

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137824.11 - phase: 0
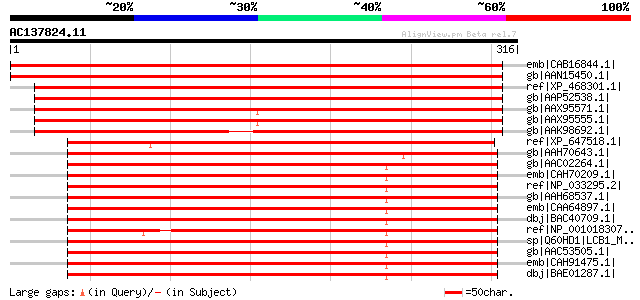
(316 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB16844.1| serine C-palmitoyltransferase like protein [Arab... 488 e-137
gb|AAN15450.1| serine C-palmitoyltransferase like protein [Arabi... 488 e-137
ref|XP_468301.1| putative serine palmitoyltransferase LCB1 subun... 443 e-123
gb|AAP52538.1| putative serine palmitoyltransferase [Oryza sativ... 431 e-119
gb|AAX95571.1| hypothetical protein [Oryza sativa (japonica cult... 417 e-115
gb|AAX95555.1| expressed protein [Oryza sativa (japonica cultiva... 417 e-115
gb|AAK98692.1| Putative serine palmitoyltransferase [Oryza sativa] 385 e-105
ref|XP_647518.1| hypothetical protein DDB0189743 [Dictyostelium ... 263 5e-69
gb|AAH70643.1| MGC81520 protein [Xenopus laevis] 257 3e-67
gb|AAC02264.1| serine palmitoyltransferase LCB1 subunit [Mus mus... 256 5e-67
emb|CAH70209.1| serine palmitoyltransferase, long chain base sub... 255 1e-66
ref|NP_033295.2| serine palmitoyltransferase subunit 1 [Mus musc... 255 1e-66
gb|AAH68537.1| SPTLC1 protein [Homo sapiens] 255 1e-66
emb|CAA64897.1| serine C-palmitoyltransferase [Mus musculus] gi|... 255 1e-66
dbj|BAC40709.1| unnamed protein product [Mus musculus] 255 1e-66
ref|NP_001018307.1| hypothetical protein LOC553981 [Danio rerio]... 253 4e-66
sp|Q60HD1|LCB1_MACFA Serine palmitoyltransferase 1 (Long chain b... 253 5e-66
gb|AAC53505.1| serine palmitoyltransferase LCB1 subunit [Cricetu... 253 5e-66
emb|CAH91475.1| hypothetical protein [Pongo pygmaeus] 253 7e-66
dbj|BAE01287.1| unnamed protein product [Macaca fascicularis] 250 3e-65
>emb|CAB16844.1| serine C-palmitoyltransferase like protein [Arabidopsis thaliana]
gi|7270596|emb|CAB80314.1| serine C-palmitoyltransferase
like protein [Arabidopsis thaliana]
gi|25407728|pir||F85430 serine C-palmitoyltransferase
like protein [imported] - Arabidopsis thaliana
Length = 475
Score = 488 bits (1256), Expect = e-137
Identities = 234/307 (76%), Positives = 269/307 (87%)
Query: 1 MASSFINFVNTTIDLVTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPP 60
MAS+ + N ++ VT L +PSAR V+FG I GH F+E LL VVI+ LL++KSYKPP
Sbjct: 1 MASNLVEMFNAALNWVTMILESPSARVVLFGVPIRGHFFVEGLLGVVIIILLTRKSYKPP 60
Query: 61 KRPLSNKEIDELCDEWVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASANYL 120
KRPL+ +EIDELCDEWVP+PLIP + ++M +EPPVLESAAGPHT VNGK+VVNFASANYL
Sbjct: 61 KRPLTEQEIDELCDEWVPEPLIPPITEDMKHEPPVLESAAGPHTTVNGKDVVNFASANYL 120
Query: 121 GLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTPDSILYSYGLS 180
GLIGH+KLL+SC+SALEKYGVGSCGPRGFYGTIDVHLDCE RISKFLGTPDSILYSYGLS
Sbjct: 121 GLIGHEKLLESCTSALEKYGVGSCGPRGFYGTIDVHLDCETRISKFLGTPDSILYSYGLS 180
Query: 181 TMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRETLENITSIYK 240
TMFS IP F KKGD+IVADEGVHWGIQNGL LSRST+VYFKHN+M+SLR TLE I + YK
Sbjct: 181 TMFSTIPCFCKKGDVIVADEGVHWGIQNGLQLSRSTIVYFKHNDMESLRITLEKIMTKYK 240
Query: 241 RTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFGVLGSSGRGLTEHY 300
R+KNLRRYIV EA+YQNSGQIAPLDEI+KLKEKY FR++LDESNSFGVLG SGRGL EH+
Sbjct: 241 RSKNLRRYIVAEAVYQNSGQIAPLDEIVKLKEKYRFRVILDESNSFGVLGRSGRGLAEHH 300
Query: 301 GVPVCRV 307
VP+ ++
Sbjct: 301 SVPIEKI 307
>gb|AAN15450.1| serine C-palmitoyltransferase like protein [Arabidopsis thaliana]
