

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119416.7 - phase: 0 /pseudo
(447 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
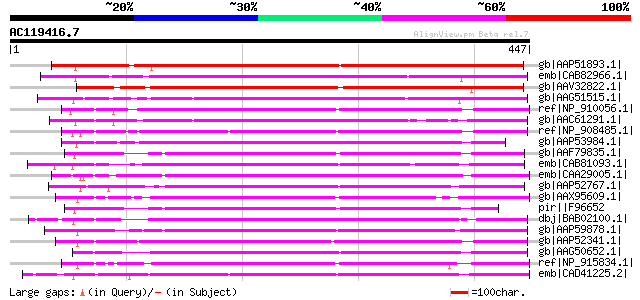
gb|AAP51893.1| putative Tam3-like transposon protein [Oryza sati... 356 9e-97
emb|CAB82966.1| putative protein [Arabidopsis thaliana] gi|11357... 315 2e-84
gb|AAV32822.1| transposase [Zea mays] 301 3e-80
gb|AAG51515.1| hAT-element transposase, putative [Arabidopsis th... 245 2e-63
ref|NP_910056.1| hypothetical protein [Oryza sativa (japonica cu... 244 3e-63
gb|AAC61291.1| Ac-like transposase [Arabidopsis thaliana] gi|254... 234 5e-60
ref|NP_908485.1| unnamed protein product [Oryza sativa (japonica... 233 1e-59
gb|AAP53984.1| putative transposase [Oryza sativa (japonica cult... 226 1e-57
gb|AAF79835.1| T6D22.19 [Arabidopsis thaliana] 223 9e-57
emb|CAB81093.1| AT4g05510 [Arabidopsis thaliana] gi|5732064|gb|A... 220 6e-56
emb|CAA29005.1| ORFa [Zea mays] gi|136125|sp|P08770|TRA1_MAIZE P... 216 2e-54
gb|AAP52767.1| putative activator-like transposable element [Ory... 212 2e-53
gb|AAX95609.1| hAT family dimerisation domain, putative [Oryza s... 212 2e-53
pir||F96652 protein F23N19.13 [imported] - Arabidopsis thaliana ... 211 4e-53
dbj|BAB02100.1| unnamed protein product [Arabidopsis thaliana] 208 3e-52
gb|AAP59878.1| Ac-like transposase THELMA13 [Silene latifolia] 207 4e-52
gb|AAP52341.1| putative transposable element [Oryza sativa (japo... 207 7e-52
gb|AAG50652.1| transposase, putative [Arabidopsis thaliana] gi|2... 202 2e-50
ref|NP_915834.1| P0003D09.25 [Oryza sativa (japonica cultivar-gr... 202 2e-50
emb|CAD41225.2| OSJNBa0010H02.12 [Oryza sativa (japonica cultiva... 201 3e-50
>gb|AAP51893.1| putative Tam3-like transposon protein [Oryza sativa (japonica
cultivar-group)] gi|37530608|ref|NP_919606.1| putative
Tam3-like transposon protein [Oryza sativa (japonica
cultivar-group)] gi|16924038|gb|AAL31650.1| Putative
Tam3-like transposon protein [Oryza sativa]
Length = 737
Score = 356 bits (913), Expect = 9e-97
Identities = 191/415 (46%), Positives = 257/415 (61%), Gaps = 13/415 (3%)
Query: 37 QKKKPQTC-SKVWRHFNRNK-----NLAVCKYCGKEYAANSSSHGTTNLGKHLKVCLKNP 90
+K+KP S VW F++ K A CK+C K + S ++GT++L HLK+C KNP
Sbjct: 53 RKRKPMALRSDVWESFSKVKLANGDERAKCKWCTKLFHCGSRTNGTSSLKAHLKICKKNP 112
Query: 91 YRVVDKKQKTLAIGMGSEDDPNSVSFKLVD--FSQEKTRLALAKMIIIDELPFKHVENEG 148
+ V Q TL + + D NS + F +K R A+MII DE PF E G
Sbjct: 113 NKPVVDNQGTLQL---TPCDGNSTLGTVTTWKFDPDKLRRCFAEMIIEDEQPFVLSERSG 169
Query: 149 FKMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSIQNMN 208
+ FM A P+F +PSR ++ R C+++Y DEKE LK N V LTTDTWT+ + N
Sbjct: 170 LRKFMTLACPRFVLPSRRTITRACVKVYEDEKEKLKKFFKDNCVRVCLTTDTWTAKNSQN 229
Query: 209 YMCVTAHYIDDEWVLKKKILSFNLIADHKGETIGIALENCMKDWGIRSICCVTVDNASAN 268
+MCVTAH+ID+EW L+KKI+ F L+ H+GE IG +LENC+ +WGI + +TVDNASAN
Sbjct: 230 FMCVTAHFIDNEWNLQKKIIGFFLVKGHRGEDIGKSLENCLAEWGIDKVFTITVDNASAN 289
Query: 269 NLAINYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVK 328
N AI Y+ R + G + GEY+HMRC AHI+NLIV DGLK+I S+QR+RAA KF++
Sbjct: 290 NNAIKYMRRVLNESKG-CVAEGEYIHMRCAAHIINLIVGDGLKEIGTSIQRVRAAVKFIR 348
Query: 329 SSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFV 388
SRL FKKCA+ V SKA + LD+ TRWNSTYLMLN AEKY+ F R D +
Sbjct: 349 CGTSRLVKFKKCAELAKVQSKAFLNLDICTRWNSTYLMLNAAEKYQKAFERYSDEDPYYK 408
Query: 389 TSLSGELG-GCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQLMEI 442
L GE G G P DWE++R FL+ FYD TL S +++++F ++ ++
Sbjct: 409 LELEGENGPGVPTRADWEKARKMADFLEHFYDLTLRVSVQSRTTSHTYFHEIADV 463
>emb|CAB82966.1| putative protein [Arabidopsis thaliana] gi|11357980|pir||T48044
hypothetical protein T12C14.220 - Arabidopsis thaliana
Length = 705
Score = 315 bits (807), Expect = 2e-84
Identities = 179/430 (41%), Positives = 252/430 (57%), Gaps = 16/430 (3%)
Query: 27 EVGAATAVSKQKKKPQTCSKVWRHFNRNK---NLAVCKYCGKEYAANSSSHGTTNLGKHL 83
E + T K K+K T S W+ F R K N C YCGK Y+ N ++ GT+N+ H+
Sbjct: 13 EPESPTVSPKAKRKLPTRSTAWKVFTRLKDDDNSCSCNYCGKIYSCNPATCGTSNMNTHM 72
Query: 84 KVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKH 143
+ C K ++ + + + G + + F Q R A KMII+DEL F
Sbjct: 73 RKC-KAYLDHLESNSQNVIVSSGGGNGAGMIK----PFDQNVCRQATVKMIIMDELSFSF 127
Query: 144 VENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTS 203
VEN+GFK F A P F IPSR ++ RD + LY EK LK++LS N Q +S TTD WTS
Sbjct: 128 VENDGFKHFCEMAVPWFTIPSRRTITRDVVGLYRAEKTALKTILSRNRQAMSFTTDIWTS 187
Query: 204 IQNMNYMCVTAHYIDDEWVLKKKILSFNLIADHKGETIGIALENCMKDWGIRSICCVTVD 263
I ++YM VTAH+ID +W L ++I+SF+ I DHKG+TI + DWGI + +TVD
Sbjct: 188 ITTLSYMVVTAHFIDMDWQLHRRIISFSPIPDHKGKTIANQFLRSLDDWGIEKVFSITVD 247
