYRSMIGS 1662
Query: 1503 LLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIG 1562
LLYLTASRPDI FSVCLCARFQ+ PR SH AVKR+FRY+K T G+ Y S +
Sbjct: 1663 LLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRMFRYIKSTLEYGIWYSCSSALSVRA 1722
Query: 1563 FCDADYAGDRIERKSTSGNCQFLGENLIS 1591
F DAD+AG +I+RKSTSG C FLG +L+S
Sbjct: 1723 FSDADFAGCKIDRKSTSGTCHFLGTSLVS 1751
Score = 118 bits (295), Expect = 2e-24
Identities = 114/498 (22%), Positives = 209/498 (41%), Gaps = 53/498 (10%)
Query: 31 APKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGV---DDLDLDEEGAAIDRRIHTPA 87
A F D F WK M +++ +W+ ++ DD D+ TPA
Sbjct: 7 AKPFVFDGHNFVIWKARMEAYLQSQGHNVWNKVKSPYTVPDDADI------------TPA 54
Query: 88 QKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVH 147
+++ R I+ I E+ ++ +A M+ +LC EG+ +++ +
Sbjct: 55 NMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQFHK 114
Query: 148 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 206
+Y+ F M ESI+ + RF ++S L+ + K + +D+ +L L W KVT+I
Sbjct: 115 EYQRFEMHPGESIDSYFKRFGEIISKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVTSIT 174
Query: 207 EAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEES 266
E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+ +
Sbjct: 175 ESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAFVYGGFSLAA 234
Query: 267 PDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIAD 326
++E + + L ARK + K K + CF C +P H +
Sbjct: 235 LHSVTEEQLE---KIPEDDLALFARKFSRAYKNVRDRKRGKTNEPFVCFECGEPNHKRVN 291
Query: 327 CPDLQKEKFKGKSKKSSFNSSKFRKQIKK----SLMATWEDLD-SESGSDKEEADDDAKA 381
CP L+K+ K +KK K +K + K ++A E++ S+ SD ++ + K
Sbjct: 292 CPKLKKKSDK-TTKKPEGQGRKGKKDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDKD 350
Query: 382 AMGLVATVSSE--------AVSEAESDSEDENEVYSKIPRQELV---------DSLKELL 424
G+ ++E A+ + + SE IP + DS+ + L
Sbjct: 351 FSGMCCLANNEDFINLCLMALEDKDDSSEHPEVCLDDIPSLDGSLCDDSCSDNDSVDDEL 410
Query: 425 SLFEHRTNELTDLKEKY------VDLMKQQKSTL-LELKASEEELKGFNLIST--TYEDR 475
S E + + ++ +KY ++ +K + + LE+ + + ST +Y
Sbjct: 411 SK-ERMAHLMIEISDKYRSSKYKIEKLKSENDGMALEIARLRSMIPEEDTCSTCASYLSE 469
Query: 476 LKSLCQKLQEKCDKGSGN 493
+ L KL + C G+GN
Sbjct: 470 INLLKDKL-KSCALGAGN 486
>gb|AAN40025.1| putative gag-pol polyprotein [Zea mays]
Length = 1892
Score = 847 bits (2189), Expect = 0.0
Identities = 456/953 (47%), Positives = 594/953 (61%), Gaps = 80/953 (8%)
Query: 669 SWYLDSGCSRHMTGEKALFLTLTMKDGGE--VKFGGNQTGKIIGTGTIGNSSISINNVWL 726
SW LDSGC+ HMTGEK +F + + + FG G + G
Sbjct: 863 SWILDSGCTNHMTGEKKMFSSYEKNKDPQRAITFGDGNQGLVKGV--------------- 907
Query: 727 VDGLKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQK 786
T+ + D SI FKG +Y + F D A+
Sbjct: 908 ----------------------------TVFRRSDDSIAFKGVLEGQLYLVVF-DRAELD 938
Query: 787 VVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKS 846