gi|21539529|gb|AAM53317.1| serine C-palmitoyltransferase
like protein [Arabidopsis thaliana]
gi|18419845|ref|NP_568005.1| aminotransferase class I
and II family protein [Arabidopsis thaliana]
gi|14488275|dbj|BAB60898.1| serine palmitoyltransferase
[Arabidopsis thaliana]
Length = 482
Score = 488 bits (1256), Expect = e-137
Identities = 234/307 (76%), Positives = 269/307 (87%)
Query: 1 MASSFINFVNTTIDLVTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPP 60
MAS+ + N ++ VT L +PSAR V+FG I GH F+E LL VVI+ LL++KSYKPP
Sbjct: 1 MASNLVEMFNAALNWVTMILESPSARVVLFGVPIRGHFFVEGLLGVVIIILLTRKSYKPP 60
Query: 61 KRPLSNKEIDELCDEWVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASANYL 120
KRPL+ +EIDELCDEWVP+PLIP + ++M +EPPVLESAAGPHT VNGK+VVNFASANYL
Sbjct: 61 KRPLTEQEIDELCDEWVPEPLIPPITEDMKHEPPVLESAAGPHTTVNGKDVVNFASANYL 120
Query: 121 GLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTPDSILYSYGLS 180
GLIGH+KLL+SC+SALEKYGVGSCGPRGFYGTIDVHLDCE RISKFLGTPDSILYSYGLS
Sbjct: 121 GLIGHEKLLESCTSALEKYGVGSCGPRGFYGTIDVHLDCETRISKFLGTPDSILYSYGLS 180
Query: 181 TMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRETLENITSIYK 240
TMFS IP F KKGD+IVADEGVHWGIQNGL LSRST+VYFKHN+M+SLR TLE I + YK
Sbjct: 181 TMFSTIPCFCKKGDVIVADEGVHWGIQNGLQLSRSTIVYFKHNDMESLRITLEKIMTKYK 240
Query: 241 RTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFGVLGSSGRGLTEHY 300
R+KNLRRYIV EA+YQNSGQIAPLDEI+KLKEKY FR++LDESNSFGVLG SGRGL EH+
Sbjct: 241 RSKNLRRYIVAEAVYQNSGQIAPLDEIVKLKEKYRFRVILDESNSFGVLGRSGRGLAEHH 300
Query: 301 GVPVCRV 307
VP+ ++
Sbjct: 301 SVPIEKI 307
>ref|XP_468301.1| putative serine palmitoyltransferase LCB1 subunit [Oryza sativa
(japonica cultivar-group)] gi|51964496|ref|XP_507033.1|
PREDICTED OJ1111_C07.25 gene product [Oryza sativa
(japonica cultivar-group)] gi|47497351|dbj|BAD19391.1|
putative serine palmitoyltransferase LCB1 subunit [Oryza
sativa (japonica cultivar-group)]
Length = 481
Score = 443 bits (1140), Expect = e-123
Identities = 211/292 (72%), Positives = 250/292 (85%)
Query: 16 VTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPPKRPLSNKEIDELCDE 75
V+ +AP ARAVVFG +I GHL +E LL+ ILF LS+KSYKPPK+PL+ +E+DELCDE
Sbjct: 18 VSAMFNAPLARAVVFGIHIDGHLVVEGLLIAAILFQLSRKSYKPPKKPLTEREVDELCDE 77
Query: 76 WVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSA 135
W P+PL P + + E P LESAAGPHTIV+GKEVVNFASANYLGLIG++K+LDSC +
Sbjct: 78 WQPEPLCPPIKEGARIEAPTLESAAGPHTIVDGKEVVNFASANYLGLIGNEKILDSCIGS 137
Query: 136 LEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDI 195
+EKYGVGSCGPRGFYGTIDVHLDCE +I+KFLGT DSILYSYG+ST+FS IPAF KKGDI
Sbjct: 138 VEKYGVGSCGPRGFYGTIDVHLDCETKIAKFLGTQDSILYSYGISTIFSVIPAFCKKGDI 197
Query: 196 IVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALY 255
IVADEGVHW +QNGL LSRSTVVYFKHN+M SL TLE +T KRT+ +RRYIV+EA+Y
Sbjct: 198 IVADEGVHWAVQNGLQLSRSTVVYFKHNDMASLASTLEKLTHGNKRTEKIRRYIVVEAIY 257
Query: 256 QNSGQIAPLDEIIKLKEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPVCRV 307
QNSGQIAPLDEI++LKEKY FR++L+ES+SFGVLG SGRGL EHYGVP+ ++
Sbjct: 258 QNSGQIAPLDEIVRLKEKYRFRVILEESHSFGVLGKSGRGLAEHYGVPIEKI 309
>gb|AAP52538.1| putative serine palmitoyltransferase [Oryza sativa (japonica
cultivar-group)] gi|37531898|ref|NP_920251.1| putative
serine palmitoyltransferase [Oryza sativa (japonica
cultivar-group)]
Length = 481
Score = 431 bits (1109), Expect = e-119
Identities = 204/292 (69%), Positives = 245/292 (83%)