Query: 264 NASANNLAINYLNRGMRVWNGRTL-FNGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRA 322
NASAN+ A L + N L NG+++H+RC AHILNLI +GL+DI S+ +R
Sbjct: 248 NASANDKANTVLKERLSNRNSDALMMNGDFMHIRCGAHILNLIANEGLEDISNSIVSVRN 307
Query: 323 ACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEH 382
A K+V++S SRL +FK+ + V ++ V LD VTRWNSTYLML A KY+ F R+
Sbjct: 308 AVKYVRTSNSRLESFKRWVE-VEKITRGSVVLDCVTRWNSTYLMLKSALKYKVAFDRMAD 366
Query: 383 ADAAF------VTSLSGELGGCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFF 436
D + V S + G P DW+ +R V+FLK FYD+TL+FS S+ V++N +
Sbjct: 367 MDKPYDAWFKEVDSNKKKRDGPPMAIDWDNARHMVRFLKAFYDSTLAFSSSIKVTSNGCY 426
Query: 437 KQLMEIQKTL 446
++ +IQ ++
Sbjct: 427 NEICKIQSSV 436
>gb|AAV32822.1| transposase [Zea mays]
Length = 674
Score = 301 bits (771), Expect = 3e-80
Identities = 164/388 (42%), Positives = 235/388 (60%), Gaps = 13/388 (3%)
Query: 58 AVCKYCGKEYAANSSSHGTTNLGKHLKVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVS-F 116
A CKYC + A + HGT++L +H VC +NP +K K + G+ +S F
Sbjct: 17 ARCKYCHRNMKAEAGRHGTSSLTRHFVVCKRNP----NKFNKDPSQGILQATHCEGISTF 72
Query: 117 KLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLY 176
K F QE RLA A+M+I DE PF E G + F+ +A P+F+ PSR + RD +R +
Sbjct: 73 K---FDQEALRLAFAEMVIEDEQPFCFGEKPGLRKFLSKACPRFQPPSRRTCTRDVVRCF 129
Query: 177 FDEKETLKSVLSANNQMVSLTTDTWTSIQNMNYMCVTAHYIDDEWVLKKKILSFNLIADH 236
F EK LK + Q V LTTD WTS Q +YM VTA +ID+ W L KK++ F ++ H
Sbjct: 130 FQEKAKLKKFFKDSCQRVCLTTDGWTSQQQDSYMTVTASFIDENWRLHKKVIGFFMVKGH 189
Query: 237 KGETIGIALENCMKDWGIRSICCVTVDNASANNLAINYLNRGMRVWNGRTLFNGEYLHMR 296
KG+ IG + CM DWG++ + VTVDNASAN+ I YL R + N + G+YLHMR
Sbjct: 190 KGDDIGRNVLRCMTDWGLQRVMPVTVDNASANDNCIGYLKRHLTKTN---VAGGKYLHMR 246
Query: 297 CCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDV 356
C AHI+NLIV DGLK++D SV+RIRAA +++++ SR+ FK+ + + +K + +DV
Sbjct: 247 CAAHIVNLIVHDGLKEVDLSVKRIRAAVRYIRNGGSRIVKFKELIEEEKLTNKQFLKIDV 306
Query: 357 VTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSLSGELG--GCPDHDDWERSRVFVKFL 414
TRWNST+LML A YE VF RL D + V LS E G P+ DW+ ++ FL
Sbjct: 307 PTRWNSTFLMLKAALVYEKVFTRLADEDMSCVNDLSEEKDRYGHPEEHDWQNAKNMADFL 366
Query: 415 KTFYDATLSFSGSLHVSANSFFKQLMEI 442
+ F+D T+ S + H+++++ F ++ E+
Sbjct: 367 EHFHDLTVRVSATNHITSHTLFHEIGEV 394
>gb|AAG51515.1| hAT-element transposase, putative [Arabidopsis thaliana]
gi|12322417|gb|AAG51228.1| Tam3-like transposon protein;
93317-95488 [Arabidopsis thaliana]
gi|25405099|pir||A96497 probable hAT-element transposase
[imported] - Arabidopsis thaliana
Length = 723
Score = 245 bits (625), Expect = 2e-63
Identities = 163/432 (37%), Positives = 232/432 (52%), Gaps = 22/432 (5%)
Query: 25 VTEVGAATAVSKQKKKPQTCSKVWRHFNR---NKNLAVCKYCGKEYAANSSSHGTTNLGK 81
V + G + K KKK QT S VW HF R + + C Y K + S+ GT+NL K
Sbjct: 37 VADDGDSANPCKPKKKLQTRSWVWDHFTRKDGDDDQCKCHYY-KRFFGCSTKSGTSNLKK 95
Query: 82 HLKVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPF 141
HL C Y +Q I + N S K+ S+E R A +M+++ +LP
Sbjct: 96 HLDCC--KHYSAWKGRQSQNVINQ----EGNLQSGKV---SKEVFREATNEMLVLGQLPL 146
Query: 142 KHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTW 201
VE+ +K +A +K SR + RD + + K +LK ++SAN + VSLTTD W
Sbjct: 147 SFVESVAWKHICSKAN-LYKPHSRRTATRDIILMNVARKASLKDLVSANKRRVSLTTDIW 205
Query: 202 TS-IQNMNYMCVTAHYIDDEWVLKKKILSFNLIADHKGETIGIALENCMKDWGIRSICCV 260
T+ +YM +T H+ID+ W L+K I+ F IADHKG TI L C+ +WGI I +
Sbjct: 206 TAQATGASYMVITVHFIDEYWRLRKFIIGFKYIADHKGATISRVLLECLTEWGIERIFTI 265
Query: 261 TVDNASANNLAINYLNRGMRVWNGRTL-FNGEYLHMRCCAHILNLIVLDGLKDIDASVQR 319
TVDNA+AN A+ R ++ +L NG+++HMRCCAHI+NLIV +GL + V+
Sbjct: 266 TVDNATANTSALRKFQRALQSQRADSLVLNGDFMHMRCCAHIINLIVKEGLHKLGNHVEA 325
Query: 320 IRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYR 379
IR +V+SS SR +F++ G ++ + LDV TRWNSTYLML A K++ F +
Sbjct: 326 IRNGVLYVRSSTSRCDSFEQKV-VTGKMTRGSLPLDVKTRWNSTYLMLTRAIKFKVAFDK 384
Query: 380 LEHADAA----FVTSLSGELG-GCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANS 434
+E D F+ + GE G P DW VKFL FY ATL S S V +
Sbjct: 385 MEAEDKLYNDHFLEVVDGEKRIGPPTTIDWREVERLVKFLGLFYTATLVVSASSIVCSYK 444
Query: 435 FFKQLMEIQKTL 446
+ +++ I+K L
Sbjct: 445 CYGEIVTIEKNL 456
>ref|NP_910056.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|27545055|gb|AAO18461.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 669
Score = 244 bits (624), Expect = 3e-63
Identities = 149/413 (36%), Positives = 219/413 (52%), Gaps = 28/413 (6%)
Query: 45 SKVWRHF-----NRNKNLAVCKYCGKEYAANSSSHGTTNLGKHLKVCLK--NPYRVVDKK 97
S+ W+ F + CK+C E A + GT++L KHL C K + ++V