CL++ + W+WH+RL H + + K+ K + + GL N+ + D +C ACQ GK V +
Sbjct: 939 T-CLIAKTNMGWLWHRRLAHVGMKNLHKLLKGEHILGLTNVHFEKDRICSACQAGKQVGT 997
Query: 847 SFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACE 906
K+I++T RPLELLH+DLFGP+ S+ GSKY LVIVDDYSR+TWV F++ K E
Sbjct: 998 HHPHKNIMTTDRPLELLHMDLFGPIAYISIGGSKYCLVIVDDYSRFTWVFFLQEKSQTQE 1057
Query: 907 VFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVER 966
F + Q+E L+I K+RSD+G EF+N E F E+ GI HEFSSP TPQQNGVVER
Sbjct: 1058 TLKGFLRRAQNEFGLRIKKIRSDNGTEFKNSQIESFLEEEGIKHEFSSPYTPQQNGVVER 1117
Query: 967 KNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISY 1026
KNRTL +MARTM+ E FWAEAVNT+CY NR+Y+ +L+KT+YEL G++PNISY
Sbjct: 1118 KNRTLLDMARTMLDEYKTPDRFWAEAVNTACYAINRLYLHRILKKTSYELLTGKKPNISY 1177
Query: 1027 FHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDR 1086
F FG C+IL + KF K G LGY ++AYRV+N T VE S V FD+
Sbjct: 1178 FRVFGSKCFILIKRGRKSKFAPKTVEGFLLGYDSNTRAYRVFNKSTGLVEVSCDVVFDET 1237
Query: 1087 EPGSKTSEQSESNAGTTDSEDAS------------ESDQPSDSEKY--TKVESSPEAEIT 1132
GS+ + G + + ES++P ++ + +++SP +
Sbjct: 1238 N-GSQVEQVDLDEIGEEQAPCIALRNMSIGDVCPKESEEPPSTQDQPPSSMQASPPTQNE 1296
Query: 1133 PEA----ESNSEAESSPIVQNESASEDFQDNTQQVIQPKFKH-------KSSHPEELIIG 1181
EA E N E + N+ + + +P+ H + HP + I+G
Sbjct: 1297 DEAQNDEEQNQEVKPPQDKSNDQGGDTNDQEKEDEEEPRPPHPRVHQAIQRDHPVDTILG 1356
Query: 1182 SKDSPRRTRS---HFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWD 1238
TRS HF + S + S IEP VEEAL D W++AMQEELN F RN+VW
Sbjct: 1357 DIHKGVTTRSRVAHFCEHYSFV---SSIEPHRVEEALQDSDWVVAMQEELNNFTRNEVWH 1413
Query: 1239 LVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETI 1298
LVP+P Q N++ TKWVFRNK +E G VTRNKARLVA+GYSQ EG+++ ET+APVARLE+I
Sbjct: 1414 LVPRPNQ-NVVGTKWVFRNKQDEHGVVTRNKARLVAKGYSQVEGLDFDETYAPVARLESI 1472
Query: 1299 RLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLK 1358
R+LL+YA HG LYQMDVKS FLNG I+EEVYV+QPPGFED ++P+HVY+L K+LYGLK
Sbjct: 1473 RILLAYATYHGFKLYQMDVKSAFLNGPIKEEVYVEQPPGFEDSEYPNHVYRLSKALYGLK 1532
Query: 1359 QAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEF 1418
QAPRAWY+ L +FLI N F+ G+ D TLF +TL+ D+ + QIYVDDIIFGSTN S C+EF
Sbjct: 1533 QAPRAWYECLRDFLIANGFKVGKADPTLFTKTLENDLFVCQIYVDDIIFGSTNESTCEEF 1592
Query: 1419 SKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHP 1478
S++M +FEMSMMGELK+FLG Q+ Q +EG ++ QTKYT+++L KF ++D K + TPM
Sbjct: 1593 SRIMTQKFEMSMMGELKYFLGFQVKQLREGTFISQTKYTQDILAKFGMKDAKPIKTPMGT 1652
Query: 1479 TCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRI 1538
L + G VDQK+YR MIGSLLYL ASRPDI+ SVC+CARFQSDP+ESHLTAVKRI
Sbjct: 1653 NGHLDLDTGGKSVDQKVYRSMIGSLLYLCASRPDIMLSVCMCARFQSDPKESHLTAVKRI 1712
Query: 1539 FRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLIS 1591
RYL T GL Y + + LIG+ DAD+AG +I RKSTSG CQFLG +L+S
Sbjct: 1713 LRYLAYTPKFGLWYPRGSTFDLIGYSDADWAGCKINRKSTSGTCQFLGRSLVS 1765
Score = 115 bits (287), Expect = 2e-23