Query: 16 VTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPPKRPLSNKEIDELCDE 75
V+ +AP ARAVVFG +I GHL +E LL+ ILF LS+KSYKPPK+PL+ +E+DELCD+
Sbjct: 18 VSAVFNAPLARAVVFGIHIDGHLVVEGLLIAAILFQLSRKSYKPPKKPLTEREVDELCDD 77
Query: 76 WVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSA 135
W P+PL P + + + P LESAAGPHT V+GKEVVNFASANYLGLIG++K++DSC +
Sbjct: 78 WQPEPLCPPIKEGARIDTPTLESAAGPHTTVDGKEVVNFASANYLGLIGNEKIIDSCVGS 137
Query: 136 LEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDI 195
+EKYGVGSCGPR FYGTIDVHLDCE +I+ FLGT DSILYSYG+ST+FS IPAF KKGDI
Sbjct: 138 VEKYGVGSCGPRSFYGTIDVHLDCESKIANFLGTQDSILYSYGISTIFSVIPAFCKKGDI 197
Query: 196 IVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALY 255
IVADEGVHW +QNGL LSRSTVVYFKHN+M SL LE +T K T+ +RRYIV+EA+Y
Sbjct: 198 IVADEGVHWAVQNGLQLSRSTVVYFKHNDMASLASILEKLTHGNKHTEKIRRYIVVEAIY 257
Query: 256 QNSGQIAPLDEIIKLKEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPVCRV 307
QNSGQIAPLDEI++LKEKY FR++L+ES+SFGVLG SGRGL EHYGVPV ++
Sbjct: 258 QNSGQIAPLDEIVRLKEKYRFRVILEESHSFGVLGKSGRGLAEHYGVPVEKI 309
>gb|AAX95571.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 508
Score = 417 bits (1071), Expect = e-115
Identities = 204/319 (63%), Positives = 245/319 (75%), Gaps = 27/319 (8%)
Query: 16 VTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPPKRPLSNKEIDELCDE 75
V+ +AP ARAVVFG +I GHL +E LL+ ILF LS+KSYKPPK+PL+ +E+DELCD+
Sbjct: 18 VSAVFNAPLARAVVFGIHIDGHLVVEGLLIAAILFQLSRKSYKPPKKPLTEREVDELCDD 77
Query: 76 WVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSA 135
W P+PL P + + + P LESAAGPHT V+GKEVVNFASANYLGLIG++K++DSC +
Sbjct: 78 WQPEPLCPPIKEGARIDTPTLESAAGPHTTVDGKEVVNFASANYLGLIGNEKIIDSCVGS 137
Query: 136 LEKYGVGSCGPRGFYGTI---------------------------DVHLDCEGRISKFLG 168
+EKYGVGSCGPR FYGTI DVHLDCE +I+ FLG
Sbjct: 138 VEKYGVGSCGPRSFYGTIGSYLLWLFSIIYILWPQKKNLAILWFADVHLDCESKIANFLG 197
Query: 169 TPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSL 228
T DSILYSYG+ST+FS IPAF KKGDIIVADEGVHW +QNGL LSRSTVVYFKHN+M SL
Sbjct: 198 TQDSILYSYGISTIFSVIPAFCKKGDIIVADEGVHWAVQNGLQLSRSTVVYFKHNDMASL 257
Query: 229 RETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFGV 288
LE +T K T+ +RRYIV+EA+YQNSGQIAPLDEI++LKEKY FR++L+ES+SFGV
Sbjct: 258 ASILEKLTHGNKHTEKIRRYIVVEAIYQNSGQIAPLDEIVRLKEKYRFRVILEESHSFGV 317
Query: 289 LGSSGRGLTEHYGVPVCRV 307
LG SGRGL EHYGVPV ++
Sbjct: 318 LGKSGRGLAEHYGVPVEKI 336
>gb|AAX95555.1| expressed protein [Oryza sativa (japonica cultivar-group)]
Length = 509
Score = 417 bits (1071), Expect = e-115
Identities = 204/319 (63%), Positives = 245/319 (75%), Gaps = 27/319 (8%)
Query: 16 VTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPPKRPLSNKEIDELCDE 75
V+ +AP ARAVVFG +I GHL +E LL+ ILF LS+KSYKPPK+PL+ +E+DELCD+
Sbjct: 19 VSAVFNAPLARAVVFGIHIDGHLVVEGLLIAAILFQLSRKSYKPPKKPLTEREVDELCDD 78
Query: 76 WVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSA 135
W P+PL P + + + P LESAAGPHT V+GKEVVNFASANYLGLIG++K++DSC +
Sbjct: 79 WQPEPLCPPIKEGARIDTPTLESAAGPHTTVDGKEVVNFASANYLGLIGNEKIIDSCVGS 138
Query: 136 LEKYGVGSCGPRGFYGTI---------------------------DVHLDCEGRISKFLG 168
+EKYGVGSCGPR FYGTI DVHLDCE +I+ FLG
Sbjct: 139 VEKYGVGSCGPRSFYGTIGSYLLWLFSIIYILWPQKKNLAILWFADVHLDCESKIANFLG 198
Query: 169 TPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSL 228
T DSILYSYG+ST+FS IPAF KKGDIIVADEGVHW +QNGL LSRSTVVYFKHN+M SL