Sbjct: 161 SEAWKEFVPILIDNEVGAGKCKHCDTEIRAKRGA-GTSSLRKHLTRCKKRISALKIVGNL 219
Query: 98 QKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQ 157
TL PNSV K F E +R L +MI++ ELPF+ VE +GF+ F
Sbjct: 220 DSTLM-------SPNSVRLKNWSFDPEVSRKELMRMIVLHELPFQFVEYDGFRSFAASLN 272
Query: 158 PKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSIQNMNYMCVTAHYI 217
P FKI SR ++ DC+ + ++K +K + N SLT D WTS Q M YMCVT H+I
Sbjct: 273 PYFKIISRTTIRNDCIAAFKEQKLAMKDMFKGANCRFSLTADMWTSNQTMGYMCVTCHFI 332
Query: 218 DDEWVLKKKILS-FNLIADHKGETIGIALENCMKDWGIR-SICCVTVDNASANNLAINYL 275
D +W ++K+I+ F + H G + A+ +C++DW I I VT+D ASAN+ L
Sbjct: 333 DTDWRVQKRIIKFFGVKTPHTGVQMFNAMLSCIQDWNIADKIFSVTLDYASANDSMAKLL 392
Query: 276 NRGMRVWNGRTL-FNGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSPSRL 334
++ +T+ G+ LH RC H++NLI DGLK ID+ V IR + K+ +S SR
Sbjct: 393 KCNLKA--KKTIPAGGKLLHNRCATHVINLIAKDGLKVIDSIVCNIRESVKYRDNSLSRK 450
Query: 335 ATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSLSGE 394
F++ G+ + T+DV TRWNSTYLMLN A + + L D + +
Sbjct: 451 EKFEEIIAQEGITCELHPTVDVCTRWNSTYLMLNAAFPFMRAYASLAVQDKNYKYA---- 506
Query: 395 LGGCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQLMEIQKTLN 447
P D WERS + LK YDAT+ SGSL+ ++N +F ++ +I+ L+
Sbjct: 507 ----PSPDQWERSTIVSGILKVLYDATMVVSGSLYPTSNLYFHEMWKIKLVLD 555
>gb|AAC61291.1| Ac-like transposase [Arabidopsis thaliana] gi|25411587|pir||C84523
Ac-like transposase [imported] - Arabidopsis thaliana
Length = 730
Score = 234 bits (596), Expect = 5e-60
Identities = 145/419 (34%), Positives = 227/419 (53%), Gaps = 29/419 (6%)
Query: 35 SKQKKKPQTCSKVWRHFNRNK---NLAVCKYCGKEYAANSSSHGTTNLGKHLKVCLK--N 89
S +KKK + S VW HF+R K N C YCGKE A S S GT+ L KHL++ K
Sbjct: 52 SSKKKKTSSRSYVWDHFSRKKGNPNKCNCHYCGKELACPSKS-GTSTLKKHLELSCKAFK 110
Query: 90 PYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKHVENEGF 149
++ + Q IG D S + S+ R A +M+I+ ELP +E+ +
Sbjct: 111 AWKSTNTDQTQTVIGRDGGDG----SLTMYKVSELVIREASIEMLILGELPLSFIESVAW 166
Query: 150 KMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSIQN-MN 208
+ F +A+ +K SR + R+ + LY +K +K +L + + +SLTTD W S +
Sbjct: 167 RHFCSKAK-SYKPVSRRTTTREIVMLYVKKKVAMKKILGKSQERMSLTTDIWVSNNTGES 225
Query: 209 YMCVTAHYIDDEWVLKKKILSFNLIADHKGETIGIALENCMKDWGIRSICCVTVDNASAN 268
YM +TAH++D +W LKK I+ F + DH TI L C+ +W IR I C+TVDNA+AN
Sbjct: 226 YMVITAHFVDVDWKLKKMIIGFKHVTDHNCGTICKVLLECLAEWDIRRIFCITVDNATAN 285
Query: 269 NLAINYLNRGMR-VWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFV 327
N A+ + M+ + + + GEY+HMRC AHILNL+V +GL ++DASV IR +FV
Sbjct: 286 NTALTKFKKTMKLIGDDALVLKGEYMHMRCDAHILNLVVKEGLTEVDASVTSIRNGIQFV 345
Query: 328 KSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAF 387
+SS +RL +F D+ G S+A+ ++ + + +K + +Y LE D
Sbjct: 346 RSSTNRLKSFDLRCDA-GTISRAV-------KFRIAFEKMEAEDKLYNDYY-LEKVDGE- 395
Query: 388 VTSLSGELGGCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQLMEIQKTL 446
+ G P DW+ + ++ L FY +TL SGS +V+++ + +++ + + L
Sbjct: 396 ------KKIGPPMSSDWDAAERLIQILAIFYKSTLVLSGSTYVTSHKMYNEIINMARNL 448
>ref|NP_908485.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 737
Score = 233 bits (593), Expect = 1e-59
Identities = 138/415 (33%), Positives = 220/415 (52%), Gaps = 26/415 (6%)
Query: 45 SKVWRHFN-------RNKNLAV----CKYCGKEYAANSSSHGTTNLGKHLKVCLKNPYRV 93
++ W+HF + + + V CK+C + Y + TT L +HL C + ++
Sbjct: 98 AECWKHFKVINVPSKKERGVMVAKAKCKFCHRSYVYHPGG-ATTTLNRHLAGCTQYQNKL 156
Query: 94 VDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFM 153
K + + G EDD NS+ +++ E TR +AKMII E F+ VE++ F + M
Sbjct: 157 TKAKAQG-TLNFGPEDD-NSLIVNPTEYNHEHTRELIAKMIIAHEYSFRMVEHKWFNILM 214
Query: 154 GEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSIQNMNYMCVT 213
++ R S+ +C+R+Y EK LK L + +SLTTD WTS QN+ YMC+
Sbjct: 215 KWMNGNYEFIGRKSIKNECMRVYESEKNQLKKSLK-EAESISLTTDLWTSNQNLQYMCLV 273
Query: 214 AHYIDDEWVLKKKILSF-NLIADHKGETIGIALENCMKDWGIRS-ICCVTVDNASANNLA 271
AHYID+ WV++ ++L+F + H G I A+ CM +W I + +T+DNA+ N+ A
Sbjct: 274 AHYIDENWVMQCRVLNFIEVDPPHTGIVIAQAVFECMVEWKIEDKVTTITLDNATNNDTA 333
Query: 272 INYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSP 331
+ L + ++F+ Y H+RC AHI+NL+V DGL+ ID + +R K+ K SP
Sbjct: 334 VTNLKAKLLA-RKNSVFDPSYFHIRCAAHIVNLVVNDGLQPIDNLISCLRNTVKYFKRSP 392
Query: 332 SRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSL 391
SR+ F + ++ V + LDV TRWNSTY ML+ Y+ F + D ++V
Sbjct: 393 SRMYKFVEVCNNYSVKVGRGLALDVKTRWNSTYKMLDTCIDYKDAFGYYKEVDTSYVWK- 451
Query: 392 SGELGGCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQLMEIQKTL 446
P DW L T +A+ +FSGSL+ +AN F+ ++++++ L
Sbjct: 452 -------PSDSDWVSFGKIRPILGTMAEASTAFSGSLYPTANCFYPYIVKVKRAL 499