Identities = 123/484 (25%), Positives = 209/484 (42%), Gaps = 59/484 (12%)
Query: 4 DEESVTTK------YTSVKHDYDTADKKTDS-----GKAPKFNGDPEEFSWWKTNMYSFI 52
D SVT+K Y+ + Y K T GK P FNG E+++ W M +
Sbjct: 95 DAASVTSKRQERKKYSKIPLRYPRVPKHTPLLSVPLGKPPTFNG--EDYAMWSNLMRFHL 152
Query: 53 MGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIPRTEYM 112
L + +WD++E GV + +E D ++ + + I++AS+ + EY
Sbjct: 153 TSLHKRIWDVVEYGVQVPSIGDEDYDTDE------VAQIEHFNSQAATILLASLSKEEYN 206
Query: 113 KMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYELFRMKDDESIEEMYSRFQTLVS 172
K+ AK ++ L EG + K K + + F ++ E ++MY+R +TLV+
Sbjct: 207 KVQGLKNAKEIWDLLKTAHEGDELTKITKRETIEGELGRFHLRQGEEPQDMYNRLKTLVN 266
Query: 173 GLQILKKSYVASDHVSK-ILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLN 231
++ L + V K ILR+L +V I ++ E+++ + E +
Sbjct: 267 QVRNLGSTKWDDHEVVKVILRALIFLNPTQVQLIRGNPRYPLMTPEEVIGNFVSFECMI- 325
Query: 232 EHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLAR 291
+ SKK + PS + A+KA+E ++E + Q + + L N+ L
Sbjct: 326 --KGSKKINELDEPSTSEAQPV--AFKATEEKKEE---STPSRQPIDASKLDNEEMALII 378
Query: 292 KQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRK 351
K + + K+ K+ K +K C+ C KPGHFIA CP + + +G KK K
Sbjct: 379 KSFRQILKQRKGKDYKPRSKKVCYKCGKPGHFIAKCP-MSSDSDRGDDKKGRRKEKKRYY 437
Query: 352 QIKKSLMATWEDLDS-ESGSDKEEADDDAKAAM--GLV-----------------ATVSS 391
+ K + DS ES SD + +D A A+ GL+ S
Sbjct: 438 KKKGGDAHVCREWDSDESSSDSSDDEDAANIAVTKGLLFPNVGHKCLMAKDGKKKVKSKS 497
Query: 392 EAVSEAESDSEDENE------VYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLM 445
E SD +D+NE +++ + E + L EL+S H ++L D +E + L+
Sbjct: 498 STKYETSSDEDDKNEEDNLRILFANL-NMEQKEKLNELISAI-HEKDDLLDSQEDF--LI 553
Query: 446 KQQK 449
K+ K
Sbjct: 554 KENK 557
>gb|AAL35396.1| Opie2a pol [Zea mays]
Length = 1048
Score = 841 bits (2172), Expect = 0.0
Identities = 446/931 (47%), Positives = 593/931 (62%), Gaps = 28/931 (3%)
Query: 680 MTGEKALFLTLTMKDGGE--VKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDGLKHNLLS 736
MTGEK +F + + + FG GK+ G G I SS SI+NV+LV+ L +N LS
Sbjct: 1 MTGEKKMFTSYVKNKDSQDSIIFGDGNQGKVKGLGKIAISSEHSISNVFLVESLGYNFLS 60
Query: 737 ISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDK 796
+SQ C+ Y+ F+ + ++ + D S+ FKG +Y ++F+ CL++
Sbjct: 61 VSQLCNMGYNCLFTNVDVSVFRRSDGSLAFKGVLDGKLYLVDFAKEEAGLDACLIAKTSM 120
Query: 797 KWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVST 856
W+WH+RL H + + K+ K + V GL N+ + D C ACQ GK V+ S +K++++T
Sbjct: 121 GWLWHRRLAHVGMKNLHKLLKGEHVIGLTNVQFKKDRPCVACQAGKQVRGSHHTKNVMTT 180
Query: 857 SRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQ 916
SRPLE+LH+DLFGPV S+ GSKYGLVIVDD+SR+TWV F++ K + + Q
Sbjct: 181 SRPLEMLHMDLFGPVAYLSIGGSKYGLVIVDDFSRFTWVFFLQDKSETQGTLKRYLRRAQ 240
Query: 917 SEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 976
+E ELK+ K+RSD+G EF+N E F + GI HEFS+P TPQQNGVVERKNRTL +MAR