Sbjct: 199 TQDSILYSYGISTIFSVIPAFCKKGDIIVADEGVHWAVQNGLQLSRSTVVYFKHNDMASL 258
Query: 229 RETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFGV 288
LE +T K T+ +RRYIV+EA+YQNSGQIAPLDEI++LKEKY FR++L+ES+SFGV
Sbjct: 259 ASILEKLTHGNKHTEKIRRYIVVEAIYQNSGQIAPLDEIVRLKEKYRFRVILEESHSFGV 318
Query: 289 LGSSGRGLTEHYGVPVCRV 307
LG SGRGL EHYGVPV ++
Sbjct: 319 LGKSGRGLAEHYGVPVEKI 337
>gb|AAK98692.1| Putative serine palmitoyltransferase [Oryza sativa]
Length = 566
Score = 385 bits (988), Expect = e-105
Identities = 192/292 (65%), Positives = 230/292 (78%), Gaps = 14/292 (4%)
Query: 16 VTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPPKRPLSNKEIDELCDE 75
V+ +AP ARAVVFG +I GHL +E LL+ ILF LS+KSYKPPK+PL+ +E+DELCDE
Sbjct: 117 VSAMFNAPLARAVVFGIHIDGHLVVEGLLIAAILFQLSRKSYKPPKKPLTEREVDELCDE 176
Query: 76 WVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSA 135
W P+PL P + + E P LESAAGPHTIV+GKEVVNFASANYLGLIG++K+L +
Sbjct: 177 WQPEPLCPPIKEGARIEAPTLESAAGPHTIVDGKEVVNFASANYLGLIGNEKILGRNLAI 236
Query: 136 LEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDI 195
L DVHLDCE +I+KFLGT DSILYSYG+ST+FS IPAF KKGDI
Sbjct: 237 LL--------------FSDVHLDCETKIAKFLGTQDSILYSYGISTIFSVIPAFCKKGDI 282
Query: 196 IVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALY 255
IVADEGVHW +QNGL LSRSTVVYFKHN+M SL TLE +T KRT+ +RRYIV+EA+Y
Sbjct: 283 IVADEGVHWAVQNGLQLSRSTVVYFKHNDMASLASTLEKLTHGNKRTEKIRRYIVVEAIY 342
Query: 256 QNSGQIAPLDEIIKLKEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPVCRV 307
QNSGQIAPLDEI++LKEKY FR++L+ES+SFGVLG SGRGL EHYGVP+ ++
Sbjct: 343 QNSGQIAPLDEIVRLKEKYRFRVILEESHSFGVLGKSGRGLAEHYGVPIEKI 394
>ref|XP_647518.1| hypothetical protein DDB0189743 [Dictyostelium discoideum]
gi|60475546|gb|EAL73481.1| hypothetical protein
DDB0189743 [Dictyostelium discoideum]
Length = 479
Score = 263 bits (672), Expect = 5e-69
Identities = 131/269 (48%), Positives = 185/269 (68%), Gaps = 3/269 (1%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLN--DEMPYEP 93
+LFI L+ V I++LL+++ +KP + L+ E DEL EW P PL P N + +
Sbjct: 25 NLFIHGLMAVFIIYLLTKRPFKPRQNDTLTKAEEDELIREWSPIPLTPKSNPLEVLNQTE 84
Query: 94 PVLESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTI 153
+++ + G H +N K+ +N A +NYLGLI + ++ +A+ KYGVGSCGPRGFYGTI
Sbjct: 85 LIIQESEGTHVTINNKKYLNLARSNYLGLINNPEINKISENAIRKYGVGSCGPRGFYGTI 144
Query: 154 DVHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLS 213
DVHLD E + + F+ TP+++LYS +T+ SAIP+FSK GDII+ D GV +Q G+ LS
Sbjct: 145 DVHLDLEKKTASFMKTPEAVLYSSAYATISSAIPSFSKIGDIIIVDRGVSQPVQVGVSLS 204
Query: 214 RSTVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEK 273
RS + YF HN+MD L+ LE + K +R+++VIE LY NSG IAPL +I+K KE+
Sbjct: 205 RSRIYYFNHNDMDDLQRVLEQTQFKGSKAKIVRKFVVIEGLYYNSGTIAPLPQILKFKEQ 264
Query: 274 YCFRILLDESNSFGVLGSSGRGLTEHYGV 302
Y FR+++DES+S GVLGS+GRGLTEHY +
Sbjct: 265 YKFRLIMDESHSVGVLGSTGRGLTEHYNI 293
>gb|AAH70643.1| MGC81520 protein [Xenopus laevis]
Length = 472
Score = 257 bits (657), Expect = 3e-67
Identities = 134/274 (48%), Positives = 188/274 (67%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ L+ K+YK +R L+ KE +EL DEW P+PL+P ++ + P
Sbjct: 23 HLILEGILILWIIRLIFFKTYKLQERSDLTEKEKEELIDEWRPEPLVPPVSKDHPALNYN 82
Query: 96 LESAAGPHTIV-NGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H IV NGKE +NFAS N+LGL+ + ++ + ++L KYGVG+CGPRGFYGT D
Sbjct: 83 IVSGPPSHKIVVNGKECINFASFNFLGLLDNARVKSAALASLRKYGVGTCGPRGFYGTFD 142