>gb|AAP53984.1| putative transposase [Oryza sativa (japonica cultivar-group)]
gi|37534790|ref|NP_921697.1| putative transposase [Oryza
sativa (japonica cultivar-group)]
Length = 693
Score = 226 bits (575), Expect = 1e-57
Identities = 142/392 (36%), Positives = 207/392 (52%), Gaps = 21/392 (5%)
Query: 45 SKVWRHFNRNKN----LAVCKYCGKEYAANSSSHGTTNLGKHLKVCLKNPYRVVDKKQKT 100
S W+ F + K A CK+C + A S + GT +L H+K C R Q
Sbjct: 89 SAAWQDFVKKKINGAWKAECKWCHNKLGAESRN-GTKHLLDHIKTCKSRQARK-GLTQSN 146
Query: 101 LAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQPKF 160
L +G+ +E V+ F QE R LA MI + E P V++ GF+ F G QP F
Sbjct: 147 LKMGIDAE---GRVTVGKYVFDQEVARKELALMICLHEYPLSIVDHVGFRRFCGALQPLF 203
Query: 161 KIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSI-QNMNYMCVTAHYIDD 219
K+ +R ++ +D + L+ K ++ + V++TTD WT+ Q YM +TAH+IDD
Sbjct: 204 KVMTRNTIRKDIIDLFGVNKISISNYFHKLQSRVAITTDLWTATHQKKGYMAITAHFIDD 263
Query: 220 EWVLKKKILSFNLI-ADHKGETIGIALENCMKDWGIRS-ICCVTVDNASANN-LAINYLN 276
EW LK +L F + A H + I + + DW + S + +T+DN S N+ L N L
Sbjct: 264 EWKLKSFLLRFIYVPAPHTADLISEIIYEVLADWNLESRLSTITLDNCSTNDKLMENLLG 323
Query: 277 RGMRVWNGRTLF-NGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSPSRLA 335
+ TL NG LHMRCCAHILNLIV DG+ +D ++++R + F ++P R
Sbjct: 324 TMLDKLPADTLMLNGSLLHMRCCAHILNLIVKDGMTILDKIIEKVRESVSFWTATPKRHE 383
Query: 336 TFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSLSGEL 395
F+K A + V + ++ LD TRWNSTYLML+ A Y+ VF +L + +
Sbjct: 384 KFEKQAQQINVKYEKVIALDCKTRWNSTYLMLSTAVLYQDVFTKLGTREKMYTPY----- 438
Query: 396 GGCPDHDDWERSRVFVKFLKTFYDATLSFSGS 427
CP +DDW+ +R LK FYDAT +FSG+
Sbjct: 439 --CPSNDDWKFARELCDRLKIFYDATEAFSGT 468
>gb|AAF79835.1| T6D22.19 [Arabidopsis thaliana]
Length = 745
Score = 223 bits (568), Expect = 9e-57
Identities = 137/404 (33%), Positives = 210/404 (51%), Gaps = 40/404 (9%)
Query: 48 WRHFNRN------KNLAVCKYCGKEYAANSSSHGTTNLGKHLKVCLKNPYRVVDKKQKTL 101
W++F+R K CKYC + Y N +GT + +H++ C K P +K
Sbjct: 147 WKNFDRGQKYPNGKTEVTCKYCEQTYHLNLRRNGTNTMNRHMRSCEKTPGSTPRISRK-- 204
Query: 102 AIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQPKFK 161
VD + +A+A ++ LP+ VE E + P +
Sbjct: 205 -----------------VDMMVFREMIAVA--LVQHNLPYSFVEYERIREAFTYVNPSIE 245
Query: 162 IPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSIQNMNYMCVTAHYIDDEW 221
SR + A D ++Y EK LK L+ + LTTD W ++ +Y+C+TAHY+D +
Sbjct: 246 FWSRNTAASDVYKIYEREKIKLKEKLAIIPGRICLTTDLWRALTVESYICLTAHYVDVDG 305
Query: 222 VLKKKILSFNLIAD-HKGETIGIALENCMKDWGI-RSICCVTVDNASANNLAINYLNRGM 279
VLK KILSF H G I + L +KDWGI + + +TVDNASAN+ + L R +
Sbjct: 306 VLKTKILSFCAFPPPHSGVAIAMKLSELLKDWGIEKKVFTLTVDNASANDTMQSILKRKL 365
Query: 280 RVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSPSRLATFKK 339
+ + +GE+ H+RC AHILNLIV DGL+ I ++++IR K+VK S +R F+
Sbjct: 366 QK---HLVCSGEFFHVRCSAHILNLIVQDGLEVISGALEKIRETVKYVKGSETRENLFQN 422
Query: 340 CADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSLSGELGGCP 399
C D++G+ ++A + LDV TRWNSTY ML+ A +++ V + L D + + P
Sbjct: 423 CMDTIGIQTEASLVLDVSTRWNSTYHMLSRAIQFKDVLHSLAEVDRGYKS--------FP 474
Query: 400 DHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQLMEIQ 443
+WER+ + LK F + T SGS + +AN +F Q+ I+
Sbjct: 475 SAVEWERAELICDLLKPFAEITKLISGSSYPTANVYFMQVWAIK 518
>emb|CAB81093.1| AT4g05510 [Arabidopsis thaliana] gi|5732064|gb|AAD48963.1| contains
similarity to transposases [Arabidopsis thaliana]
gi|25407345|pir||C85069 hypothetical protein AT4g05510
[imported] - Arabidopsis thaliana
Length = 604
Score = 220 bits (561), Expect = 6e-56
Identities = 140/439 (31%), Positives = 222/439 (49%), Gaps = 46/439 (10%)
Query: 16 DGEVTEAGAVTEVGAATAVSKQ-----KKKPQTCSKVWRHFN----RNKNLAVCKYCGKE 66
+ E T G + A T+ SK + K S +W +F + +A CK C K
Sbjct: 4 EDECTYGGEGGQTSADTSQSKSLVSASRFKRSRTSDMWDYFTLEDENDGKIAYCKKCLKP 63
Query: 67 YAANSSSHGTTNLGKHLKVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKT 126
Y ++ GT+NL +H + C MG + V K +
Sbjct: 64 YPILPTT-GTSNLIRHHRKC-----------------SMGLD-----VGRKTTKIDHKVV 100
Query: 127 RLALAKMIIIDELPFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKSV 186
R +++II +LPF VE E + F+ P +K +R + A D ++ + EK+ LKS
Sbjct: 101 REKFSRVIIRHDLPFLCVEYEELRDFISYMNPDYKCYTRNTAAADVVKTWEKEKQILKSE 160
Query: 187 LSANNQMVSLTTDTWTSIQNMNYMCVTAHYIDDEWVLKKKILSF-NLIADHKGETIGIAL 245
L + LT+D WTS+ Y+ +TAHY+D W+L KILSF +++ H G+ + +
Sbjct: 161 LERIPSRICLTSDCWTSLGGDGYIVLTAHYVDTRWILNSKILSFSDMLPPHTGDALASKI 220
Query: 246 ENCMKDWGI-RSICCVTVDNASANNLAINYLNRGMRVWNGRTLFNGEYLHMRCCAHILNL 304
C+K+WGI + + +T+DNA+ANN L +++ N + GE+ H+RCCAH+LN
Sbjct: 221 HECLKEWGIEKKVFTLTLDNATANNSMQEVLIDRLKLDN-NLMCKGEFFHVRCCAHVLNR 279