Sbjct: 241 NEFELKVKKIRSDNGSEFKNLQVEEFLVEEGIKHEFSAPYTPQQNGVVERKNRTLMDMAR 300
Query: 977 TMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYI 1036
TM+ E + FW+EAVNT+C+ NR+Y+ +L+ T+YEL G +PN+SYF FG CYI
Sbjct: 301 TMLGEFKTPERFWSEAVNTACHSINRVYLHRLLKNTSYELLTGNKPNVSYFRVFGSKCYI 360
Query: 1037 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPGSKTSEQS 1096
L K KF KA G LGY +KAYRV+N + VE S V FD E EQ
Sbjct: 361 LVKKGRTSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSSDVVFD--ETNGSLREQ- 417
Query: 1097 ESNAGTTDSEDASESDQPSDSEKYTKV-ESSPEAEITPEAESNSEAESSPIVQNESA--- 1152
+ +D E D P+ + + + + P+ + + S++ P +E A
Sbjct: 418 -----VVNLDDVDEEDVPTAAMRTMAIGDVRPQEHLEQDQPSSTTMVHPPTQDDEQAPQV 472
Query: 1153 -------SEDFQDNTQQV-----IQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLI 1200
++D Q ++ Q + + +HP I+G TRS
Sbjct: 473 GAHDQGGAQDVQVEEEEAPQAPPTQVRATIQRNHPVNQILGDISKGVTTRSRLVNFCEHY 532
Query: 1201 GLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLN 1260
+S IEP VEE L D W+LAMQEELN F+RN+VW LVP+P Q N++ TKWVFRNK +
Sbjct: 533 SFVSSIEPFRVEEVLLDPDWVLAMQEELNNFKRNEVWTLVPRPKQ-NVVGTKWVFRNKQD 591
Query: 1261 EQGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSV 1320
E G VTRNKARLVA+GY+Q G+++ ETFAPVARL++IR+ L+YA +H LYQMDVKS
Sbjct: 592 EHGVVTRNKARLVAKGYAQVAGLDFEETFAPVARLKSIRIWLAYAAHHSFRLYQMDVKSA 651
Query: 1321 FLNGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERG 1380
FLNG I+EEVYV+QPPGFED + PDHV KL K+LYGLKQAPRAWY+ L +FLI N F+ G
Sbjct: 652 FLNGPIKEEVYVEQPPGFEDERFPDHVCKLSKALYGLKQAPRAWYECLRDFLIANAFKVG 711
Query: 1381 QVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGI 1440
+ D TLF +T D+ + QIYVDDIIFGSTN + C+EFS++M +FEMSMMGEL +FLG
Sbjct: 712 KADPTLFTKTCDGDLFVCQIYVDDIIFGSTNQNSCEEFSRVMTQKFEMSMMGELSYFLGF 771
Query: 1441 QINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMI 1500
Q+ Q K+G ++ QTKYT++L+K+F ++D K TPM G VDQK YR I
Sbjct: 772 QVRQLKDGTFISQTKYTQDLIKRFGMKDAKPAKTPMGTDGHTDLNKGGKSVDQKAYRSTI 831
Query: 1501 GSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKL 1560
GSLLYL ASRPDI+ SVC+CARFQSDPRE HL AVKRI RYL T G+ Y K + L
Sbjct: 832 GSLLYLCASRPDIMLSVCMCARFQSDPRECHLVAVKRILRYLVATPCFGIWYPKGSTFDL 891
Query: 1561 IGFCDADYAGDRIERKSTSGNCQFLGENLIS 1591
IG+ D+DYA +++RKSTS CQFLG +L+S
Sbjct: 892 IGYSDSDYARCKVDRKSTSRMCQFLGRSLVS 922
>emb|CAE04884.2| OSJNBa0042I15.6 [Oryza sativa (japonica cultivar-group)]
Length = 1510
Score = 813 bits (2099), Expect = 0.0
Identities = 445/953 (46%), Positives = 584/953 (60%), Gaps = 73/953 (7%)
Query: 669 SWYLDSGCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSISINNVWLVD 728
SW +DSGC+ HMTGE+++F +L K G S N+ D
Sbjct: 470 SWVVDSGCTNHMTGERSMFTSLDEKGG------------------------SRENIVFGD 505
Query: 729 GLKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVV 788
K L I + + +DD SI FKG ++Y ++F