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHL+ E R++KF+ T ++I+YSYG +T+ SAIPA+SK+GDI+ ADE + IQ GL SR
Sbjct: 143 VHLELEERLAKFMKTEEAIIYSYGFATVASAIPAYSKRGDIVFADEAACFAIQKGLQASR 202
Query: 215 STVVYFKHNNMDSLRETL-ENITSIYKRTKN---LRRYIVIEALYQNSGQIAPLDEIIKL 270
S++ YFKHN+MD L L E K + RR+IV E LY N+G I PL ++++L
Sbjct: 203 SSIKYFKHNDMDDLERLLKEQELEDQKNPRKACVTRRFIVAEGLYMNTGDICPLPKLVEL 262
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEH+G+ +
Sbjct: 263 KYKYKVRIFLEESLSFGVLGEHGRGVTEHFGINI 296
>gb|AAC02264.1| serine palmitoyltransferase LCB1 subunit [Mus musculus]
Length = 473
Score = 256 bits (655), Expect = 5e-67
Identities = 134/274 (48%), Positives = 186/274 (66%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ L+ K+YK +R L+ KE +EL +EW P+PL+P ++ P
Sbjct: 24 HLILEGILILWIIRLVFSKTYKLQERSDLTAKEKEELIEEWQPEPLVPPVSKNHPALNYN 83
Query: 96 LESAAGPHTIV-NGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ES H IV NGKE VNFAS N+LGL+ + ++ + S+L+KYGVG+CGPRGFYGT D
Sbjct: 84 IESGPPTHNIVVNGKECVNFASFNFLGLLANPRVKAAAFSSLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+Y+YG ST+ SAIPA+SK+GDII D + IQ GL SR
Sbjct: 144 VHLDLEERLAKFMKTEEAIIYTYGFSTVASAIPAYSKRGDIIFVDSAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN++ L L+ ++ + RR+IV+E LY N+G I PL E++KL
Sbjct: 204 SDIKLFKHNDVADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTICPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGISI 297
>emb|CAH70209.1| serine palmitoyltransferase, long chain base subunit 1 [Homo
sapiens] gi|55662329|emb|CAH69924.1| serine
palmitoyltransferase, long chain base subunit 1 [Homo
sapiens] gi|5454084|ref|NP_006406.1| serine
palmitoyltransferase subunit 1 isoform a [Homo sapiens]
gi|13540158|gb|AAK29328.1| serine palmitoyltransferase
[Homo sapiens] gi|2564247|emb|CAA69941.1| serine
palmitoyltransferase, subunit I [Homo sapiens]
gi|6685579|sp|O15269|LCB1_HUMAN Serine
palmitoyltransferase 1 (Long chain base biosynthesis
protein 1) (LCB 1) (Serine-palmitoyl-CoA transferase 1)
(SPT 1) (SPT1)
Length = 473
Score = 255 bits (651), Expect = 1e-66
Identities = 132/274 (48%), Positives = 186/274 (67%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ LL K+YK +R L+ KE +EL +EW P+PL+P + + P
Sbjct: 24 HLILEGILILWIIRLLFSKTYKLQERSDLTVKEKEELIEEWQPEPLVPPVPKDHPALNYN 83
Query: 96 LESAAGPH-TIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H T+VNGKE +NFAS N+LGL+ + ++ + ++L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPSHKTVVNGKECINFASFNFLGLLDNPRVKAAALASLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG +T+ SAIPA+SK+GDI+ D + IQ GL SR
Sbjct: 144 VHLDLEDRLAKFMKTEEAIIYSYGFATIASAIPAYSKRGDIVFVDRAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN+M L L+ ++ + RR+IV+E LY N+G I PL E++KL
Sbjct: 204 SDIKLFKHNDMADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTICPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGINI 297
>ref|NP_033295.2| serine palmitoyltransferase subunit 1 [Mus musculus]
gi|28374372|gb|AAH46323.1| Serine palmitoyltransferase
subunit 1 [Mus musculus] gi|26351097|dbj|BAC39185.1|
unnamed protein product [Mus musculus]
gi|26351071|dbj|BAC39172.1| unnamed protein product [Mus
musculus] gi|26348084|dbj|BAC37690.1| unnamed protein
product [Mus musculus] gi|26343307|dbj|BAC35310.1|
unnamed protein product [Mus musculus]