Query: 305 IVLDGLKDIDASVQRIRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTY 364
IV +GL I ++ +IR K+VK S SR +C + G + +++LDV TRWNSTY
Sbjct: 280 IVQNGLDVISDALSKIRETVKYVKGSTSRRLALAECVEGKG---EVLLSLDVQTRWNSTY 336
Query: 365 LMLNIAEKYEHVFYRLEHADAAFVTSLSGELGGCPDHDDWERSRVFVKFLKTFYDATLSF 424
LML+ A KY+ R + D + CP ++W+R++ + L FY T
Sbjct: 337 LMLHKALKYQRALNRFKIVDKNY--------KNCPSSEEWKRAKTIHEILMPFYKITNLM 388
Query: 425 SGSLHVSANSFFKQLMEIQ 443
SG + ++N +F + +IQ
Sbjct: 389 SGRSYSTSNLYFGHVWKIQ 407
>emb|CAA29005.1| ORFa [Zea mays] gi|136125|sp|P08770|TRA1_MAIZE Putative AC
transposase (ORFA)
Length = 807
Score = 216 bits (549), Expect = 2e-54
Identities = 141/428 (32%), Positives = 214/428 (49%), Gaps = 32/428 (7%)
Query: 37 QKKKPQTCSKVWRHFNRNKNLAV-------------CKY--CGKEYAANSSSHGTTNLGK 81
QK+ + S VW+HF + K + V C + C +Y A HGT+
Sbjct: 133 QKRAKKCTSDVWQHFTK-KEIEVEVDGKKYVQVWGHCNFPNCKAKYRAEGH-HGTSGFRN 190
Query: 82 HLKVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPF 141
HL+ + K Q L D N + D +L LA II+ E PF
Sbjct: 191 HLRTS-----HSLVKGQLCLKSEKDHGKDINLIEPYKYDEVVSLKKLHLA--IIMHEYPF 243
Query: 142 KHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTW 201
VE+E F F+ +P F I SRV+ + + LY +EKE L L S T D W
Sbjct: 244 NIVEHEYFVEFVKSLRPHFPIKSRVTARKYIMDLYLEEKEKLYGKLKDVQSRFSTTMDMW 303
Query: 202 TSIQNMNYMCVTAHYIDDEWVLKKKILSF-NLIADHKGETIGIALENCMKDWGI-RSICC 259
TS QN +YMCVT H+IDD+W L+K+I+ F ++ H G+ + M W I + +
Sbjct: 304 TSCQNKSYMCVTIHWIDDDWCLQKRIVGFFHVEGRHTGQRLSQTFTAIMVKWNIEKKLFA 363
Query: 260 VTVDNASANNLAINYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQR 319
+++DNASAN +A++ + ++ + + +G + H+RC HILNL+ DGL I ++++
Sbjct: 364 LSLDNASANEVAVHDIIEDLQDTDSNLVCDGAFFHVRCACHILNLVAKDGLAVIAGTIEK 423
Query: 320 IRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYR 379
I+A VKSSP + KCA + ++ DV TRWNSTYLML A Y+ R
Sbjct: 424 IKAIVLAVKSSPLQWEELMKCASECDLDKSKGISYDVSTRWNSTYLMLRDALYYKPALIR 483
Query: 380 LEHADAAFVTSLSGELGGCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQL 439
L+ +D ++ CP ++W+ + K LK F+D T SG+ + +AN F+K
Sbjct: 484 LKTSDPRRYDAI------CPKAEEWKMALTLFKCLKKFFDLTELLSGTQYSTANLFYKGF 537
Query: 440 MEIQKTLN 447
EI+ ++
Sbjct: 538 CEIKDLID 545
>gb|AAP52767.1| putative activator-like transposable element [Oryza sativa
(japonica cultivar-group)] gi|37532356|ref|NP_920480.1|
putative activator-like transposable element [Oryza
sativa (japonica cultivar-group)]
gi|20270084|gb|AAM18172.1| Putative activator-like
transposable element [Oryza sativa (japonica
cultivar-group)]
Length = 737
Score = 212 bits (540), Expect = 2e-53
Identities = 144/428 (33%), Positives = 214/428 (49%), Gaps = 37/428 (8%)
Query: 34 VSKQKKKPQTCSKVWRHFNRNKNLAV-------------CKYCGKEYAANSSSHGTTNLG 80
V KK + S+VW HF++ + V CK C + S+ GTT
Sbjct: 58 VPPTKKAKKCSSEVWSHFDKYEKKVVGDDGTEIVELWAKCKKCSYT-SRRESNRGTTIFW 116
Query: 81 KHL--KVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDE 138
HL K +K+ ++++ K+ + + +VS K K LA II+ E
Sbjct: 117 SHLDKKHQIKSGQQLLNLKKSESGTSVETYRYDEAVSLK-------KFYLA----IIMHE 165
Query: 139 LPFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTT 198
PF VE+E F F+ +P F I SR++V +D L ++ +EK+ + + T
Sbjct: 166 YPFNIVEHEYFVDFIKSLRPTFPIKSRITVRKDILNMFLEEKKKMYEYFKTLSCRFCTTM 225
Query: 199 DTWTSIQNMNYMCVTAHYIDDEWVLKKKILSF-NLIADHKGETIGIALENCMKDWGI-RS 256
D WTS QN YMC+T H+IDD W ++K+I+ F ++ H G + + + DW + R
Sbjct: 226 DMWTSNQNKCYMCITVHWIDDNWCMQKRIIKFMHVEGHHSGNNLCKVFYDSVLDWNLDRK 285
Query: 257 ICCVTVDNASANNLAINYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDAS 316
+ +T+DNASAN++ L + + + +G + H+RC HI NL+ DGLK I +S
Sbjct: 286 LLALTLDNASANDVCARGLVQKLNKIQ-PLICDGAFFHVRCFNHIFNLVAQDGLKQISSS 344
Query: 317 VQRIRAACKFVKSSPSRLATFKKCADSV-GVCSKAMVTLDVVTRWNSTYLMLNIAEKYEH 375
+ IR VK+SP + F KCA G+ ++LDV TRWNST+LML A Y
Sbjct: 345 ISNIRNTVWIVKNSPLQWEEFMKCASECDGLDITCGLSLDVPTRWNSTFLMLKQAIHYRL 404
Query: 376 VFYRLEHADAAFVTSLSGELGGCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSF 435
F RL F+ L P DDW ++ LK FYDATL FSGS + +AN F
Sbjct: 405 AFNRL------FLRHRHKYLKCAPTDDDWTMAKSLCTCLKRFYDATLIFSGSSYPTANLF 458
Query: 436 FKQLMEIQ 443
F + +I+
Sbjct: 459 FTKFCQIK 466
>gb|AAX95609.1| hAT family dimerisation domain, putative [Oryza sativa (japonica
cultivar-group)] gi|50901008|ref|XP_462937.1| putative
transposase [Oryza sativa (japonica cultivar-group)]
Length = 687
Score = 212 bits (540), Expect = 2e-53
Identities = 141/417 (33%), Positives = 214/417 (50%), Gaps = 33/417 (7%)
Query: 40 KPQTCSKVWRHFNRNKNL------AVCKYCGKEYAANSSSHGTTNLGKHLKVCLKNPYRV 93
+P+ +KVW HF + + A CKYC A S GT++L H VC P
Sbjct: 68 QPRKRAKVWEHFEQELVMVDGVPKAQCKYCRLMLTATRKS-GTSHLINH--VCDSCPLVE 124