Sbjct: 506 DGKEKLQFIIRVS---------------IVRDDSSIAFKGVLKGDLYLVDFDVDRVNPEA 550
Query: 789 CLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSF 848
CL++ + W+WH+RL H R ++ + K + + GL N+ + D +C ACQ GK V S
Sbjct: 551 CLIAKSSMGWLWHRRLAHVGMRNLASLLKGEHILGLSNVSFEMDRVCSACQAGKQVGSPH 610
Query: 849 KSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVF 908
K+I++T+RPLELLH+DLFGPV S+ G+KYG VIVDD+S +TWV F+ K A +VF
Sbjct: 611 PIKNIMTTTRPLELLHMDLFGPVAYISIGGNKYGFVIVDDFSCFTWVYFLHDKSEAQDVF 670
Query: 909 SSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKN 968
F Q Q+ +L I +VRSD+GGEF+N E F ++ GI HEFS+P P QNG+VERKN
Sbjct: 671 KRFTKQAQNLYDLTIKRVRSDNGGEFKNTQVEEFLDEEGIKHEFSAPYDPPQNGIVERKN 730
Query: 969 RTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFH 1028
RTL E AR M+ E + FWAEAV+T+C+ NR+Y+ +L+KT+YEL G++PN+SYF
Sbjct: 731 RTLIEAARAMLDEYKTSDVFWAEAVSTACHAINRLYLHKILKKTSYELLSGKKPNVSYFR 790
Query: 1029 QFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREP 1088
FG +IL+ KF K G LGY + AYRV+N +T + +HV D +P
Sbjct: 791 VFGSKFFILSKMPRSSKFSPKVDEGFLLGYESNAHAYRVFN-KTSGEQVVIHV-VRDVDP 848
Query: 1089 GSKTSEQSESNAGTTDSEDASES-DQP--SDSEKYTKVESSPEAEITPEAESNSEAESSP 1145
++ + +++D E DQP S S T V S+ E E+ + N P
Sbjct: 849 SQAIGTKAIGDIRPVETQDDQEDRDQPPSSTSNSPTSVVSA-EPEVPGPIDRNLRTSPGP 907
Query: 1146 IVQNE-------SASEDF------------------QDNTQQVIQPKFKH--KSSHPEEL 1178
V S SED Q V P+ H + HP +
Sbjct: 908 EVPGSTVRNLRTSGSEDVPTAQVDGIDAAGTLGHTDQAQVPLVHHPRIHHTVQRDHPVDN 967
Query: 1179 IIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWD 1238
I+G TRS +S +EP VE+AL D W++AMQEELN F RN VW+
Sbjct: 968 ILGDIRKGVTTRSRVASFCQHYSFVSSLEPTRVEDALGDSDWVMAMQEELNNFARNQVWN 1027
Query: 1239 LVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETI 1298
LV +P Q N+I TKW+FRNK +E V RNKARLV QG++Q EG+++ ETFAPVARLE+I
Sbjct: 1028 LVERPKQ-NVIGTKWIFRNKQDEHVVVVRNKARLVTQGFTQVEGLDFGETFAPVARLESI 1086
Query: 1299 RLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLK 1358
R+LL+YA +H L+QMDVKS FLNG I E VYV+QPPGFED K P+HVYKL K+LYGLK
Sbjct: 1087 RILLAYAAHHDFRLFQMDVKSAFLNGPISELVYVEQPPGFEDPKLPNHVYKLHKALYGLK 1146
Query: 1359 QAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEF 1418
QAPRAWY+ L +FL+KN FE G DTTLF + K D+ I QIYVDDIIFGSTNAS C+EF
Sbjct: 1147 QAPRAWYECLRDFLLKNGFEIGNADTTLFTKKFKSDLFICQIYVDDIIFGSTNASFCEEF 1206
Query: 1419 SKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHP 1478
S +M FEMSMMGEL FFL +Q+ Q++EG ++ QTKY K++LKKF +ED K + TPM
Sbjct: 1207 SSIMTKRFEMSMMGELTFFLWLQVKQAQEGTFISQTKYVKDILKKFGMEDAKPIKTPMPT 1266
Query: 1479 TCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRI 1538
L +D G VDQK+YR MIGSLLYL ASRPDI+ SVC+CARFQ++P+E HL AVKRI
Sbjct: 1267 NGHLDLDDNGKCVDQKVYRSMIGSLLYLCASRPDIMLSVCMCARFQAEPKECHLIAVKRI 1326
Query: 1539 FRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLIS 1591
RYL T NLGL Y K D++L+G+ D+DYAG +++RKS +G CQFLG +L+S