Length = 473
Score = 255 bits (651), Expect = 1e-66
Identities = 134/274 (48%), Positives = 185/274 (66%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ L+ K+YK +R L+ KE +EL +EW P+PL+P ++ P
Sbjct: 24 HLILEGILILWIIRLVFSKTYKLQERSDLTAKEKEELIEEWQPEPLVPPVSKNHPALNYN 83
Query: 96 LESAAGPHTIV-NGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H IV NGKE VNFAS N+LGL+ + ++ + S+L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPTHNIVVNGKECVNFASFNFLGLLANPRVKATAFSSLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG ST+ SAIPA+SK+GDII D + IQ GL SR
Sbjct: 144 VHLDLEERLAKFMKTEEAIIYSYGFSTIASAIPAYSKRGDIIFVDSAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN++ L L+ ++ + RR+IV+E LY N+G I PL E++KL
Sbjct: 204 SDIKLFKHNDVADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTICPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGISI 297
>gb|AAH68537.1| SPTLC1 protein [Homo sapiens]
Length = 513
Score = 255 bits (651), Expect = 1e-66
Identities = 132/274 (48%), Positives = 186/274 (67%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ LL K+YK +R L+ KE +EL +EW P+PL+P + + P
Sbjct: 24 HLILEGILILWIIRLLFSKTYKLQERSDLTVKEKEELIEEWQPEPLVPPVPKDHPALNYN 83
Query: 96 LESAAGPH-TIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H T+VNGKE +NFAS N+LGL+ + ++ + ++L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPSHKTVVNGKECINFASFNFLGLLDNPRVKAAALASLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG +T+ SAIPA+SK+GDI+ D + IQ GL SR
Sbjct: 144 VHLDLEDRLAKFMKTEEAIIYSYGFATIASAIPAYSKRGDIVFVDRAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN+M L L+ ++ + RR+IV+E LY N+G I PL E++KL
Sbjct: 204 SDIKLFKHNDMADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTICPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGINI 297
>emb|CAA64897.1| serine C-palmitoyltransferase [Mus musculus]
gi|6685592|sp|O35704|LCB1_MOUSE Serine
palmitoyltransferase 1 (Long chain base biosynthesis
protein 1) (LCB 1) (Serine-palmitoyl-CoA transferase 1)
(SPT 1) (SPT1)
Length = 473
Score = 255 bits (651), Expect = 1e-66
Identities = 134/274 (48%), Positives = 185/274 (66%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ L+ K+YK +R L+ KE +EL +EW P+PL+P ++ P
Sbjct: 24 HLILEGILILWIIRLVFSKTYKLQERSDLTAKEKEELIEEWQPEPLVPPVSKNHPALNYN 83
Query: 96 LESAAGPHTIV-NGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H IV NGKE VNFAS N+LGL+ + ++ + S+L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPTHNIVVNGKECVNFASFNFLGLLANPRVKAAAFSSLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG ST+ SAIPA+SK+GDII D + IQ GL SR
Sbjct: 144 VHLDLEERLAKFMKTEEAIIYSYGFSTVASAIPAYSKRGDIIFVDSAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN++ L L+ ++ + RR+IV+E LY N+G I PL E++KL
Sbjct: 204 SDIKLFKHNDVADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTICPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGISI 297
>dbj|BAC40709.1| unnamed protein product [Mus musculus]
Length = 473
Score = 255 bits (651), Expect = 1e-66
Identities = 134/274 (48%), Positives = 185/274 (66%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ L+ K+YK +R L+ KE +EL +EW P+PL+P ++ P
Sbjct: 24 HLILEGILILWIIRLVFSKTYKLQERSDLTAKEKEELIEEWQPEPLVPPVSKNHPALNYN 83
Query: 96 LESAAGPHTIV-NGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H IV NGKE VNFAS N+LGL+ + ++ + S+L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPTHNIVVNGKECVNFASFNFLGLLANPRVKAAAFSSLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG ST+ SAIPA+SK+GDII D + IQ GL SR