Query: 94 VDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFM 153
V+ + + +AI + N V F ++ + K I DE+PF+ +E+ F +M
Sbjct: 125 VEVRNRFIAIAR-KQPVENFV------FYPRRSEELMIKFFIHDEIPFRKIEDPYFAEWM 177
Query: 154 GEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSIQNMNYMCVT 213
QP FK+ R ++ Y K+ L L + V LT+D WTS QN+ YM VT
Sbjct: 178 ESMQPTFKVVGRQTIRDKIYTCYIRMKQELHDELQNIDSRVCLTSDMWTSNQNLRYMVVT 237
Query: 214 AHYIDDEWVLKKKILSFNLIA-DHKGETIGIALENCMKDWGIR-SICCVTVDNASANNLA 271
AHYID E+ LKKKI+S + H I A+ C+ DWG+R + +T+DNAS N A
Sbjct: 238 AHYIDAEFSLKKKIISLKPVKYPHTSFAIEEAMMRCLTDWGLRGKLFTLTLDNASNNTAA 297
Query: 272 INYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSP 331
+ + + LF G + H+RCCAHILNL+V DG++ I ++ +I K++++S
Sbjct: 298 CEEMVKNQK---NELLFEGRHFHVRCCAHILNLLVQDGMRLIRGAIHKIHELLKYIENSA 354
Query: 332 SRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSL 391
SR+ F A+S +CSK +T+DV RWN+T+ M+ E + YR A + S
Sbjct: 355 SRIQAFNSIANSSCLCSKFGLTVDVPNRWNTTFKMV-----LEALVYR------AVLNSY 403
Query: 392 SGELGG-CPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQLMEIQKTLN 447
+ E G P ++W + +FLK F +AT S +A+SF ++ I+ L+
Sbjct: 404 ANENGEVAPSDEEWLTAESICEFLKAFEEATRLVSADRKPTAHSFLHLVLCIRHALS 460
>pir||F96652 protein F23N19.13 [imported] - Arabidopsis thaliana
gi|6630458|gb|AAF19546.1| F23N19.13 [Arabidopsis
thaliana]
Length = 633
Score = 211 bits (537), Expect = 4e-53
Identities = 131/382 (34%), Positives = 198/382 (51%), Gaps = 40/382 (10%)
Query: 48 WRHFNRN------KNLAVCKYCGKEYAANSSSHGTTNLGKHLKVCLKNPYRVVDKKQKTL 101
W++F+R K CKYC + Y N +GT + +H++ C K P +K
Sbjct: 57 WKNFDRGQKYPNGKTEVTCKYCEQTYHLNLRRNGTNTMNRHMRSCEKTPGSTPRISRK-- 114
Query: 102 AIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQPKFK 161
VD + +A+A ++ LP+ VE E + A P +
Sbjct: 115 -----------------VDMMVFREMIAVA--LVQHNLPYSFVEYERIREAFTYANPSIE 155
Query: 162 IPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSIQNMNYMCVTAHYIDDEW 221
SR + A D ++Y EK LK L+ + LTTD W ++ +Y+C+TAHY+D +
Sbjct: 156 FWSRNTAASDVYKIYEREKIKLKEKLAIIPGRICLTTDLWRALTVESYICLTAHYVDVDG 215
Query: 222 VLKKKILSFNLIAD-HKGETIGIALENCMKDWGI-RSICCVTVDNASANNLAINYLNRGM 279
VLK KILSF+ H G I + L +KDWGI + I +TVDNASAN+ + L R +
Sbjct: 216 VLKTKILSFSAFPPPHSGVAIAMKLSELLKDWGIEKKIFTLTVDNASANDTMQSILKRKL 275
Query: 280 RVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSPSRLATFKK 339
+ + +GE+ H+RC AHILNLIV DGL+ I ++++IR K+VK S +R F+
Sbjct: 276 QK---DLVCSGEFFHVRCSAHILNLIVQDGLEVISGALEKIRETVKYVKGSETRENLFQN 332
Query: 340 CADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSLSGELGGCP 399
C D++G+ ++A + LDV TRWNSTY ML+ A +++ V L D + + P
Sbjct: 333 CMDTIGIQTEASLVLDVSTRWNSTYHMLSRAIQFKDVLRSLAEVDRVYKS--------FP 384
Query: 400 DHDDWERSRVFVKFLKTFYDAT 421
+WER+ + LK F + T
Sbjct: 385 SAVEWERAELICDLLKPFAEIT 406
>dbj|BAB02100.1| unnamed protein product [Arabidopsis thaliana]
Length = 463
Score = 208 bits (529), Expect = 3e-52
Identities = 142/441 (32%), Positives = 223/441 (50%), Gaps = 45/441 (10%)
Query: 17 GEVTEAGAVTEVGAATAVSKQKKKPQTCSKVWRHFNR-------NKNLAVCKYCGKEYAA 69
GE T++ A AAT V K+ + S VW+ F K C + K+
Sbjct: 35 GEATQS-ATQSTQAATTVYNSNKRLK--SDVWKEFRPILELEEDGKQRGRCIHYDKKLII 91
Query: 70 NSSSHGTTNLGKHLKVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLA 129
+S GT+ L +HL++C K P + +K + + + R
Sbjct: 92 ENSQ-GTSALKRHLQICQKRPQVLSEK----------------------IVYDHKVDREM 128
Query: 130 LAKMIIIDELPFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSA 189
++++I+ +LPF++VE E + P + R + D + Y EK LK
Sbjct: 129 VSEIIVYHDLPFRYVEYEKVRARDKYLNPNCQPICRQTAGNDVFKRYELEKGKLKKFFEQ 188
Query: 190 NNQMVSLTTDTWTSIQNMN-YMCVTAHYIDDEWVLKKKILSF-NLIADHKGETIGIALEN 247
V T D WT+ + Y+C+TAHY+DDEW L KIL+F ++ H GE + + +
Sbjct: 189 FRGRVCCTADLWTARGIVTGYICLTAHYVDDEWRLNNKILAFCDMKPPHTGEELANKILS 248
Query: 248 CMKDWGI-RSICCVTVDNASANNLAINYLNRGMRVWNGRTLF-NGEYLHMRCCAHILNLI 305
C+K+WG+ + I +T+DNA N+ + L +++ +G L +G++ H+RCCAH+LNLI
Sbjct: 249 CLKEWGLEKKIFSLTLDNARNNDSMQSILKHRLQMISGNGLLCDGKFFHVRCCAHVLNLI 308
Query: 306 VLDGLKDIDASVQRIRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYL 365
V +GL ++ IR + +FVK+S SR F C +SVG+ S A ++LDV TRWNSTY
Sbjct: 309 VQEGLSIATELLENIRESVRFVKASESRKDAFAACVESVGIRSGAGLSLDVPTRWNSTYD 368
Query: 366 MLNIAEKYEHVFYRLEHADAAFVTSLSGELGGCPDHDDWERSRVFVKFLKTFYDATLSFS 425
ML A K+ F L+ D + SL+ E ++W+R LK F T FS
Sbjct: 369 MLARALKFRKAFASLKECDRNY-KSLTSE-------NEWDRGERICDLLKPFSTITTYFS 420
Query: 426 GSLHVSANSFFKQLMEIQKTL 446
G + +AN +F Q+ +I++ L
Sbjct: 421 GVKYPTANVYFLQVWKIERLL 441
>gb|AAP59878.1| Ac-like transposase THELMA13 [Silene latifolia]
Length = 682
Score = 207 bits (528), Expect = 4e-52