Sbjct: 1327 QRYLVHTPNLGLWYPKGCDFELLGYSDSDYAGCKVDRKSITGTCQFLGPSLVS 1379
Score = 96.3 bits (238), Expect = 8e-18
Identities = 108/436 (24%), Positives = 177/436 (39%), Gaps = 87/436 (19%)
Query: 52 IMGLDEELWDILEDGVD----DLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIP 107
++ L +W ++ GVD D++L E Q++L ++ + I++++
Sbjct: 5 LISLHPSIWKVVCTGVDVPHDDMELTSE------------QEQLIHRNAQASNAILSTLS 52
Query: 108 RTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYELFRMKDDESIEEMYSRF 167
E+ K+ AK + +L EGS V+EAK +L + F M D E+ +EMY R
Sbjct: 53 LEEFNKVDGLEEAKEICDTLQLAHEGSPAVREAKIELLEGRLGRFVMDDKETPQEMYDRM 112
Query: 168 QTLVSGLQILKKSYVASDHVSK-ILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVH 226
LV+ ++ L + + V K +LR R V+ I E KD L++ D++ + H
Sbjct: 113 MILVNKIKGLGSEDMTNHFVVKRLLREFGPRNPTLVSMIREKKDFKRLTLSDILGRIVSH 172
Query: 227 EMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKL 286
EM E KT + K +K + G D+D+
Sbjct: 173 EMQEEEAR--------------KTRRRVKRFKHFLRKSGYGKGRKDDDK----------- 207
Query: 287 EYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKFKGKSKKSSFNS 346
KK+ ++ CFNC + GHFIAD P + K KG KK
Sbjct: 208 -------------------GKKQSKRACFNCGEYGHFIADFPKSNEAKAKGGKKKPE--- 245
Query: 347 SKFRKQIKKSLM-ATWEDLDSESGSDKEEADDDAKA-AMGLVATVSSEAVSEAE------ 398
R + ++ M W D E K + K G VATV+ ++ S ++
Sbjct: 246 ---RAHVAEAHMPEVWYSGDEEDPEVKPKPKPKDKVDGEGGVATVTFKSSSSSKERLFNN 302
Query: 399 -SDSEDENEVYS-------KIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKS 450
SD +D++ YS K+ Q+ + ++ S E NEL D+ + + Q +
Sbjct: 303 LSDDDDDSYHYSCFMAQGRKVMTQKPSHTSLDVDSSDEESDNELDDVLKSFSKPAMQHLA 362
Query: 451 TLLE----LKASEEEL 462
L+ LK E L
Sbjct: 363 KLMRALDTLKKENERL 378
>gb|AAW57789.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1799
Score = 791 bits (2042), Expect = 0.0
Identities = 431/935 (46%), Positives = 580/935 (61%), Gaps = 45/935 (4%)
Query: 666 KQRSWYLDSGCSRHMTGEKALFLTLTMKDGGE-VKFGGNQTGKIIGTGTIG-NSSISINN 723
+ SW +DSGCSRHMTGE F +LT G E + FG +G+++ GTI N + +
Sbjct: 775 RSNSWLVDSGCSRHMTGEAKWFTSLTRASGDETITFGDASSGRVMAKGTIKVNDKFMLKD 834
Query: 724 VWLVDGLKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLA 783
V LV LK+NLLS+SQ CD +V F K +++ + + F RV V+ NF A
Sbjct: 835 VALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFDSSA 893
Query: 784 DQKVVCLL-SMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGK 842
CL+ S N + WH+RLGH + +S+IS + L++GLP + D +C C+ GK
Sbjct: 894 PGPSRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVPKDLVCAPCRHGK 953
Query: 843 IVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYG------SKYGLVIVDDYSRWTWVK 896
+ SS K +V T P +LLH+D GP S+ G + + L++V W W
Sbjct: 954 MTSSSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWRKHLASFSLLLV----VWLWSF 1009
Query: 897 FIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPR 956