Sbjct: 144 VHLDLEERLAKFMKTEEAIIYSYGFSTIASAIPAYSKRGDIIFVDSAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN++ L L+ ++ + RR+IV+E LY N+G I PL E++KL
Sbjct: 204 SDIKLFKHNDVADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTICPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGISI 297
>ref|NP_001018307.1| hypothetical protein LOC553981 [Danio rerio]
gi|63102002|gb|AAH95705.1| Hypothetical protein
LOC553981 [Danio rerio]
Length = 472
Score = 253 bits (647), Expect = 4e-66
Identities = 134/280 (47%), Positives = 188/280 (66%), Gaps = 18/280 (6%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLI-------PSLNDE 88
HL +E L++ I+ LL K+YK +R L+ KE +EL +EW P+PL+ PSLN +
Sbjct: 23 HLILEGFLILWIIRLLFSKTYKLQERSDLTEKEKEELIEEWQPEPLVSPVSKDHPSLNYD 82
Query: 89 MPYEPPVLESAAGPHTIVNGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRG 148
+ PP IVNGKE +NFAS N+LGL+ ++++ ++L+KYGVG+CGPRG
Sbjct: 83 VVTGPP------SHKIIVNGKECINFASFNFLGLLDNERVKLKALASLKKYGVGTCGPRG 136
Query: 149 FYGTIDVHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQN 208
FYGT DVHL+ E R++KF+ T ++I+YSYG +T+ SAIPA+SK+GDII DE + IQ
Sbjct: 137 FYGTFDVHLELEERLAKFMRTEEAIIYSYGFATIASAIPAYSKRGDIIFVDEAACFSIQK 196
Query: 209 GLYLSRSTVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPL 264
GL SRS + YFKHN+M+ L L+ ++ + +RR+I++E LY N+ I PL
Sbjct: 197 GLQASRSFIKYFKHNDMEDLERLLKEQEIEDQKNPRKARVIRRFILVEGLYINTADICPL 256
Query: 265 DEIIKLKEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
E++KLK KY RI L+ES SFGVLG GRG+TEH+GV +
Sbjct: 257 PELVKLKYKYKVRIFLEESMSFGVLGEHGRGVTEHFGVNI 296
>sp|Q60HD1|LCB1_MACFA Serine palmitoyltransferase 1 (Long chain base biosynthesis protein
1) (LCB 1) (Serine-palmitoyl-CoA transferase 1) (SPT 1)
(SPT1) gi|52782275|dbj|BAD51984.1| serine
palmitoyltransferase, long chain base subunit 1 [Macaca
fascicularis]
Length = 473
Score = 253 bits (646), Expect = 5e-66
Identities = 132/274 (48%), Positives = 185/274 (67%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ LL K+YK +R L+ KE +EL +EW P+PL+P + + P
Sbjct: 24 HLILEGILILWIIRLLFSKTYKLQERSDLTVKEKEELIEEWQPEPLVPPVPKDHPALNYN 83
Query: 96 LESAAGPHTIV-NGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H IV NGKE +NFAS N+LGL+ + ++ + ++L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPSHKIVVNGKECINFASFNFLGLLDNPRVKAAALASLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG +T+ SAIPA+SK+GDI+ D + IQ GL SR
Sbjct: 144 VHLDLEDRLAKFMKTEEAIIYSYGFATIASAIPAYSKRGDIVFVDRAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN+M L L+ ++ + RR+IV+E LY N+G I PL E++KL
Sbjct: 204 SDIKLFKHNDMADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTICPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGINI 297
>gb|AAC53505.1| serine palmitoyltransferase LCB1 subunit [Cricetulus griseus]
gi|6685595|sp|O54695|LCB1_CRIGR Serine
palmitoyltransferase 1 (Long chain base biosynthesis
protein 1) (LCB 1) (Serine-palmitoyl-CoA transferase 1)
(SPT 1) (SPT1)
Length = 473
Score = 253 bits (646), Expect = 5e-66
Identities = 131/274 (47%), Positives = 185/274 (66%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ L+ K+YK +R L+ KE +EL +EW P+PL+P ++ P
Sbjct: 24 HLILEGILILWIIRLVFSKTYKLQERSDLTAKEKEELIEEWQPEPLVPPVSKNHPALNYN 83