Identities = 144/426 (33%), Positives = 214/426 (49%), Gaps = 28/426 (6%)
Query: 31 ATAVSKQKKKPQTCSKVWRHFNR-NKNL-------AVCKYCGKEYAANSSSHGTTNLGKH 82
A +VS + + + S VW+H+ + +L A+CKYC S +GT+N +H
Sbjct: 44 APSVSSETRNRKWTSPVWQHYKLFDASLFPDGIARAICKYCDGGPTLAYSGNGTSNFKRH 103
Query: 83 LKVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFK 142
+ C K P +G+ S K +D K R+ALA +I PF
Sbjct: 104 TETCPKRPL-----------LGVAHLTSDGSF-IKKMDPLVYKERVALA--VIRHAFPFS 149
Query: 143 HVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWT 202
+ E +G + +K SR ++ C++++ EK+ LK LS + LTTD WT
Sbjct: 150 YAEYDGNRWLHEGLNESYKPISRNTLRNYCMKIHKREKQILKESLSNLPGKICLTTDMWT 209
Query: 203 SIQNMNYMCVTAHYIDDEWVLKKKILSF-NLIADHKGETIGIALENCMKDWGIRS-ICCV 260
+ M Y+ +TAHYID EW L KIL+F +L H ++ ++ +K+W IRS I +
Sbjct: 210 AFVGMGYISLTAHYIDSEWNLHSKILNFCHLEPPHDAPSLHDSIYAKLKEWDIRSKIFTI 269
Query: 261 TVDNASANNLAINYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQRI 320
T+DNA N+ + L + + + L +GEY H+RC AHILNLIV DGLK ID+ V+++
Sbjct: 270 TLDNARCNDNMQDLLMNSLSL-HSPILCDGEYFHVRCAAHILNLIVQDGLKVIDSGVRKL 328
Query: 321 RAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRL 380
R + S RL FK A ++GV + + LD VTRWNSTY ML A Y +VF +
Sbjct: 329 RMVVAHIVGSERRLIKFKGNASALGVDTSKKLCLDCVTRWNSTYNMLERAMIYRNVFPTM 388
Query: 381 EHADAAFVTSLSGELGGCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQLM 440
+ + P +W R V+ LK F T SG + +AN +FK +
Sbjct: 389 RGPE---MKKFDPHFPEPPSEAEWIRIVKIVELLKPFDHITTLISGRKYPTANLYFKSVW 445
Query: 441 EIQKTL 446
+IQ L
Sbjct: 446 KIQYLL 451
>gb|AAP52341.1| putative transposable element [Oryza sativa (japonica
cultivar-group)] gi|37531504|ref|NP_920054.1| putative
transposable element [Oryza sativa (japonica
cultivar-group)] gi|21671885|gb|AAM74247.1| Putative
transposable element [Oryza sativa (japonica
cultivar-group)]
Length = 772
Score = 207 bits (526), Expect = 7e-52
Identities = 135/414 (32%), Positives = 210/414 (50%), Gaps = 19/414 (4%)
Query: 40 KPQTCSKVWRHFNRNKNL----AVCKYCGKEYAANSSSHGTTNLGKHLKVCLKNPYRVVD 95
K + S VW+ F + K A C +C K SS+ GT++L HLK+C ++
Sbjct: 134 KRKLTSVVWKDFKKVKVCGDVKAECLHCHKRLGGKSSN-GTSHLHDHLKICTLRKIKMGP 192
Query: 96 KKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGE 155
K ++ S + VS F R LA MII+ E P V++ GF+ F+
Sbjct: 193 KTLAQSSLRFNSIEG-GKVSVDTYTFDPAVARRELAAMIILHEYPLSIVDHIGFRRFVSA 251
Query: 156 AQPKFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSI-QNMNYMCVTA 214
QP FK+ +R ++ +D + Y +EK+ + ++ V +TTD WTS Q YM +TA
Sbjct: 252 LQPLFKMVTRNTIRKDIMDTYEEEKKRALAYMAGAKSRVGITTDLWTSDNQKRGYMAITA 311
Query: 215 HYIDDEWVLKKKILSFNLI-ADHKGETIGIALENCMKDWGI-RSICCVTVDNASANNLAI 272
H+IDD W L+ I+ F + A H E I L + +W + I +T+DN + N+ I
Sbjct: 312 HFIDDSWTLRSIIMRFIYVPAPHTAEVICEHLYEALVEWNLDEKISTLTLDNCTTNDKVI 371
Query: 273 NYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSPS 332
+ L + ++ + + G+ LHMRC AHILNLIV DGL I S+ ++R + F ++P
Sbjct: 372 SELIK--KIGKRKLMLEGKLLHMRCAAHILNLIVRDGLDVIKDSIAKVRESVAFWTATPK 429
Query: 333 RLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSLS 392
R+ F++ A V V + + LD TRWNSTY ML+IA F R + F +
Sbjct: 430 RVEKFEEIAKHVKVKMENKLGLDCKTRWNSTYRMLSIALPCARAFDRAIRVEKLFDCA-- 487
Query: 393 GELGGCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQLMEIQKTL 446
P ++W + V LK F D T FSG+ +V++N ++ E ++ +
Sbjct: 488 ------PSEEEWAFASEVVDRLKLFNDITAVFSGTNYVTSNIQLLKICEAKEQI 535
>gb|AAG50652.1| transposase, putative [Arabidopsis thaliana]
gi|25405068|pir||B96493 probable transposase [imported]
- Arabidopsis thaliana
Length = 659
Score = 202 bits (513), Expect = 2e-50
Identities = 133/395 (33%), Positives = 198/395 (49%), Gaps = 36/395 (9%)
Query: 55 KNLAVCKYCGKEYAANSSSHGTTNLGKHLKVCLKNPYRVVDKKQKTLAIGMGSEDDPNSV 114
K A C +CG + S +GT+ + +HL +C + P K
Sbjct: 50 KERARCHHCGIKLVVEKS-YGTSTMNRHLTLCPERPQPETRPK----------------- 91
Query: 115 SFKLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCLR 174
+ + R +++II ++PF++VE E + P K R + A D +
Sbjct: 92 ------YDHKVDREMTSEIIIYHDMPFRYVEYEKVRARDKFLNPDCKPICRQTAALDVFK 145
Query: 175 LYFDEKETLKSVLSANNQMVSLTTDTWTSIQNMN-YMCVTAHYIDDEWVLKKKILSF-NL 232
+ EK L V + +N V LT D W+S + Y+CVT+HYID+ W L KIL+F +L
Sbjct: 146 RFEIEKAKLIDVFAKHNGQVCLTADLWSSRSTVTGYICVTSHYIDESWRLNNKILAFCDL 205
Query: 233 IADHKGETIGIALENCMKDWGI-RSICCVTVDNASANNLAINYLNRGMRVWNGRTLFNGE 291
H GE I + +C+K+WG+ + I +T+DNASAN L ++ NG L G
Sbjct: 206 KPPHNGEEIAKKVYDCLKEWGLEKKILTITLDNASANTSMQTILKHRLQSGNG-LLCGGN 264
Query: 292 YLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSPSRLATFKKCADSVGVCSKAM 351
+LH+RCCAHILNLIV GL+ ++ I + KFVK+S SR +F C + VG+ S A