+ + +A V + ++I L ++ + S+ H+FSSP
Sbjct: 1010 LVLFEPFA--VIMALNSKI-LHLNLSVILLESN--------------------HQFSSPY 1046
Query: 957 TPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYEL 1016
PQQNGVVERKNRTL EMARTM+ E + FW EA++ +C+I NR+++R +L KT YEL
Sbjct: 1047 VPQQNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYEL 1106
Query: 1017 FKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVE 1076
GRRP +S+ FGC C++L + + L KF++++ GIFLGY+ S+AYRVY T +
Sbjct: 1107 RFGRRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIV 1165
Query: 1077 ESMHVKFDDREPGSKTSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAE 1136
E+ V FD+ PG++ + ED+ + D S S ++S+P + T
Sbjct: 1166 ETCEVTFDEASPGARPEISGVPDESIFVDEDSDDDDDDSIS---PPLDSTPPVQETGSPL 1222
Query: 1137 SNSEAESSPIVQNESASEDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQE 1196
+ S + +P + SA+E+ T P+ P+ +I G + R RS+
Sbjct: 1223 TTSPSGDAP-TTSSSAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYELVN 1281
Query: 1197 ESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFR 1256
+ + + EPK V ALSD+ W+ AM EEL F+RN VW LV P ++I TKWVF+
Sbjct: 1282 SAFV---ASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFSVIGTKWVFK 1338
Query: 1257 NKLNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMD 1316
NKL E G + RNKARLVAQG++Q EG+++ ETFAPVARLE IR+LL++A + G L+QMD
Sbjct: 1339 NKLGEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMD 1398
Query: 1317 VKSVFLNGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKND 1376
VKS FLNGVIEEEVYVKQPPGFE+ K P+HV+KL K+LYGLKQAPRAWY+RL FL++N
Sbjct: 1399 VKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLDKALYGLKQAPRAWYERLKTFLLQNG 1458
Query: 1377 FERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKF 1436
FE G VD TLF D L+VQIYVDDIIFG ++ +L +F +M EFEMSMMGEL F
Sbjct: 1459 FEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFFDVMSREFEMSMMGELTF 1518
Query: 1437 FLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLY 1496
FLG+QI Q+KEG++VHQTKY+KELLKKF + DCK + TPM T +L ++ G VDQ+ Y
Sbjct: 1519 FLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREY 1578
Query: 1497 RGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSL 1556
R MIGSLLYLTASRPDI FSVCLCARFQ+ PR SH AVKRIFRY+K T G+ Y S
Sbjct: 1579 RSMIGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRIFRYIKSTLEYGIWYSCSS 1638
Query: 1557 DYKLIGFCDADYAGDRIERKSTSGNCQFLGENLIS 1591
+ F DAD+AG +I+RKSTSG C FLG +L+S
Sbjct: 1639 ALSVRAFSDADFAGCKIDRKSTSGTCHFLGTSLVS 1673
Score = 115 bits (289), Expect = 9e-24
Identities = 157/678 (23%), Positives = 270/678 (39%), Gaps = 87/678 (12%)
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGV---DDLDLDEEGAAIDRRIHTPAQKKLYK 93
D F WK M +++ +W+ ++ DD+D+ TPA
Sbjct: 13 DGHNFVIWKARMEAYLPSQGHNVWNKVKSPYTVPDDVDI------------TPANMAQVD 60
Query: 94 KHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVH-QYELF 152