Query: 96 LESAAGPHTIV-NGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H IV NGKE VNFAS N+LGL+ + ++ + ++L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPTHNIVVNGKECVNFASFNFLGLLANPRVKAAALASLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG ST+ SAIPA+SK+GDI+ D + IQ GL SR
Sbjct: 144 VHLDLEERLAKFMRTEEAIIYSYGFSTIASAIPAYSKRGDIVFVDSAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN++ L L+ ++ + RR+IV+E LY N+G + PL E++KL
Sbjct: 204 SDIKLFKHNDVADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTVCPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGISI 297
>emb|CAH91475.1| hypothetical protein [Pongo pygmaeus]
Length = 473
Score = 253 bits (645), Expect = 7e-66
Identities = 132/274 (48%), Positives = 185/274 (67%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ LL K+YK +R L+ KE +EL +EW P+PL+P + + P
Sbjct: 24 HLILEGILILWIIRLLFSKTYKLQERSDLTVKEKEELIEEWQPEPLVPLVPKDHPALNYN 83
Query: 96 LESAAGPHTIV-NGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H IV NGKE +NFAS N+LGL+ + ++ + ++L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPSHKIVVNGKECINFASFNFLGLLDNPRVKAAALASLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG +T+ SAIPA+SK+GDI+ D + IQ GL SR
Sbjct: 144 VHLDLEDRLAKFMKTEEAIIYSYGFATIASAIPAYSKRGDIVFVDRAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN+M L L+ ++ + RR+IV+E LY N+G I PL E++KL
Sbjct: 204 SDIKLFKHNDMADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTICPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGINI 297
>dbj|BAE01287.1| unnamed protein product [Macaca fascicularis]
Length = 473
Score = 250 bits (639), Expect = 3e-65
Identities = 132/274 (48%), Positives = 184/274 (66%), Gaps = 6/274 (2%)
Query: 37 HLFIEVLLLVVILFLLSQKSYKPPKRP-LSNKEIDELCDEWVPQPLIPSLNDEMPYEPPV 95
HL +E +L++ I+ LL K+YK +R L+ KE +EL +EW P+PL P + + P
Sbjct: 24 HLILEGILILWIIRLLFSKTYKLQERSDLTVKEKEELIEEWQPEPLGPPVPKDHPALNYN 83
Query: 96 LESAAGPHTIV-NGKEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTID 154
+ S H IV NGKE +NFAS N+LGL+ + ++ + ++L+KYGVG+CGPRGFYGT D
Sbjct: 84 IVSGPPSHKIVVNGKECINFASFNFLGLLDNPRVKAAALASLKKYGVGTCGPRGFYGTFD 143
Query: 155 VHLDCEGRISKFLGTPDSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSR 214
VHLD E R++KF+ T ++I+YSYG +T+ SAIPA+SK+GDI+ D + IQ GL SR
Sbjct: 144 VHLDLEDRLAKFMKTEEAIIYSYGFATIASAIPAYSKRGDIVFVDRAACFAIQKGLQASR 203
Query: 215 STVVYFKHNNMDSLRETLE----NITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKL 270
S + FKHN+M L L+ ++ + RR+IV+E LY N+G I PL E++KL
Sbjct: 204 SDIKLFKHNDMADLERLLKEQEIEDQKNPRKARVTRRFIVVEGLYMNTGTICPLPELVKL 263
Query: 271 KEKYCFRILLDESNSFGVLGSSGRGLTEHYGVPV 304
K KY RI L+ES SFGVLG GRG+TEHYG+ +
Sbjct: 264 KYKYKARIFLEESLSFGVLGEHGRGVTEHYGINI 297
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 555,810,778
Number of Sequences: 2540612
Number of extensions: 24094896
Number of successful extensions: 85127
Number of sequences better than 10.0: 708
Number of HSP's better than 10.0 without gapping: 666
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 83551
Number of HSP's gapped (non-prelim): 728
length of query: 316
length of database: 863,360,394
effective HSP length: 128
effective length of query: 188
effective length of database: 538,162,058
effective search space: 101174466904
effective search space used: 101174466904
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC137824.11