Sbjct: 265 FLHVRCCAHILNLIVQAGLELASGLLENITESVKFVKASESRKDSFATCLECVGIKSGAG 324
Query: 352 VTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSLSGELGGCPDHDDWERSRVFV 411
++LDV TRWNSTY ML A K+ F L + + + P ++ +R
Sbjct: 325 LSLDVSTRWNSTYEMLARALKFRKAFAILNLYERGYCS--------LPTEEECDRGEKIC 376
Query: 412 KFLKTFYDATLSFSGSLHVSANSFFKQLMEIQKTL 446
LK F T FSG + +AN +F Q+ +I+ L
Sbjct: 377 DLLKPFNTITTYFSGVKYPTANIYFIQVWKIELLL 411
>ref|NP_915834.1| P0003D09.25 [Oryza sativa (japonica cultivar-group)]
Length = 639
Score = 202 bits (513), Expect = 2e-50
Identities = 134/413 (32%), Positives = 208/413 (49%), Gaps = 35/413 (8%)
Query: 45 SKVWRHFNRNKNL------AVCKYCGKEYAANSSSHGTTNLGKHLKVCLKNPYRVVDKKQ 98
+KVW HF + + A CKYCG +A S GT++L H +C P +VD +
Sbjct: 26 AKVWEHFEQELVMIDGLPKAQCKYCGLMLSATRKS-GTSHLINH--ICESCP--LVDGEV 80
Query: 99 KTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQP 158
+ I + + F +KT + K I E+PF +E+ +M QP
Sbjct: 81 RNRFISTVRKQPVENFVF-----DPKKTEELMIKYFIHAEIPFHKIEDPYLDDWMASMQP 135
Query: 159 KFKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSIQNMNYMCVTAHYID 218
FK+ R S+ Y K+ L + L + V LT+D WTS QN+ YM VTAHY+D
Sbjct: 136 TFKLVGRQSIRDKIYNYYNRLKQELHAELENLDSRVCLTSDMWTSNQNLGYMVVTAHYVD 195
Query: 219 DEWVLKKKILSFNLIA-DHKGETIGIALENCMKDWGIRS-ICCVTVDNASANNLAINYLN 276
E+ +KKKI+S + H I A+ C+ +WG+ S + +T+DNAS N A L
Sbjct: 196 AEFKMKKKIISLKPVKYPHTSFAIEEAMMRCLTEWGLSSKLFTLTLDNASNNTAACQELV 255
Query: 277 RGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSPSRLAT 336
+ ++ + G++ H+RCCAHILNL+V DG++ I A++ +IR K+++ SPSR+
Sbjct: 256 KTLK---DELVLEGKHFHVRCCAHILNLLVQDGMRVIRAAIDKIREILKYIEHSPSRIQA 312
Query: 337 FKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVF--YRLEHADAAFVTSLSGE 394
F A S + K+ TLDV TRWNST+ M+ Y+ + Y E+ +
Sbjct: 313 FNSIASSKSLPPKSGFTLDVPTRWNSTFKMIREILPYKAILNSYASENCEL--------- 363
Query: 395 LGGCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQLMEIQKTLN 447
P ++W + +FLK F +AT S +A++F ++ I+ L+
Sbjct: 364 ---LPTDEEWLHAESICEFLKAFEEATRGVSAHRTPTAHTFLPHVLCIRHALS 413
>emb|CAD41225.2| OSJNBa0010H02.12 [Oryza sativa (japonica cultivar-group)]
gi|50927821|ref|XP_473438.1| OSJNBa0010H02.12 [Oryza
sativa (japonica cultivar-group)]
Length = 899
Score = 201 bits (512), Expect = 3e-50
Identities = 148/453 (32%), Positives = 217/453 (47%), Gaps = 36/453 (7%)
Query: 12 QQGKDGEVTEAGAVTEVGAATAVSKQKKKPQTCSKVWRHFNR-----------NKNLAVC 60
QQ GEV G + G T +++KK P +W F +N C
Sbjct: 79 QQLVPGEVR--GEEGQEGNPTKATRKKKSP-----LWDFFEECTVPSKKKKGEMENKVKC 131
Query: 61 KYCGKEYAANSSSHGTTNLGKHLKVCLKNPYRVVDKKQKTL---AIGMGSED-DPNSVSF 116
K C NSS TT+ +HL+ C + + KQ+ + ++ G D D VS
Sbjct: 132 KACETLLTKNSSGT-TTHWRRHLEQCDYHKLQQKSIKQQNINFPSLDEGDVDLDAPCVSV 190
Query: 117 KLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCLRLY 176
+ K R + KMII+ ELPF VE F + + P +K SR ++ DC+RLY
Sbjct: 191 PGY-YDPTKIRELICKMIIVHELPFYLVEYTWFNVLLKRLNPSYKKVSRNTIRSDCMRLY 249
Query: 177 FDEKETLKSVLSANNQMVSLTTDTWTSIQNMNYMCVTAHYIDDEWVLKKKILSF-NLIAD 235
EKE LK + + +SLT D WTS Q + YM + AHYID +W + ++++F L
Sbjct: 250 ESEKEKLKRTFK-DVRKISLTCDLWTSNQTICYMSLVAHYIDADWSMHCRVINFLELEPP 308
Query: 236 HKGETIGIALENCMKDWGIRS-ICCVTVDNASANNLAINYLNRGMRVWNGRTLFNGEYLH 294
H G I A+ +C+ W I I +T DNAS+N+ A N L G F G++LH
Sbjct: 309 HTGVVIANAISDCLASWRIEDKIASITFDNASSNDSAANLLLAKFTK-RGSLWFYGKFLH 367
Query: 295 MRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTL 354
+RCCAHILNLIV DGL I + ++R K++K S +R F DS+ + S + +
Sbjct: 368 IRCCAHILNLIVQDGLAVIKHIIDKVRDTIKYIKKSNNRAYKFSADIDSLNLKSDMGLAI 427
Query: 355 DVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSLSGELGGCPDHDDWERSRVFVKFL 414
D TRW ST+ ML A Y DA + P ++W L
Sbjct: 428 DSCTRWGSTFKMLQSAFFYRSALDVYAAGDANY--------RWLPTPEEWNLYCEVNDAL 479
Query: 415 KTFYDATLSFSGSLHVSANSFFKQLMEIQKTLN 447
+ AT FSGS + ++N F+ +++I++ L+
Sbjct: 480 SVIHAATEEFSGSTYPTSNLFYSHIVDIKRVLD 512
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.133 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 724,801,722
Number of Sequences: 2540612
Number of extensions: 28843437
Number of successful extensions: 69256
Number of sequences better than 10.0: 202
Number of HSP's better than 10.0 without gapping: 121
Number of HSP's successfully gapped in prelim test: 82
Number of HSP's that attempted gapping in prelim test: 68679
Number of HSP's gapped (non-prelim): 263
length of query: 447
length of database: 863,360,394
effective HSP length: 131
effective length of query: 316
effective length of database: 530,540,222
effective search space: 167650710152
effective search space used: 167650710152
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC119416.7