+++ R I+ I E+ ++ +A M+ +LC EG+ +++ L H +Y+ F
Sbjct: 61 FNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQ----LNQFHKEYQRF 116
Query: 153 RMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIEEAKDL 211
M ESI+ + RF +VS L+ + K + +D+ +L L W KVT+I E+ L
Sbjct: 117 EMHPGESIDSYFKRFGEIVSKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVTSITESAPL 176
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ L+++ L S LK HEM + + K S ++ G TS + A+ + +
Sbjct: 177 SELTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAFVCGGFSLAALHSVT 236
Query: 272 DEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIADCPDLQ 331
+E + + L ARK + K K + CF C +P H +CP L+
Sbjct: 237 EEQLE---KIPEDDLALFARKFSRAYKNVRDRKRGKTNEPFVCFECGEPNHIRVNCPKLK 293
Query: 332 KEKFKGKSKKSSFNSSKFRKQIKK----SLMATWEDLD-SESGSDKEEADDDAKAAMGLV 386
K+ K +KK K +K + K ++A E++ S+ SD ++ + K G+
Sbjct: 294 KKSDK-TTKKPEGQGRKGKKDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDKDFSGMC 352
Query: 387 ATVSSE--------AVSEAESDSEDENEVYSKIPRQE--LVDSLKELLSLFEHRTNELTD 436
++E A+ + + SE IP + L D S ++ T +
Sbjct: 353 CLANNEDFINLCLMALEDKDDSSEHPEVCLDDIPSLDGSLCDD-----SCSDNDTVDDEL 407
Query: 437 LKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKHE 496
KE+ LM + K E+LK N RL+S+ + ++ C + E
Sbjct: 408 SKERMAHLMIEISDKYRSSKYKIEKLKSENDGMALEIARLRSMIPE-EDTCSTCASYLSE 466
Query: 497 I-ALDDFIMA---GIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKS 552
I L D + + G AS ST +G E + KE LK
Sbjct: 467 INLLKDKLKSCTLGAGNPSSASAACSTCYEMKVDMGLLEMELKE-------------LKE 513
Query: 553 TFVPEGTNAVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQN 612
FV + + ++ P + ++ + L+ TK+D ++ EP+H +
Sbjct: 514 KFVHD---RIGRCENCPILTSDNDELRQQVAMLR----TKND----LLESFATKEPIHSS 562
Query: 613 LIKPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRSWYL 672
I K + + K S K+I D I KV L + +K+ S YL
Sbjct: 563 C--ANCAILKTELKDAKTVIDSIKSI----------DSCSSCISLKVDLESAKKENS-YL 609
Query: 673 DSGCSRHMTGEKALFLTL 690
R G+K L + L
Sbjct: 610 QQSLERFAQGKKKLNMIL 627
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.131 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,632,398,003
Number of Sequences: 2540612
Number of extensions: 113943361
Number of successful extensions: 443953
Number of sequences better than 10.0: 7268
Number of HSP's better than 10.0 without gapping: 3556
Number of HSP's successfully gapped in prelim test: 3897
Number of HSP's that attempted gapping in prelim test: 400936
Number of HSP's gapped (non-prelim): 30474
length of query: 1591
length of database: 863,360,394
effective HSP length: 142
effective length of query: 1449
effective length of database: 502,593,490
effective search space: 728257967010
effective search space used: 728257967010
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 82 (36.2 bits)
Medicago: description of AC139344.2


